Abstract
Drought is one of the major environmental stresses limiting crop growth and production. MYB family transcription factors play crucial roles in response to abiotic stresses. Previous studies found that TaMYB31 is transcriptionally induced by drought stress. However, the biological functions of TaMYB31 in drought stress responses remained unknown. In this study, three TaMYB31 homoeologous genes from hexaploid wheat, designated TaMYB31-A, TaMYB31-B, and TaMYB31-D, were cloned and characterized. Expression analysis showed that TaMYB31 genes have different tissue expression patterns, and TaMYB31-B has relatively high expression levels in most tested tissues. All the three homoeologs were up-regulated by polyethylene glycol (PEG) 6000 and abscisic acid (ABA) treatments. Subcellular localization analyses revealed that TaMYB31 is localized to the nucleus. Ectopic expression of the TaMYB31-B gene in Arabidopsis affected plants growth and enhanced drought tolerance. In addition, seed germination and seedling root growth of TaMYB31-B transgenic plants were more sensitive to exogenous ABA treatment compared to wild type control. RNA-seq analysis indicated that TaMYB31 functions through up-regulation of wax biosynthesis genes and drought-responsive genes. These results provide evidence that TaMYB31 acts as a positive regulator of drought resistance, and justify its potential application in genetic modification of crop drought tolerance.
Keywords: wheat, MYB, drought, transgenic, Arabidopsis, RNA-seq
Introduction
Drought is a major environmental stress that limits crop growth and production. A large proportion of agriculturally productive areas worldwide have been seriously affected by enduring droughts, which have led to serious damage to crops (Kudo et al., 2017). It was calculated that drought has caused an average of 13.7% reduction in cereal production worldwide over the past few decades (Lesk et al., 2016). Moreover, the world population continues to grow rapidly, therefore, to sustain such a large population, it is essential to enhance crop drought tolerance. Unraveling the molecular and biochemical mechanisms which enable plants to cope with environmental challenges is of great importance to improve stress tolerance and yield (Hrmova and Lopato, 2014).
The molecular mechanisms underlying plant drought tolerance have been investigated intensively. In fact, plants respond to drought via various dynamic regulatory networks, in which transcription factors (TFs) play important roles (Krasensky and Jonak, 2012). A single TF could activate and/or repress the expression of a number of target genes through direct binding to its specific DNA recognition sequence, also known as cis-acting element, present in the promoters of its downstream genes. So far, several TF families, such as MYB, WRKY, AP2/ERF, NAC, and HD-ZIP, have been reported to act as master regulators in response to drought stress in plants (Sakuma et al., 2006; Ding et al., 2009; Nuruzzaman et al., 2013; Banerjee and Roychoudhury, 2017).
MYB TFs constitute one of the largest TF families and have conserved MYB DNA-binding domain containing 52 amino acids. The MYB TF family consists of four major subfamilies differing in the number of imperfect repeat of R1-R3 DNA-binding domains: 1R-MYB or MYB-related proteins, R2R3-MYB, 3R-MYB (R1R2R3-MYB), and 4R-MYB. In particular, R2R3-MYB TFs regulate various plant developmental processes, such as epidermal cell, cuticle, trichome and stomata formation, as well as a series of environmental stresses including drought (Dubos et al., 2010). For example, increased expression of Arabidopsis MYB15 resulted in enhanced drought and salt tolerance and hypersensitivity of transgenic Arabidopsis to exogenous ABA (Ding et al., 2009). Overexpression of the drought-induced AtMYB41 improved drought tolerance in transgenic Arabidopsis (Cominelli et al., 2008; Lippold et al., 2009). AtMYB96 was demonstrated to play important roles in increasing cuticular wax deposition and improving drought tolerance in Arabidopsis (Seo et al., 2009, 2011). Overexpression of the rice OsMYB48-1 conferred drought tolerance to transgenic rice plants (Xiong et al., 2014). Heterologous expression of OsMYB55 led to improved tolerance to drought and heat stresses in transgenic maize (Casaretto et al., 2016).
In our previous work, six cuticle biosynthesis-related wheat MYB genes were cloned. As an ortholog of AtMYB96, the drought inducible TaMYB31 has been suggested to be involved in the regulation of drought responses (Bi et al., 2016). However, the biological function of TaMYB31 has not yet been investigated. In this study, we cloned TaMYB31 homoeologous genes (TaMYB31-A, TaMYB31-B, and TaMYB31-D) from hexaploid wheat, analyzed their expression patterns, and investigated their functions via heterologous overexpression and RNA-seq analysis. The three homoeologous genes displayed differential expression in various wheat organs, and ectopic overexpression of TaMYB31-B in Arabidopsis increased drought tolerance. The improved drought tolerance was at least partly due to up-regulation of wax biosynthesis genes and drought responsive genes. Our results shed light on the potential value of TaMYB31 in genetic improvement of crop drought tolerance.
Materials and methods
Plant materials and growth conditions
The hexaploid wheat Chinese Spring and three diploid ancestor wheats, the Triticum urartu accession UR206, the Aegilops tauschii accession Y2282 and the Aegilops speltoides accession Y2006, were used to isolate the genomic and ORF sequences of TaMYB31 genes. For gene expression analyses under abiotic stresses, 10-day-old wheat seedlings were treated with 20% polyethylene glycol (PEG) 6000 and 200 μM ABA, and harvested at 0, 0.5, 1, 2, 4, 6, 12, and 24 h. Each sampled material was immediately rinsed in sterile distilled water, dried with filter papers, then frozen in liquid nitrogen and stored at −80°C for further analyses. To analyze tissue-specific expression patterns, roots, stems, leaves, young spikes, and flowering spikes were harvested from Chinese Spring wheat and stored at −80°C.
The Arabidopsis genotype Col-0 was used to generate TaMYB31-B transgenic plants. The plants were grown in soil in a greenhouse with a constant 22°C, 50–70% relative humidity and a 16/8 h light/dark cycle at 100 μmol m−2s−1 or on 1/2MS medium. Images of rosette leaves were taken after 4 weeks grown in soil and rosette diameter was measured using ImageJ software (http://imagej.nih.gov/ij/). Plant height and fresh weight were recorded at mature period.
