Abstract
Genetic variation in FGFR2 is a newly described risk factor for breast cancer. We estimated the relative risk and contribution of FGFR2 polymorphisms to breast cancer risk in diverse ethnic groups within Jewish and other Middle Eastern populations. We genotyped four FGFR2 single nucleotide polymorphisms (SNP) and tested for association of these SNPs and haplotypes with breast cancer risk in a population-based case-control study of 1,529 women with breast cancer and 1,528 controls. We found significant associations between breast cancer risk and all four studied SNPs in FGFR2 (Ptrend for all SNPs < 0.0001). In ethnicity-specific analysis, all four SNPs were significantly associated with breast cancer risk in Ashkenazi and Sephardi Jews, with a similar but not significant trend in Arabs. Haplotype analysis identified five common haplotypes (>1%). The previously described AAGT risk haplotype was significantly associated with breast cancer risk in Ashkenazi [odds ratio (OR), 1.25; 95% confidence interval (95% CI), 1.07-1.45; P = 0.0059] and Sephardi Jews (OR, 1.46; 95% CI, 1.17-1.80; P = 0.0006) compared with the reference GGAC haplotype. The AAAC haplotype was significantly associated with breast cancer risk in Sephardi Jews (OR, 1.97; 95% CI, 1.16-3.35; P = 0.0125) but not in Ashkenazi Jews (OR, 0.83; 95% CI, 0.41-1.62; P = 0.5613) or in Arabs (OR, 1.31; 95% CI, 0.80-2.14; P = 0.2881). Genetic variation in FGFR2, identified by rs1219648, may account for a substantial fraction of breast cancer in Arab (12%), Ashkenazi (15%), and Sephardi Jewish (22%) populations. The identification of population-specific risk haplotypes in FGFR2 is likely to help identify causal variants for breast cancer.
Introduction
FGFR2 is a tyrosine kinase receptor that is a member of the family of individually distinct fibroblast growth factor receptors involved in tumorigenesis. FGFR2 is amplified and overexpressed in breast cancer (1-3), and data suggest that FGFR2 is a tumor suppressor gene in prostate cancer and urothelial cancer (4). Aberrant expression of alternatively spliced isoforms of FGFR2 transforms breast cancer cells by sustained signal transduction, and one splice variant of FGFR2 is overexpressed in ~5% of breast cancers (1-6). Two recent genome-wide association (GWA) studies demonstrated an association of FGFR2 single nucleotide polymorphisms (SNP) with breast cancer risk (7, 8). The association was restricted to SNPs in the linkage disequilibrium block covering intron 2. The association was found with four SNPs (rs11200014, rs2981579, rs1219648, and rs2420946; ref. 8) in one study and one SNP (rs2981582) in another (7). All five SNPs are in high linkage disequilibrium, indicating that both studies found the same association. Efforts to identify the causal allele by resequencing the FGFR2 intron 2 region and fine-mapping with haplotype-tagging SNPs in European and Asian populations have not yet revealed the causal sequence variant (7).
In Israel, ~60% of Jews are Ashkenazi (Eastern European) and others are Sephardi (Middle Eastern/African and Iraqi) or of mixed ancestry. Other major populations include Muslim Arabs, Christian Arabs, Druze, and Bedouin. Three Ashkenazi Jewish founder mutations in BRCA1 (185delAG and 5382insC) and BRCA2 (6174delT) genes have combined frequency of 2.5% in general Ashkenazi population and appear in ~10% (9-11) of breast cancer cases in Ashkenazi Jewish women. In Ashkenazi Jewish breast cancer patients with family history of breast and ovary cancer, 45% carry a mutation in one of these genes (11). The mutations in other breast cancer susceptibility genes (PTEN, ATM, and p53) are not frequent enough in Israel to explain the rest of BRCA-negative familial breast cancers (12, 13). Extrapolating estimates of the population attributable fraction of 16% that was calculated from the Nurses′ Health Study (8) suggest that genetic variation in FGFR2 may make a considerable contribution to breast cancer risk in Jewish and other Middle Eastern populations.
In the present study we validated GWA study findings of FGFR2 association with breast cancer and estimated the contribution of FGFR2 polymorphisms to breast cancer risk in diverse ethnic groups within the Israeli population.
Materials and Methods
Study Population.
