Abstract
Aberrant DNA methylation is the most common type of epigenetic alteration and is associated with many types of cancer. Although previous studies have provided a few novel DNA methylation markers in hepatocellular carcinoma (HCC), specific DNA methylation patterns and comparisons of the aberrant alterations in methylation between HCC and normal liver cell lines have not yet been reported. Therefore, in the present study the Illumina Infinium HumanMethylation 450K BeadChip was employed to identify the genome-wide aberrant DNA methylation profiles of Huh7 and L02 cells. Following Bonferroni adjustment, 102,254 differentially methylated CpG sites (covering 26,511 genes) were detected between Huh7 and L02 cells. Of those CpG sites, 62,702 (61.3%) sites were hypermethylated (covering 12,665 genes) and 39,552 (38.7%) sites were hypomethylated (covering 13,846 genes). The results of the present study indicated that 40.3% of the CpG sites were in CpG island regions, 20.7% were in CpG shores and 8.8% were in shelf regions. A total of 57.3% hypermethylated CpG sites and 39.4% of the hypomethylated CpG sites had a |β-Difference| ≥50%. Within the significant differentially methylated CpG sites, 490 genes were located within 598 differentially methylated regions. Gene Ontology enrichment analysis revealed that 2,107 differentially methylated genes were associated with ‘biological process’, 13,351 differentially methylated genes were associated with ‘molecular function’, and 18,041 differentially methylated genes were associated with ‘cellular component’. Kyoto Encyclopedia of Genes and Genomes pathway-based analysis revealed 43 signaling pathways that were associated with 5,195 differentially methylated genes. These results demonstrated that aberrant DNA methylation may be a key and common event underlying the tumorigenesis of Huh7 cells. The present study also identified many subsets of hypo- or hyper-methylated CpG sites, genes and signaling pathways, which have an importance in the occurrence and development of HCC.
Keywords: DNA methylation, genome-wide, hepatocellular carcinoma, BeadChip
Introduction
Worldwide, primary liver cancer (PLC) is the second leading cause of cancer-associated mortality in poorly developed countries and the sixth in more developed countries (1). Of the ~782,500 new annual cases of PLC, China accounts for >50% of the associated incidence (2). In 2015, PLC was the fourth most common type of cancer and the third most common cause of cancer-associated mortality in China (3). The majority of PLC cases occurring worldwide are cases of hepatocellular carcinoma (HCC) (1). Many risk factors can induce the development of HCC, including chronic hepatitis B virus or hepatitis C virus infections, chronic alcoholic cirrhosis and high doses of aflatoxin B1 (4). Although a number of studies have identified a few of the molecular alterations associated with the pathogenesis of HCC, the main mechanism underlying HCC is still unclear.
Previous studies have demonstrated that epigenetic alterations are one of the many early events that occur during tumorigenesis (5,6). DNA methylation is the main epigenetic feature in regulating gene transcriptional regulation and preserving genome stability; however, aberrant DNA methylation can lead to the inactivation of tumor suppressor genes or activation of oncogenes, which eventually induces the development of many types of cancer (7,8). A number of studies have also reported alterations in one or several genes at one time; the abnormal methylation of genes, including Ras association domain family member 1 (9), p16, postmeiotic segregation increased 2, MutL homolog 1, MutS homolog 2 (10), Adenomatosis polyposis coli (11) and glutathione S-transferase Pi 1 (12,13), has also been associated with HCC. Shen et al (14) used Illumina Infinium HumanMethylation 27K arrays to analyze 27,578 CpG sites covering 14,495 genes in paired HCC tumor and adjacent non-tumor tissues. The Illumina Infinium HumanMethylation 450K BeadChip represents a significant improvement in the detection of CpG sites (482,421 CpG and 3,091 non-CpG sites), covers 99% of RefSeq genes with multiple sites in annotated promoters (1,500 or 200 bp upstream of the transcription start site), 5′-untranslated regions (UTRs), first exons, gene body, and 3′-UTRs (15). Previously, aberrant DNA hypermethylation of CpG islands was reported to induce the inactivation of tumor suppressor genes (16), which was thought to contribute to tumorigenesis (17). Recently, previous studies have revealed that cancer-associated aberrant DNA methylation not only occurs within CpG islands but may also be detected within CpG shores or CpG shelves (18–20).