Isolation of ORF and genomic sequences
Genomic sequences of TaMYB31 homoeologous genes were retrieved from IWGSC database1 using TaMYB31 ORF sequence (GenBank: KU674897.1) as a query. The acquired sequences were then used to design primers. Genomic DNA was extracted from young leaves using the CTAB method. The PCR conditions for amplifying TaMYB31s were as follows: 5 min at 98°C; 36 cycles of 20 s at 98°C, 20 s at 60°C, and 90 s at 72°C; and then 5 min at 72°C. The obtained purified PCR products were cloned into the pEASY-Blunt (TransGen, Beijing, China) cloning vector for sequencing. Total RNA was isolated from leaves using the TRIzol reagent following the manufacturer's protocol (TransGen, Beijing, China) and followed by treatment with DNase I (TaKaRa, Dalian, China). cDNA synthesis was conducted with 2 micrograms of purified total RNA using M-MLV reverse transcriptase (Promega, Madison, USA) according to the manufacturer's instructions. Based on the acquired genomic sequences, primers were designed to amplify the TaMYB31 ORF sequences from wheat cv. Chinese Spring. All of the primers used are listed in Table S1.
Primers were designed using DNAMAN software2 Multiple sequence alignment of MYB proteins obtained in this study was performed with ClustalW (Larkin et al., 2007). Phylogenetic tree was constructed using MEGA 6.0 software (Tamura et al., 2013) with the neighbor-joining (NJ) method.
Quantitative RT-PCR
The quantitative RT-PCR analyses were performed using the SYBR Mix ExTaq II kit (TaKaRa, Dalian, China) and the Bio-Rad CFX96 real-time system3 The PCR parameters were as follows: 95°C for 4 min, followed by 40 cycles of 94°C for 20 s, 60°C for 20 s and 72°C for 20 s. For expression analysis of TaMYB31 genes in wheat, the wheat β-actin gene was selected as an internal control (Hu et al., 2013). AtActin2 gene was used as an internal control for gene expression in Arabidopsis. Three biological replicates and three technical replicates were used in all gene expression experiments. The relative gene expression were calculated based on the comparative CT method (Livak and Schmittgen, 2001). The primers for quantitative RT-PCR are listed in Table S1.
Subcellular localization analysis
The GFP ORF sequence was introduced in frame to the 3′ end of the TaMYB31-B gene sequence, and the resulting gene fusion was subcloned into the pCAMBIA1300 vector for transient expression in leaf epidermal cells of Nicotiana benthamiana. After agro-infiltration, plants were allowed to grow for 2 days at 22°C with a photoperiod of 16/8 h light/dark before visualization under a confocal laser-scanning microscope (FluoView™ FV1000; Olympus).
Arabidopsis transformation and stress treatments
TaMYB31-B ORF was amplified using primers TaMYB31-OE-F and TaMYB31-OE-R (containing attB sites) and cloned into the Gateway plant expression vector pB2GW7 to generate p35S:: TaMYB31-B. The transformation of the generated construct into Arabidopsis was performed via A. tumefaciens (strain GV3101) mediated floral-dip method (Clough and Bent, 1998). The transgenic seedlings were confirmed by PCR and quantitative RT-PCR.
For the drought stress treatment, water was withheld for 2 weeks from 3-week-old plants with similar size grown under normal conditions in soil in a growth room, afterwards, rewatering was applied and survival rates were measured 3 days after rewatering. For water loss rate assays, leaves were detached from 4-week-old plants grown under normal conditions, and placed on filter paper at room temperature for 1, 2, 3, 4, 5, and 6 h, and then fresh weights of the leaves at each time point were measured as described previously (Zhao et al., 2017b). Three independent experiments were performed.
For the PEG stress treatment, the seeds of wild type and three homozygous T3 TaMYB31-B transgenic lines were grown on 1/2MS media in a growth chamber. After 7 days, seedlings were transferred onto 1/2MS media with or without 10% PEG6000. After another 7 days, the plates with seedlings were photographed and the fresh weights and root lengths were measured. Seedlings were grown vertically during the entire experiment. Three independent experiments were performed.
For the seed germination assay in response to ABA, WT and transgenic seeds were sterilized and germinated on 1/2MS agar plates without or with 0.3, 0.5, or 1.0 μM ABA. After 4 days of growth, the plates with seeds/seedlings were photographed and seed germination rates were calculated. Seed germination was defined as emergence of the radicle tip through the endosperm. At least 60 seeds were used for each replicate of three independent replicates. Root elongation rate assay was performed on 1/2MS media supplemented with 0 or 10 μM ABA as described in Ling et al. (2017).
RNA-Seq analysis
Leaves were collected from 4-week-old transgenic L2 and WT plants under drought conditions (water was withheld for 1 week from 3-week-old plants). Total RNA was extracted using Trizol reagent (TransGen, Beijing, China) and RNA integrity was checked using a Bioanalyzer 2100 (Agilent, California, USA). cDNA synthesis, libraries construction, sequencing, and raw reads analysis were performed as previously described (Zhao et al., 2017a). Approximately 2 GB clean data were produced for each sample. The clean reads generated were aligned against the reference Arabidopsis genome using TopHat2 (Kim et al., 2013). Gene expression was calculated using the Reads Per Kilobase per Million mapped reads (RPKM) method (Mortazavi et al., 2008). Gene Ontology enrichment was analyzed using the agriGO analysis tool (Tian et al., 2017). RNA-Seq data were submitted to NCBI and can be accessed under the SRA accession number SRP157487.
Results
Molecular characterization of the TaMYB31 homoeologous genes
To comprehensively understand the TaMYB31 gene, the genomic and ORF sequences of MYB31 were isolated from the diploid wheat UR206 (AA), Y2006 (SS), and Y2282 (DD) as well as hexaploid wheat Chinese Spring (BBAADD). Three homoeologs of TaMYB31 were amplified from the hexaploid wheat, and sequence alignment indicated that the six MYB31s (three from diploid wheat and three from hexaploid wheat) obtained shared 98.4–100% identity at the amino acid level (Figure 1A). The GenBank accession numbers of the six MYB31s are MH428951-MH428956.
Figure 1.
