Abstract
Gastrointestinal stromal tumors (GISTs) are the most common type of mesenchymal tumor in the gastrointestinal tract. The present study aimed to identify the potential candidate biomarkers that may be involved in the pathogenesis and progression of v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog (KIT)/platelet-derived growth factor receptor α (PDGFRA) wild-type GISTs. A joint bioinformatics analysis was performed to identify the differentially expressed genes (DEGs) in wild-type GIST samples compared with KIT/PDGFRA mutant GIST samples. Gene Ontology function and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of DEGs was conducted using Database for Annotation, Visualization and Integrated Discovery and KEGG Orthology-Based Annotation System (KOBAS) online tools, respectively. Protein-protein interaction (PPI) networks of the DEGs were constructed using Search Tool for the Retrieval of Interacting Genes online tool and Cytoscape, and divided into sub-networks using the Molecular Complex Detection (MCODE) plug-in. Furthermore, enrichment analysis of DEGs in the modules was analyzed with KOBAS. In total, 546 DEGs were identified, including 238 upregulated genes primarily enriched in ‘cell adhesion’, ‘biological adhesion’, ‘cell-cell signaling’, ‘PI3K-Akt signaling pathway’ and ‘ECM-receptor interaction’, while the 308 downregulated genes were predominantly involved in ‘inflammatory response’, ‘sterol metabolic process’ and ‘fatty acid metabolic process’, ‘small GTPase mediated signal transduction’, ‘cAMP signaling pathway’ and ‘proteoglycans in cancer’. A total of 25 hub genes were obtained and four modules were mined from the PPI network, and sub-networks also revealed these genes were primarily involved in significant pathways, including ‘PI3K-Akt signaling pathway’, ‘proteoglycans in cancer’, ‘pathways in cancer’, ‘Rap1 signaling pathway’, ‘ECM-receptor interaction’, ‘phospholipase D signaling pathway’, ‘ras signaling pathway’ and ‘cGMP-PKG signaling pathway’. These results suggested that several key hub DEGs may serve as potential candidate biomarkers for wild-type GISTs, including phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit γ, insulin like growth factor 1 receptor, hepatocyte growth factor, thrombospondin 1, Erb-B2 receptor tyrosine kinase 2 and matrix metallopeptidase 2. However, further experiments are required to confirm these results.
Keywords: gastrointestinal stromal tumors, v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog, PDGFRA, wild type, bioinformatics analysis, microarray
Introduction
Gastrointestinal stromal tumors (GISTs) are the most common type of mesenchymal tumor in the gastrointestinal tract, which account for 20% of all soft-tissue sarcomas (1,2). GISTs originate from the interstitial cells of Cajal, and may occur in any part of the gastrointestinal tract; the most frequent sites of origin are the stomach (50–60%), followed by the small intestine (30–35%), the colon and rectum (5%), and finally the esophagus (<1%) (3–5). The number of newly diagnosed GISTs is increasing yearly, since the identification of v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog (KIT) and platelet-derived growth factor receptor α (PDGFRA) proteins as reliable biomarkers of these tumors (6,7).
Approximately 75–80% of GISTs have KIT gene mutations in exons 9, 11, 13, 14 and 17. Of the remaining GISTs with no detected KIT mutations, ~1/3 have mutations in the PDGFRA gene, in exons 12, 14 and 18 (8). While the majority of GISTs are characterized by KIT/PDGFRA gene mutations, 10–15% of GISTs lack such mutations and are defined as KIT/PDGFRA wild-type GISTs (9,10). KIT-also known as CD117 or C-kit receptor-encodes the c-KIT type III receptor tyrosine kinase, which is a cytokine receptor located on the surface of hematopoietic stem cells, as well as other cell types (11). PDGFRA-also known as PDGFRα - is also a type III receptor tyrosine kinase that is expressed on the surfaces of a wide range of cell types (12). Mutations in the PDGFRA gene may induce activation of constitutive ligand-independent kinases and are mutually exclusive with KIT gene mutations, i.e., KIT and PDGFRA mutations do not coexist in patients with GISTs (8). Imatinib, a small molecule selective tyrosine kinase inhibitor, has been used to treat KIT/PDGFRA mutated GISTs (1); however, the efficacy of imatinib depends on the mutated domains of KIT/PDGFRA. It has been reported that ~10% of patients with GISTs are resistant to imatinib, and 40–50% of imatinib-sensitive patients will develop secondary resistance in 2 years (13). In addition, wild-type GISTs are resistant to imatinib treatment and the genetic alterations in wild-type GISTs remain unclear (8). Therefore, it is necessary to identify new target molecules that may be involved in the development and progression of wild-type GISTs.
Currently, gene profiling is widely used in the field of cancer genetics research, which is particularly suitable for the differentially expressed gene (DEG) screening. A large amount of gene profile data has been generated, and most of the data has been shared in public databases. Reintegrating these public data may provide valuable clues for further research. Although many gene profile studies have been performed on GISTs in recent years, research regarding wild-type GISTs is limited and the results are not consistent. Therefore, a joint bioinformatics analysis will be innovative and may provide valuable clues for further research.
In the present study, a joint bioinformatics analysis of two gene expression profiles was performed, in order to identify potential genetic changes in wild-type GIST samples compared to KIT/PDGFRA-mutant GIST samples. Subsequently, functional and pathway enrichment analyses were performed on the DEGs to identify potential biological functions and signaling pathways. Furthermore, a protein-protein interaction (PPI) network was constructed to identify key hub genes. The aim of the current study was to investigate the underlying biological functions and pathways involved in the development and progression of GISTs, and to identify potential candidate biomarkers for these tumors.
Materials and methods
Microarray data
The raw gene expression profiles (GSE17743 and GSE20708) were downloaded from the public Gene Expression Omnibus database (www.ncbi.nlm.nih.gov/geo), which were based on the GPL570 platform (Affymetrix Human Genome U133 Plus 2.0 Array; Thermo Fisher Scientific, Inc., Waltham, MA, USA), and were submitted by Ostrowski et al (14) and Astolfi et al (15), respectively. The GSE17743 dataset contained 29 GIST samples, including 15 with KIT mutations detected, 11 with PDGFRA mutations detected, and three with no mutations detected. The GSE20708 dataset included 22 GIST tumor samples, including 13 with KIT mutations detected, five with PDGFRA mutations detected, and four with no mutations detected. Thus, a total of 51 GIST tumor samples were used for further analysis in the present study.
Data processing
Samples (n=51) were divided into two groups, including wild-type GIST groups (n=7) and KIT/PDGFRA mutant GIST groups (n=44). The CEL files were first converted into probe expression values and were preprocessed for background adjustment and quantile normalization by robust multiarray average algorithm using the ‘affy’ package in R (version 3.4.2) (16,17). The ‘sva’ package in R was used to remove batch effects between two gene expression profiles (18). The ‘Hclust’ method of R was used to perform cluster analysis for gene expression alterations at two batch levels (19). Following this, the probe-level data were transformed to the expression values of genes according to the latest version of annotation file (HG-U133_Plus_2; release 35) for Affymetrix Human Genome U133 Plus 2.0 Array, which was obtained from the official website (www.affymetrix.com/support/technical/byproduct.affx?product=hg-u133-plus). If one gene symbol was matched by multiple probes, then the average expression value was calculated for this gene.
Identification of DEGs
The ‘limma’ package (version 3.26.9) in R language was used to identify DEGs between two groups (20). Fold change (FC) of the gene expression was also observed and log2 FC was calculated. The threshold was defined as a |log2 FC| of >1 and an adjusted P-value of <0.05. Hierarchical clustering analysis was subsequently performed using the ‘pheatmap’ package in R (21).
Functional and pathway enrichment analysis
Gene Ontology (GO) enrichment analysis and functional annotation of DEGs were performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) network software version 6.8 (https://david.ncifcrf.gov/) (22), and enriched GO terms were visualized using the BiNGO plug-in of Cytoscape software (version 3.5.1) (23). Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis and functional annotation were processed by KEGG Orthology-Based Annotation System (KOBAS) network software version 3.0 (kobas.cbi.pku.edu.cn/download.php) (24). An adjusted P-value <0.05 was set as the cut-off criterion.
PPI network construction and modules selection
The PPI networks of DEGs were identified using the Search Tool for the Retrieval of Interacting Genes (STRING) database (string-db.org; release 10) (25). Interactions with confidence scores of ≥0.4 were selected as significant and visualized using Cytoscape software (www.cytoscape.org) (26). The hub genes were selected by the cytoHubba plug-in, with ≥10 degrees for each gene (27), and also mapped into ClueGO to visualize functionally grouped GO terms and KEGG pathway annotation networks (28). The Molecular Complex Detection (MCODE) plug-in was applied to screen modules of the PPI network with degree cutoff=2, node score cutoff=0.2, k-core=2, and max. depth=100 (29). Subsequently, the functional and pathway enrichment analysis of genes in each module (MCODE score ≥6 and number of nodes ≥6) was performed by KOBAS.
