Abstract
Idiopathic pulmonary fibrosis (IPF) is considered to be an ailment of the lungs that cannot be cured, wherein the lung tissues are characterized by increased thickness and stiffness, and/or scars. Despite the fact that extensive success has been achieved regarding the molecular diagnostics and pathobiology, the basic pathogenesis associated with IPF has not yet been fully elucidated and requires further clarification. In the current research, the changes in microRNA (miRNA) and mRNA expression in IPF were investigated through an integrative network technique. The authentic miRNA and mRNA expression profiling datasets were downloaded from Gene Expression Omnibus, followed by identification of differentially expressed miRNAs and mRNAs with use of the Significance Analysis of Microarrays algorithm. Expansion of the molecular targets associated with miRNAs was performed with the use of CyTargetLinker in Cytoscape, which was succeeded by validation with the use of mRNA array expression profiling. The incorporated miRNA-mRNA network covered 27 genes, in addition to 22 miRNAs that were associated with IPF development. As revealed by the functional enrichment analysis, the cytokine-cytokine receptor interaction and glycine, serine and threonine metabolism signalling pathways were extensively associated with IPF development. Overall, the present incorporated network illustrated the key link between miRNA and genes in IPF; in particular, it was elucidated that miR-409-5p and has-miR-376c, together with their target genes (C-C motif chemokine ligand 20 and oncostatin M), are likely candidates involved in the promotion of IPF initiation and progression.
Keywords: idiopathic pulmonary fibrosis, microRNA, mRNA, expression profiles, network
Introduction
Idiopathic pulmonary fibrosis (IPF) is a chronic and eventually fatal ailment, characterized by a progressive decrease in the function of the lungs, and is regarded as a rarely occurring ailment that influences almost 5 million individuals worldwide (1). As revealed by previous research, the 3- and 5-year mortality rates associated with IPF are below 50 percent. However, these estimations were made well before the identification of nonspecific interstitial pneumonia, which is classed as a pathological subtype of IPF in its medicinal presentation (2). Clinical phenotypes of IPF exhibit associations with old age, a history of smoking, serious physiological damage, an extensive degree of radiological damage, and pulmonary hypertension (3). Despite the fact that the diagnostics for IPF are considered to have reached an adequate level of efficacy (4), the comprehensive molecular mechanisms associated with IPF remain poorly defined.
MicroRNAs (miRNAs) are classed as a subset of small, non-coding RNA molecules of ~22 nucleotides in length, comprised of RNA and DNA, which make up one percent of genes in the animal genome; these molecules are capable of repressing gene expression through interaction with the three prime untranslated regions of target mRNAs. To date, several studies have suggested that miRNAs represent a key topic in the field of biomedical research into pulmonary fibrosis (5). Despite the fact that there have been reports on modified miRNA expression profiles in numerous respiratory ailments, including asthma, chronic obstructive pulmonary disease and cystic fibrosis in addition to IPF, comprehensive information on IPF regarding potential miRNA-mRNA interaction networks from a systematic biological perspective remains inadequate.
In order to better address this topic, in the present study, the authors depicted an miRNA-mRNA interaction network through use of publicly accessible databases based around high-throughput screening data (6). It has been hypothesized that the interaction network between miRNA and mRNA associated with IPF may provide a novel approach for the determination of gene functions and mechanisms involved in the pathogenesis of IPF.
Materials and methods
Methods
The workflow of the integrative methodology, in general, comprised four methodologies, namely data processing and miRNA/mRNA identification, miRNA target prediction, miRNA-mRNA network construction, and target validation.
Patient samples and data processing
Raw microarray datasets were downloaded from Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/). The microarrays GSE27430 for miRNA isoform and GSE24206 for mRNA isoform expression profiling that stemmed from a cluster of patients with IPF (7,8) were selected. Both of these individual datasets were produced on Agilent (Agilent Technologies, Inc., Santa Clara, CA, USA) and Affymetrix (Thermo Fisher Scientific, Inc., Waltham, MA, USA) platforms, followed by normalization with the use of quantile normalization (9) and Robust Multi-Array (10) methodology, respectively, as described previously (11).