The series of 1,529 women with breast cancer and 1,528 controls was derived from a large ongoing population-based case-control study of the molecular and environmental etiology of breast cancer in Israel. In this study, which was initiated in 2000, all incident breast cancer cases in a defined geographic region in northern Israel are invited to participate. All participants signed an informed consent approved by Carmel Medical Center Institutional Review Board Committee. Controls are randomly sampled from the list of all women enrolled in the health-care program provided by CHS (the largest nonprofit health services provider in Israel, covering the majority of the Israeli population). Controls are individually matched by age (within 1 year), residence, and Jewish/ Arab status. The study includes an interview by trained nurses (to evaluate risk factors, including a detailed three-generation family history of cancer) and venous blood collection. DNA extracted from the blood is stored at −20°C for a variety of molecular studies, among them genotyping for each of the Jewish founder mutations in the BRCA1 and BRCA2 genes.
DNA Extraction and Genotyping.
Genomic DNA was extracted from whole blood using a commercially available kit according to the manufacturer′s protocol (Puregene DNA extraction kit; Gentra Systems). Genetic testing for BRCA1 (185delAG and 5382insC) and BRCA2 (6174delT) was done using the Pronto BRCA kit (Pronto Diagnostics). Genomic DNA from patients with breast cancer known to carry one of the three mutations (185delAG, 5382insC, and 6174delT) served as positive controls for each assay, and genomic DNA samples from patients who did not carry one of these mutations were used as negative control. The Pronto kit testing of all samples was confirmed by TaqMan Assay-by-Design for each Ashkenazi founder mutation.
Taking into account the redundant nature of recent GWA study findings (7, 8), we genotyped four SNPs from the Hunter et al. study, which allowed us to reconstruct the haplotypes. The genotyping of rs11200014 (SNP Assay-on-Demand C_31019228_10), rs2981579 (SNP Assay-on-Demand C_15885469_10), rs1219648 (SNP Assay-on-Demand C_2917314_20), rs2420946 (SNP Assay-on-Demand C_2917305_10) was done by allelic discrimination using the 5′-nuclease Assay-on-Demand on 7900HT sequence detection system (Applied Biosystems). The assay was done in 5 μL reaction volume containing 1× TaqMan PCR core reagents (Applied Biosystems), 5 mmol/L MgCl2, 200 nmol/L each PCR primer, 100 nmol/L MGB probes (Applied Biosystems), 0.5 units AmpliTaq Gold, 0.2 units AmpErase UNG, and 5 ng genomic DNA. Approximately 1% of duplicated samples were used as internal control. No discrepancies were observed. Genotyping failures were repeated at least once, and the final genotyping failure rate ranged between 3.4% and 3.8% [rs11200014 (3.6%), rs2981579 (3.8%), rs1219648 (3.4%), and rs2420946 (3.8%)].
Statistical Analysis.
The allelic distribution of FGFR2 was determined for each ethnic group (Ashkenazi, Sephardi, and other) and tested for deviation from Hardy-Weinberg equilibrium among controls by SNP Hardy-Weinberg equilibrium within the R statistical package (version 2.4.1). Odds ratios (OR) were calculated by unconditional and conditional logistic regression as implemented in R and SAS (version 9.1). For haplotype reconstruction and the analysis of the association of haplotypes with breast cancer, the “haplo.stats” package for R language was used (14). Based on genotype data, haplotype frequencies were compared using age and BRCA mutation status adjusted global and haplotype-specific score tests (“haplo.score” function). Rare haplotypes (combined frequency less than 1%) were pooled. Unmatched results are reported in the tables for consistency between genotype and haplotype analyses. Matched multivariate estimates are reported in the text and were similar to unmatched analyses. All analyses were adjusted for age and BRCA mutations status, and measures of association were stratified by ethnicity (Ashkenazi Jews, Sephardi Jews, and Arabs) and by family history, using reported history of breast cancer in any first-degree relative as a binary variable, to test for effect modification. Effect modification was evaluated by a likelihood ratio test for multivariate interaction terms. A meta-analysis of previously published data (8) and results from the present study included the construction of a Forestplot with “metaplot” function from “rmeta” package for R (version 2.4.1).
Results
The demographic characteristics of cases and controls are shown in Table 1. The response rate from this ongoing case-control study is derived from 4,075 detected cases, where 3,372 were approached and invited to participate at the time of the analysis. The refusal rate was 14% (474/3,372), and 82.7% (2,788/3,372) of those approached were interviewed. Blood samples and DNA were available from 1,529 matched cases for this analysis, and these samples were representative of the larger group of interviewed cases. Among controls, 3,190 potential controls were approached and 73.2% (2,335/3,190) agreed to participate, and 1,528 matched controls with blood samples were included in the present analysis. Family history of breast cancer was reported more often among first-degree relatives of cases than controls. The distribution of BRCA carriers among cases and controls is shown in Table 1.