Results
Identification of DEGs
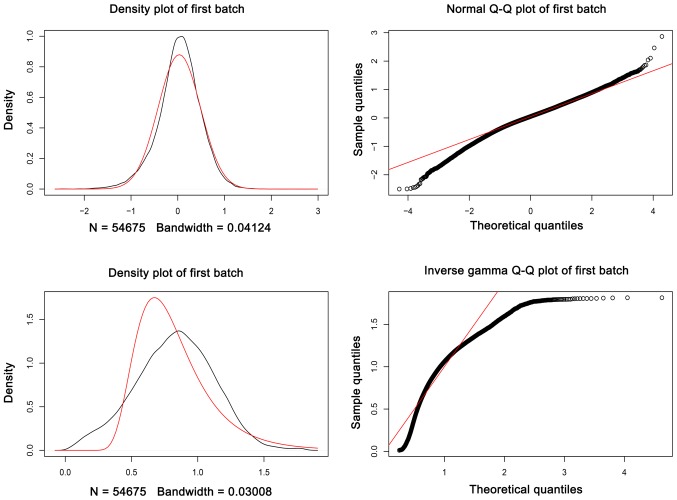
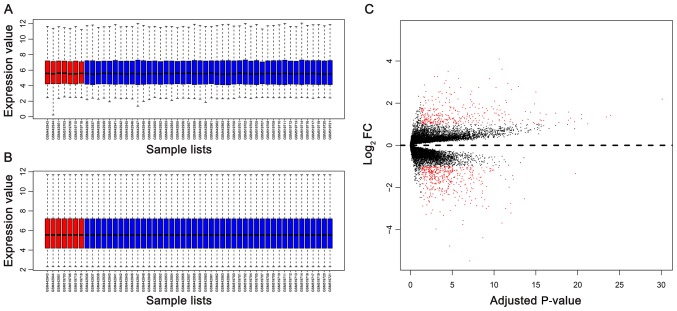
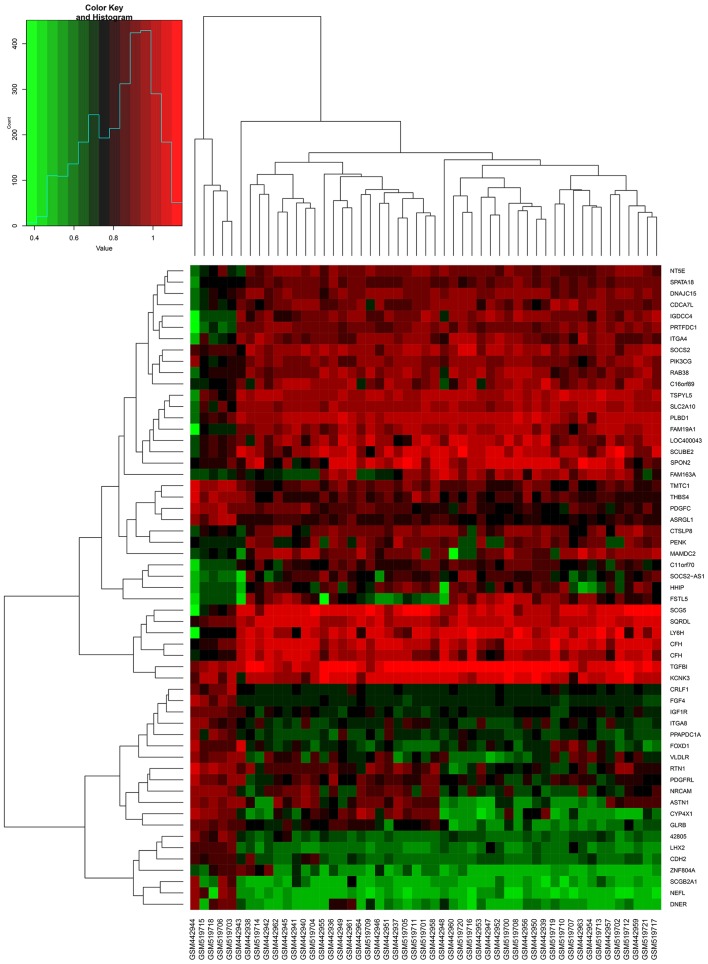
As presented in Fig. 1, the batch effects between two gene expression profiles datasets were removed. The data was normalized prior to further analysis (Fig. 2A and B). In total, 546 DEGs (238 upregulated DEGs and 308 downregulated DEGs) were identified in wild-type GIST samples, compared with KIT/PDGFRA mutant GIST samples, based on the cut-off criteria. The volcano plot (Fig. 2C) and heatmap (Fig. 3) demonstrated the distribution and cluster of DEGs, respectively.
Figure 1.
Batch effect plots between the two gene expression profile datasets.
Figure 2.
Box plots of gene expression profiles of GIST samples and the distribution of DEGs. (A) Gene expression profile of each sample prior to data normalization. (B) Gene expression profile of each sample following data normalization. (C) Volcano plot of gene distribution in wild-type GIST samples compared with KIT/PDGFRA mutant GIST samples. In (A) and (B), the horizontal axis and vertical axis represent samples and gene expression values, respectively; the red boxes and the blue boxes represent the wild-type GIST samples and the KIT/PDGFRA mutant GIST samples, respectively. In (C), the red plot represents statistically significant DEGs (FC≥1 and P<0.05). The black plot represents genes with no significant expression changes. DEGs, differentially expressed genes; GISTs, gastrointestinal stromal tumors; FC, fold change; KIT, v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog; PDGFRA, platelet-derived growth factor receptor α.
Figure 3.
Hierarchical clustering heatmap of fractional DEGs. The horizontal axis indicates the sample, and the vertical axis indicates the DEGs. Red represents the upregulated DEGs and green represents the downregulated DEGs. DEGs, differentially expressed genes.
Functional and pathway enrichment analysis
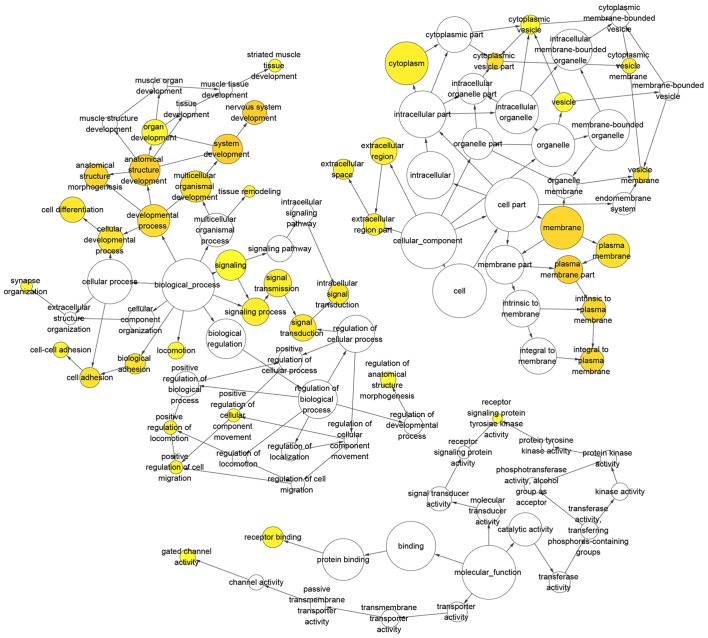
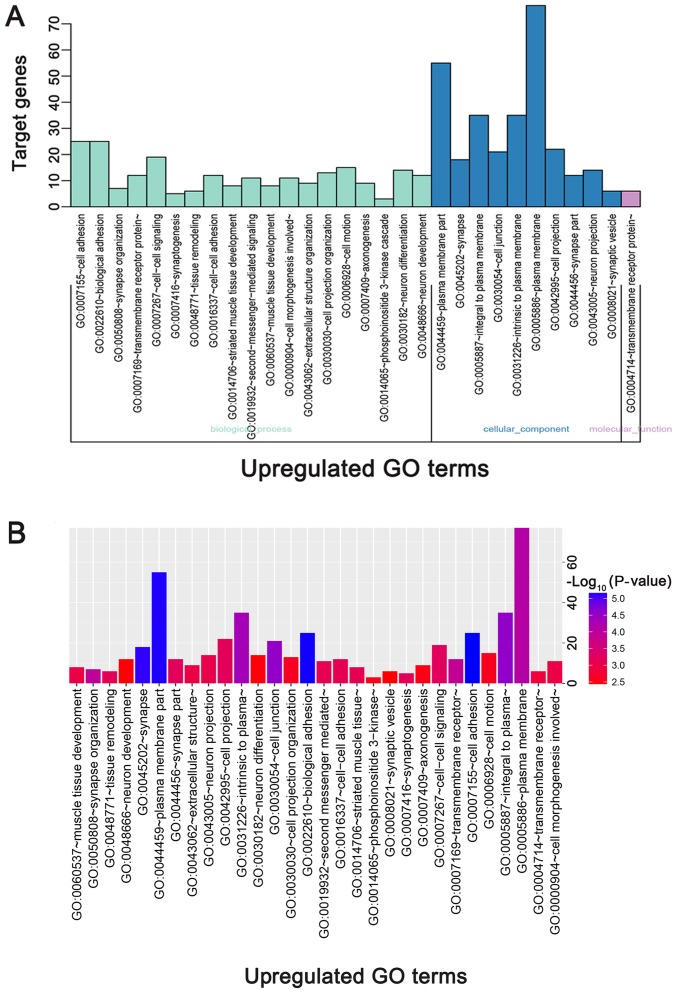
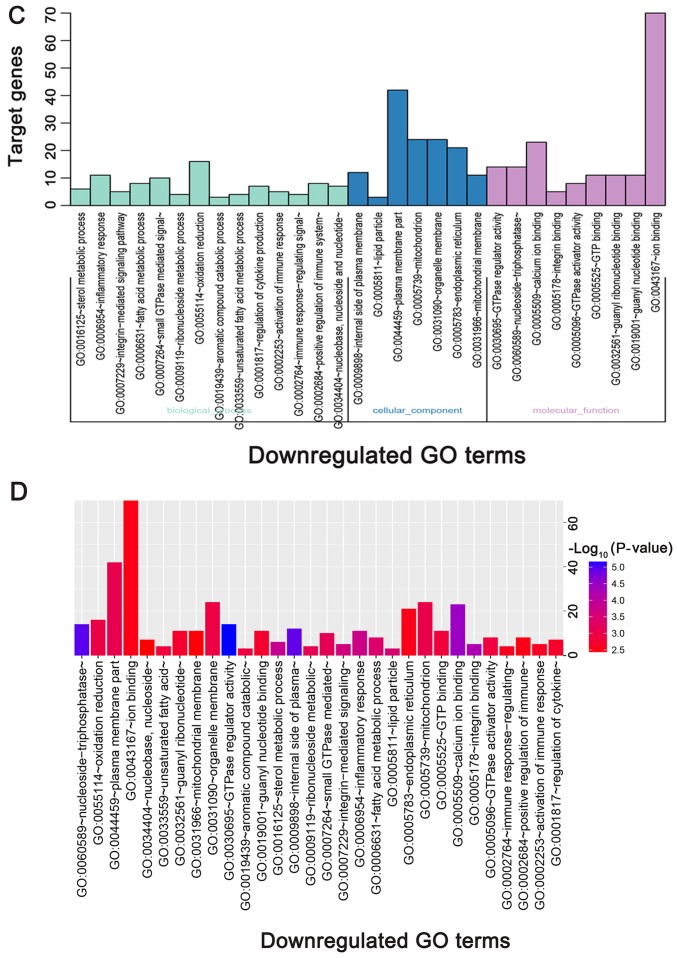
The directed acyclic graph of GO enrichment analysis and functional annotation of DEGs is depicted in Fig. 4. For biological processes (BP), the upregulated DEGs were primarily enriched in ‘cell adhesion’, ‘biological adhesion’ and ‘synapse organization’ (Table I; Fig. 5A and B); the downregulated DEGs were primarily enriched in ‘sterol metabolic process’, ‘inflammatory response’, and ‘integrin-mediated signaling pathway’ (Table I; Fig. 5C and D). For cellular component (CC), the upregulated DEGs were primarily enriched in ‘plasma membrane part’, ‘synapse’ and ‘integral to plasma membrane’ (Table I; Fig. 5A and B); the downregulated DEGs were primarily enriched in ‘internal side of plasma membrane’, ‘lipid particle’, and ‘plasma membrane part’ (Table I; Fig. 5C and D). For molecular function (MF), the upregulated DEGs were primarily enriched in ‘transmembrane receptor protein tyrosine kinase activity’, ‘calcium ion binding’ and ‘transmembrane receptor protein tyrosine kinase signaling pathway’ (Table I; Fig. 5A and B); the downregulated DEGs were primarily enriched in ‘GTPase regulator activity’, ‘nucleoside-triphosphatase regulator activity’, and ‘calcium ion binding’ (Table I; Fig. 5C and D).
Figure 4.
Directed acyclic graph of GO enrichment analysis and DEG functional annotation. Circles represent GO function, the upstream GO function includes the downstream GO function (arrows) and the color intensity represents the degree of GO function enrichment. DEGs, differentially expressed genes; GO, gene ontology.
Table I.
GO enrichment analysis and functional annotation of DEGs.
| A, Upregulated | |||||
|---|---|---|---|---|---|
| Category | Term | Description | Count | % | P-value |
| GOTERM_BP_FAT | GO:0007155 | Cell adhesion | 25 | 11.1 | 7.08×10−6 |