Differentially expressed miRNA/mRNA identification
Identification of differentially expressed miRNAs/mRNAs between IPF cases and their normal controls was performed with the use of Significance Analysis of Microarrays (SAM, http://statweb.stanford.edu/~tibs/SAM/) in accordance with previously detailed methodology (12). miRNAs/mRNAs considered to be significantly differentially expressed when the fold change in expression was ≥2, together with a False Discovery Rate ≤0.05.
miRNA target prediction
For the purpose of improving understanding of the association between miRNAs and gene expression, molecular targets linked to the aberrant miRNAs were identified and mapped with the use of CyTargetLinker (http://projects.bigcat.unimaas.nl/cytargetlinker/), which is regarded as an easy-to-use Cytoscape application, providing an immediate and widespread presentation of biological networks with regards to particular miRNAs, which in the present study was visualized in the Cytoscape environment (http://www.cytoscape.org/) (13).
miRNA target validation and co-expression network integration
For the purpose of further validating the molecular targets of the aberrant miRNAs linked to IPF, the present study examined the differentially expressed genes determined from the mRNA microarray expression profiling data combined with the in silico forecasted targets from CyTargetLinker, in a bid to additionally optimize and authenticate the miRNA/mRNA co-regulatory network linked to IPF.
Enrichment analysis
Signalling pathway enrichment analysis was carried out with the use of the Database for Annotation, Visualization and Integrated Discovery (DAVID; http://david.ncifcrf.gov/) version 6.8, utilizing the Kyoto Encyclopaedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg/) annotations (14,15).
Results
A total of 22 miRNAs and 27 genes are aberrantly expressed in IPF tissues
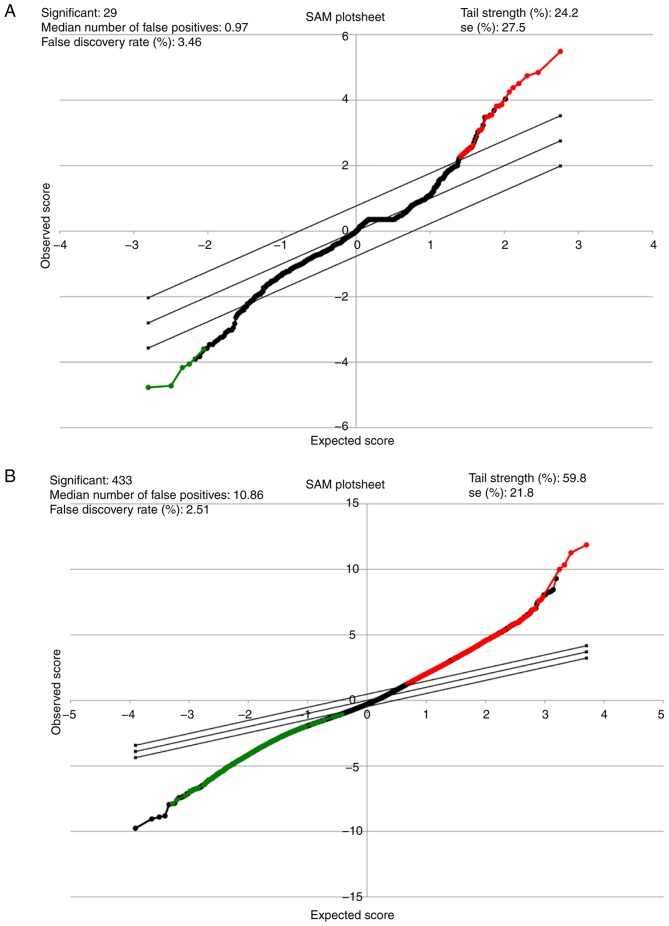
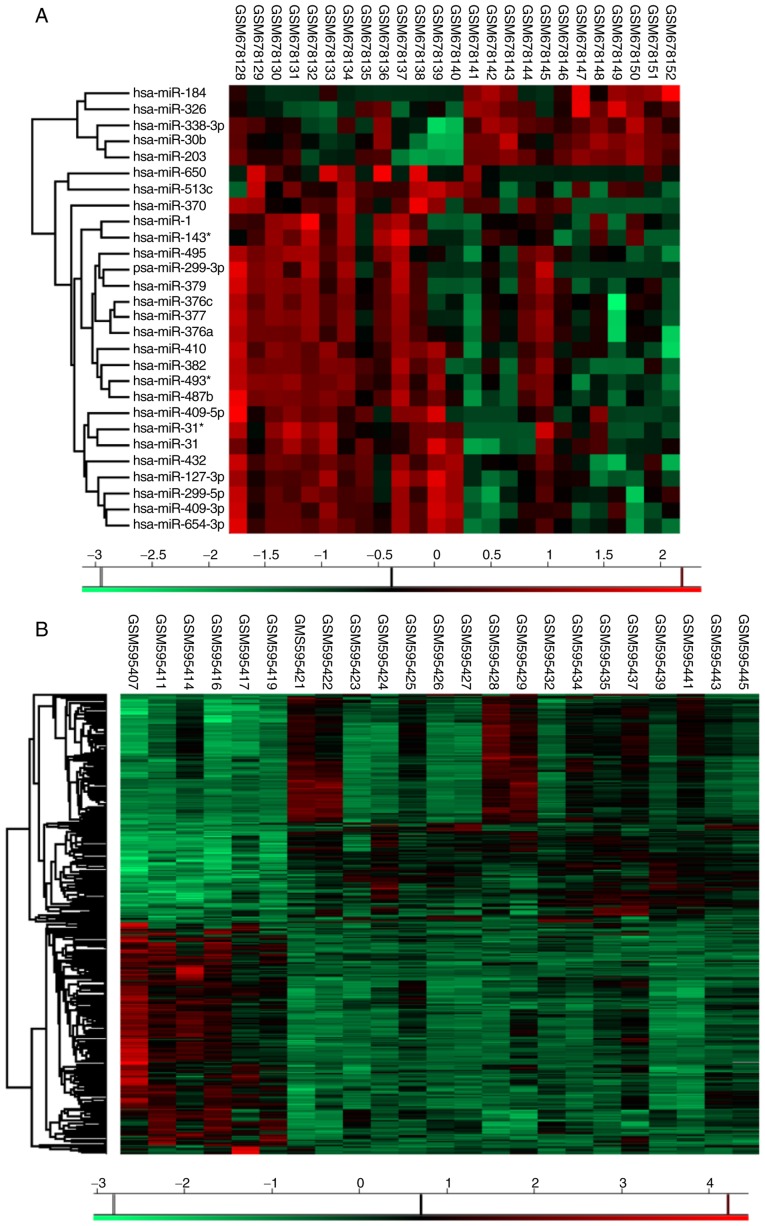