Table 1. Overview of study population.
| Characteristics | Cases (%) | Controls (%) |
|---|---|---|
| All subjects | 1,529 | 1,528 |
| Age* | 58 | 58.8 |
| Family history† | 214 (14.0) | 97 (6.4) |
| BRCA1/2 carriers‡ | 56 (3.7) | 12 (0.8) |
| Ethnicity | ||
| Jewish | ||
| Ashkenazi | 729 (47.7) | 685 (44.8) |
| Sephardi | 332 (21.7) | 395 (25.9) |
| Both | 31 (2.0) | 32 (2.1) |
| Unknown | 57 (3.7) | 62 (4.1) |
| Arabs | 337 (22.0) | 331 (21.7) |
| Other | 43 (2.8) | 23 (1.5) |
Mean age in years.
Self-reported breast cancer history in first-degree relatives.
Ashkenazi founder BRCA1/2 mutations only.
We genotyped four FGFR2 SNPs (rs11200014, rs2981579, rs1219648, and rs2420946) in 1,529 breast cancer cases and 1,528 controls. The minor allele frequencies of all tested SNPs in Ashkenazi Jews are consistent with the corresponding frequencies in Caucasian population of HapMap project and roughly similar in Sephardi Jews and Arabs (Table 2). FGFR2 genotypes did not deviate from the Hardy-Weinberg equilibrium in controls.
Table 2. Minor allele frequency distribution in Caucasians from HapMap study and studied populations (Ashkenazi and Sephardi Jews and Arabs) without breast cancer.
| rs1219648(G) | rs2981579(A) | rs11200014(A) | rs2420946(T) | |
|---|---|---|---|---|
| Caucasians* | 0.42 | 0.42 | 0.42 | 0.41 |
| Ashkenazi Jews | 0.39 | 0.39 | 0.39 | 0.40 |
| Sephardi Jews | 0.48 | 0.50 | 0.48 | 0.49 |
| Arabs | 0.45 | 0.51 | 0.48 | 0.45 |
According to the HapMap project.
We found evidence of significant associations between risk of breast cancer and all four studied SNPs in FGFR2 (Table 3). The analysis in different ethnic groups showed significant association of all four studied SNPs with breast cancer risk in Ashkenazi and Sephardi Jews, with consistent trends within Arab women. The codominant and log-additive models are presented for consistency with prior published reports (8). The adjusted OR for the associations of individual SNPs with breast cancer risk from multivariate models are shown in the Table 4.
Table 3. Association of FGFR2 intron 2 SNPs (rs11200014, rs2981579, rs1219648, rs2420946) with breast cancer risk in different ethnic groups from Israel.
| Ethnicity | Genotype | Cases | Controls | OR (95% CI) | P | Ptrend | |
|---|---|---|---|---|---|---|---|
| All | rs11200014 | G/G | 417 | 488 | Reference | <0.0001 | |
| G/A | 698 | 701 | 1.19 (1.01-1.41) | 0.0425 | |||
| A/A | 366 | 288 | 1.52 (1.24-1.86) | <0.0001 | |||
| rs2981579 | G/G | 377 | 460 | Reference | <0.0001 | ||
| G/A | 722 | 710 | 1.27 (1.07-1.51) | 0.006 | |||