To the best of our knowledge, no research analyzing the genome-wide DNA methylation status within a HCC cell line using the Illumina Infinium HumanMethylation 450K BeadChip has been conducted. Therefore, in the present study, the Illumina 450K Methylation BeadChip was employed to screen promoter DNA methylation and the expression profiles of methylated genes in a human hepatocellular carcinoma cell line (Huh7 cells) and in a human normal liver cell line (L02 cells). The results may aid the characterization of differentially methylated CpG sites, regions and genes associated with the pathogenesis of HCC, thereby improving our current understanding of the methylation mechanisms underlying the development and progression of HCC.
Materials and methods
Cell culture
The human HCC cell line, Huh7 and the human normal liver cell line, L02, were purchased from the Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China). Huh7 cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA), while L02 cells were maintained in RPMI-1640 (Invitrogen; Thermo Fisher Scientific, Inc.). The cell lines were supplemented with 100 U/ml penicillin and 100 g/ml streptomycin in the presence of 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) and incubated in a humidified atmosphere containing 5% CO2 at 37°C.
DNA preparation and Illumina Infinium HumanMethylation 450K BeadChip assay
DNA was extracted from the two cell lines (Huh7 and L02) using a QIAamp DNA Micro kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer's protocol. Bisulfite modification of 1 µg DNA was conducted using an EZ DNA Methylation kit (Zymo Research Corp., Irvine, CA, USA) according to the manufacturer's protocol. The Illumina Infinium HumanMethylation 450K BeadChip assay was performed according to Illumina's standard protocol (Illumina, Inc., San Diego, CA, USA). Experiments with Huh7 and L02 cells were performed in triplicate to avoid false positive and false negative results.
Statistical analysis
The methylation data were processed with the Methylation Module of GenomeStudio software (Methylation v1.9; Illumina, Inc.). The methylation levels of the CpG sites were calculated as β-values: β=intensity of the methylated allele (M)/[intensity of the unmethylated allele (U) + M + 100] (15). A t-test, in addition to analysis of variance with Bonferroni correction for multiple comparisons was used to compare differentially methylated CpG sites between Huh7 and L02 cells. The differentially methylated CpG sites were defined as sites with Adjusted P-values of ≤0.05 and |β-Difference|≥0.2. Methylation measures with a detection P>0.05 and CpG coverage <95% were excluded (14). For the selection of candidate CpG sites that had significant differences between Huh7 and L02 cell methylation levels, the following additional filtering criteria were applied: i) Adjusted P≤0.05, which corresponds to a raw P-value of ≤1.06×10−7; ii) for significantly hypermethylated CpG sites, the |β-Difference| in the methylation levels between Huh7 and L02 cells was >20%, and the mean methylation level for L02 was <25%; and iii) for significantly hypomethylated CpG sites, the methylation level |β-Difference| between L02 and Huh7 cells was >20%, and the mean methylation level for Huh7 cells was <25% (14).
Functional annotation of differentially methylated genes
The genes for which the CpG sites corresponded with differential methylation levels were determined using Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway (www.genome.jp/kegg/) and Gene Ontology (GO; www.geneontology.org/) databases to analyze the pathway and enrichment information of these genes.
Results
Global DNA methylation in Huh7 and LO2 cells
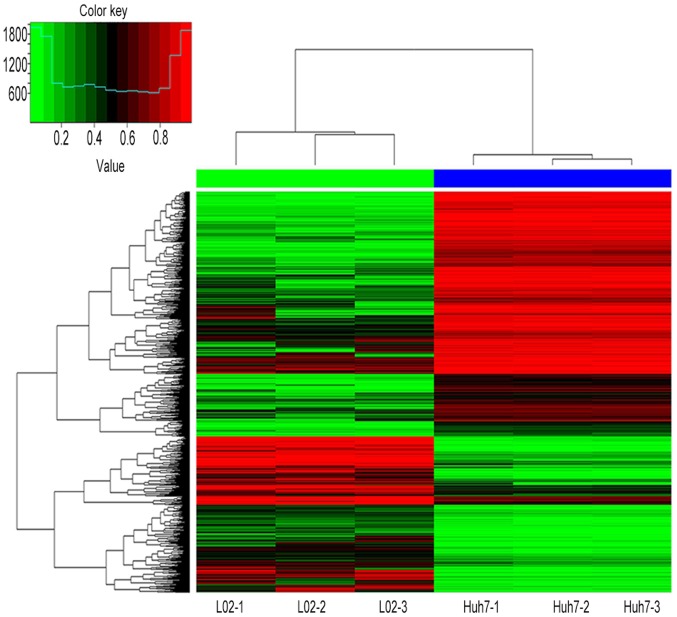
Following a t-test with Bonferroni correction for multiple comparisons, 102,254 differentially methylated CpG sites (covering 26,511 genes) were detected between Huh7 and L02 cells. Within these CpG sites, 62,702 (61.3%) sites were hypermethylated (covering 12,665 genes), and 39,552 (38.7%) sites were hypomethylated (covering 13,846 genes). The results suggested that aberrant DNA methylation may a very common event in Huh7 cells, and alterations in hypermethylation were more frequently observed than hypomethylation. Figs. 1 and 2 present hierarchical cluster analysis of the differentially methylated CpG sites and genes that distinguish Huh7 from L02 cells.