Overall, 22 miRNAs, together with 27 genes, were determined be differentially expressed in the IPF patient specimens compared with in the vehicle control tissues (Tables I and II). Making use of the SAM Plot Controller, the present study constructed a list of up-(red) and down-(green) regulated miRNAs and mRNA associated with IPF, which has been presented in Fig. 1A and B. Unsupervised hierarchical clustering analysis of these miRNAs (Fig. 2A), together with the genes (Fig. 2B) revealed a notable segregation between the fibrosis and normal lung tissues, in that the transcription profiles of miRNAs as well as mRNAs had the capability of discriminating the IPF group from the vehicle cohort, in addition to a possible application as a phenotypic discriminator.
Table I.
List of idiopathic pulmonary fibrosis associated miRNAs in the integrated networks.
| miRNAs associated with idiopathic pulmonary fibrosis | Fold change |
|---|---|
| hsa-miR-299-5p | 2.091 |
| hsa-miR-410 | 2.463 |
| hsa-miR-495 | 2.124 |
| hsa-miR-338-3p | 0.374 |
| hsa-miR-31 | 4.434 |
| hsa-miR-1 | 2.089 |
| hsa-miR-326 | 0.442 |
| hsa-miR-143* | 2.089 |
| hsa-miR-203 | 0.347 |
| hsa-miR-370 | 2.074 |
| hsa-miR-513c | 2.061 |
| hsa-miR-376a | 2.206 |
| hsa-miR-382 | 2.440 |
| hsa-miR-30b | 0.491 |
| hsa-miR-376c | 2.274 |
| hsa-miR-31* | 4.434 |
| hsa-miR-409-3p | 2.234 |
| hsa-miR-299-3p | 2.146 |
| hsa-miR-409-5p | 2.028 |
| hsa-miR-379 | 2.193 |
| hsa-miR-377 | 2.269 |
| hsa-miR-650 | 2.830 |
Table II.
List of idiopathic pulmonary fibrosis associated genes in the integrated networks.
| Genes associated with idiopathic pulmonary fibrosis | Fold change |
|---|---|
| WISP1 | 0.242 |
| FLJ36031 | 0.267 |
| CXCL14 | 6.770 |
| TTN | 0.166 |
| CASC1 | 3.076 |
| PSPH | 0.303 |
| CCL20 | 0.146 |
| CD69 | 0.297 |
| LPPR4 | 5.542 |
| NFKBIZ | 0.272 |
| CP | 3.309 |
| SIX1 | 4.568 |
| CBS | 0.286 |
| PSD3 | 4.656 |
| OSM | 0.295 |
| ATF3 | 0.216 |
| CHIT1 | 9.515 |
| APOBEC3A | 0.183 |
| COL10A1 | 5.493 |
| BMPR1B | 3.042 |
| YOD1 | 0.315 |
| PTGFRN | 3.658 |
| IGHM | 3.055 |
| AFF3 | 0.202 |
| C10orf81 | 6.874 |
| CAPS2 | 3.019 |
| SLC25A37 | 0.318 |
Figure 1.