| A/A | 381 | 301 | 1.59 (1.30-1.96) | <0.0001 | |||
| rs1219648 | A/A | 420 | 499 | Reference | <0.0001 | ||
| A/G | 717 | 701 | 1.24 (1.05-1.47) | 0.0116 | |||
| G/G | 350 | 277 | 1.53 (1.25-1.89) | <0.0001 | |||
| rs2420946 | C/C | 409 | 489 | Reference | <0.0001 | ||
| C/T | 715 | 700 | 1.25 (1.06-1.48) | 0.009 | |||
| T/T | 356 | 285 | 1.53 (1.25-1.89) | <0.0001 | |||
| Ashkenazi | rs11200014 | G/G | 241 | 251 | Reference | 0.0169 | |
| G/A | 340 | 334 | 1.10 (0.87-1.40) | 0.4146 | |||
| A/A | 145 | 100 | 1.51 (1.10-2.07) | 0.0109 | |||
| rs2981579 | G/G | 228 | 242 | Reference | 0.0055 | ||
| G/A | 348 | 342 | 1.13 (0.89-1.43) | 0.3289 | |||
| A/A | 149 | 98 | 1.62 (1.18-2.22) | 0.0031 | |||
| rs1219648 | A/A | 233 | 250 | Reference | 0.0037 | ||
| A/G | 350 | 340 | 1.14 (0.90-1.45) | 0.2614 | |||
| G/G | 146 | 95 | 1.65 (1.20-2.28) | 0.0021 | |||
| rs2420946 | C/C | 225 | 241 | Reference | 0.0039 | ||
| C/T | 350 | 341 | 1.15 (0.91-1.46) | 0.2530 | |||
| T/T | 151 | 100 | 1.64 (1.19-2.25) | 0.0022 | |||
| Sephardi | rs11200014 | G/G | 64 | 114 | Reference | 0.0006 | |
| G/A | 157 | 184 | 1.54 (1.06-2.24) | 0.0230 | |||
| A/A | 108 | 97 | 2.06 (1.37-3.10) | 0.0005 | |||
| rs2981579 | G/G | 54 | 103 | Reference | 0.0002 | ||
| G/A | 162 | 189 | 1.66 (1.12-2.45) | 0.0113 | |||
| A/A | 114 | 100 | 2.27 (1.49-3.47) | 0.0002 | |||
| rs1219648 | A/A | 70 | 112 | Reference | 0.0037 | ||
| A/G | 156 | 185 | 1.37 (0.95-1.97) | 0.0936 | |||
| G/G | 106 | 97 | 1.82 (1.21-2.73) | 0.0038 | |||
| rs2420946 | C/C | 69 | 108 | Reference | 0.0052 | ||
| C/T | 155 | 189 | 1.30 (0.90-1.89) | 0.1588 | |||
| T/T | 106 | 97 | 1.78 (1.19-2.68) | 0.0055 | |||
| Arabs | rs11200014 | G/G | 87 | 95 | Reference | 0.2472 | |
| G/A | 155 | 153 | 1.11 (0.77-1.60) | 0.5792 | |||
| A/A | 95 | 82 | 1.27 (0.84-1.93) | 0.2573 | |||
| rs2981579 | G/G | 71 | 88 | Reference | 0.1997 | ||
| G/A | 166 | 149 | 1.39 (0.94-2.04) | 0.0959 | |||
| A/A | 99 | 93 | 1.35 (0.88-2.06) | 0.1703 | |||
| rs1219648 | A/A | 91 | 107 | Reference | 0.2590 | ||
| A/G | 166 | 148 | 1.33 (0.93-1.90) | 0.1197 | |||
| G/G | 80 | 76 | 1.24 (0.82-1.89) | 0.3138 | |||
| rs2420946 | C/C | 87 | 110 | Reference | 0.1323 | ||
| C/T | 167 | 144 | 1.48 (1.03-2.12) | 0.0329 | |||
| T/T | 81 | 77 | 1.33 (0.88-2.03) | 0.18 |
NOTE: Adjusted for age and BRCA mutation status. Different sample sizes for each SNP reflect slightly different genotyping failure rates.
Table 4. Multivariate modeling of breast cancer risk for four SNPs in FGFR2, adjusted for family history of breast cancer in a first-degree relative, age, ethnicity, and interaction terms.