Figure 1.
Hierarchical cluster analysis of all of the differentially methylated CpG sites between Huh7 and L02 cells.
Figure 2.
Hierarchical cluster analysis of all of the differentially methylated genes between Huh7 and L02 cells.
In addition, the results revealed that there were 41,178 (40.3%) CpG sites located in CpG island regions, 21,150 (20.7%) CpG sites were within CpG shores, 9,030 (8.8%) CpG sites were in shelves, and 30,896 (30.2%) CpG sites were in open sea (Table I). Furthermore, the results also demonstrated that within different regions, the distribution of hypo- or hypermethylated CpG sites differed: In CpG island regions, 19,462 (47.3%) CpG sites were hypermethylated and 21,716 (52.7%) CpG sites were hypomethylated. In CpG shore regions, 13,729 (64.9%) CpG sites were hypermethylated and 7,421 (35.1%) CpG sites were hypomethylated. In shelf regions, 7,340 (81.3%) CpG sites were hypermethylated and 1,690 (18.7%) CpG sites were hypomethylated.
Table I.
Distribution of all of the differentially methylated CpG sites.
| Type | All methylated CpG sites, n (%) | Hypermethylated CpG sites, n (%) | Hypomethylation CpG sites, n (%) |
|---|---|---|---|
| CpG island | 41,178 (40.3) | 19,462 (47.3) | 21,716 (52.7) |
| CpG shores | 21,150 (20.7) | 13,729 (64.9) | 7,421 (35.1) |
| CpG shelves | 9,030 (8.8) | 7,340 (81.3) | 1,690 (18.7) |
| Open sea | 30,896 (30.2) | 22,171 (71.8) | 8,725 (28.2) |
| Total | 102,254 | 62,702 | 39,552 |
Frequency distribution of differentially methylated CpG sites between Huh7 and L02 cells. In the results of the present study, 35,937 (57.3%) hypermethylated CpG sites and 15,587 (39.4%) hypomethylated CpG sites were observed to have an |β-Difference|≥50%. A total of 18,529 (29.5%) of hypermethylated CpG sites and 14,177 (35.9%) hypomethylated CpG sites had an |β-Difference|≥30% but <50%. A total of 8,236 (13.1%) hypermethylated CpG sites and 9,788 (24.7%) of hypomethylated CpG sites had an |β-Difference|<30% but ≥20% (Table II). Collectively, these results revealed that DNA aberrant hypermethylation in Huh7 cells was more frequent than in L02 cells, which may serve a potential role in genomic instability.
Table II.
Frequency distribution of all of the differentially methylated CpG sites in Huh7 and L02 cells by methylation status.