Sequence analysis of TaMYB31 homologs. (A) Comparison of amino acid sequences of seven MYB31 proteins, i.e., hexaploid wheat TaMYB31-A/B/D (GenBank: MH428951-53), Triticum urartu TuMYB31 (GenBank: MH428954), Aegilops speltoides AsMYB31 (GenBank: MH428955), Aegilops tauschii AetMYB31 (GenBank: MH428956), and AtMYB96 (AT5G62470). The SANT domain is underlined. (B) Phylogenetic and gene structure analysis of TaMYB31 homoeologs, three diploid ancestral wheat MYB31 genes and AtMYB96. Exons and introns are represented by black boxes and lines, respectively.
Based on the sequence alignment, the three TaMYB sequences were assigned to the A, B, and D genomes and accordingly named as TaMYB31-A, TaMYB31-B, and TaMYB31-D, respectively (Figure 1B). Gene structure analyses using the genomic sequences and the cDNA sequences of TaMYB31s revealed that the three TaMYB31 homoeologs, TaMYB31-A, TaMYB31-B, and TaMYB31-D, contain three exons and two introns (Figure 1B).
Expression analysis of TaMYB31 genes in wheat
TaMYB31 was demonstrated to be induced by drought stress (Bi et al., 2016), implying that this gene might play a role in the regulation of plant responses to drought stress. To unravel the biological function of the TaMYB31 homoeologous genes, we first examined the tissue-specific expression of TaMYB31 genes in various tissues of hexaploid wheat Chinese Spring, including roots, stems, leaves, young spikes and flowering spikes. As shown in Figure 2A, TaMYB31 genes were strongly expressed in leaves, young spikes, and flowering spikes. Notably, TaMYB31-A, TaMYB31-B, and TaMYB31-D displayed different expression patterns, suggesting that they may play diverse roles at various developmental stages. TaMYB31-B had the highest transcript level in most of the tissues examined (Figure 2A), indicating that it might play a more significant regulatory role on plant development than TaMYB31-A and TaMYB31-D.
Figure 2.

Expression patterns of TaMYB31s. (A) TaMYB31s expression in various wheat tissues by quantitative RT-PCR. YS, young spikes and FS, flowering spikes. Data are presented as means of three replicates ± SD. (B,C) Expression of TaMYB31s under PEG (B) and ABA treatment (C) for the indicated time points by quantitative RT-PCR. Data are presented as means of three replicates ± SD.
Next, the expression patterns of TaMYB31 genes were analyzed by quantitative RT-PCR under stress conditions. Under a PEG treatment, the expression of the TaMYB31 genes increased gradually and TaMYB31-A and TaMYB31-B peaked at 12 h, while TaMYB31-D peaked at 6 h (Figure 2B). As for ABA treatment, the expression of TaMYB31-A and TaMYB31-D were gradually up-regulated and peaked at 12 h (Figure 2C). The transcript of TaMYB31-B under ABA treatment reached a high level at 2 h and then declined rapidly and reached the untreated level at 6 h; however, TaMYB31-B expression increased again and reached the highest level at 12 h. In addition, the expression levels of the genes after ABA treatment appeared to be lower than that after PEG treatment. Based on the expression analyses of TaMYB31s in wheat tissues and under stress treatments, the TaMYB31-B gene was selected for further functional analysis.
Subcellular location of the TaMYB31-B protein
Nuclear localization is necessary for transcription factors to execute their functions. Subcellular localization of TaMYB31-B was investigated through transient transformation in Nicotiana benthamiana leaf epidermal cells. We constructed the fusion protein of TaMYB31-B with green florescent protein (GFP), and the resulting constructs containing TaMYB31-B-GFP and GFP, respectively, were introduced to Nicotiana benthamiana leaf epidermal cells. As shown in Figure 3, TaMYB31-B protein was localized primarily in the nucleus, while GFP was observed in both nucleus and cytosol (Figure 3).
Figure 3.
Subcellular localization of TaMYB31-B in Nicotiana benthamiana leaf epidermal cells. 35S::GFP and 35S::TaMYB31-B-GFP plasmids were transformed into Nicotiana benthamiana leaf epidermal cells and GFP fluorescence were visualized under a confocal laser-scanning microscope. From left to right, GFP fluorescence (A,D), bright-field (B,E), and overlays of the GFP fluorescence and bright-field (C,F). Bar = 40 μm.
Overexpression of TaMYB31-B in Arabidopsis
To further dissect the biological function of TaMYB31-B, transgenic Arabidopsis plants overexpressing TaMYB31-B driven by 35S promoter were generated (Figure 4A); A total of 10 independent transgenic plants were obtained, and various levels of TaMYB31-B gene expression were detected in T3 homozygous lines (Figure 4B). Three transgenic lines designated L1, L2, and L3 with different expression levels of TaMYB31-B were selected for further phenotypic assays. Under normal growth conditions, the TaMYB31-B transgenic plants were smaller than the wild-type plants (Figure 4C). The rosette diameter after 4 weeks of growth in soil was smaller for L1, L2, and L3 compared with the wild type (Figure 4D). Plant height and fresh weight of mature plants were reduced in the transgenic lines (Figure 4D). Notably, line L1, which had the highest expression level of TaMYB31-B among the three examined lines, had the smallest plant size, while L3 with the lowest transcript level of TaMYB31-B had the largest plant size. These results indicated that altered TaMYB31-B expression affects plant growth and development under normal growth conditions, possibly in a dose-dependent manner.
Figure 4.
Generation of transgenic Arabidopsis plants overexpressing TaMYB31-B. (A) Schematic diagram of the 35S::TaMYB31-B construct. LB left border, p35S cauliflower mosaic virus 35S promoter, Tnos nopaline synthase gene (NOS) terminator, Bar bialaphos-resistance gene, RB right border. (B) Transcript levels of TaMYB31-B in the leaves of T3 transgenic Arabidopsis lines via quantitative RT-PCR analysis. Bars indicate standard deviations of three replicates. WT non-transgenic plants, N.D, Not Detected. (C) Phenotypes of TaMYB31-B transgenic plants. Bar = 2 cM. (D) Statistical analyses of rosette diameter, plant height and fresh weight of mature plants grown in soil. * and ** indicate significant differences at P < 0.05 and P < 0.01 levels, respectively, by Student's t-test.