| GOTERM_BP_FAT | GO:0022610 | Biological adhesion | 25 | 11.1 | 7.26×10−6 |
| GOTERM_BP_FAT | GO:0050808 | Synapse organization | 7 | 3.1 | 1.13×10−4 |
| GOTERM_BP_FAT | GO:0007169 | Transmembrane receptor protein tyrosine kinase signaling pathway | 12 | 5.3 | 1.24×10−4 |
| GOTERM_BP_FAT | GO:0007267 | Cell-cell signaling | 19 | 8.4 | 5.50×10−4 |
| GOTERM_CC_FAT | GO:0044459 | Plasma membrane part | 55 | 24.4 | 7.52×10−6 |
| GOTERM_CC_FAT | GO:0045202 | Synapse | 18 | 8.0 | 8.07×10−6 |
| GOTERM_CC_FAT | GO:0005887 | Integral to plasma membrane | 35 | 15.6 | 2.90×10−5 |
| GOTERM_CC_FAT | GO:0030054 | Cell junction | 21 | 9.3 | 3.00×10−5 |
| GOTERM_CC_FAT | GO:0031226 | Intrinsic to plasma membrane | 35 | 15.6 | 4.59×10−5 |
| GOTERM_MF_FAT | GO:0004714 | Transmembrane receptor protein tyrosine kinase activity | 6 | 2. 7 | 1.36×10−3 |
| GOTERM_MF_FAT | GO:0005509 | Calcium ion binding | 22 | 9.8 | 4.10×10−3 |
| GOTERM_MF_FAT | GO:0022843 | Voltage-gated cation channel activity | 7 | 3.1 | 9.44×10−3 |
| GOTERM_MF_FAT | GO:0022836 | Gated channel activity | 10 | 4.4 | 1.41×10−2 |
| GOTERM_MF_FAT | GO:0008083 | Growth factor activity | 7 | 3.1 | 1.43×10−2 |
| B, Downregulated | |||||
| GOTERM_BP_FAT | GO:0016125 | Sterol metabolic process | 6 | 2.2 | 1.22×10−2 |
| GOTERM_BP_FAT | GO:0006954 | Inflammatory response | 11 | 4.1 | 1.34×10−2 |
| GOTERM_BP_FAT | GO:0007229 | Integrin-mediated signaling pathway | 5 | 1.9 | 1.51×10−2 |
| GOTERM_BP_FAT | GO:0006631 | Fatty acid metabolic process | 8 | 3.0 | 1.87×10−2 |
| GOTERM_BP_FAT | GO:0007264 | Small GTPase mediated signal transduction | 10 | 3.7 | 2.38×10−2 |
| GOTERM_CC_FAT | GO:0009898 | Internal side of plasma membrane | 12 | 4.5 | 4.55×10−2 |
| GOTERM_CC_FAT | GO:0005811 | Lipid particle | 3 | 1.1 | 1.78×10−2 |
| GOTERM_CC_FAT | GO:0044459 | Plasma membrane part | 42 | 15.7 | 2.57×10−2 |
| GOTERM_CC_FAT | GO:0005739 | Mitochondrion | 24 | 9.0 | 2.68×10−2 |
| GOTERM_CC_FAT | GO:0031090 | Organelle membrane | 24 | 9.0 | 2.91×10−2 |
| GOTERM_MF_FAT | GO:0030695 | GTPase regulator activity | 14 | 5.2 | 3.60×10−3 |
| GOTERM_MF_FAT | GO:0060589 | Nucleoside-triphosphatase regulator activity | 14 | 5.2 | 4.34×10−3 |
| GOTERM_MF_FAT | GO:0005509 | Calcium ion binding | 23 | 8.6 | 6.95×10−3 |
| GOTERM_MF_FAT | GO:0005178 | Integrin binding | 5 | 1.9 | 8.49×10−3 |
| GOTERM_MF_FAT | GO:0005096 | GTPase activator activity | 8 | 3.0 | 3.12×10−2 |
Count indicates the enriched gene number in the category. GO, gene ontology; DEGs, differentially expressed genes BP, biological process; CC, cellular component; MF, molecular function.
Figure 5.
GO functional and KEGG pathway enrichment analysis of DEGs. (A) GO functional enrichment analysis of upregulated DEGs based on the three types of sub-ontologies and (B) P-value. (C) GO functional enrichment analysis of downregulated DEGs (top 30 terms) based on the three types of sub-ontologies and (D) P-value. The horizontal axis and vertical axis indicate the names of GO terms and the number of target genes, respectively. KEGG functional enrichment analysis of (E) upregulated and (F) downregulated DEGs (top 30 terms) are presented, based on P-value. The color of the pillars represents the size of the P-value, with the red color representing the smallest P-value. DEGs, differentially expressed genes; GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
According to the KEGG pathway enrichment analysis, upregulated genes were primarily enriched in ‘PI3K-Akt signaling pathway’, ‘ras signaling pathway’, ‘Rap1 signaling pathway’, ‘calcium signaling pathway’ and ‘ErbB signaling pathway’ (Table II; Fig. 5E). Downregulated genes were primarily enriched in ‘insulin signaling pathway’, ‘cAMP signaling pathway’, ‘PPAR signaling pathway’, and ‘NF-kappa B signaling pathway’ (Table II; Fig. 5F).