| |β-difference|, % | Hypermethylated CpG sites, n (%) | Cumulative, % | Hypomethylated CpG sites, n (%) | Cumulative, % | Total CpG sites, n (%) | Cumulative, % |
|---|---|---|---|---|---|---|
| ≥60 | 25,585 (40.8) | 40.8 | 10,796 (27.3) | 27.3 | 36,381 (35.6) | 35.6 |
| 50≤x<60 | 10,352 (16.5) | 57.3 | 4,791 (12.1) | 39.4 | 15,134 (14.8) | 50.4 |
| 40≤x<50 | 9,681 (15.4) | 72.7 | 6,159 (15.6) | 54.0 | 15,840 (15.5) | 65.9 |
| 30≤x<40 | 8,848 (14.1) | 86.8 | 8,018 (20.3) | 75.3 | 16,866 (16.5) | 82.4 |
| 20≤x<30 | 8,236 (13.1) | 100.0 | 9,788 (24.7) | 100.0 | 18,024 (17.6) | 100.0 |
| Total | 62,702 | – | 39,552 | – | 102,254 | – |
Significant differentially methylated CpG sites and genes
To reduce the potential impact of an extreme β value on methylation differences, the present study applied stringent criteria to select potentially biologically important CpG sites (14). A total of 5,285 significantly hypermethylated CpG sites (covering 3,222 genes) and 2,659 significantly hypomethylated CpG sites (covering 2,204 genes) were observed. For the significantly hypermethylated CpG sites, there were 1,544 sites in CpG islands, 1,137 sites in CpG shores, 655 sites within CpG shelves and 1,949 sites in open sea regions. By contrast, for the significantly hypomethylated CpG sites, there were 1,201 sites in CpG islands, 632 sites in CpG shores, 133 sites in CpG shelves and 693 sites in open sea regions (Table III). The top 20 differentially hypermethylated and hypomethylated sites and genes are presented in the Tables IV and V.
Table III.
Distribution of genomic regions for significant differentially methylated CpG sites in Huh7 cells when compared with L02 cells.
| Type | Hypermethylated CpG sites, n | Hypomethylated CpG sites, n |
|---|---|---|
| CpG island | 1,544 | 1,201 |
| CpG shores | 1,137 | 632 |
| CpG shelves | 655 | 133 |
| Open sea | 1,949 | 693 |
| Total | 5,285 | 2,659 |
Table IV.
Top 20 significant hypermethylated CpG sites and genes within differentially methylated regions in Huh7 cells when compared with L02 cells.
| CpG sites | Adjust P-value | |β-difference| | Mean Huh | Mean L02 | Hypermethylated genes |
|---|---|---|---|---|---|
| cg11058366 | 2.09×10−6 | 0.924 | 0.934 | 0.010 | ERBB4 |
| cg13245152 | 1.69×10−6 | 0.863 | 0.876 | 0.013 | PAX6 |
| cg25758545 | 1.69×10−6 | 0.863 | 0.876 | 0.013 | SALL4 |
| cg14950829 | 1.69×10−6 | 0.925 | 0.940 | 0.015 | PCDH8 |
| cg09260089 | 1.69×10−6 | 0.952 | 0.968 | 0.016 | NKX6-2 |
| cg11459773 | 1.69×10−6 | 0.876 | 0.891 | 0.015 | BCL3 |
| cg12989574 | 1.69×10−6 | 0.965 | 0.983 | 0.018 | GPC6 |
| cg03129384 | 1.69×10−6 | 0.956 | 0.974 | 0.018 | FAM196A; DOCK1 |
| cg03396151 | 1.69×10−6 | 0.929 | 0.947 | 0.018 | MEIS2 |
| cg04556126 | 1.69×10−6 | 0.920 | 0.938 | 0.018 | ZIC4 |
| cg20317123 | 1.69×10−6 | 0.947 | 0.966 | 0.019 | TCF21 |
| cg21062760 | 1.69×10−6 | 0.876 | 0.894 | 0.018 | ZBTB32 |
| cg09454560 | 1.99×10−6 | 0.624 | 0.636 | 0.013 | LRFN2 |
| cg12090740 | 1.69×10−6 | 0.892 | 0.911 | 0.020 | BCL2 |
| cg24249411 | 1.69×10−6 | 0.887 | 0.907 | 0.020 | BDNF |
| cg00057722 | 1.69×10−6 | 0.929 | 0.950 | 0.021 | – |
| cg08640046 | 1.69×10−6 | 0.810 | 0.828 | 0.018 | – |
| cg03283124 | 2.03×10−6 | 0.898 | 0.920 | 0.021 | PCDH9 |
| cg13087076 | 1.69×10−6 | 0.890 | 0.912 | 0.021 | DYDC2 |
| cg25453154 | 2.03×10−6 | 0.820 | 0.839 | 0.020 | ZCCHC24 |
Data are presented to 3 decimal places.
Table V.
