Abstract
We investigated whether machine learning methods could potentially identify a subgroup of persons with autism spectrum disorder (ASD) who show vitamin B6 responsiveness by selected phenotype variables. We analyzed the existing data from our intervention study with 17 persons. First, we focused on signs and biomarkers that have been identified as candidates for vitamin B6 responsiveness indicators. Second, we conducted hypothesis testing among these selected variables and their combinations. Finally, we further investigated the results by conducting cluster analyses with two different algorithms, affinity propagation and k-medoids. Statistically significant variables for vitamin B6 responsiveness, including combination of hypersensitivity to sound and clumsiness, and plasma glutamine level, were included. As an a priori variable, the Pervasive Developmental Disorders Autism Society Japan Rating Scale (PARS) scores was also included. The affinity propagation analysis showed good classification of three potential vitamin B6-responsive persons with ASD. The k-medoids analysis also showed good classification. To our knowledge, this is the first study to attempt to identify subgroup of persons with ASD who show specific treatment responsiveness using selected phenotype variables. We applied machine learning methods to further investigate these variables’ ability to identify this subgroup of ASD, even when only a small sample size was available.
Introduction
Autism spectrum disorder (ASD) is a disorder characterized by difficulties in social interaction and communication, and repetitive behaviours1. In recent years, ASD has been investigated using advanced technologies such as machine learning to improve diagnosis and prognosis prediction2,3. Machine learning employs artificial intelligence techniques to discover useful masked patterns.
The effects of vitamin B6 in ASD are controversial. Previous attempts to evaluate the efficacy of vitamin B6 in treating persons with ASD have failed to produce consistent findings4–8. ASD is highly heterogeneous9, and identifying a subgroup of persons who may show responsiveness to vitamin B6, if any, is important.
We previously identified similarities between ASD and pyridoxine-dependent epilepsy (PDE)10, which are generally considered quite different. Pyridoxine is one form of vitamin B6. Many of the signs of PDE parallel those of ASD. These signs include epilepsy, autistic tendency, hypersensitivity to sound, expressive verbal disorders, and clumsiness1,11,12. Therefore, we hypothesized that some persons with ASD share, even in part, some etiologies with persons with PDE and that this subgroup of persons with ASD also responds to vitamin B6 intervention. We previously conducted a preliminary randomized controlled trial to evaluate the efficacy of a high-dose vitamin B6 treatment in persons with ASD who exhibited hypersensitivity to sound, expressive verbal disorders, and clumsiness, and we found that the intervention improved verbal IQ10,13. Our preliminary results indicate that hypersensitivity to sound, expressive verbal disorders, and clumsiness might predict responsiveness to vitamin B6.
In addition to signs, biomarkers might also be candidates contributing to the classification of ASD. Several previous studies have consistently suggested the presence of increased plasma amino acids of glutamate and decreased glutamine among persons with some ASD14–16. These findings suggest that plasma glutamate and glutamine levels might contribute to the classification of ASD in addition to signs.
In the present study, we explored whether the signs and biomarkers described above can be used to classify vitamin B6-responsive persons with ASD from nonresponsive ASD persons. We apply machine learning methods of affinity propagation (AP)17 and k-medoids18 to further investigate these variables’ ability to identify a subgroup of persons with ASD.
Results
Characteristics of participants
The participants’ pre-intervention characteristics are shown in Table 1. The 17 participants included 13 boys and 4 girls. The mean age was 8.8 years (standard deviation, 4.1 years). Sixteen of 17 participants showed expressive verbal disorders.
Table 1.
Characteristics and selected signs of participants before intervention.
| Boys/Girls (No.) | 13/4 |
| Age (years) (SD) | 8.8 (4.1), range; 5–19 |
| Body weight (kg) (SD) | 32.8 (15.6), range; 15.6–62.0 |
| Body height (cm) (SD) | 130.6 (21.8), range; 98.2–170.0 |
| Signs (No.) | |
| Hypersensitivity to sound | 8/17 |
| Expressive verbal disorders | 16/17 |
| Clumsiness | 10/17 |
No., number. SD, standard deviation.