Table II.
KEGG pathways analysis results of DEGs.
| A, Upregulated | ||||
|---|---|---|---|---|
| Pathway ID | Description | Count | P-value | Genes |
| hsa05218 | Melanoma | 7 | 4.22×10−7 | CDK6, HGF, FGF3, FGF10, IGF1R, FGF4, PDGFC |
| hsa04151 | PI3K-Akt signaling pathway | 12 | 1.53×10−6 | CDK6, HGF, FGF3, MYB, EFNA2, FGF10, IGF1R, FGF4, ITGA8, PPP2R2B, PDGFC, THBS4 |
| hsa04514 | Cell adhesion molecules (CAMs) | 8 | 4.06×10−6 | PVRL3, NRXN1, CDH2, NRCAM, CNTNAP2, ITGA8, NLGN4X, CADM1 |
| hsa04014 | Ras signaling pathway | 9 | 1.30×10−5 | HGF, SHC3, FGF3, EFNA2, FGF10, IGF1R, HTR7, FGF4, PDGFC |
| hsa01521 | EGFR tyrosine kinase inhibitor resistance | 6 | 1.34×10−5 | HGF, SHC3, ERBB3, ERBB2, IGF1R, PDGFC |
| hsa04512 | ECM-receptor interaction | 6 | 1.43×10−5 | CD44, SV2B, CD36, ITGA8, THBS4, SV2C |
| hsa04510 | Focal adhesion | 8 | 3.98×10−5 | ARHGAP5, HGF, SHC3, ERBB2, IGF1R, ITGA8, PDGFC, THBS4 |
| hsa04015 | Rap1 signaling pathway | 8 | 5.17×10−5 | HGF, FGF3, EFNA2, FGF10, IGF1R, FGF4, ADCY2, PDGFC |
| hsa04020 | Calcium signaling pathway | 7 | 1.31×10−4 | ATP2B1, ERBB3, ERBB2, HTR7, TACR1, CAMK4, ADCY2 |
| hsa05200 | Pathways in cancer | 10 | 1.70×10−4 | CDK6, HGF, DAPK1, ERBB2, FGF3, FGF10, IGF1R, FZD2, FGF4, ADCY2 |
| hsa05014 | Amyotrophic lateral sclerosis (ALS) | 4 | 3.18×10−4 | GRIA2, MAP3K5, GRIA1, NEFL |
| hsa04080 | Neuroactive ligand-receptor interaction | 8 | 3.22×10−4 | GRIA2, PRLHR, PTH1R, HTR7, TACR1, GRIA1, OPRK1, GLRB |
| hsa04730 | Long-term depression | 4 | 5.69×10−4 | GRIA2, IGF1R, GRIA1, PPP1R17 |
| hsa05205 | Proteoglycans in cancer | 6 | 1.66×10−3 | HGF, CD44, ERBB3, ERBB2, IGF1R, FZD2 |
| hsa04012 | ErbB signaling pathway | 4 | 2.20×10−3 | SHC3, ERBB3, ERBB2, NRG3 |
| B, Downregulated | ||||
| Pathway ID | Description | Count | P-value | Genes |
| hsa05145 | Toxoplasmosis | 7 | 5.50×10−5 | PIK3CG, SOCS1, HLA-DPA1, MYD88, CD40, LDLR, MAPK10 |
| hsa04925 | Aldosterone synthesis and secretion | 6 | 5.68×10−5 | KCNK3, PDE2A, CREB3L1, LDLR, ORAI1, CACNA1H |
| hsa05144 | Malaria | 5 | 5.85×10−5 | VCAM1, THBS1, GYPC, MYD88, CD40 |
| hsa04810 | Regulation of actin cytoskeleton | 9 | 6.24×10−5 | ITGA4, PIK3CG, CD14, F2R, FGFR4, MRAS, SSH3, VAV2, ITGAE |
| hsa04910 | Insulin signaling pathway | 7 | 1.39×10−4 | PIK3CG, SOCS2, SOCS1, SREBF1, PYGM, PDE3B, MAPK10 |
| hsa04024 | cAMP signaling pathway | 8 | 2.08×10−4 | HHIP, PIK3CG, F2R, CREB3L1, PDE3B, ORAI1, MAPK10, VAV2 |
| hsa05205 | Proteoglycans in cancer | 8 | 2.52×10−4 | MMP2, CTSL, PIK3CG, THBS1, WNT5B, WNT9A, MRAS, VAV2 |
| hsa04931 | Insulin resistance | 6 | 2.65×10−4 | PIK3CG, CREB3L1, SREBF1, PYGM, CPT1A, MAPK10 |
| hsa03320 | PPAR signaling pathway | 5 | 3.17×10−4 | ME1, PLIN2, CPT1A, ACSL5, CYP27A1 |
| hsa00071 | Fatty acid degradation | 4 | 4.98×10−4 | ACAA2, CPT1A, ACSL5, ALDH7A1 |
| hsa01212 | Fatty acid metabolism | 4 | 6.77×10−4 | PTPLAD2, ACAA2, CPT1A, ACSL5 |
| hsa04930 | Type II diabetes mellitus | 4 | 6.77×10−4 | PIK3CG, SOCS2, SOCS1, MAPK10 |
| hsa04064 | NF-kappa B signaling pathway | 5 | 9.57×10−4 | VCAM1, CD14, SYK, MYD88, CD40 |
| hsa05166 | HTLV-I infection | 8 | 1.11×10−3 | JAK3, PIK3CG, VCAM1, WNT5B, HLA-DPA1, WNT9A, MRAS, CD40 |
| hsa04510 | Focal adhesion | 7 | 1.22×10−3 | ITGA4, PIK3CG, THBS1, PARVA, MAPK10, VAV2, PARVB |
Count indicates the enriched gene number in the pathway. KEGG, Kyoto Encyclopedia of Genes and Genomes; DEGs, differentially expressed genes.
PPI network construction and modules selection
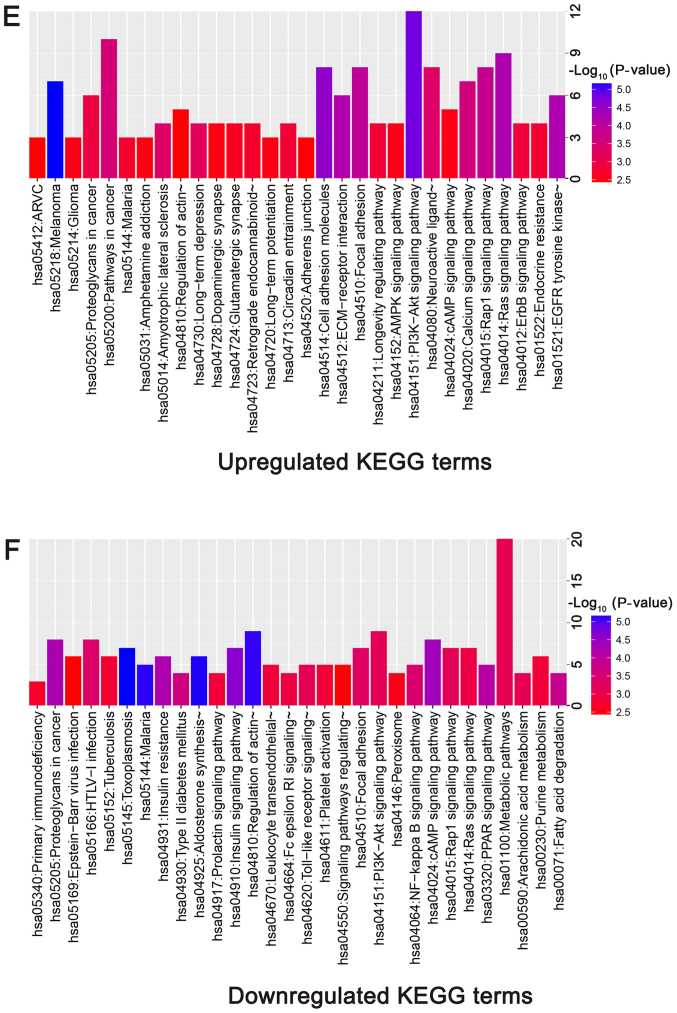
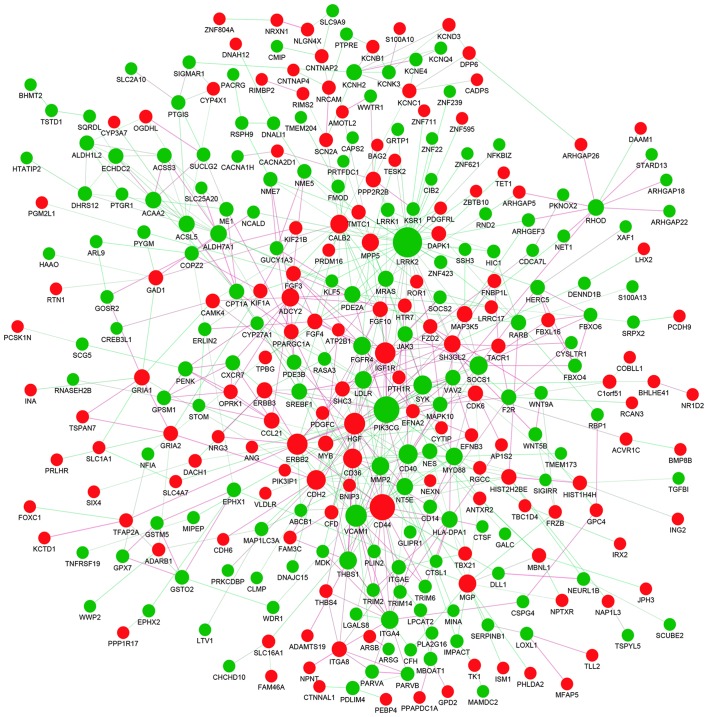
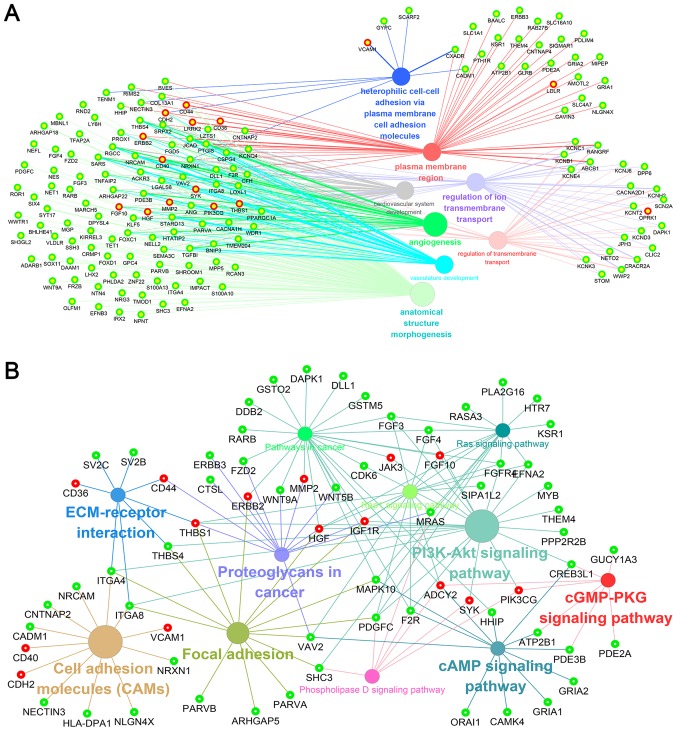