| OR (95% CI) | P | |
|---|---|---|
| rs1219648 | ||
| rs1219648 | 1.23 (1.11-1.37) | <0.0001 |
| First-degree family history | 2.13 (1.04-4.36) | 0.039 |
| Age <50 y | 0.93 (0.56-1.54) | 0.777 |
| Ethnicity | 1.03 (0.79-1.33) | 0.854 |
| rs1219648 with family history interaction | 1.04 (0.73-1.48) | 0.822 |
| rs1219648 with age <50 interaction | 1.02 (0.81-1.28) | 0.895 |
| rs1219648 with ethnicity interaction | 0.96 (0.84-1.09) | 0.522 |
| rs11200014 | ||
| rs11200014 | 1.23 (1.11-1.36) | <0.0001 |
| First-degree family history | 1.84 (0.88-3.81) | 0.104 |
| Age <50 y | 0.88 (0.53-1.47) | 0.623 |
| Ethnicity | 0.96 (0.74-1.25) | 0.753 |
| rs11200014 with family history interaction | 1.12 (0.78-1.61) | 0.528 |
| rs11200014 with age <50 interaction | 1.05 (0.84-1.32) | 0.671 |
| rs11200014 with ethnicity interaction | 0.99 (0.87-1.13) | 0.894 |
| rs2420946 | ||
| rs2420946 | 1.23 (1.11-1.37) | <0.0001 |
| First-degree family history | 2.58 (1.26-5.29) | 0.009 |
| Age <50 y | 0.94 (0.57-1.56) | 0.813 |
| Ethnicity | 0.99 (0.76-1.29) | 0.941 |
| rs2420946 with family history interaction | 0.94 (0.66-1.33) | 0.730 |
| rs2420946 with age <50 interaction | 1.01 (0.80-1.27) | 0.941 |
| rs2420946 with ethnicity interaction | 0.98 (0.86-1.11) | 0.704 |
| rs2981579 | ||
| rs2981579 | 1.26 (1.13-1.39) | <0.0001 |
| First-degree family history | 1.71 (0.82-3.59) | 0.154 |
| Age <50 y | 0.92 (0.54-1.54) | 0.338 |
| Ethnicity | 0.96 (0.73-1.26) | 0.770 |
| rs2981579 with family history interaction | 1.16 (0.81-1.66) | 0.413 |
| rs2981579 with age <50 interaction | 1.03 (0.82-1.30) | 0.807 |
| rs2981579 with ethnicity interaction | 0.99 (0.87-1.12) | 0.828 |
NOTE: All variable estimates shown are adjusted for each element within the SNP table.
We then considered whether young age-of-onset (designated as <50 for this sample) or first-degree family history of breast cancer modified the association of FGFR2 SNPs with risk of breast cancer. There was no significant modification effect of age, first-degree family history of breast cancer, or ethnicity on the association of FGFR2 SNPs with risk of breast cancer (Table 4). In rs1219648, the OR per allele in Ashkenazi Jews compared with OR per allele in Sephardi Jews was not significantly different [ratio of OR, 1.07; 95% confidence interval (95% CI), 0.83-1.38; P = 0.607], suggesting no important heterogeneity of risks for this SNP between Ashkenazi and Sephardi Jews. Similarly, no meaningful difference was observed in the per-allele risk between Ashkenazi Jews and Arabs (ratio of OR, 0.90; 95% CI, 0.70-1.17; P = 0.4354).
Haplotype analysis identified four common haplotypes (five in Sephardi Jews and Arabs) with frequencies greater than 0.01 (Table 5). The common AAGT risk haplotype, identified previously in White non-Hispanic populations, was significantly associated with breast cancer risk in Ashkenazi (OR, 1.25; 95% CI, 1.07-1.45; P = 0.0059) and Sephardi Jews (OR, 1.46; 95% CI, 1.17-1.80; P = 0.0006) compared with the reference GGAC haplotype. Another haplotype with low frequency (AAAC) was significantly associated with breast cancer risk in Sephardi Jews only (OR, 1.97; 95% CI, 1.16-3.35; P = 0.0125). The haplotype AAAC is about four times more frequent in Sephardi Jews and Arabs than in Ashkenazi Jews. The reference GGAC haplotype is the most frequent in Ashkenazi Jews and Arabs but not in Sephardi Jews. None of the haplotypes showed a significant association with breast cancer risk in Arabs, although power is limited in this group and the direction of the associations was consistent with those observed in other populations. The GAGT haplotype, equally frequent among Sephardi Jews and Arabs (0.016 and 0.020, respectively) but less frequent in Ashkenazi Jews (0.008), was also significantly associated with risk of breast cancer in combined data (Table 5).
Table 5. FGFR2 intron 2 haplotypes and association with breast cancer in different ethnic groups.