Significance analysis of microarrays (SAM) for miRNA and mRNA from patients with IPF. (A) Representative SAM plot sheet for GSE24206 (n=23) and (B) GSE27430 (n=25) in patients with IPF compared with the normal lung tissues. In the SAM plot sheet, red represents miRNAs or genes upregulated, while green represents miRNAs or genes downregulated. miRNA, microRNA; SAM, significance analysis of microarrays; IPF, idiopathic pulmonary fibrosis.
Figure 2.
Unsupervised hierarchical clustering analysis. Differentially expressed (A) miRNAs and (B) genes in idiopathic pulmonary fibrosis patients. In these heatmaps, red represents gene upregulated and green represents genes downregulated. miRNA, microRNA; SAM, significance analysis of microarrays; IPF, idiopathic pulmonary fibrosis.
Potential miRNA targets forecast using CyTargetLinker
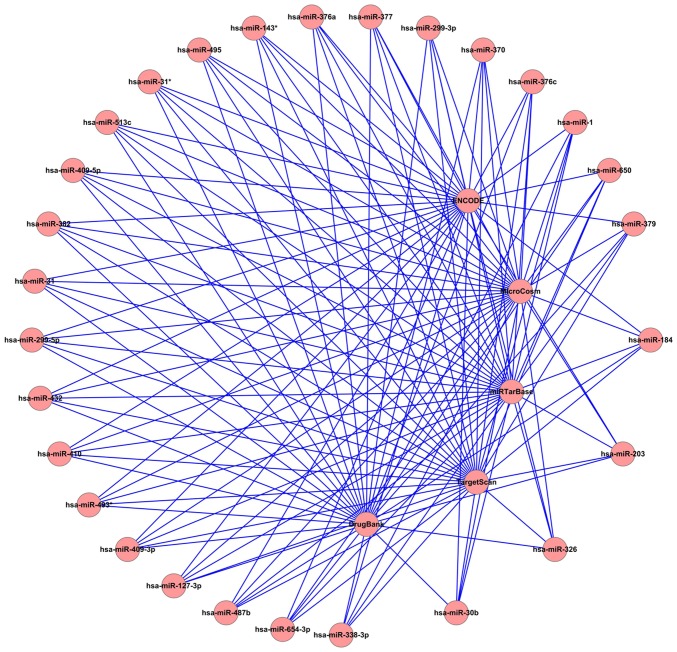
Mapping of the molecular targets that were linked to the aberrantly expressed miRNAs was conducted with the use of CyTargetLinker, by means of integration of Homo sapiens data from different microRNA-target, transcription factor-target and drug-target databases, including MicroCosm (http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/), TargetScan (http://www.targetscan.org/), miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/), ENCODE (https://genome.ucsc.edu/ENCODE/) and DrugBank (http://www.drugbank.ca/). A representative genetic interaction network of miRNAs associated with idiopathic pulmonary fibrosis is presented in Fig. 3.
Figure 3.
Representative genetic interaction network of miRNAs associated with idiopathic pulmonary fibrosis in the Cytoscape environment. In this diagrammatic sketch, the outer circles represent the differently expressed miRNAs screened from the microarray (GSE24206). The central circles represent the databases of MicroCosm, TargetScan, miRTarBase, ENCODE and DrugBank. A total of 17,879 targets from MicroCosm, 9,378 targets from TargetScan and 1,137 targets from miRTarBase were subsequently identified.