Overexpression of TaMYB31-B increases drought tolerance
To evaluate the impact of TaMYB31-B overexpression on the drought tolerance of plants, watering was terminated for 3-week-old transgenic (L2 and L3) and wild type (WT) plants with similar size grown in soil. After 2 weeks of water deprivation, the two TaMYB31-B transgenic lines exhibited better growth status than WT plants (Figure 5A). The plants were then rewatered and allowed to grow for 3 days when plant survival rates were measured. The transgenic plants had significantly higher survival rates than WT plants (Figure 5B).
Figure 5.
Drought tolerance responses of TaMYB31-B overexpressing transgenic lines. (A) Images of drought tolerance assays and (B) survival rates of WT and TaMYB31-B overexpressing lines of the T3 generation. Watering was withheld for two weeks and re-applied for 3 days. (C) Water loss assays in detached leaves of WT and transgenic lines. **Indicates significant differences at P < 0.01 level by Student's t-test.
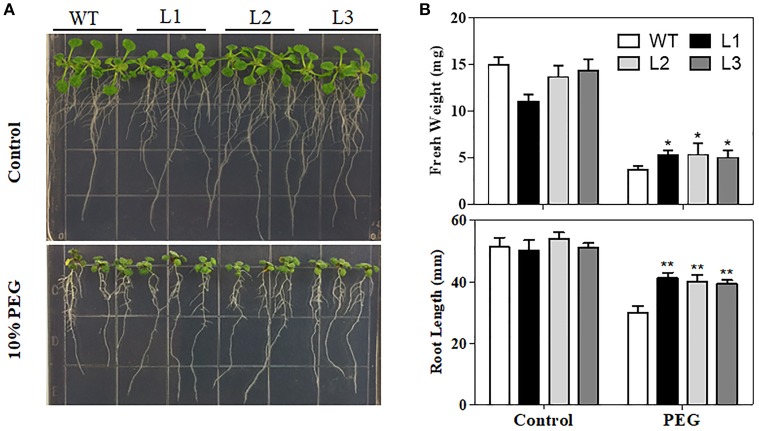
Water loss rates of detached leaves have been widely used to reflect drought tolerance in plants. Water loss rates were assessed every hour at room temperature for both transgenic plants and control plants. As shown in Figure 5C, the two transgenic lines exhibited lower rates of water loss than the control plants. Furthermore, PEG treatment, as a routine dehydration treatment, was applied; transgenic plants displayed a more tolerant phenotype in seedling growth compared to WT plants under PEG treatment (Figure 6A). The three TaMYB31-B transgenic lines showed higher fresh weight and longer root length than the WT under PEG conditions (Figure 6B). Taken together, these results demonstrated that TaMYB31-B positively regulates plant response to drought stress.
Figure 6.
Phenotypes of WT and TaMYB31-B transgenic plants under PEG stress. (A) Images of WT and transgenic lines under PEG treatment. (B) Fresh weight and root lengths of Arabidopsis seedlings. Data are presented as means ± SD of three replicates (*P < 0.05, **P < 0.01).
Transgenic Arabidopsis plants are hypersensitive to exogenous ABA
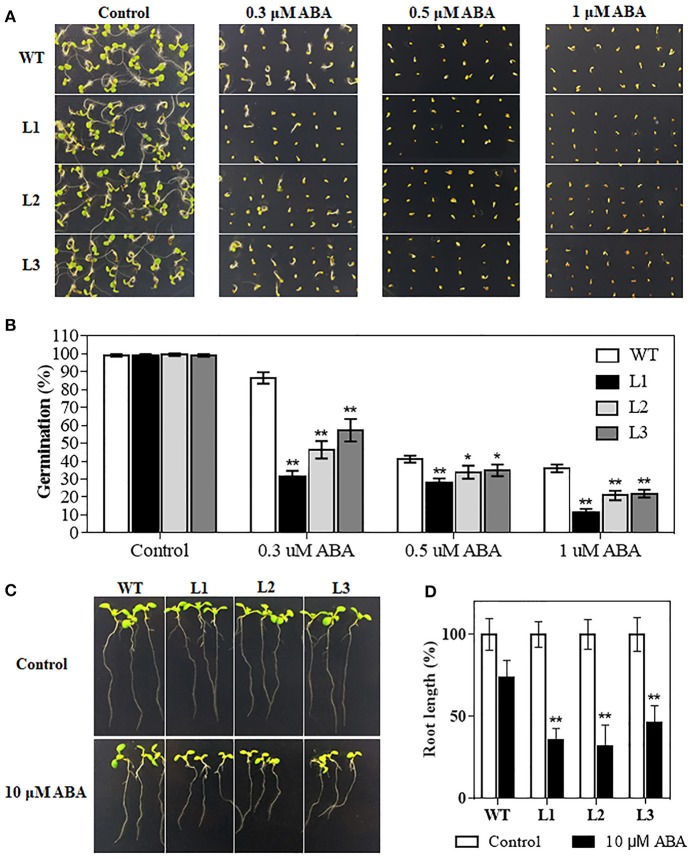
The plant hormone ABA mediates plant development and various abiotic stresses. TaMYB31-B expression was significantly induced by ABA (Figure 2C). We therefore investigated whether TaMYB31 plays a role in ABA signaling pathway. Homozygous transgenic lines (L1, L2, and L3) and WT plants were selected and treated with exogenous ABA. As shown in Figures 7A,B, germination of the transgenic lines and WT was similar on ABA-free medium (Control), whereas transgenic lines had inferior germination on media containing 0.3, 0.5, or 1μM ABA compared to WT plants. Furthermore, the transgenic seedlings grown on 10 μM ABA showed reduced root elongation compared with that of the WT seedlings (Figures 7C,D). These observations indicated that TaMYB31-B overexpressing plants are hypersensitive to ABA during seed germination and seedling root growth.
Figure 7.
Seed germination rates analysis of WT and TaMYB31-B transgenic lines under ABA treatment. (A) Seeds of WT and three T3 TaMYB31-B transgenic lines grown on 1/2MS agar plates with the indicated concentrations of ABA. The photographs were taken 4 d after sowing. (B) Seed germination rates recorded before photographing. (C) Morphology of WT and transgenic seedlings on plates without or with 10 μM ABA. Images were taken 3 d after seedlings were transferred to the shown plates. (D) Root elongation rates of WT and transgenic seedlings at 3 d after being transferred to the plates without or with 10 μM ABA. Error bars indicate SD of three independent experiments (*P < 0.05, **P < 0.01).