| Haplotype | Frequency |
OR (95% CI) | P | ||
|---|---|---|---|---|---|
| Total | Cases | Controls | |||
| All | |||||
| GGAC | 0.505 | 0.479 | 0.532 | 1.00 | |
| AAGT | 0.420 | 0.444 | 0.397 | 1.25 (1.12-1.39) | <0.0001 |
| AAAC | 0.029 | 0.031 | 0.027 | 1.28 (0.94-1.73) | 0.1113 |
| GAGT | 0.013 | 0.015 | 0.010 | 1.75 (1.09-2.80) | 0.0210 |
| GGGT | 0.010 | 0.010 | 0.010 | 1.11 (0.66-1.89) | 0.6872 |
| GGAT | 0.010 | 0.009 | 0.011 | 0.91 (0.55-1.52) | 0.7303 |
| Rare <1% | 0.023 | 0.010 | 0.011 | 0.98 (0.68-1.41) | 0.9160 |
| Ashkenazi | |||||
| GGAC | 0.556 | 0.535 | 0.580 | 1.00 | |
| AAGT | 0.393 | 0.417 | 0.367 | 1.25 (1.07-1.45) | 0.0059 |
| GGAT | 0.014 | 0.013 | 0.016 | 0.92 (0.49-1.71) | 0.7841 |
| AAAC | 0.013 | 0.011 | 0.014 | 0.82 (0.41-1.62) | 0.5613 |
| GAGT | 0.008 | 0.011 | 0.005 | 2.26 (0.92-5.54) | 0.0740 |
| Rare <1% | 0.016 | 0.014 | 0.017 | 1.18 (0.72-1.95) | 0.5072 |
| Sephardi | |||||
| GGAC | 0.476 | 0.383 | 0.477 | 1.00 | |
| AAGT | 0.434 | 0.516 | 0.442 | 1.46 (1.17-1.80) | 0.0006 |
| AAAC | 0.041 | 0.051 | 0.032 | 1.97 (1.16-3.35) | 0.0125 |
| GGGT | 0.020 | 0.021 | 0.019 | 1.29 (0.60-2.81) | 0.5160 |
| GAGT | 0.016 | 0.019 | 0.014 | 1.88 (0.80-4.43) | 0.1463 |
| Rare <1% | 0.015 | 0.011 | 0.015 | 0.88 (0.33-2.32) | 0.7964 |
| Arabs | |||||
| GGAC | 0.464 | 0.443 | 0.485 | 1.00 | |
| AAGT | 0.432 | 0.445 | 0.418 | 1.15 (0.92-1.43) | 0.2211 |
| AAAC | 0.051 | 0.055 | 0.046 | 1.31 (0.80-2.14) | 0.2881 |
| GAGT | 0.020 | 0.024 | 0.016 | 1.66 (0.74-3.75) | 0.2212 |
| A-A-G-C | 0.011 | 0.006 | 0.016 | 0.41 (0.12-1.32) | 0.1345 |
| Rare <1% | 0.023 | 0.024 | 0.022 | 1.62 (0.74-3.54) | 0.2255 |
NOTE: The significant association of AAGT haplotype with breast cancer risk was found in Ashkenazi and Sephardi Jews but not in Arabs. The AAAC haplotype was significantly associated with breast cancer in Sephardi Jews only. The reference haplotype GGAC is not the most frequent haplotype in Sephardi Jews.
Adjusted for age and BRCA status. Haplotypes are shown in the physical order: rs11200014, rs2981579, rs1219648, and rs2420946. GAGT haplotype in Ashkenazi Jews is shown for comparison with other ethnic groups.
Discussion
The present study estimates the contribution of FGFR2 polymorphisms to breast cancer risk in different Israeli populations. For this purpose, we genotyped four SNPs (rs11200014, rs2981579, rs1219648, and rs2420946) in intron 2 of FGFR2 gene that were found to be most significantly associated with breast cancer in a previous GWA study (8). In the Nurses′ Health Study, more than 500,000 SNPs were genotyped in a GWA scan of sporadic postmenopausal breast cancer in matched 1,145 cases and 1,142 controls. This study identified an association of four SNPs in intron 2 of FGFR2 gene (rs11200014, rs2981579, rs1219648, and rs2420946) with postmenopausal breast cancer. The SNPs lying outside the intron 2 were not in linkage disequilibrium with these four polymorphisms and were not associated with risk of breast cancer. The authors replicated the results in 1,776 cases and 2,072 controls from three additional studies. The most strongly associated SNP across all four studies was rs1219648 (Ptrend = 1.1 × 10−10 with population attributable fraction = 16%). The most frequent risk haplotype (AAGT) was found significantly associated with breast cancer risk in all populations.