Target validation and miRNA target gene co-expression network construction
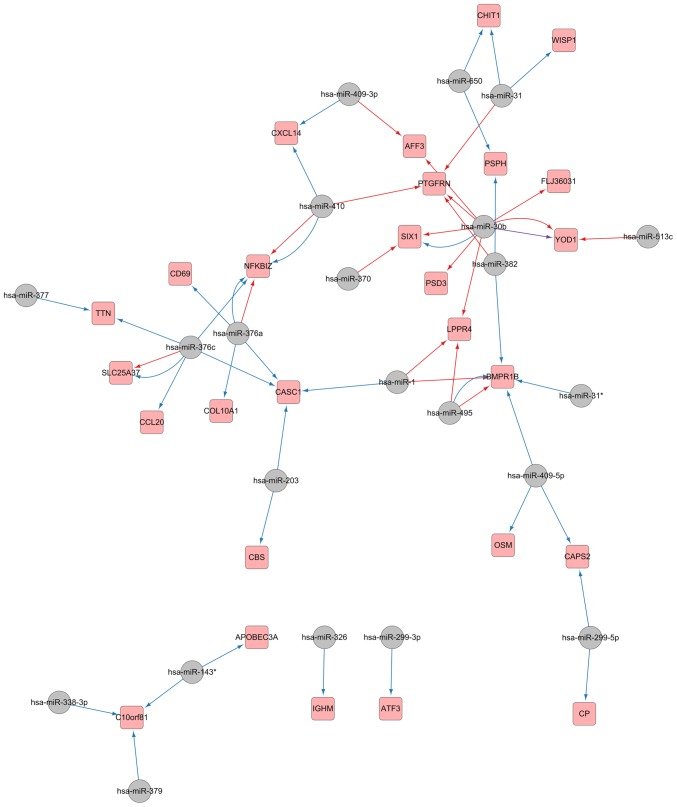
In order to further understanding of the regulatory network linked to pulmonary fibrosis, the confirmed target genes attained from the mRNA array were mapped into the forecasted miRNA-target network; furthermore, optimization and visualization of the miRNA-mRNA regulatory network were performed with the use of Cytoscape. As evident in Fig. 4, 22 miRNAs were eventually observed as having an association with IPF initiation by means of their regulation of 27 validated targets.
Figure 4.
Validated microRNA-mRNA interaction obtained from the predicted network and the mRNA microarray of idiopathic pulmonary fibrosis. Grey circles represent microRNAs, and pink squares represent their targets. Blue arrows represent regulatory interaction network identified from MicroCosm, red from TargetScan and purple from miRTarBase.
Signalling pathway enrichment analysis
For the purpose of obtaining an improved understanding of the biological function of the miRNA-mRNA interaction network that was implicated in IPF, the enriched KEGG pathways of the target genes established were assessed with the use of DAVID. With the use of an adjusted P-value cut-off for correction, cytokine-cytokine receptor interaction [P=6.9E−3, genes C-C motif chemokine ligand 20 (CCL20), oncostatin M (OSM), C-X-C motif chemokine ligand 14 and bone morphogenetic protein receptor type 1B] and glycine, serine and threonine metabolism (P=7.0E−2, genes cystathionine-β-synthase and phosphoserine phosphatase) were determined as being substantially enriched pathways in IPF.
Discussion
Owing to the development of high-throughput screening approaches, the integration of miRNA-mediated regulation into biological network analyses provides a novel perspective with regard to the pathogenesis and progression of diseases. Previously, Maimaiti et al (16) mapped a miRNA-mRNA network that demonstrated the malignant commencement and transformation of oral leukoplakia. In particular, they shed light on the significance of miR-129-5p, miR-339-5p and miR-31 in mediating the evolution of oral leukoplakia from the non-malignant to the aggressive form. Furthermore, through microRNA-mRNA regulatory network analyses, Li et al (17) demonstrated the underlying molecular mechanism of glioma malignant progression. Other similar studies on miRNAs as well as their target gene interaction networks in conditions including leukaemia (18), prostate cancer (19), premature ovarian failure (20) and pancreatic ductal adenocarcinoma (21) also identified target miRNAs, as well as mRNAs that appeared likely to aid in diagnosing and treating the respective ailments.
As single-stranded RNAs, it has been demonstrated that miRNAs perform regulation of gene expression in pulmonary fibrosis. Previously, let-7d has been observed to be downregulated in IPF lung tissues, whereas let-7d inhibition gives rise to a substantial epithelial to mesenchymal shift in a bleomycin-stimulated lung fibrosis mouse model (22,23). As reported by a number of other research studies, miR-200 family members may serve key roles in IPF. In particular, it has been documented that downregulation of miR-200a and miR-200c is evident in the lungs of mice with empirical pulmonary fibrosis, and notably, in the lungs of the patients with IPF (24). Additionally, there are reports suggesting that miR-29 (25), miR-375 (26), miR-145 (27), miR-96 (28), miR-26a (29) and the miR-17-92 cluster (30) have crucial importance in lung epithelial cell homeostasis and IPF evolution.