Transcriptome analysis of TaMYB31-B transgenic Arabidopsis
To decipher the possible mechanism of TaMYB31 in regulating Arabidopsis drought tolerance, comparative gene expression analysis was conducted in transgenic and WT plants using RNA-seq. A total of 359 genes were found to be differentially expressed (fold change >2, false discovery rate <0.01). Of these, 238 genes were up-regulated and 121 were down-regulated in transgenic plants (Figure 8A; Table S2). To obtain the functional information of these genes, we searched for gene ontology (GO) terms using agriGO web-based tool and database. Most of the differentially expressed genes were associated with developmental process, lipid metabolism, and responses to stress (Figure 8B). In particular, a group of genes associated with wax biosynthesis in plants, including WIN1/SHN1, CYP96A15, FAR3, CER1-L1, and WSD1 (Table 1) (Broun et al., 2004; Rowland et al., 2006; Greer et al., 2007; Li et al., 2008; Wang et al., 2018), and those that are involved in water stress responses, including LTP3, WIN1/SHN1, KIN1, and LEA were up-regulated in transgenic plants (Table 1). Interestingly, CYP707A3 was found to be down-regulated in transgenic plants, which is involved in the abscisic acid degradation pathway.
Figure 8.
Transcriptome analysis of the 35S::TaMYB31-B transgenic Arabidopsis under drought conditions. (A) Number of up- or down-regulated genes in transgenic plants compared with WT plants using a significant cutoff of false discovery rate (FDR) < 0.01, and a fold change (FC) > 2. (B) Gene ontology classification for differentially expressed genes (DEGs) between WT and TaMYB31-B transgenic plants. (C) Confirmation of DEGs expression by quantitative RT-PCR analysis. Error bars represent SD of three replicates.
Table 1.
The list of DEGs in 35S::TaMYB31-B transgenic Arabidopsis that are highlighted in this paper.
| Gene ID | Gene symbol | log2FC | Profile | Description | Function |
|---|---|---|---|---|---|
| AT5G59310 | LTP4 | 5.29 | UP | Non-specific lipid-transfer protein 4 | lipid metabolism, drought/ABA responses |
| AT2G21490 | LEA | 4.87 | UP | Probable dehydrin LEA | drought/ABA responses |
| AT5G59320 | LTP3 | 4.60 | UP | Non-specific lipid-transfer protein 3 | lipid metabolism, drought/ABA responses |
| AT4G33355 | LTP11 | 3.84 | UP | Non-specific lipid-transfer protein 11 | lipid metabolism |
| AT1G57750 | CYP96A15 | 3.35 | UP | Alkane hydroxylase MAH1 | wax biosynthesis |
| AT1G15360 | WIN1/SHN1 | 2.84 | UP | WAX INDUCER1/SHINE1 | wax biosynthesis, drought/ABA responses |
| AT4G33790 | FAR3 | 1.81 | UP | Fatty acyl-CoA reductase 3 | wax biosynthesis |
| AT3G51590 | LTP12 | 1.70 | UP | Non-specific lipid-transfer protein 12 | lipid metabolism |
| AT5G37300 | WSD1 | 1.64 | UP | O-acyltransferase WSD1 | wax biosynthesis |
| AT4G12470 | AZI1 | 1.60 | UP | pEARLI1-like lipid transfer protein 1 | lipid metabolism |
| AT1G02190 | CER1-L1 | 1.28 | UP | CER1-like 1 | wax biosynthesis |
| AT5G15960 | KIN1 | 1.06 | UP | Stress-induced protein KIN1 | drought/ABA responses |
| AT2G44080 | ARL | −1.17 | DOWN | ARGOS-like protein | regulates lateral organ size |
| AT1G73330 | ATDR4 | −1.24 | DOWN | drought-repressed 4 | drought response |
| AT5G45340 | CYP707A3 | −1.88 | DOWN | Abscisic acid 8'-hydroxylase 3 | drought/ABA responses |
Several wax biosynthesis and stress-related genes differentially expressed in our transcriptome data were further validated using quantitative RT-PCR. As shown in Figure 8C, the expression levels of LTP3, WIN1, and FAR3 in the transgenic lines were significantly higher than that in control; in contrast, the expression levels of CYP707A3 in the transgenic lines were significantly lower than that in control. The fold changes in gene expression determined by quantitative RT-PCR were in accordance with the fold changes of normalized expression level (FPKM) by RNA-Seq.
Discussion
Common wheat (Triticum aestivum, BBAADD) is an allohexaploid species originating from two independent hybridization events involving three diploid species, Triticum urartu (AA), Aegilops speltoides (SS≈BB), and Aegilops tauschii (DD) (Gill and Friebe, 2002). Thus, there are generally three homoeologous genes present in common wheat; a large quantity of these homoeologs exhibit expression partitioning (Liu et al., 2015). In the course of evolution, the triplicate homoeologs have three possible fates: preservation of original or similar function, functional diversification, and gene silencing (Wendel, 2000). Functional diversification of homoeologous genes is one of the driving factors leading to the successful evolution of polyploidy species (Shitsukawa et al., 2007). In the present study, the three TaMYB31s were differentially expressed; TaMYB31-B displayed the highest transcript level in almost all of the tissues examined, especially in flowering spikes, while TaMYB31-A and TaMYB31-D were less expressed, especially in wheat roots (Figure 2A). Temporal and spatial gene expression is generally associated with specific developmental and physiological functions. Our results suggested that TaMYB31-B may play a role in the development of flowering spikes. In addition, we found that TaMYB31-A has a rapid response to ABA treatment. These results indicated that functional diversification might have occurred in TaMYB31s. Future investigation should focus on analyzing the promoters of the three TaMYB31 genes to unravel the regulatory mechanism underlying their specific expression patterns.
Many MYBs participate in various stress-related responses in plants. Though more than 84 MYB genes have been found in wheat, only a few of them had been functionally analyzed, such as TaMYB1 (Lee et al., 2007), TaMYB2A (Mao et al., 2011), TaMYB19-B (Zhang et al., 2014), TaMYB30-B (Zhang et al., 2012), TaMYB33 (Qin et al., 2012), TaMYB74 (Bi et al., 2016), and TaMYB80 (Zhao et al., 2017b). In this study, we found that TaMYB31 genes were up-regulated in wheat leaves by PEG and ABA treatments (Figures 2B,C), implying a potential role of TaMYB31 genes in stress responses. Indeed, TaMYB31-B transgenic Arabidopsis plants exhibited enhanced tolerance to drought stress in soil as well as to dehydration stress simulated by PEG treatment (Figures 5, 6).