Another large three-stage GWA study in 4,398 breast cancer cases and 4,316 controls followed by confirmation in 21,860 cases and 22,578 controls from 22 studies used more than 200,000 SNPs and found an association of six SNPs in five novel independent loci with risk of breast cancer (7). The most significantly associated SNP (rs2981582) lies within a 25-kb linkage disequilibrium block in intron 2 of FGFR2. This SNP has a r2 of 1.0 with rs1219648 and rs2420946, r2 of 0.97 with rs2981579, and r2 of 0.96 with rs11200014 in the HapMap CEU samples. This high degree of linkage disequilibrium shows close correspondence between the results of both studies. Both studies genotyped White non-Hispanic women of European descent. Four of 22 validation studies in the larger GWA study included women of Asian descent. The association of SNPs in intron 2 of FGFR2 with breast cancer risk was essentially confirmed in all populations participated in these GWA studies but at different degrees of risk for European and Asian populations.
Our study confirms this strong and consistent association and extends these results to other population groups. FGFR2 is a breast cancer susceptibility gene in Ashkenazi and Sephardi Jews and appears to confer modest risk in Arab populations as well. Although the increased risk of breast cancer associated with FGFR2 polymorphisms is modest (rs1219648 OR per allele, 1.25 and 1.32 in Ashkenazi and Sephardi Jews, respectively; Fig. 1), the population attributable fraction is substantial for Ashkenazi Jews (15%) and Sephardi Jews (22%), because the susceptibility alleles differ in prevalence and are very common. The population attributable fraction for FGFR2 among Arabs is also likely to be meaningful (12%), although the relative risk does not quite reach the usual threshold of statistical significance. Genetic variation in FGFR2 appears to be implicated in a considerable portion of breast cancers in Israel and throughout the world.
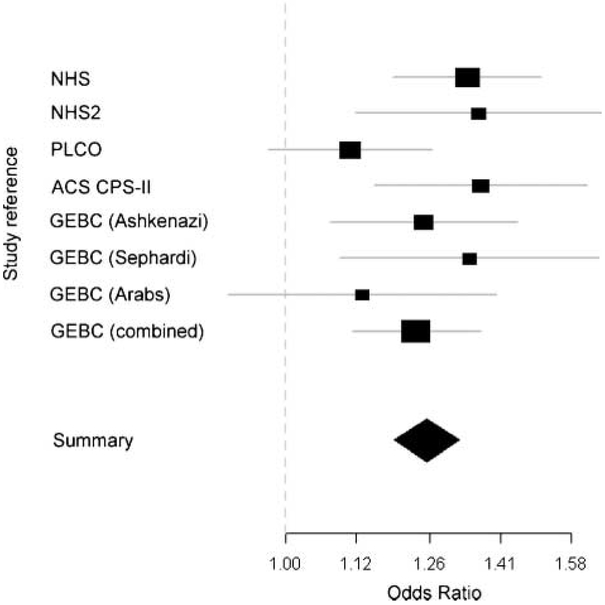
Figure 1.
Meta-analysis forest plot representing per-allele OR for rs1219648 in different studies. The Nurses’ Health Study I and II, PLCO, and ACS CPS-II were used for validation in recently published GWA scan (8). ORs for different ethnic groups are shown for the present study of Genetic Epidemiology of Breast Cancer. The square’s area is proportional to the inverse of the variance of the estimate. Flat lines, 95% CI; diamond, summary OR with 95% CI for all studies.
The haplotype-specific risks are interesting to consider. We unequivocally replicated the association between the AAGT haplotype observed previously in White non-Hispanic women (8). We also evaluated the risk associated with the GAGT haplotype that is reasonably common in Sephardi Jews and Arabs. Although analyses that restrict consideration to each ethnic group do not show significant association with GAGT, there is no evidence of risk heterogeneity by ethnicity for this haplotype (P = 0.1845 interaction for haplotype by Sephardi Jews versus Ashkenazi Jews). Therefore, the summary estimate for GAGT suggests that this haplo-type also may harbor a causal risk variant. There may be value to sequencing these risk haplotypes in Sephardi Jews and Arabs to help identify a causal variant that is not yet appreciated in White non-Hispanic populations.
Given that Arab and Sephardi Jewish populations have comparable size in our study (668 and 727, respectively), we had 80% power to detect OR as low as 1.3 at confidence level of 0.05 in both populations. A statistically significant association with breast cancer risk for this haplotypes was found in Sephardi Jews, with a similar trend toward association in Arabs. Resequencing of this region in different ethnic groups may help to isolate causal alleles. The causal allele linked to the risk haplotypes (AAGT, GAGT, and AAAC) in Ashkenazi and Sephardi Jews may be easier to identify with rare haplotypes in Arabs, and comparison of polymorphic sequences of intron 2 may reveal the functional variant underlying changes in FGFR2 function.