Hypothesis testing
Three participants were classified as vitamin B6 possible responders, whereas 14 were classified as less responders. All three possible responders showed coexisting hypersensitivity to sound and clumsiness, compared with only two of the 14 less responders, representing a statistically significant difference (P = 0.01) (Table 2).
Table 2.
Number of participants according to signs and potential vitamin B6 responsiveness.
| Signs | Potential vitamin B6 responsivenessa | P-valueb | |
|---|---|---|---|
| “possible responders” | “less responders” | ||
| Hypersensitivity to sound | |||
| Yes | 3 | 5 | 0.08 |
| No | 0 | 9 | |
| Clumsiness | |||
| Yes | 3 | 7 | 0.18 |
| No | 0 | 7 | |
| Hypersensitivity to sound and clumsiness | |||
| Yes | 3 | 2 | 0.01 |
| No | 0 | 12 | |
aAccording to the Clinical Global Impression-Improvement scale.
bFisher’s exact test.
A comparison of plasma amino acids levels between the possible responders and less responders showed large differences in glutamine levels (reference range: 420–700 nmol/mL). The mean glutamine levels measured before the vitamin B6 treatment were 400.5 nmol/mL (standard error (SE), 11.9 nmol/mL) in the possible responders and 481.4 nmol/mL (SE, 10.4 nmol/mL) in the less responders; this difference was significant (P = 0.004) (Table 3).
Table 3.
Differences in selected plasma amino acids levels according to potential vitamin B6 responsiveness.
| Variables | Potential vitamin B6 responsivenessa | Plasma levels (SE) | P-valueb |
|---|---|---|---|
| Glutamate (nmol/mL) | “possible responders” | 21.5 (3.0) | 0.86 |
| “less responders” | 20.5 (2.4) | ||
| Glutamine (nmol/mL) | “possible responders” | 400.5 (11.9) | 0.004 |
| “less responders” | 481.4 (10.4) |
SE, standard error.
aAccording to the Clinical Global Impression-Improvement scale.
bStudent t-test.
Cluster analysis
Using the presence of hypersensitivity to sound concomitant with the presence of clumsiness, plasma glutamine levels, and the Pervasive Developmental Disorders Autism Society Japan Rating Scale (PARS)19 scores, the AP analysis showed good classification of potential vitamin B6-responsive persons with ASD (cluster 1). All the participants were relatively well classified into five groups, and clusters 2 to 5 consisted of persons who exhibited a low response to vitamin B6. A graphical representation of the AP clustering results using principal component analysis (PCA)20 is presented in Fig. 1a. The mean glutamine levels before the vitamin B6 treatment were 495.6 nmol/mL in cluster 2, 476.3 nmol/mL in cluster 3, 417.8 nmol/mL in cluster 4, and 501.2 nmol/mL in cluster 5, respectively.
Figure 1.
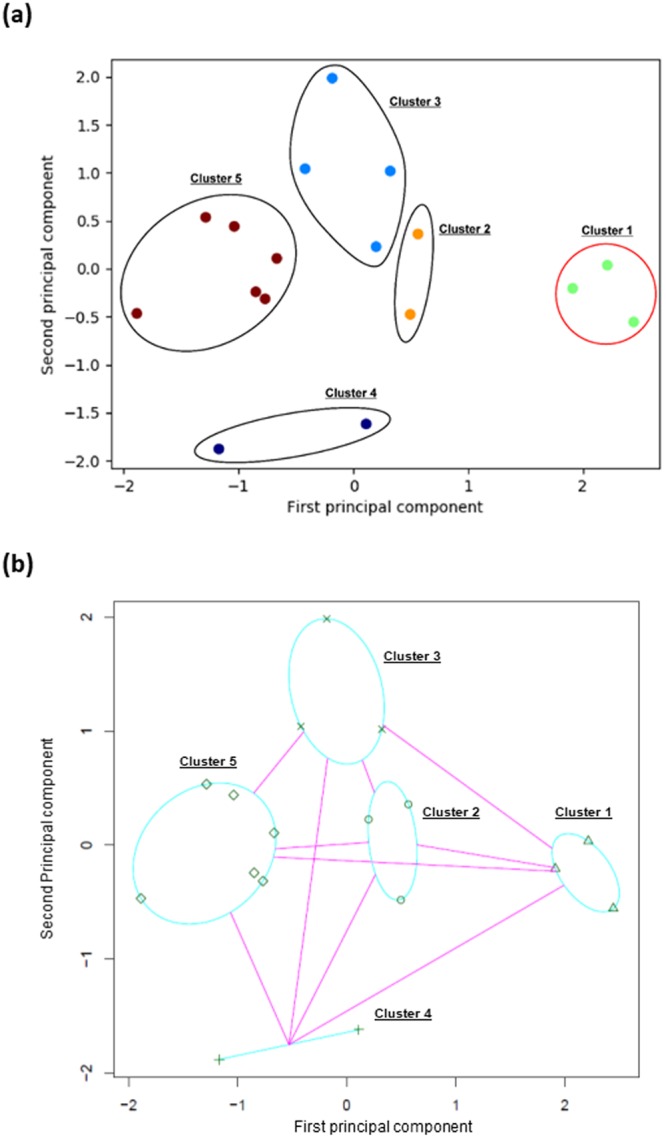
A graphical representation of the affinity propagation (a) and k-medoids (b) clustering results using principal component analysis. The affinity propagation (AP) analysis showed good classifying of potential vitamin B6-responsive persons with ASD (cluster 1). All the participants were relatively well classified into five groups, and clusters 2 to 5 consisted of persons who exhibited a low response to vitamin B6. A graphical representation of the AP clustering results using principal component analysis (PCA) is presented in 1a. The k-medoids analysis also showed good classification . The selected number of clusters by k-medoids was also five and the result was identical to that by the AP except for one participants who was classified in Cluster 2 by the AP was classified in Cluster 3 by the k-medoids method. A graphical representation of the k-medoids clustering results using PCA is presented in 1b.