To reveal possible regulatory mechanisms of TaMYB31-B in response to drought stress, transcriptome analysis of WT and L2 was conducted using RNA-Seq. The results showed that the genes involved in lipid metabolism were up-regulated in TaMYB30-B overexpressing lines under drought treatment, such as lipid-transfer protein 3 (LTP3), LTP4, LTP11, and LTP12 (Table 1). Plant LTPs participate in the transportation of cuticular waxes during the assembly of plant cuticle (Yeats and Rose, 2008). Arabidopsis LTP3 was shown to be the direct target of the AtMYB96; overexpression of LTP3 resulted in a constitutively enhanced tolerance to drought and freezing stresses (Guo et al., 2013). The authors speculated that LTP3 might bind and deliver lipids from cytosol to the cell wall to synthesize cuticular waxes, which protect plants against biotic and abiotic stresses, including drought. In addition, the expression levels of several wax biosynthesis genes, including WIN1/SHN1, CYP96A15, FAR3, CER1-L1, and WSD1, were up-regulated in TaMYB31-B overexpressing plants under drought treatments (Table 1). Arabidopsis WIN1/SHN1 was the first TF whose role in the regulation of cuticle biosynthesis was experimentally demonstrated; overexpression of WIN1/SHN1 genes activates cuticular wax biosynthesis, reduces water loss and increases drought tolerance in transgenic Arabidopsis (Aharoni et al., 2004), rice (Wang et al., 2012) and wheat (Bi et al., 2018). The above results suggested that TaMYB31 shared similar functions with its homolog AtMYB96. It was shown that overexpression of the ABA- and drought-inducible AtMYB96 activates the expression of wax biosynthesis genes and increases the deposition of cuticular waxes, which consequently leads to improved drought tolerance in Arabidopsis (Seo et al., 2009, 2011).
It is noteworthy that overexpression of TaMYB31-B causes severe morphological defects such as dwarfed stature under normal conditions. Previous research showed that ARGOS-LIKE (AtARL) regulates lateral organ size (Hu et al., 2006). Increased AtARL expression was shown to enlarge organ size, while reduced AtARL expression led to organ reduction. We found that AtARL was down-regulated in TaMYB31-B transgenic plants (Table 1), suggesting that dwarfed stature observed here might result from the deceased expression of AtARL by TaMYB31-B. In addition, ABA regulates many developmental processes and mediates adaptive responses to environmental stresses. Among others, ABA plays a negative role in seed germination and seedling growth (Koornneef et al., 2002), while promoting tolerance to abiotic stresses including drought (Fujita et al., 2011). It is commonly accepted that the balance between ABA biosynthesis and catabolism determines its action (Seiler et al., 2011). In this study, the expression levels of TaMYB31s were increased in response to ABA treatment. Moreover, overexpression of the TaMYB31-B gene increased ABA sensitivity in seed germination. RNA-seq data revealed that the expression of CYP707A3, a cytochrome P450 CYP707A family gene involved in ABA catabolism and determining threshold levels of ABA during dehydration-rehydration response (Umezawa et al., 2006), is markedly reduced in TaMYB31-B transgenic plants (Table 1). Taken together, our results suggest that the growth retardation of TaMYB31-B, as well as the improved tolerance to drought are at least partly attributed to ABA.
Although a variety of transgenic crops with drought tolerance have been generated, many of these transgenics show growth retardation (Todaka et al., 2015). Growth retardation is commonly present in transgenic plants which tremendously limits the potential utilization of candidate genes in crop breeding (Mao et al., 2011). To reduce such negative effects, stress inducible promoters have been utilized to subtly control the expression of target genes in transgenic plants (Pino et al., 2007). Recently, it has been reported that application of a gene stacking approach with two transcription factor genes DREB1A and OsPIL1 overcomes growth retardation in drought-tolerant transgenic plants (Kudo et al., 2017). In the present study, TaMYB31-B overexpression plants in Arabidopsis also showed growth retardation under normal growth conditions. This inspires us to use stress-inducible promoters or the gene stacking approach when utilization of TaMYB31-B in crop drought improvement.
Author contributions
YZ carried out the experiments, analyzed the results, and drafted the manuscript. XC and HX provided the idea and instructed the research work. XL and HW provided assistance to perform experiments and collect data. HB designed the experiments and revised the manuscript. All authors have read and approved the final manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Professor Zhongfu Ni and Professor Huiru Peng from China Agricultural University for kindly offering the wild diploid wheat seeds. This work was supported by the National Transgenic Research Project (2016ZX08002003-004), National Key Basic Research Program (973 Program, 2014CB138105), and PhD Scientific Research Start-up Program (30500653) of Henan Agricultural University.