To investigate the interactions between DEGs, a PPI network for the DEGs was constructed. The PPI network consisted of 338 nodes and 628 edges with a confidence score of ≥0.4 (Fig. 6). A total of 25 hub genes were selected from the PPI network with a degree of ≥10 (Table III), including leucine-rich repeat kinase 2 (LRRK2), phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit γ (PIK3CG), CD44, vascular cell adhesion molecule 1 (VCAM1) and hepatocyte growth factor (HGF). GO enrichment analysis revealed that hub genes were primarily enriched in ‘plasma membrane region’, ‘extracellular exosome’, ‘angiogenesis’, ‘regulation of transmembrane transport’ and ‘protein binding’ (Fig. 7A; Table IV). The KEGG analysis indicated that hub genes were predominantly enriched in ‘PI3K-Akt signaling pathway’, ‘proteoglycans in cancer’, ‘focal adhesion’, ‘Rap1 signaling pathway’, ‘pathways in cancer’ and ‘ECM-receptor interaction’ (Fig. 7B; Table IV).
Figure 6.
Protein-protein interaction network of identified differentially expressed genes. Red nodes represent the upregulated genes, while green nodes represent the downregulated genes. The magnitude of each node represents the extent to which each gene is linked, and the lines represent the interaction relationship between nodes.
Table III.
Top 25 hub genes identified in PPI network of DEGs.
| Affy ID | Gene symbol | Gene name | Degree | log FC | P-value | Regulation |
|---|---|---|---|---|---|---|
| 229584_at | LRRK2 | Leucine rich repeat kinase 2 | 36 | −1.78 | 1.04×10−4 | Down |
| 206369_s_at | PIK3CG | Phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma | 29 | −1.89 | 5.37×10−6 | Down |
| 204489_s_at | CD44 | CD44 molecule (Indian blood group) | 28 | 1.11 | 1.14×10−3 | Up |
| 203868_s_at | VCAM1 | Vascular cell adhesion molecule 1 | 20 | −1.76 | 8.36×10−4 | Down |
| 209960_at | HGF | Hepatocyte growth factor | 19 | 1.05 | 2.25×10−3 | Up |
| 216836_s_at | ERBB2 | Erb-b2 receptor tyrosine kinase 2 | 18 | 1.21 | 6.69×10−5 | Up |
| 243358_at | IGF1R | Insulin-like growth factor 1 receptor | 18 | 1.39 | 1.72×10−11 | Up |
| 203441_s_at | CDH2 | Cadherin 2 | 15 | 1.41 | 2.63×10−8 | Up |
| 206488_s_at | CD36 | CD36 molecule | 15 | 1.49 | 2.97×10−3 | Up |
| 205153_s_at | CD40 | CD40 molecule | 15 | −1.12 | 5.87×10−4 | Down |
| 207540_s_at | SYK | Spleen associated tyrosine kinase | 14 | −1.48 | 2.52×10−3 | Down |
| 205428_s_at | CALB2 | Calbindin 2 | 13 | 1.25 | 3.93×10−6 | Up |
| 210001_s_at | SOCS1 | Suppressor of cytokine signaling 1 | 13 | −1.46 | 4.87×10−5 | Down |
| 202291_s_at | MGP | Matrix Gla protein | 12 | 1.52 | 5.01×10−6 | Up |
| 213217_at | ADCY2 | Adenylate cyclase 2 | 12 | 1.96 | 7.75×10−4 | Up |
| 201069_at | MMP2 | Matrix metallopeptidase 2 | 12 | −2.29 | 2.25×10−4 | Down |
| 205884_at | ITGA4 | Integrin subunit alpha 4 | 12 | −1.94 | 6.88×10−4 | Down |
| 201108_s_at | THBS1 | Thrombospondin 1 | 12 | −1.65 | 2.30×10−3 | Down |
| 204579_at | FGFR4 | Fibroblast growth factor receptor 4 | 12 | −1.49 | 2.41×10−3 | Down |
| 219321_at | MPP5 | Membrane protein, palmitoylated 5 | 11 | 1.07 | 6.57×10−5 | Up |
| 209124_at | MYD88 | Myeloid differentiation primary response 88 | 11 | −1.15 | 9.07×10−6 | Down |
| 217173_s_at | LDLR | Low density lipoprotein receptor | 11 | −1.06 | 8.12×10−5 | Down |
| 227486_at | NT5E | 5′-nucleotidase ecto | 10 | −1.64 | 1.19×10−4 | Down |
| 205751_at | SH3GL2 | SH3-domain GRB2-like 2 | 10 | 1.48 | 1.32×10−3 | Up |
| 208951_at | ALDH7A1 | Aldehyde dehydrogenase 7 family, member A1 | 10 | −1.21 | 5.77×10−5 | Down |
PPI, protein-protein interaction; DEGs, differentially expressed genes; FC, fold change.
Figure 7.
Nodes linking the enriched (A) Gene Ontology function and (B) Kyoto Encyclopedia of Genes and Genomes pathway by hub genes. Solid circles and hollow circles represent each annotation and the DEGs, respectively. Red hollow circles represent hub genes in the protein-protein interaction network.
Table IV.
Enriched function and pathways of hub genes and selected modules of PPI network.