The k-medoids analysis also showed good classification. The selected number of clusters by k-medoids was also five and the result was identical to that by AP except for one participants who was classified into cluster 2 by AP and into cluster 3 by the k-medoids method. A graphical representation of the k-medoids clustering results using PCA is presented in Fig. 1b.
Discussion
Using the machine learning clustering algorithms of AP and k-medoids, we successfully classified an ASD subgroup that responded to vitamin B6 from others by selected phenotype variables. The additional k-medoids analysis reflected the robustness of our clustering result because the k-medoids method is completely different from AP. Our data indicate that common variables used in previous studies may be able to identify subgroup that exhibit responsiveness to specific treatment.
Our sample size is relatively small. Because we selected candidate phenotype variables according to evidence from previous studies, they are free from the multiplicity of hypothesis tests. Furthermore, contrary to hypothesis testing, cluster analysis does not necessarily require a large sample size because it separates data according to structures. In cluster analyses, distance between clusters is more important for stable clustering than sample sizes.
Variable selection is critical issues in clustering analysis. In this study, we focused on signs and biomarkers that have been identified in previous studies as candidates and conducted hypothesis testing among these selected variables and their combinations to further select relevant variables and to reduce the dimension. Recent medical studies have frequently used the method of reducing dimension and subsequent clustering. Based on the clinical and/or statistical approach, researchers identify subsets or patterns of variables to address their study aims21,22. We believe that our protocol is one appropriate strategy to identify subgroup of diseases including ASD.
AP is an unsupervised clustering algorithm that identifies clusters of similar points using a set of points and a set of similarity values between the points and provides a representative example, called an exemplar, for each cluster17. We identified five clusters from 17 persons with ASD using the AP analysis. Of the five clusters, cluster 1 consisted of all persons who responded to vitamin B6. Although we could not identify meaningful characteristics of the other four clusters (clusters 2 to 5), the AP analysis suggested that persons with ASD may be highly heterogeneous, as previously reported9.
Cluster analysis classifies data into groups according to their structure. However, cluster analysis itself does not predict a sample as a specific cluster. Although our cluster analyses could potentially identify a subgroup of ASD, their predictive value requires further analyses utilizing different algorithms, such as support vector machine23,24, and evaluation of the accuracy of these results. Based on our data, further studies to determine the predictive value are warranted, presumably with a larger sample size and validation data sets.