In the current research, it was indicated that the cytokine-cytokine receptor interaction and glycine, serine and threonine metabolism signalling pathways are involved in IPF, which was consistent with the results of a preliminary bioinformatics analysis (31). Specifically, both the CCL20 and OSM genes were observed to be downregulated in IPF patient specimens in comparison with normal lung specimens, and appeared to be the fundamental hubs regulated in the cytokine-cytokine receptor interaction pathway according to the enrichment analysis (P≤0.05). As one of the genes present in the integrated network, CCL20 has been observed to be involved in a number of physiological and pathological mechanisms, including in cancer and inflammatory ailments (32,33). Being a minor cytokine associated with the CC chemokine family, CCL20 has been found to be a critical inflammatory regulator associated with chronically inflamed fibrosis, in addition to being produced in the regions of epithelial hyperplasia, which results in the development and maintenance of the ectopic lymphoid aggregates (34). In accordance with the network analysis, CCL20 was observed as being a direct target of has-miR-376c in IPF commencement. Nevertheless, comprehensive information concerning whether and how miR-376c regulates IPF remains to be elucidated.
Another gene identified in the interaction network was OSM, which has been observed to be downregulated in IPF tissues, and serves as a predictor of inflammation (35). OSM is regarded as a pleiotropic cytokine, associated with the interleukin 6 cohort of cytokines. As an IL-6 family cytokine, OSM reportedly is involved in inflammation as well as modulation of the extracellular matrix. Additionally, OSM has been observed to be upregulated in the bronchoalveolar lavage fluid of patients with idiopathic pulmonary fibrosis, wherein it stimulates the pulmonary inflammation and fibrosis (36). Notably, OSM has been observed as a hub gene regulated by miR-409-5p. As an oncomiR, miR-409-5p has been identified to be capable of promoting tumorigenesis, in addition to epithelial-to-mesenchymal transition and even bone metastasis in prostate cancer (37). Nevertheless, the potential function of miR-409-5p in lung fibrosis has not been defined to date.
In conclusion, by application of publicly accessible datasets obtained from the GEO database, the present study developed an miRNA-mRNA interaction network for IPF, and identified certain miRNA candidates (including miR-409-5p as well as has-miR-376c), in addition to genes (CCL20 and OSM) implicated in IPF commencement and evolution. Overall, the findings suggest potential targets for application in IPF diagnostics and therapeutics.
Acknowledgements
The authors would like to thank Gene Expression Omnibus database for making their data readily available to the scientific community.
Glossary
Abbreviations
- PF
idiopathic pulmonary fibrosis
- miRNA
microRNA
- GEO
Gene Expression Omnibus
- SAM
significance analysis of microarrays
- DAVID
Database for Annotation, Visualization and Integrated Discovery
- KEGG
Kyoto Encyclopaedia of Genes and Genomes
Funding
This study was supported by Hunan Provincial Science and Technology Department Project (grant no. 2015SK20402). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
LW, LZ, QC and HZ developed the concept of the paper, designed the study, analyzed the data and interpreted the data. LW, LZ, WH and HZ performed the experiments. WH and HZ provided valuable support and input of the project, and wrote the manuscript. All the authors approved the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Meltzer EB, Noble PW. Idiopathic pulmonary fibrosis. Orphanet J Rare Dis. 2008;3:8. doi: 10.1186/1750-1172-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cai M, Zhu M, Ban C, Su J, Ye Q, Liu Y, Zhao W, Wang C, Dai H. Clinical features and outcomes of 210 patients with idiopathic pulmonary fibrosis. Chin Med J (Engl) 2014;127:1868–1873. [PubMed] [Google Scholar]
- 3.Ley B, Collard HR, King TE., Jr Clinical course and prediction of survival in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2011;183:431–440. doi: 10.1164/rccm.201006-0894CI. [DOI] [PubMed] [Google Scholar]