Because the association is restricted to intronic sequence only, possible mechanisms of FGFR2 involvement in breast cancer development include variable regulation of FGFR2 expression and alternative splicing of the gene. The first hypothesis is plausible to consider based on the presence of several transcription-binding sites that lie in the intron 2 of FGFR2 (according to PReMod database (15)). Aberrant expression of nine different isoforms of FGFR2 that arise from alternative splicing has been shown to activate signal transduction and lead to transformation in breast cancer cells (3, 16). Genetic variation in FGFR2 is an important marker of susceptibility to breast cancer at the population level, although its clinical utility has not yet been examined. Along with known susceptibility alleles in BRCA1, BRCA2, PTEN, p53, and possibly CHEK2, FGFR2 polymorphisms may have a clinical predictive value in conjunction with a panel of other markers and should be explored as potential modifiers of high penetrance risk alleles.
Acknowledgments
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Grant support: NCI R01 81488 and Research grants from the Israeli Ministry of Health and Israel Cancer Association.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Grose R, Dickson C. Fibroblast growth factor signaling in tumorigenesis. Cytokine Growth Factor Rev 2005;16:179–86. [DOI] [PubMed] [Google Scholar]
- 2.Moffa AB, Ethier SP. Differential signal transduction of alternatively spliced FGFR2 variants expressed in human mammary epithelial cells. J Cell Physiol 2007;210:720–31. [DOI] [PubMed] [Google Scholar]
- 3.Moffa AB, Tannheimer SL, Ethier SP. Transforming potential of alternatively spliced variants of fibroblast growth factor receptor 2 in human mammary epithelial cells. Mol Cancer Res 2004;2:643−52. [PubMed] [Google Scholar]
- 4.Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev 2005;16:139–49. [DOI] [PubMed] [Google Scholar]
- 5.Adnane J, Gaudray P, Dionne CA, et al. BEK and FLG, two receptors to members of the FGF family, are amplified in subsets of human breast cancers. Oncogene 1991;6:659–63. [PubMed] [Google Scholar]
- 6.Jang JH, Shin KH, Park JG. Mutations in fibroblast growth factor receptor 2 and fibroblast growth factor receptor 3 genes associated with human gastric and colorectal cancers. Cancer Res 2001;61:3541–3. [PubMed] [Google Scholar]
- 7.Easton DF, Pooley KA, Dunning AM, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature 2007; 447:1087–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hunter DJ, Kraft P, Jacobs KB, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet 2007;39:870–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abeliovich D, Kaduri L, Lerer I, et al. The founder mutations 185delAG and 5382insC in BRCA1 and 6174delT in BRCA2 appear in 60% of ovarian cancer and 30% of early-onset breast cancer patients among Ashkenazi women. Am J Hum Genet 1997;60:505–14. [PMC free article] [PubMed] [Google Scholar]
- 10.Rennert G, Bisland-Naggan S, Barnett-Griness O, et al. Clinical outcomes of breast cancer in carriers of BRCA1 and BRCA2 mutations. N Engl J Med 2007;357:115–23. [DOI] [PubMed] [Google Scholar]
- 11.Tonin P, Weber B, Offit K, et al. Frequency of recurrent BRCA1 and BRCA2 mutations in Ashkenazi Jewish breast cancer families. Nat Med 1996;2:1179–83. [DOI] [PubMed] [Google Scholar]
- 12.Figer A, Kaplan A, Frydman M, et al. Germline mutations in the PTEN gene in Israeli patients with Bannayan-Riley-Ruvalcaba syndrome and women with familial breast cancer. Clin Genet 2002; 62:298–302. [DOI] [PubMed] [Google Scholar]
- 13.Koren M, Kimmel G, Ben-Asher E, et al. ATM haplotypes and breast cancer risk in Jewish high-risk women. Br J Cancer 2006;94:1537–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet 2002;70:425–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Blanchette M, Bataille AR, Chen X, et al. Genome-wide computational prediction of transcriptional regulatory modules reveals new insights into human gene expression. Genome Res 2006;16:656–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwabi-Addo B, Ropiquet F, Giri D, Ittmann M. Alternative splicing of fibroblast growth factor receptors in human prostate cancer. Prostate 2001;46:163–72. [DOI] [PubMed] [Google Scholar]