Why low levels of plasma glutamine levels predict vitamin B6 responsiveness is unclear. Glutamine plays a central role in nitrogen metabolism in many cell systems and in the central nervous system, and glutamine synthesis has a neuroprotective function because it removes ammonia and glutamate25. Vitamin B6 also plays a predominant role as a coenzyme for these substances26. Therefore, we speculate that consuming a high dose of vitamin B6 may improve the activity of the metabolic pathways described above.
The prevalence of pyridoxine responsiveness is unknown. Although three of the 17 participants exhibited a possible response to vitamin B6 in our intervention study, the study population was not randomly recruited from among persons with ASD, rather, we restrictively recruited the participants with potential responsiveness. Therefore, the proportion of vitamin B6 responders in the general population with ASD may be relatively small. Unfortunately, in daily practice, clinically used ASD assessment tools generally do not include hypersensitivity to sound or clumsiness, and clinical tests do not typically include serum or plasma glutamine levels. Therefore, more data may be necessary to assess vitamin B6 responsiveness and estimate its prevalence.
We might be able to identify the genetic factors responsible for vitamin B6-responsive ASD. Although the high heritability of ASD is supported by high concordance rates (from 36% to 95%) in monozygotic twins and a higher recurrence risk of 11% and 19% with single-sibling involvement27–29, no susceptibility genes have been specified in ASD30. Based on the results of the present study, the clustering of some signs and biomarkers might be informative and provide the best model for identifying etiologically similar and medically treatable predictive cases of ASD.
The possibility that a low proportion of vitamin B6 responders with ASD and the small sample size were limitations of the present study. Therefore, the present findings should be interpreted with caution, and external validity should be confirmed by further studies.
To our knowledge, this is the first study to attempt to identify subgroup of persons with ASD who show vitamin B6 responsiveness using selected phenotype variables. We applied the machine learning methods of AP and k-medoids to further investigate these variables’ ability to identify a subgroup of ASD, even when only a small sample size was available.
Methods
Study design
We analyzed the existing data from our intervention study as follows: 1) we focused on signs and biomarkers that have been identified in previous studies10,13–16 as candidates for vitamin B6 responsiveness indicators; 2) we conducted hypothesis testing among these selected variables and their combinations to further select variables and to reduce the dimension; and 3) we conducted cluster analysis to further investigate the variables’ predictive ability using two different algorithms.
Data characteristics
Intervention study
We conducted a single-arm intervention from October 24, 2007 to September 2, 2009. Seventeen persons with ASD were recruited from seven medical institutions in Japan. Medical doctors invited potential vitamin B6 responders who showed hypersensitivity to sound, expressive verbal disorders, and/or clumsiness. The inclusion criteria for participation in this intervention study were as follows: (1) a diagnosis of ASD (code F84) determined by a doctor according to the International Classification of Diseases, 10th Revision (ICD-10)31; (2) 5 to 20 years of age; (3) no history of epilepsy; (4) no diagnosis of homocystinuria or fragile X syndrome; and (5) not currently receiving vitamin B6 treatments.
We selected the above age range to attempt to avoid an unstable phenotype for younger children and to exclude the effects of deuteropathy of more elderly persons.
Vitamin B6 responsiveness
Participants received 5 mg vitamin B6/kg body weight per day for two weeks followed by 10 mg vitamin B6/kg body weight per day for two weeks, resulting in a total treatment period of four weeks. Using the Clinical Global Impression-Improvement (CGI-I) scale32, medical doctors evaluated responsiveness to vitamin B6 as “very much improved,” “much improved,” “minimally improved,” or “no change.” The CGI Scale is a measure of global clinical change with strong validity that has been widely used as an outcome measure in clinical trials of central nervous system disorders33. We divided participants into two groups: “possible responders,” who were “very much improved” or “much improved”, and “less responders,” who were “minimally improved” or showed “no change”.
Variable measurements
Before the intervention, participants were assessed for signs including hypersensitivity to sound, expressive verbal disorders, and clumsiness, and plasma amino acids including glutamate and glutamine. A person with ASD was considered to show a sign, such as hypersensitivity to sound, when both his/her medical doctor and guardian independently assessed the sign as present. The PARS was used to evaluate autistic traits19.