Funding. This work was supported by the National Transgenic Research Project (2016ZX08002003-004), National Key Basic Research Program (973 Program, 2014CB138105), and PhD Scientific Research Start-up Program (30500653) of Henan Agricultural University.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01426/full#supplementary-material
References
- Aharoni A., Dixit S., Jetter R., Thoenes E., van Arkel G., Pereira A. (2004). The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 16, 2463–2480. 10.1105/tpc.104.022897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A., Roychoudhury A. (2017). Abscisic-acid-dependent basic leucine zipper (bZIP) transcription factors in plant abiotic stress. Protoplasma 254, 3–16. 10.1007/s00709-015-0920-4 [DOI] [PubMed] [Google Scholar]
- Bi H., Luang S., Li Y., Bazanova N., Morran S., Song Z., et al. (2016). Identification and characterization of wheat drought-responsive MYB transcription factors involved in the regulation of cuticle biosynthesis. J. Exp. Bot. 67, 5363–5380. 10.1093/jxb/erw298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi H., Shi J., Kovalchuk N., Luang S., Bazanova N., Chirkova L., et al. (2018). Overexpression of the TaSHN1 transcription factor in bread wheat leads to leaf surface modifications, improved drought tolerance and no yield penalty under controlled growth conditions. Plant Cell Environ. [Epub ahead of print]. 10.1111/pce.13339 [DOI] [PubMed] [Google Scholar]
- Broun P., Poindexter P., Osborne E., Jiang C. Z., Riechmann J. L. (2004). WIN1, a transcriptional activator of epidermal wax accumulation in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 101, 4706–4711. 10.1073/pnas.0305574101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casaretto J. A., El-Kereamy A., Zeng B., Stiegelmeyer S. M., Chen X., Bi Y. M., et al. (2016). Expression of OsMYB55 in maize activates stress-responsive genes and enhances heat and drought tolerance. BMC Genomics 17:312. 10.1186/s12864-016-2659-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. 10.1046/j,1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Cominelli E., Sala T., Calvi D., Gusmaroli G., Tonelli C. (2008). Over-expression of the Arabidopsis AtMYB41 gene alters cell expansion and leaf surface permeability. Plant J. 53, 53–64. 10.1111/j.1365-313X.2007.03310.x [DOI] [PubMed] [Google Scholar]
- Ding Z., Li S., An X., Liu X., Qin H., Wang D. (2009). Transgenic expression of MYB15 confers enhanced sensitivity to abscisic acid and improved drought tolerance in Arabidopsis thaliana. J. Genet. Genomics 36, 17–29. 10.1016/S1673-8527(09)60003-5 [DOI] [PubMed] [Google Scholar]
- Dubos C., Stracke R., Grotewold E., Weisshaar B., Martin C., Lepiniec L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15, 573–581. 10.1016/j.tplants.2010.06.005 [DOI] [PubMed] [Google Scholar]
- Fujita Y., Fujita M., Shinozaki K., Yamaguchi-Shinozaki K. (2011). ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 124, 509–525. 10.1007/s10265-011-0412-3 [DOI] [PubMed] [Google Scholar]
- Gill B. S., Friebe B. (2002). Cytogenetics, phylogeny and evolution of cultivated wheats, in Bread Wheat Improvement and Production, eds Curtis B. C., Rajaram S., Gómez Macpherson H. (Rome: FAO; ), 71–88. [Google Scholar]
- Greer S., Wen M., Bird D., Wu X., Samuels L., Kunst L., et al. (2007). The cytochrome P450 enzyme CYP96A15 is the midchain alkane hydroxylase responsible for formation of secondary alcohols and ketones in stem cuticular wax of Arabidopsis. Plant Physiol. 145, 653–667. 10.1104/pp.107.107300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Yang H., Zhang X., Yang S. (2013). Lipid transfer protein 3 as a target of MYB96 mediates freezing and drought stress in Arabidopsis. J. Exp. Bot. 64, 1755–1767. 10.1093/jxb/ert040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrmova M., Lopato S. (2014). Enhancing Abiotic Stress Tolerance in Plants by Modulating Properties of Stress Responsive Transcription Factors, in Genomics of Plant Genetic Resources, eds Tuberosa R., Graner A., Frison E. (Dordrecht: Springer; ), 291–316. [Google Scholar]
- Hu Y., Poh H. M., Chua N. H. (2006). The Arabidopsis ARGOS-LIKE gene regulates cell expansion during organ growth. Plant J. 47, 1–9. 10.1111/j.1365-313X.2006.02750.x [DOI] [PubMed] [Google Scholar]
- Hu Z., Han Z., Song N., Chai L., Yao Y., Peng H., et al. (2013). Epigenetic modification contributes to the expression divergence of three TaEXPA1 homoeologs in hexaploid wheat (Triticum aestivum). New Phytol. 197, 1344–1352. 10.1111/nph.12131 [DOI] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S. L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36. 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M., Bentsink L., Hilhorst H. (2002). Seed dormancy and germination. Curr. Opin. Plant Biol. 5, 33–36. 10.1016/S1369-5266(01)00219-9 [DOI] [PubMed] [Google Scholar]
- Krasensky J., Jonak C. (2012). Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J. Exp. Bot. 63, 1593–1608. 10.1093/jxb/err460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo M., Kidokoro S., Yoshida T., Mizoi J., Todaka D., Fernie A. R., et al. (2017). Double overexpression of DREB and PIF transcription factors improves drought stress tolerance and cell elongation in transgenic plants. Plant Biotechnol. J. 15, 458–471. 10.1111/pbi.12644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Lee T. G., Jang C. S., Kim J. Y., Kim D. S., Park J. H., Kim D. Y., et al. (2007). A MYB transcription factor (TaMYB1) from wheat roots is expressed during hypoxia: roles in response to the oxygen concentration in root environment and abiotic stresses. Physiol. Planta. 129, 375–385. 10.1111/j.1399-3054.2006.00828.x [DOI] [Google Scholar]
- Lesk C., Rowhani P., Ramankutty N. (2016). Influence of extreme weather disasters on global crop production. Nature 529, 84–87. 10.1038/nature16467 [DOI] [PubMed] [Google Scholar]
- Li F., Wu X., Lam P., Bird D., Zheng H., Samuels L., et al. (2008). Identification of the wax ester synthase/acyl-coenzyme A: diacylglycerol acyltransferase WSD1 required for stem wax ester biosynthesis in Arabidopsis. Plant Physiol. 148, 97–107. 10.1104/pp.108.123471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling Y., Alshareef S., Butt H., Lozano-Juste J., Li L., Galal A. A., et al. (2017). Pre-mRNA splicing repression triggers abiotic stress signaling in plants. Plant J. 89, 291–309. 10.1111/tpj.13383 [DOI] [PubMed] [Google Scholar]