| A, Hub genes | ||||
|---|---|---|---|---|
| Term | Description | Count | P-value | Hub genes |
| GO:0005886 | Plasma membrane | 19 | 7.42×10−8 | PIK3CG, FGFR4, ADCY2, LDLR, ERBB2, MPP5, CD40, ITGA4, CDH2, MMP2, VCAM1, IGF1R, MYD88, CD36, CD44, LRRK2, NT5E, SH3GL2, SYK |
| GO:0016020 | Membrane | 11 | 2.53×10−4 | PIK3CG, VCAM1, IGF1R, ADCY2, CD36, LDLR, ERBB2, ITGA4, HGF, CDH2, NT5E |
| GO:0070062 | Extracellular exosome | 12 | 4.07×10−4 | VCAM1, ALDH7A1, CD44, MPP5, MGP, ITGA4, CD40, CDH2, THBS1, LRRK2, NT5E, SH3GL2 |
| GO:0005737 | Cytoplasm | 16 | 5.71×10−4 | PIK3CG, FGFR4, ADCY2, ERBB2, SOCS1, MPP5, CD40, CDH2, CALB2, ALDH7A1, MYD88, CD44, LRRK2, NT5E, SH3GL2, SYK |
| GO:0005515 | Protein binding | 21 | 1.47×10−3 | PIK3CG, FGFR4, LDLR, ERBB2, SOCS1, MPP5, MGP, HGF, CD40, ITGA4, CDH2, MMP2, IGF1R, ALDH7A1, MYD88, CD36, CD44, THBS1, LRRK2, SH3GL2, SYK |
| hsa05205 | Proteoglycans in cancer | 7 | 4.89×10−11 | PIK3CG, IGF1R, CD44, ERBB2, HGF, THBS1, MMP2 |
| hsa04151 | PI3K-Akt signaling pathway | 7 | 1.58×10−9 | PIK3CG, IGF1R, FGFR4, ITGA4, HGF, THBS1, SYK |
| hsa04510 | Focal adhesion | 6 | 3.19×10−9 | PIK3CG, IGF1R, ERBB2, ITGA4, HGF, THBS1 |
| hsa04015 | Rap1 signaling pathway | 6 | 3.99×10−9 | PIK3CG, IGF1R, FGFR4, ADCY2, HGF, THBS1 |
| hsa05200 | Pathways in cancer | 6 | 1.57×10−7 | PIK3CG, IGF1R, ADCY2, ERBB2, HGF, MMP2 |
| B, Module 1 | ||||
| Term | Description | Count | P-value | Genes |
| hsa04151 | PI3K-Akt signaling pathway | 4 | 6.16×10−5 | IGF1R, FGFR4, JAK3, SYK |
| hsa04550 | Signaling pathways regulating pluripotency of stem cells | 3 | 1.03×10−4 | IGF1R, FGFR4, JAK3 |
| hsa05203 | Viral carcinogenesis | 2 | 7.47×10−3 | FGFR4, SYK |
| hsa04015 | Rap1 signaling pathway | 2 | 7.89×10−3 | IGF1R, JAK3 |
| hsa04014 | Ras signaling pathway | 2 | 9.15×10−3 | IGF1R, JAK3 |
| C, Module 2 | ||||
| Term | Description | Count | P-value | Genes |
| hsa04668 | TNF signaling pathway | 2 | 5.09×10−4 | PIK3CG, MAP3K5 |
| hsa04071 | Sphingolipid signaling pathway | 2 | 6.14×10−4 | PIK3CG, MAP3K5 |
| hsa04210 | Apoptosis | 2 | 8.16×10−4 | PIK3CG, MAP3K5 |
| hsa04072 | Phospholipase D signaling pathway | 2 | 8.62×10−4 | MRAS, PIK3CG |
| hsa05205 | Proteoglycans in cancer | 2 | 1.72×10−3 | MRAS, PIK3CG |
| D, Module 3 | ||||
| Term | Description | Count | P-value | Genes |
| hsa05200 | Pathways in cancer | 7 | 5.55×10−8 | ADCY2, ERBB2, FGF10, FZD2, MMP2, FGF3, FGF4 |
| hsa05205 | Proteoglycans in cancer | 5 | 1.11×10−6 | CD44, ERBB2, FZD2, THBS1, MMP2 |
| hsa04015 | Rap1 signaling pathway | 5 | 1.28×10−6 | ADCY2, FGF10, THBS1, FGF3, FGF4 |
| hsa04512 | ECM-receptor interaction | 3 | 5.86×10−5 | CD36, CD44, THBS1 |
| hsa05166 | HTLV-I infection | 4 | 8.31×10−5 | VCAM1, ADCY2, FZD2, CD40 |
| E, Module 4 | ||||
| Term | Description | Count | P-value | Genes |
| hsa05205 | Proteoglycans in cancer | 4 | 3.03×10−5 | WNT5B, ERBB3, WNT9A, HGF |
| hsa04810 | Regulation of actin cytoskeleton | 4 | 3.64×10−5 | ITGAE, ITGA4, CD14, F2R |
| hsa04151 | PI3K-Akt signaling pathway | 4 | 2.12×10−4 | MYB, ITGA4, CD14, WNT5B |
| hsa05200 | Pathways in cancer | 4 | 3.71×10−4 | F2R, ERBB3, WNT9A, WNT5B |
| hsa01100 | Metabolic pathways | 6 | 5.98×10−4 | CYP3A7, ACSL5, NT5E, PTGIS, ACSS3, ACAA2 |
Count indicates the enriched gene number in the category. PPI, protein-protein interaction; GO, gene ontology.
Four significant modules were screened from the PPI network of DEGs using MCODE plug-in, and enrichment analysis revealed that the module genes in the sub-networks were mainly associated with ‘PI3K-Akt signaling pathway’, ‘Rap1 signaling pathway’, ‘proteoglycans in cancer’ and ‘pathways in cancer’ (Table IV).
Discussion
KIT/PDGFRA mutations are the major genetic alterations that occur in the development and progression of GISTs, and have been the only targets of molecular-based therapies in the last decade (4,30). However, other genetic alterations may also be associated with the development and progression of GISTs (15). Currently, little is known about differences in gene expression levels between wild-type GISTs and KIT/PDGFRA mutant GISTs. Therefore, joint bioinformatics analysis was performed in the present study, to obtain other potential candidate biomarkers that may be involved in the development and progression of wild-type GISTs. Ultimately, a total of 546 DEGs were identified, including 238 upregulated DEGs and 308 downregulated DEGs. GO functional and KEGG pathway enrichment analysis of DEGs was subsequently performed. Traditionally, distant metastasis is determined to be the leading cause of morbidity and mortality in patients with cancer, and genes encoding adhesion proteins, inflammatory factors, cytokines, growth factors and transduction molecules are considered major mediators of metastasis (31,32). Enrichment analysis in the present study revealed that the identified DEGs may be involved in the aforementioned process and signaling pathway, and were associated with proliferation, differentiation, apoptosis and distant metastasis in GISTs.
Furthermore, PPI networks of DEGs were constructed and 25 hub genes with degrees of ≥10 were identified. Functional enrichment analysis of hub genes determined that these genes were significantly associated with heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules, regulation of transmembrane transport, cardiovascular system development and angiogenesis. Additionally, pathway analysis indicated that these genes were primarily associated with the following pathways: ‘PI3K-Akt signaling pathway’, ‘proteoglycans in cancer’, ‘focal adhesion’, ‘pathways in cancer’, ‘Rap1 signaling pathway’, ‘ECM-receptor interaction’, ‘cell adhesion molecules’, ‘phospholipase D signaling pathway’, ‘cAMP signaling pathway’, ‘Ras signaling pathway’ and ‘cGMP-PKG signaling pathway’. Furthermore, pathway analyses of four significant modules filtered from the PPI network was performed, and the results revealed that the genes in these modules were also primarily involved in the aforementioned pathways. Specifically, several high-frequency hub genes were identified that may be involved in the progression of GISTs by combining these results, including PIK3CG, insulin like growth factor 1 receptor (IGF1R), HGF, thrombospondin 1 (THBS1), Erb-B2 receptor tyrosine kinase (ERBB) 2 and matrix metallopeptidase 2 (MMP2).
The PIK3CG gene is located on chromosome 7 long arm q22.3, contains 12 exons, and encodes phosphoinositide 3-kinase (PI3K)γ, which phosphorylates inositol lipids and is involved in the immune response (33). Semba et al (34) reported that the PIK3CG gene is downregulated in colorectal cancer and is involved in tumorigenesis and progression, mainly through the PI3K-protein kinase B (Akt) signaling pathway. Li et al (35) determined that the PI3K-Akt signaling pathway is partially activated following imatinib secondary resistance, and PI3K activation may occur at an early stage of secondary resistance. In the present study, PIK3CG was downregulated in wild-type GISTs. PIK3CG was identified as a hub gene with 29 degrees in the PPI network, and as the core gene of module 2. The enrichment analyses demonstrated that PIK3CG was associated with cardiovascular and vasculature development, regulation of transmembrane transport, angiogenesis and anatomical structure morphogenesis, as well as the PI3K-Akt signaling pathway, phospholipase D signaling pathway and cAMP signaling pathway. This suggested that PIK3CG may be a key molecule associated with wild-type GISTs.
IGF1R, a transmembrane receptor tyrosine kinase, serves a critical role in tumor transformation and malignant cell survival. IGF1R is predominantly involved in two signaling pathways: The PI3K-Akt pathway and the Ras-mitogen-activated protein kinase pathway (36). Ludovini et al (37) identified that activation of IGF1R is a necessary condition for mediating tumor cell proliferation and invasion, and is an independent poor prognostic factor in early stage non-small cell lung cancer. In addition, previous studies have demonstrated that IGF1R is overexpressed in wild-type GISTs, and inhibition of IGF1R signaling may be an effective therapeutic strategy (38–41). In the present study, IGF1R was upregulated in wild-type GIST samples and was associated with fibroblast growth factor receptor (FGFR) 4, Janus kinase 3 and tyrosine-protein kinase SYK in module 1. Pathway analysis revealed that IGF1R was associated with the PI3K-Akt signaling pathway, proteoglycans in cancer, and ras signaling pathway. These findings were consistent with the results of the aforementioned studies and indicated that IGF1R overexpression may act as an alternative genetic alteration event to the KIT/PDGFRA mutations in GISTs. Therefore, further research is necessary to clarify underlying mechanisms of IGF1R in wild-type GISTs.