- 4.King TE, Jr, Pardo A, Selman M. Idiopathic pulmonary fibrosis. Lancet. 2011;378:1949–1961. doi: 10.1016/S0140-6736(11)60052-4. [DOI] [PubMed] [Google Scholar]
- 5.Rajasekaran S, Rajaguru P, Sudhakar Gandhi PS. MicroRNAs as potential targets for progressive pulmonary fibrosis. Front Pharmacol. 2015;6:254. doi: 10.3389/fphar.2015.00254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu W, Chen YP. Computational developments in microRNA-regulated protein-protein interactions. BMC Syst Biol. 2014;8:14. doi: 10.1186/1752-0509-8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meltzer EB, Barry WT, D'Amico TA, Davis RD, Lin SS, Onaitis MW, Morrison LD, Sporn TA, Steele MP, Noble PW. Bayesian probit regression model for the diagnosis of pulmonary fibrosis: Proof-of-principle. BMC Med Genomics. 2011;4:70. doi: 10.1186/1755-8794-4-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Milosevic J, Pandit K, Magister M, Rabinovich E, Ellwanger DC, Yu G, Vuga LJ, Weksler B, Benos PV, Gibson KF, et al. Profibrotic role of miR-154 in pulmonary fibrosis. Am J Respir Cell Mol Biol. 2012;47:879–887. doi: 10.1165/rcmb.2011-0377OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qiu X, Wu H, Hu R. The impact of quantile and rank normalization procedures on the testing power of gene differential expression analysis. BMC Bioinformatics. 2013;14:124. doi: 10.1186/1471-2105-14-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan S, Pan Z, Geng Q, Li X, Wang Y, An Y, Xu Y, Tie L, Pan Y, Li X. Layered signaling regulatory networks analysis of gene expression involved in malignant tumorigenesis of non-resolving ulcerative colitis via integration of cross-study microarray profiles. PLoS One. 2013;8:e67142. doi: 10.1371/journal.pone.0067142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan S, Li X, Tie L, Pan Y, Li X. KIAA0101 is associated with human renal cell carcinoma proliferation and migration induced by erythropoietin. Oncotarget. 2016;7:13520–13537. doi: 10.18632/oncotarget.5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan S, Geng Q, Pan Z, Li X, Tie L, Pan Y, Li X. Clarifying off-target effects for torcetrapib using network pharmacology and reverse docking approach. BMC Syst Biol. 2012;6:152. doi: 10.1186/1752-0509-6-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kutmon M, Kelder T, Mandaviya P, Evelo CT, Coort SL. CyTargetLinker: A cytoscape app to integrate regulatory interactions in network analysis. PLoS One. 2013;8:e82160. doi: 10.1371/journal.pone.0082160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 15.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maimaiti A, Abudoukeremu K, Tie L, Pan Y, Li X. MicroRNA expression profiling and functional annotation analysis of their targets associated with the malignant transformation of oral leukoplakia. Gene. 2015;558:271–277. doi: 10.1016/j.gene.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 17.Li Y, Xu J, Chen H, Bai J, Li S, Zhao Z, Shao T, Jiang T, Ren H, Kang C, Li X. Comprehensive analysis of the functional microRNA-mRNA regulatory network identifies miRNA signatures associated with glioma malignant progression. Nucleic Acids Res. 2013;41:e203. doi: 10.1093/nar/gkt1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong L, Bi KH, Huang N, Chen CY. Biological analysis of chronic lymphocytic leukemia: Integration of mRNA and microRNA expression profiles. Genet Mol Res. 2016;15 doi: 10.4238/gmr.15017170. [DOI] [PubMed] [Google Scholar]
- 19.Zhu J, Wang S, Zhang W, Qiu J, Shan Y, Yang D, Shen B. Screening key microRNAs for castration-resistant prostate cancer based on miRNA/mRNA functional synergistic network. Oncotarget. 2015;6:43819–43830. doi: 10.18632/oncotarget.6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J, Fan S, Han D, Xie J, Kuang H, Ge P. Microarray gene expression profiling and bioinformatics analysis of premature ovarian failure in a rat model. Exp Mol Pathol. 2014;97:535–541. doi: 10.1016/j.yexmp.2014.10.015. [DOI] [PubMed] [Google Scholar]
- 21.Liu PF, Jiang WH, Han YT, He LF, Zhang HL, Ren H. Integrated microRNA-mRNA analysis of pancreatic ductal adenocarcinoma. Genet Mol Res. 2015;14:10288–10297. doi: 10.4238/2015.August.28.14. [DOI] [PubMed] [Google Scholar]