Ethical issues
We conducted this intervention study in accordance with the guidelines of the Declaration of Helsinki34 and all other applicable guidelines. The protocol was reviewed and approved by the institutional review board of Tohoku University Graduate School of Medicine. The board also reviewed and approved the study protocol for Miyagi Psychiatric Center, Kakuta Child & Allergy Clinic, NTT Medical Center Tokyo, Fujimoto Shinjuku Hospital, Yasuhara Children’s Clinic, and National Center of Neurology and Psychiatry. For participants less than 16 years of age, we obtained informed assent from the individuals and informed consent from their guardians. For participants aged 16 to 19 years, we obtained informed consent from both the individuals and their guardians.
Hypothesis testing
Regarding the signs’ variables, we excluded expressive verbal disorders in the present analyses because they were observed in 16 of 17 participants. Signs and their combination of hypersensitivity to sound and clumsiness, and also biomarkers of plasma glutamate level, glutamine level were compared between vitamin B6 possible responders and less responders. Signs of the participants were compared using Fisher’s exact test. Student’s t-test was used to investigate the significance of the differences in the biomarkers.
The above statistical analyses were performed using SAS version 9.4 (SAS Inc., Cary, NC, USA). We used approximate variance formulas to calculate 95% confidence intervals (CIs). A P value < 0.05 was accepted as a statistically significant value. All P values were two-tailed.
Cluster analysis
In the cluster analysis, statistically significant variables for vitamin B6 responsiveness, including hypersensitivity to sound combined with clumsiness, and plasma glutamine level, were included in the preprocessed dataset. As an a priori variable, the PARS score was also included. A relatively recently developed algorithm, AP17, was first applied to the preprocessed dataset to divide the participants into vitamin B6 possible-responsive and less-responsive groups. AP is an unsupervised (i.e. without using the information of possible responders and less responders) clustering analysis method using a message-passing-based algorithm. In the present analysis, AP was performed without diagonal components using a dumping factor of 0.9. These analyses were performed with the scikit-learn toolkit in Python 2.635,36.
To examine the robustness of the clustering by AP, we additionally conducted a completely different clustering analysis using k-medoids18 implemented in the “fpc” package for R version 3.3.237. It is a fully automated method similar to AP in which the optimal number of clusters is automatically determined by the average silhouette width.
To create a graphical representation of the clustering results, we adopted a PCA20.
Acknowledgements
The present study was supported by the Ministry of Education, Culture, Sports, Science and Technology (MEXT) KAKENHI Grant Numbers 19390171 and 16H05242. MEXT had no role in the design or execution of the study.
Author Contributions
Study conception and design: T.O., M.I., M.K., M.W., M.I., M.K., S.K. and S.K. Data acquisition: M.K., M.W., K.K., M.O., A.K., N.F., A.Y., M.I., M.K. and S.K. Data analysis and interpretation: T.O., M.I., G.T., M.U., C.Y., S.M., M.K., H.M., H.M., M.N., T.K. and S.K. Drafting of the manuscript: T.O., M.I., G.T., S.M. and S.K. Critical revision of the manuscript for important intellectual content: T.O., M.I., G.T., M.U., C.Y., S.M., M.K., H.M., H.M., M.N., T.K., M.K., M.W., K.K., M.O., A.K., N.F., A.Y., M.I., M.K., S.K. and S.K. Statistical analysis: T.O., M.I., G.T., M.U., S.M. and S.K. Obtaining funding: S.K. Administrative, technical, or material support: C.Y., S.M., M.K., H.M., H.M., M.N. and T.K. Study supervision: S.K.
Data Availability Statement
The data used in this study will be shared upon request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Taku Obara and Mami Ishikuro contributed equally.