Top 20 significant hypomethylated CpG sites and genes within differentially methylated regions in Huh7 cells when compared with L02 cells.
| CpG sites | Adjust P-value | |β-difference| | Mean Huh | Mean L02 | Hypomethylated gene |
|---|---|---|---|---|---|
| cg10739344 | 1.70×10−6 | 0.894 | 0.019 | 0.913 | WDR76 |
| cg00618865 | 1.69×10−6 | 0.945 | 0.023 | 0.967 | PLXND1 |
| cg16267343 | 1.69×10−6 | 0.898 | 0.024 | 0.922 | NPR3 |
| cg00138041 | 1.69×10−6 | 0.943 | 0.029 | 0.972 | PRDM8 |
| cg01529365 | 1.69×10−6 | 0.930 | 0.031 | 0.961 | – |
| cg09564253 | 1.69×10−6 | 0.902 | 0.031 | 0.933 | LASP1 |
| cg08176368 | 1.69×10−6 | 0.926 | 0.033 | 0.959 | MMP9 |
| cg08812555 | 2.05×10−6 | 0.784 | 0.029 | 0.813 | DKK1 |
| cg25612391 | 1.77×10−6 | 0.741 | 0.028 | 0.769 | SLC25A42 |
| cg22417879 | 1.70×10−6 | 0.908 | 0.035 | 0.943 | SDCBP2 |
| cg15019790 | 1.69×10−6 | 0.924 | 0.036 | 0.960 | SIX2 |
| cg07407787 | 1.69×10−6 | 0.922 | 0.036 | 0.958 | ARSG; SLC16A6 |
| cg08361684 | 1.69×10−6 | 0.911 | 0.038 | 0.949 | FJX1 |
| cg16195157 | 1.69×10−6 | 0.900 | 0.038 | 0.938 | DNAJB1 |
| cg15842502 | 1.69×10−6 | 0.913 | 0.040 | 0.953 | RB1 |
| cg13848566 | 1.95×10−6 | 0.934 | 0.041 | 0.975 | GAS1 |
| cg27454412 | 1.81×10−6 | 0.781 | 0.034 | 0.815 | C7orf50 |
| cg02152578 | 1.69×10−6 | 0.931 | 0.041 | 0.972 | AHCYL1 |
| cg13355248 | 1.69×10−6 | 0.792 | 0.038 | 0.829 | NPTX1 |
| cg16443866 | 1.69×10−6 | 0.878 | 0.042 | 0.920 | STC2 |
Data are presented to 3 decimal places.
Significant differentially methylated regions (DMRs)
The results of the present study revealed that 390 significantly hypermethylated CpG sites (covering 287 genes) and 208 significantly hypomethylated CpG sites (covering 203 genes) were in DMRs. For the significantly hypermethylated CpG sites, 64 sites were in cancer-specific (c)-DMRs, 125 sites were in reprogramming-specific (r)-DMRs and 201 sites were in DMRs; for the significantly hypomethylated CpG sites, 30 were located within cDMRs, 74 were located within rDMRs and 104 were located within DMRs (Table VI; Fig. 3).
Table VI.
Significant differentially methylated regions.
| DMRs | Significantly hypermethylated sites (n) | Significantly hypermethylated sites (n) |
|---|---|---|
| cDMRs | 64 | 30 |
| Island | 16 | 4 |
| Shores | 36 | 18 |
| Shelves | 5 | 3 |
| Open sea | 7 | 5 |
| rDMRs | 125 | 74 |
| Island | 38 | 13 |
| Shores | 70 | 50 |
| Shelves | 7 | 3 |
| Open sea | 10 | 8 |
| DMRs | 201 | 104 |
| Island | 179 | 70 |
| Shores | 13 | 21 |
| Shelves | 3 | 0 |
| Open sea | 6 | 13 |
| Total | 390 | 208 |
DMRs, differentially methylated regions; cDMRs, cancer-specific-DMRs; rDMRs, reprogramming-specific-DMRs.
Figure 3.
Significantly hypermethylated and hypomethylated genes located in DMRs. DMRs, differentially methylated regions.
GO enrichment and KEGG pathway analysis
GO enrichment and the KEGG Pathway database were employed to analyze information regarding the differentially methylated genes. The results of GO enrichment revealed that there were 2,107 differentially methylated genes associated with ‘biological process’, and the most enriched groups included negative regulators of cell proliferation, negative regulators of transcription such as RNA polymerase II promoter, and synaptic transmission. A total of 13,351 differentially methylated genes were associated with ‘molecular function’, and the most enriched groups included protein binding, DNA binding and metal ion binding. A total of 18,041 differentially methylated genes were associated with ‘cellular component’, and the most enriched groups included the nucleus, cytoplasm and cytosol (Fig. 4). The top 20 significant differentially methylated genes obtained from GO enrichment analysis are listed in Table VII.