HGF, also known as scatter factor, is a member of the endothelium-specific growth factor family, and regulates cell growth and motility, migration, and angiogenesis through binding to its receptor c-Met (42). Hack et al (43) revealed that aberrant activation of the HGF/MET signaling pathway occurs in the malignant transformation and progression of gastroesophageal cancer, and consistently correlates with an aggressive metastatic phenotype and poor prognosis. However, there is a lack of research on the role of HGF in GISTs. The results of enrichment analysis in the present study demonstrated that HGF was associated with angiogenesis, cardiovascular and vasculature development, proteoglycans in cancer, focal adhesion, pathways in cancer, and PI3K-Akt signaling pathway. Furthermore, it was associated with Wnt family member (WNT) 5B, ERBB3, and WNT9A in module 4, revealing that HGF may be involved in progression of wild-type GISTs.
THBS1, also known as TSP-1, encodes an adhesive glycoprotein that is involved in platelet aggregation, angiogenesis and tumorigenesis (44). In the present study, THBS1 was associated with proteoglycans in cancer, ECM-receptor interaction, PI3K-Akt signaling pathway, and rap1 signaling pathway. In addition, functional enrichment analysis identified that THBS1 was associated with angiogenesis, protein binding, cell adhesion, regulation of transmembrane transport and inflammatory response. Kashihara et al (45) revealed that THBS1 is associated with carcinogenesis occurring in patients with intestinal inflammation, and may serve an important role in gastric carcinogenesis. Huang et al (46) reported that upregulation of THBS1 is induced by FGF7/FGFR2 via the PI3K/Akt/mechanistic target of rapamycin signaling pathway, and is associated with the regulation of invasion and migration in gastric cancer. However, no studies have elucidated the mechanism of THBS1 in GISTs. In module 3, THBS1 was associated with adenyl cyclase type 2 (ADCY2), FGF10, FGF3, FGF4, CD36 and CD44, indicating that THBS1 may also be involved in GISTs by mediating these genes. Therefore, further research is necessary to clarify the underlying mechanism of THBS1 in wild-type GISTs.
ERBB2, also known as Her2 or Neu, is a member of the ERBB family of receptor tyrosine kinases. It encodes a transmembrane tyrosine kinase receptor of the ERBB family that has important roles in many aspects of various human cancers (47,48). However, the oncogenic role and clinical significance of ERBB2 in GISTs has not been investigated in detail. In the present study, ERBB2 was upregulated and identified as the core gene of module 3. Enrichment analyses revealed that ERBB2 was associated with plasma membrane region, cardiovascular system development, angiogenesis, focal adhesion and proteoglycans in cancer. Furthermore, it was associated with ADCY2, FGF10, frizzled-2 (FZD2), FGF3, FGF4, CD44, THBS1, and MMP2, revealing these genes may have joint function in GISTs. Hence, further studies are also required to determine the role of ERBB2 in wild-type GISTs.
MMP2, a member of the matrix metalloproteinase (MMP) gene family, is involved in many cancer pathways and exists in several proteoglycans in cancer. MMP family proteins are zinc-dependent enzymes capable of cleaving components of the extracellular matrix and molecules involved in signal transduction. MMP activity has been implicated in a number of key pathological processes, including tumor growth, progression, metastasis and dysregulated angiogenesis (49). Sebastiano et al (50) reported that MMP2 increases platelet activation by cleaving PAR1 at a noncanonical extracellular site, which induces biased receptor signaling through certain signaling pathways, usually only activated by full PAR1 agonism. Furthermore, it has been reported that the gene interaction between MMP2 and PARP1 may increase the incidence of gastric cancer development and lymph node metastasis (51). In the current study, MMP2 was identified as a downregulated hub gene, enriched in pathways and proteoglycans in cancer of module 3 and associated with ADCY2, ERBB2, FGF10, FZD2, FGF3, FGF4, CD44 and THBS1.
There are several limitations of the present study that require acknowledgement. First, due to the limitations of the gene chip itself, the differentially expressed genes between GIST tissue and normal gastrointestinal tract tissue could not be identified. Second, all predicted results still require confirmation by laboratory data. Finally, a limited number of samples were used in the present study, which should be increased in future studies to improve the reliability of the conclusions drawn.
In conclusion, 546 DEGs were identified in wild-type GISTs, compared with the mutant GIST samples, which may be closely associated with GIST progression. In addition, several key hub DEGs were selected as potential candidate biomarkers for wild-type GISTs, including PIK3CG, IGF1R, HGF, THBS1, ERBB2 and MMP2. However, further verification experiments are required to confirm these results.
Acknowledgements
The authors are grateful to medical staff of Department of General Surgery, Lanzhou General Hospital of Chinese People's Liberation Army for their support.
Funding
This study was supported by the Huimin Plan of Ministry of Science and Technology & Ministry of Finance, P.R. China (grant no. 2012GS620101). the Major Projects of Science and Technology in Gansu Province of P.R. China (grant no. 2011GS04390), the Natural Science Foundation of in Gansu Province of P.R. China (grant no. 1506RJZA309), and the Postdoctoral Research Foundation of P.R. China (grant no. 2015M572710).
Availability of data and materials
The datasets analyzed in the present study are available from the GEO repository, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE17743 and https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE20708.
Authors' contributions
The original study design was undertaken by HBL, WJW and ZYJ. Data collection was undertaken JPY and YML. Microarray data were analyzed by WJW and HTL, and appraised by XPH, PC and WWY. Functional and pathway enrichment analysis was undertaken by XPH, PC, WWY and WKC. The draft manuscript was written by WJW, and was reviewed and edited by HTL, JPY, WKC, ZYJ and HBL. All authors have read and approved the final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Valsangkar N, Sehdev A, Misra S, Zimmers TA, O'Neil BH, Koniaris LG. Current management of gastrointestinal stromal tumors: Surgery, current biomarkers, mutations, and therapy. Surgery. 2015;158:1149–1164. doi: 10.1016/j.surg.2015.06.027. [DOI] [PubMed] [Google Scholar]
- 2.Ducimetière F, Lurkin A, Ranchère-Vince D, Decouvelaere AV, Péoc'h M, Istier L, Chalabreysse P, Muller C, Alberti L, Bringuier PP, et al. Incidence of sarcoma histotypes and molecular subtypes in a prospective epidemiological study with central pathology review and molecular testing. PLoS One. 2011;6:e20294. doi: 10.1371/journal.pone.0020294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patil DT, Rubin BP. Gastrointestinal stromal tumor: Advances in diagnosis and management. Arch Pathol Lab Med. 2011;135:1298–1310. doi: 10.5858/arpa.2011-0022-RA. [DOI] [PubMed] [Google Scholar]
- 4.Joensuu H, Hohenberger P, Corless CL. Gastrointestinal stromal tumour. Lancet. 2013;382:973–983. doi: 10.1016/S0140-6736(13)62398-3. [DOI] [PubMed] [Google Scholar]
- 5.Joensuu H, Vehtari A, Riihimaki J, Nishida T, Steigen SE, Brabec P, Plank L, Nilsson B, Cirilli C, Braconi C, et al. Risk of recurrence of gastrointestinal stromal tumour after surgery: An analysis of pooled population-based cohorts. Lancet Oncol. 2012;13:265–274. doi: 10.1016/S1470-2045(11)70299-6. [DOI] [PubMed] [Google Scholar]
- 6.Pedroso FE, Raut CP, Xiao H, Yeo CJ, Koniaris LG. Has the survival rate for surgically resected gastric gastrointestinal stromal tumors improved in the tyrosine kinase inhibitor era? Ann Surg Oncol. 2012;19:1748–1758. doi: 10.1245/s10434-012-2222-9. [DOI] [PubMed] [Google Scholar]
- 7.Artinyan A, Kim J, Soriano P, Chow W, Bhatia S, Ellenhorn JD. Metastatic gastrointestinal stromal tumors in the era of imatinib: Improved survival and elimination of socioeconomic survival disparities. Cancer Epidemiol Biomarkers Prev. 2008;17:2194–2201. doi: 10.1158/1055-9965.EPI-08-0237. [DOI] [PubMed] [Google Scholar]
- 8.Corless CL, Barnett CM, Heinrich MC. Gastrointestinal stromal tumours: Origin and molecular oncology. Nat Rev Cancer. 2011;11:865–878. doi: 10.1038/nrc3143. [DOI] [PubMed] [Google Scholar]
- 9.Vernuccio F, Taibbi A, Picone DLA, Grutta L, Midiri M, Lagalla R, Lo Re G, Bartolotta TV. Imaging of gastrointestinal stromal tumors: From diagnosis to evaluation of therapeutic response. Anticancer Res. 2016;36:2639–2648. [PubMed] [Google Scholar]
- 10.Heinrich MC, Corless CL, Duensing A, McGreevey L, Chen CJ, Joseph N, Singer S, Griffith DJ, Haley A, Town A, et al. PDGFRA activating mutations in gastrointestinal stromal tumors. Science. 2003;299:708–710. doi: 10.1126/science.1079666. [DOI] [PubMed] [Google Scholar]