References
- 1.Rapin I. Current concepts: autism. N Engl J Med. 1997;337:97–104. doi: 10.1056/NEJM199707103370206. [DOI] [PubMed] [Google Scholar]
- 2.Anzulewicz A, Sobota K, Delafield-Butt JT. Toward the autism motor signature: gesture patterns during smart tablet gameplay identify persons with autism. Sci Rep. 2016;6:31107. doi: 10.1038/srep31107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thabtah, F. Machine learning in autistic spectrum disorder behavioral research: a review and ways forward. Inform Health Soc Care. Feb 13, 1–20, 10.1080/17538157.2017.139913210.1080/17538157.2017.1399132 [Epub ahead of print] (2018). [DOI] [PubMed]
- 4.Rimland B, Callaway E, Dreyfus P. The effect of high doses of vitamin B6 on autistic children: a double blind crossover study. Am J Psychiatry. 1978;135:472–475. doi: 10.1176/ajp.135.4.472. [DOI] [PubMed] [Google Scholar]
- 5.Findling RL, et al. High-dose pyridoxine and magnesium administration in children with autistic disorder: an absence of salutary effects in a double-blind, placebo- controlled study. J Autism Dev Disord. 1997;27:467–478. doi: 10.1023/A:1025861522935. [DOI] [PubMed] [Google Scholar]
- 6.Nye C, Brice A. Combined vitamin B6-magnesium treatment in autism spectrum disorder. Cochrane Database Syst Rev. 2005;4:CD003497. doi: 10.1002/14651858.CD003497.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kummer A, Harsányi E. Comments on “vitamin supplementation reduces the level of homocysteine in the urine of autistic children”. Nutr Res. 2011;31:873. doi: 10.1016/j.nutres.2011.09.019. [DOI] [PubMed] [Google Scholar]
- 8.Gogou M, Kolios G. The effect of dietary supplements on clinical aspects of autism spectrum disorder: a systematic review of the literature. Brain Dev. 2017;39:656–664. doi: 10.1016/j.braindev.2017.03.029. [DOI] [PubMed] [Google Scholar]
- 9.David MM, et al. Comorbid analysis of genes associated with autism spectrum disorders reveals differential evolutionary constraints. PLoS One. 2016;11:e0157937. doi: 10.1371/journal.pone.0157937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuriyama S, et al. Pyridoxine treatment in a subgroup of children with pervasive developmental disorders. Dev Med Child Neurol. 2002;44:284–286. doi: 10.1111/j.1469-8749.2002.tb00809.x. [DOI] [PubMed] [Google Scholar]
- 11.Baxter P, Griffiths P, Kelly T, Gardner-Medwin D. Pyridoxine-dependent seizures: demographic, clinical MRI and psychometric features, and effect of dose on intelligence quotient. Dev Med Child Neurol. 1996;38:998–1006. doi: 10.1111/j.1469-8749.1996.tb15060.x. [DOI] [PubMed] [Google Scholar]
- 12.Burd L, Stenehjem A, Franceschini LA, Kerbeshian J. A 15-year follow-up of a boy with pyridoxine (vitamin B6)-dependent seizures with autism, breath holding, and severe mental retardation. J Child Neurol. 2000;15:763–765. doi: 10.1177/088307380001501111. [DOI] [PubMed] [Google Scholar]
- 13.Kamiyama M, Kuriyama S, Watanabe M. A clinical study of pyridoxine treatment for pervasive developmental disorders with hypersensitivity to sound. No To Hattatsu. 2006;38:277–282. [PubMed] [Google Scholar]
- 14.Shimmura C, et al. Alteration of plasma glutamate and glutamine levels in children with high-functioning autism. PLoS One. 2011;6:e25340. doi: 10.1371/journal.pone.0025340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghanizadeh A. Increased glutamate and homocysteine and decreased glutamine levels in autism: a review and strategies for future studies of amino acids in autism. Dis Markers. 2013;35:281–286. doi: 10.1155/2013/536521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Al-Otaish Hanoof, Al-Ayadhi Laila, Bjørklund Geir, Chirumbolo Salvatore, Urbina Mauricio A., El-Ansary Afaf. Relationship between absolute and relative ratios of glutamate, glutamine and GABA and severity of autism spectrum disorder. Metabolic Brain Disease. 2018;33(3):843–854. doi: 10.1007/s11011-018-0186-6. [DOI] [PubMed] [Google Scholar]
- 17.Frey BJ, Dueck D. Clustering by passing messages between data points. Science. 2007;315:972–976. doi: 10.1126/science.1136800. [DOI] [PubMed] [Google Scholar]
- 18.Kaufman, L. & Rousseeuw, P. J. Finding groups in data: an introduction to cluster analysis (John Wiley & Sons, 2009).