- Lippold F., Sanchez D. H., Musialak M., Schlereth A., Scheible W. R., Hincha D. K., et al. (2009). AtMYB41 regulates transcriptional and metabolic responses to osmotic stress in Arabidopsis. Plant Physiol. 149, 1761–1772. 10.1104/pp.108.134874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Xin M., Qin J., Peng H., Ni Z., Yao Y., et al. (2015). Temporal transcriptome profiling reveals expression partitioning of homeologous genes contributing to heat and drought acclimation in wheat (Triticum aestivum L.). BMC Plant Biol. 15:152. 10.1186/s12870-015-0511-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Mao X., Jia D., Li A., Zhang H., Tian S., Zhang X., et al. (2011). Transgenic expression of TaMYB2A confers enhanced tolerance to multiple abiotic stresses in Arabidopsis. Funct. Integr. Genomics 11, 445–465. 10.1007/s10142-011-0218-3 [DOI] [PubMed] [Google Scholar]
- Mortazavi A., Williams B. A., McCue K., Schaeffer L., Wold B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat. Methods 5, 621–628. 10.1038/nmeth.1226 [DOI] [PubMed] [Google Scholar]
- Nuruzzaman M., Sharoni A. M., Kikuchi S. (2013). Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front. Microbiol. 4:248. 10.3389/fmicb.2013.00248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pino M. T., Skinner J. S., Park E. J., Jeknic Z., Hayes P. M., Thomashow M. F., et al. (2007). Use of a stress inducible promoter to drive ectopic AtCBF expression improves potato freezing tolerance while minimizing negative effects on tuber yield. Plant Biotechnol. J. 5, 591–604. 10.1111/j.1467-7652.2007.00269.x [DOI] [PubMed] [Google Scholar]
- Qin Y., Wang M., Tian Y., He W., Han L., Xia G. (2012). Over-expression of TaMYB33 encoding a novel wheat MYB transcription factor increases salt and drought tolerance in Arabidopsis. Mol. Biol. Rep. 39, 7183–7192. 10.1007/s11033-012-1550-y [DOI] [PubMed] [Google Scholar]
- Rowland O., Zheng H., Hepworth S. R., Lam P., Jetter R., Kunst L. (2006). CER4 encodes an alcohol-forming fatty acyl-coenzyme A reductase involved in cuticular wax production in Arabidopsis. Plant Physiol. 142, 866–877. 10.1104/pp.106.086785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma Y., Maruyama K., Osakabe Y., Qin F., Seki M., Shinozaki K., et al. (2006). Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 18, 1292–1309. 10.1105/tpc.105.035881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler C., Harshavardhan V. T., Rajesh K., Reddy P. S., Strickert M., Rolletschek H., et al. (2011). ABA biosynthesis and degradation contributing to ABA homeostasis during barley seed development under control and terminal drought-stress conditions. J. Exp. Bot. 62, 2615–2632. 10.1093/jxb/erq446 [DOI] [PubMed] [Google Scholar]
- Seo P. J., Lee S. B., Suh M. C., Park M. J., Go Y. S., Park C. M. (2011). The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. Plant Cell 23, 1138–1152. 10.1105/tpc.111.083485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo P. J., Xiang F., Qiao M., Park J., Lee Y. N., Kim S. G., et al. (2009). The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol. 151, 275–289. 10.1104/pp.109.144220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shitsukawa N., Tahira C., Kassai K., Hirabayashi C., Shimizu T., Takumi S., et al. (2007). Genetic and epigenetic alteration among three homoeologous genes of a class E MADS box gene in hexaploid wheat. Plant Cell 19, 1723–1737. 10.1105/tpc.107.051813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T., Liu Y., Yan H., You Q., Yi X., Du Z., et al. (2017). agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 45, W122–W129. 10.1093/nar/gkx382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todaka D., Shinozaki K., Yamaguchi-Shinozaki K. (2015). Recent advances in the dissection of drought-stress regulatory networks and strategies for development of drought-tolerant transgenic rice plants. Front. Plant Sci. 6:84. 10.3389/fpls.2015.00084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T., Okamoto M., Kushiro T., Nambara E., Oono Y., Seki M., et al. (2006). CYP707A3, a major ABA 8'-hydroxylase involved in dehydration and rehydration response in Arabidopsis thaliana. Plant J. 46, 171–182. 10.1111/j.1365-313x.2006.02683.x [DOI] [PubMed] [Google Scholar]
- Wang T., Xing J., Liu X., Yao Y., Hu Z., Peng H., et al. (2018). GCN5 contributes to stem cuticular wax biosynthesis by histone acetylation of CER3 in Arabidopsis. J. Exp. Bot. 69, 2911–2922. 10.1093/jxb/ery077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wan L., Zhang L., Zhang Z., Zhang H., Quan R., et al. (2012). An ethylene response factor OsWR1 responsive to drought stress transcriptionally activates wax synthesis related genes and increases wax production in rice. Plant Mol. Biol. 78, 275–288. 10.1007/s11103-011-9861-2 [DOI] [PubMed] [Google Scholar]
- Wendel J. F. (2000). Genome evolution in polyploids. Plant Mol. Biol. 42, 225–249. 10.1023/A:1006392424384 [DOI] [PubMed] [Google Scholar]
- Xiong H., Li J., Liu P., Duan J., Zhao Y., Guo X., et al. (2014). Overexpression of OsMYB48-1, a novel MYB-related transcription factor, enhances drought and salinity tolerance in rice. PLoS ONE 9:e92913. 10.1371/journal.pone.0092913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeats T. H., Rose J. K. (2008). The biochemistry and biology of extracellular plant lipid-transfer proteins (LTPs). Protein Sci. 17, 191–198. 10.1110/ps.073300108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Liu G., Zhao G., Xia C., Jia J., Liu X., et al. (2014). Characterization of a wheat R2R3-MYB transcription factor gene, TaMYB19, involved in enhanced abiotic stresses in Arabidopsis. Plant Cell Physiol. 55, 1802–1812. 10.1093/pcp/pcu109 [DOI] [PubMed] [Google Scholar]
- Zhang L., Zhao G., Xia C., Jia J., Liu X., Kong X. (2012). A wheat R2R3-MYB gene, TaMYB30-B, improves drought stress tolerance in transgenic Arabidopsis. J. Exp. Bot. 63, 5873–5885. 10.1093/jxb/ers237 [DOI] [PubMed] [Google Scholar]
- Zhao Y., Tian X., Li Y., Zhang L., Guan P., Kou X., et al. (2017a). Molecular and functional characterization of wheat ARGOS genes influencing plant growth and stress tolerance. Front. Plant Sci. 8:170. 10.3389/fpls.2017.00170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Tian X., Wang F., Zhang L., Xin M., Hu Z., et al. (2017b). Characterization of wheat MYB genes responsive to high temperatures. BMC Plant Biol. 17:208. 10.1186/s12870-017-1158-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.