- 11.Ricci R. Syndromic gastrointestinal stromal tumors. Hered Cancer Clin Pract. 2016;14:15. doi: 10.1186/s13053-016-0055-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joensuu H, DeMatteo RP. The management of gastrointestinal stromal tumors: A model for targeted and multidisciplinary therapy of malignancy. Annu Rev Med. 2012;63:247–258. doi: 10.1146/annurev-med-043010-091813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gramza AW, Corless CL, Heinrich MC. Resistance to tyrosine kinase inhibitors in gastrointestinal stromal tumors. Clin Cancer Res. 2009;15:7510–7518. doi: 10.1158/1078-0432.CCR-09-0190. [DOI] [PubMed] [Google Scholar]
- 14.Ostrowski J, Polkowski M, Paziewska A, Skrzypczak M, Goryca K, Rubel T, Kokoszyñska K, Rutkowski P, Nowecki ZI, Vel Dobosz AJ, et al. Functional features of gene expression profiles differentiating gastrointestinal stromal tumours according to KIT mutations and expression. BMC Cancer. 2009;9:413. doi: 10.1186/1471-2407-9-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Astolfi A, Nannini M, Pantaleo MA, Di Battista M, Heinrich MC, Santini D, Catena F, Corless CL, Maleddu A, Saponara M, et al. A molecular portrait of gastrointestinal stromal tumors: An integrative analysis of gene expression profiling and high-resolution genomic copy number. Lab Invest. 2010;90:1285–1294. doi: 10.1038/labinvest.2010.110. [DOI] [PubMed] [Google Scholar]
- 16.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 17.R Core Team, corp-author. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna: 2017. [Google Scholar]
- 18.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paradis E, Claude J, Strimmer K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 20.Smyth GK, Michaud J, Scott HS. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 2005;21:2067–2075. doi: 10.1093/bioinformatics/bti270. [DOI] [PubMed] [Google Scholar]
- 21.Metsalu T, Vilo J. ClustVis: A web tool for visualizing clustering of multivariate data using principal component analysis and heatmap. Nucleic Acids Res. 2015;43:W566–W570. doi: 10.1093/nar/gkv468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 23.Maere S, Heymans K, Kuiper M. BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 24.Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li CY, Wei L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39(Web Server Issue):W316–W322. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. CytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8(Suppl 4):S11. doi: 10.1186/1752-0509-8-S4-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z, Galon J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahmad F, Lad P, Bhatia S, Das BR. Molecular spectrum of c-KIT and PDGFRA gene mutations in gastro intestinal stromal tumor: Determination of frequency, distribution pattern and identification of novel mutations in Indian patients. Med Oncol. 2015;32:424. doi: 10.1007/s12032-014-0424-7. [DOI] [PubMed] [Google Scholar]
- 31.Eccles SA, Welch DR. Metastasis: Recent discoveries and novel treatment strategies. Lancet. 2007;369:1742–1757. doi: 10.1016/S0140-6736(07)60781-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malanchi I, Santamaria-Martínez A, Susanto E, Peng H, Lehr HA, Delaloye JF, Huelsken J. Interactions between cancer stem cells and their niche govern metastatic colonization. Nature. 2011;481:85–89. doi: 10.1038/nature10694. [DOI] [PubMed] [Google Scholar]
- 33.Wain LV, Verwoert GC, O'Reilly PF, Shi G, Johnson T, Johnson AD, Bochud M, Rice KM, Henneman P, Smith AV, et al. Genome-wide association study identifies six new loci influencing pulse pressure and mean arterial pressure. Nat Genet. 2011;43:1005–1011. doi: 10.1038/ng.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Semba S, Itoh N, Ito M, Youssef EM, Harada M, Moriya T, Kimura W, Yamakawa M. Down-regulation of PIK3CG, a catalytic subunit of phosphatidylinositol 3-OH kinase, by CpG hypermethylation in human colorectal carcinoma. Clin Cancer Res. 2002;8:3824–3831. [PubMed] [Google Scholar]
- 35.Li J, Dang Y, Gao J, Li Y, Zou J, Shen L. PI3K/AKT/mTOR pathway is activated after imatinib secondary resistance in gastrointestinal stromal tumors (GISTs) Med Oncol. 2015;32:111. doi: 10.1007/s12032-015-0554-6. [DOI] [PubMed] [Google Scholar]
- 36.Yamaguchi R, Harada H, Hirota K. VHL-deficient renal cancer cells gain resistance to mitochondria-activating apoptosis inducers by activating AKT through the IGF1R-PI3K pathway. Tumour Biol. 2016;37:13295–13306. doi: 10.1007/s13277-016-5260-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ludovini V, Flacco A, Bianconi F, Ragusa M, Vannucci J, Bellezza G, Chiari R, Minotti V, Pistola L, Tofanetti FR, et al. Concomitant high gene copy number and protein overexpression of IGF1R and EGFR negatively affect disease-free survival of surgically resected non-small-cell-lung cancer patients. Cancer Chemother Pharmacol. 2013;71:671–680. doi: 10.1007/s00280-012-2056-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee EJ, Kang G, Kang SW, Jang KT, Lee J, Park JO, Park CK, Sohn TS, Kim S, Kim KM. GSTT1 copy number gain and ZNF overexpression are predictors of poor response to imatinib in gastrointestinal stromal tumors. PLoS One. 2013;8:e77219. doi: 10.1371/journal.pone.0077219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tarn C, Rink L, Merkel E, Flieder D, Pathak H, Koumbi D, Testa JR, Eisenberg B, von Mehren M, Godwin AK. Insulin-like growth factor 1 receptor is a potential therapeutic target for gastrointestinal stromal tumors. Proc Natl Acad Sci USA. 2008;105:8387–8392. doi: 10.1073/pnas.0803383105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Belinsky MG, Rink L, Flieder DB, Jahromi MS, Schiffman JD, Godwin AK, Mehren Mv. Overexpression of insulin-like growth factor 1 receptor and frequent mutational inactivation of SDHA in wild-type SDHB-negative gastrointestinal stromal tumors. Genes Chromosomes Cancer. 2013;52:214–224. doi: 10.1002/gcc.22023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pantaleo MA, Ravegnini G, Astolfi A, Simeon V, Nannini M, Saponara M, Urbini M, Gatto L, Indio V, Sammarini G, et al. Integrating miRNA and gene expression profiling analysis revealed regulatory networks in gastrointestinal stromal tumors. Epigenomics. 2016;8:1347–1366. doi: 10.2217/epi-2016-0030. [DOI] [PubMed] [Google Scholar]
- 42.Patel MB, Pothula SP, Xu Z, Lee AK, Goldstein D, Pirola RC, Apte MV, Wilson JS. The role of the hepatocyte growth factor/c-MET pathway in pancreatic stellate cell-endothelial cell interactions: Antiangiogenic implications in pancreatic cancer. Carcinogenesis. 2014;35:1891–1900. doi: 10.1093/carcin/bgu122. [DOI] [PubMed] [Google Scholar]
- 43.Hack SP, Bruey JM, Koeppen H. HGF/MET-directed therapeutics in gastroesophageal cancer: A review of clinical and biomarker development. Oncotarget. 2014;5:2866–2880. doi: 10.18632/oncotarget.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soto-Pantoja DR, Sipes JM, Martin-Manso G, Westwood B, Morris NL, Ghosh A, Emenaker NJ, Roberts DD. Dietary fat overcomes the protective activity of thrombospondin-1 signaling in the Apc(Min/+) model of colon cancer. Oncogenesis. 2016;5:e230. doi: 10.1038/oncsis.2016.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kashihara H, Shimada M, Yoshikawa K, Higashijima J, Tokunaga T, Nishi M, Takasu C, Ishikawa D. Correlation between thrombospondin-1 expression in non-cancer tissue and gastric carcinogenesis. Anticancer Res. 2017;37:3547–3552. doi: 10.21873/anticanres.11724. [DOI] [PubMed] [Google Scholar]
- 46.Huang T, Wang L, Liu D, Li P, Xiong H, Zhuang L, Sun L, Yuan X, Qiu H. FGF7/FGFR2 signal promotes invasion and migration in human gastric cancer through upregulation of thrombospondin-1. Int J Oncol. 2017;50:1501–1512. doi: 10.3892/ijo.2017.3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moasser MM. Targeting the function of the HER2 oncogene in human cancer therapeutics. Oncogene. 2007;26:6577–6592. doi: 10.1038/sj.onc.1210478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Cutsem E, Sagaert X, Topal B, Haustermans K, Prenen H. Gastric cancer. Lancet. 2016;388:2654–2664. doi: 10.1016/S0140-6736(16)30354-3. [DOI] [PubMed] [Google Scholar]
- 49.Roy R, Yang J, Moses MA. Matrix metalloproteinases as novel biomarkers and potential therapeutic targets in human cancer. J Clin Oncol. 2009;27:5287–5297. doi: 10.1200/JCO.2009.23.5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sebastiano M, Momi S, Falcinelli E, Bury L, Hoylaerts MF, Gresele P. A novel mechanism regulating human platelet activation by MMP-2-mediated PAR1 biased signaling. Blood. 2017;129:883–895. doi: 10.1182/blood-2016-06-724245. [DOI] [PubMed] [Google Scholar]
- 51.Kim J, Pyun JA, Cho SW, Lee K, Kwack K. Lymph node metastasis of gastric cancer is associated with the interaction between poly(ADP-ribose) polymerase 1 and matrix metallopeptidase 2. DNA Cell Biol. 2011;30:1011–1017. doi: 10.1089/dna.2011.1250. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets analyzed in the present study are available from the GEO repository, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE17743 and https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE20708.