- 19.Ito H, et al. Validation of an interview-based rating scale developed in Japan for pervasive developmental disorders. Res Autism Spectr Disord. 2012;6:1265–1272. doi: 10.1016/j.rasd.2012.04.002. [DOI] [Google Scholar]
- 20.B.Y. Christophe. Analytical Chemistry. 2012. PCA: The Basic Building Block of Chemometrics. [Google Scholar]
- 21.Terada A, Okada-Hatakeyama M, Tsuda K, Sese J. Statistical significance of combinatorial regulations. Proc Natl Acad Sci USA. 2013;110:12996–13001. doi: 10.1073/pnas.1302233110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zampetaki A, et al. Prospective study on circulating MicroRNAs and risk of myocardial infarction. J Am Coll Cardiol. 2012;60:290–299. doi: 10.1016/j.jacc.2012.03.056. [DOI] [PubMed] [Google Scholar]
- 23.Raudys S. How good are support vector machines? Neural Netw. 2000;13:17–19. doi: 10.1016/S0893-6080(99)00097-0. [DOI] [PubMed] [Google Scholar]
- 24.Bruining H, et al. Behavioral signatures related to genetic disorders in autism. Mol Autism. 2014;5:11. doi: 10.1186/2040-2392-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Häberle J, et al. Inborn error of amino acid synthesis: human glutamine synthetase deficiency. J Inherit Metab Dis. 2006;29:352–358. doi: 10.1007/s10545-006-0256-5. [DOI] [PubMed] [Google Scholar]
- 26.Sato K. Why is vitamin B6 effective in alleviating the symptoms of autism? Med Hypotheses. 2018;115:103–106. doi: 10.1016/j.mehy.2018.04.007. [DOI] [PubMed] [Google Scholar]
- 27.Rosenberg RE, et al. Characteristics and concordance of autism spectrum disorders among 277 twin pairs. Arch Pediatr Adolesc Med. 2009;163:907–914. doi: 10.1001/archpediatrics.2009.98. [DOI] [PubMed] [Google Scholar]
- 28.Constantino JN, Zhang Y, Frazier T, Abbacchi AM, Law P. Sibling recurrence and the genetic epidemiology of autism. Am J Psychiatry. 2010;167:1349–1356. doi: 10.1176/appi.ajp.2010.09101470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ozonoff S, et al. Recurrence risk for autism spectrum disorders: a baby siblings research consortium study. Pediatrics. 2011;128:e488–495. doi: 10.1542/peds.2010-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sfari, G. Gene scoring. https://gene.sfari.org/database/gene-scoring/ (2018).
- 31.World Health Organization. The ICD-10 classification of mental and behavioural disorders: clinical descriptions and diagnostic guidelines. http://www.who.int/classifications/icd/en/bluebook.pdf (1992).
- 32.Busner J, Targum SD. The clinical global impressions scale: applying a research tool in clinical practice. Psychiatry (Edgmont). 2007;4:28–37. [PMC free article] [PubMed] [Google Scholar]
- 33.Neul JL, et al. Improving treatment trial outcomes for rett syndrome: the development of rett-specific anchors for the clinical global impression scale. J Child Neurol. 2015;30:1743–1748. doi: 10.1177/0883073815579707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.World Medical Association. WMA Declaration of Helsinki – Ethical principles for medical research involving human subjects. https://www.wma.net/policies-post/wma-declaration-of-helsinki-ethical-principles-for-medical-research-involving-human-subjects/ (2018).
- 35.Pedregosa F, et al. Scikit-learn: Machine learning in Python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- 36.Scikit-learn. Machine learning in python (Version 0.18.1). http://scikit-learn.org/stable/# (2018).
- 37.Mente S, Kuhn M. The use of the R language for medicinal chemistry applications. Curr Top Med Chem. 2012;12:1957–1964. doi: 10.2174/156802612804910322. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used in this study will be shared upon request.