- 22.Pandit KV, Corcoran D, Yousef H, Yarlagadda M, Tzouvelekis A, Gibson KF, Konishi K, Yousem SA, Singh M, Handley D, et al. Inhibition and role of let-7d in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2010;182:220–229. doi: 10.1164/rccm.200911-1698OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pandit KV, Milosevic J, Kaminski N. MicroRNAs in idiopathic pulmonary fibrosis. Transl Res. 2011;157:191–199. doi: 10.1016/j.trsl.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 24.Yang S, Banerjee S, de Freitas A, Sanders YY, Ding Q, Matalon S, Thannickal VJ, Abraham E, Liu G. Participation of miR-200 in pulmonary fibrosis. Am J Pathol. 2012;180:484–493. doi: 10.1016/j.ajpath.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cushing L, Kuang PP, Qian J, Shao F, Wu J, Little F, Thannickal VJ, Cardoso WV, Lü J. miR-29 is a major regulator of genes associated with pulmonary fibrosis. Am J Respir Cell Mol Biol. 2011;45:287–294. doi: 10.1165/rcmb.2010-0323OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, Huang C, Reddy Chintagari N, Bhaskaran M, Weng T, Guo Y, Xiao X, Liu L. miR-375 regulates rat alveolar epithelial cell trans-differentiation by inhibiting Wnt/β-catenin pathway. Nucleic Acids Res. 2013;41:3833–3844. doi: 10.1093/nar/gks1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang S, Cui H, Xie N, Icyuz M, Banerjee S, Antony VB, Abraham E, Thannickal VJ, Liu G. miR-145 regulates myofibroblast differentiation and lung fibrosis. FASEB J. 2013;27:2382–2391. doi: 10.1096/fj.12-219493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nho RS, Im J, Ho YY, Hergert P. MicroRNA-96 inhibits FoxO3a function in IPF fibroblasts on type I collagen matrix. Am J Physiol Lung Cell Mol Physiol. 2014;307:L632–L642. doi: 10.1152/ajplung.00127.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang H, Xu C, Pan Z, Zhang Y, Xu Z, Chen Y, Li T, Li X, Liu Y, Huangfu L, et al. The antifibrotic effects and mechanisms of microRNA-26a action in idiopathic pulmonary fibrosis. Mol Ther. 2014;22:1122–1133. doi: 10.1038/mt.2014.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dakhlallah D, Batte K, Wang Y, Cantemir-Stone CZ, Yan P, Nuovo G, Mikhail A, Hitchcock CL, Wright VP, Nana-Sinkam SP, et al. Epigenetic regulation of miR-17~92 contributes to the pathogenesis of pulmonary fibrosis. Am J Respir Crit Care Med. 2013;187:397–405. doi: 10.1164/rccm.201205-0888OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang G, Yang L, Wang W, Wang J, Wang J, Xu Z. Discovery and validation of extracellular/circulating microRNAs during idiopathic pulmonary fibrosis disease progression. Gene. 2015;562:138–144. doi: 10.1016/j.gene.2015.02.065. [DOI] [PubMed] [Google Scholar]
- 32.Frick VO, Rubie C, Keilholz U, Ghadjar P. Chemokine/chemokine receptor pair CCL20/CCR6 in human colorectal malignancy: An overview. World J Gastroenterol. 2016;22:833–841. doi: 10.3748/wjg.v22.i2.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheluvappa R. Experimental appendicitis and appendectomy modulate the CCL20-CCR6 axis to limit inflammatory colitis pathology. Int J Colorectal Dis. 2014;29:1181–1188. doi: 10.1007/s00384-014-1936-5. [DOI] [PubMed] [Google Scholar]
- 34.Marchal-Sommé J, Uzunhan Y, Marchand-Adam S, Kambouchner M, Valeyre D, Crestani B, Soler P. Dendritic cells accumulate in human fibrotic interstitial lung disease. Am J Respir Crit Care Med. 2007;176:1007–1014. doi: 10.1164/rccm.200609-1347OC. [DOI] [PubMed] [Google Scholar]
- 35.O'Hara KA, Kedda MA, Thompson PJ, Knight DA. Oncostatin M: An interleukin-6-like cytokine relevant to airway remodelling and the pathogenesis of asthma. Clin Exp Allergy. 2003;33:1026–1032. doi: 10.1046/j.1365-2222.2003.01714.x. [DOI] [PubMed] [Google Scholar]
- 36.Mozaffarian A, Brewer AW, Trueblood ES, Luzina IG, Todd NW, Atamas SP, Arnett HA. Mechanisms of oncostatin M-induced pulmonary inflammation and fibrosis. J Immunol. 2008;181:7243–7253. doi: 10.4049/jimmunol.181.10.7243. [DOI] [PubMed] [Google Scholar]
- 37.Josson S, Gururajan M, Hu P, Shao C, Chu GY, Zhau HE, Liu C, Lao K, Lu CL, Lu YT, et al. miR-409-3p/-5p promotes tumorigenesis, epithelial-to-mesenchymal transition, and bone metastasis of human prostate cancer. Clin Cancer Res. 2014;20:4636–4646. doi: 10.1158/1078-0432.CCR-14-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.