Figure 4.
GO enrichment analysis of all of the differentially methylated genes. Green bars indicate ‘biological process, blue bars indicate ‘molecular function’, and red bars indicate ‘cellular component’. GO, Gene Ontology.
Table VII.
Top 20 significant differentially methylated genes in Gene Ontology enrichment.
| GO enrichment | Top 20 significantly hypermethylated genes in GO enrichment | Top 20 significantly hypomethylated genes in GO enrichment |
|---|---|---|
| Biological process | ||
| Positive regulation of protein phosphorylation | ERBB4 | – |
| Negative regulation of cell proliferation | ERBB4 | – |
| Epidermal growth factor receptor signaling | ERBB4 | – |
| pathway | ||
| Synaptic transmission | PCDH8 | – |
| Negative regulation of transcription from RNA | SALL4; NKX6-2; | – |
| polymerase II promoter | MEIS2; TCF21; ZBTB32; | |
| Fibroblast growth factor receptor signaling pathway | ERBB4 | – |
| Molecular function | ||
| Protein binding | ERBB4; PAX6; BCL3; DOCK1; ZBTB32; BCL2 | PLXND1; LASP1; MMP9; DKK1; DNAJB1; RB1; GAS1; AHCYL1 |
| ATP binding | ERBB4 | – |
| Sequence-specific DNA binding transcription factor activity | PAX6; NKX6-2; BCL3 | SIX2; RB1 |
| DNA binding | PAX6; SALL4; BCL3; ZIC4; ZBTB32 | PRDM8; RB1 |
| Metal ion binding | SALL4; ZIC4; ZBTB3 | PRDM8; ARSG; NPTX1 |
| Protein heterodimerization activity | BCL2 | NPR3; SDCBP2 |
| Sequence-specific DNA binding | BCL2 | SIX2 |
| Protein kinase binding | PAX6 | – |
| Transcription regulatory region DNA binding | ERBB4; TCF21 | – |
| Protein complex binding | – | SIX2 |
| Protein dimerization activity | TCF21 | – |
| Identical protein binding | BCL2 | MMP9; RB1 |
| Protein homodimerization activity | ERBB4; BCL2 | SDCBP2 |
| Protein C-terminus binding | – | SDCBP2 |
| Cellular component | ||
| Nucleus | ERBB4; PAX6; SALL4; NKX6-2; BCL3; DOCK1; MEIS2; ZIC4; TCF21; ZBTB32; BCL2 | PRDM8; SIX2; RB1 |
| Cytosol | ERBB4 | – |
| Cytoplasm | ERBB4; PAX6; SALL4; BCL3; DOCK1; BCL2; BDNF | SDCBP2; DNAJB1 |
| Nucleoplasm | ERBB4; ZBTB32 | RB1 |
| Golgi apparatus | – | STC2 |
| Perinuclear region of cytoplasm | BCL3; BDNF | NPTX1 |
| Mitochondrion | ERBB4; BCL2 | SLC25A42 |
| Nucleolus | ERBB4; PAX6 | DNAJB1; RB1 |
| Transcription factor complex | LRFN2 | – |
| Cell junction | PCDH | – |
| Membrane | ERBB4; DOCK1; BCL2 | SLC16A6 |
| Nuclear chromatin | PAX6 | – |
GO, Gene Ontology.
KEGG Pathway-based analyses revealed that 43 signaling pathways involved 5,195 differentially methylated genes, and these genes were significantly enriched in specific pathways, including the cancer, metabolic, mitogen-activated protein kinase (MAPK), calcium, Wnt, hepatitis C, Erb-B2 receptor tyrosine kinase (ErbB), transforming growth factor (TGF)-β, vascular endothelial growth factor (VEGF), p53 and Notch signaling pathways (Table VIII).
Table VIII.
Kyoto encyclopedia of genes and genomes pathway analysis of differentially methylated genes.
| Pathway | Number of differentially methylated genes (n) |
|---|---|
| Pathways in cancer | 309 |
| Focal adhesion | 186 |
| MAPK signaling pathway | 251 |
| Wnt signaling pathway | 143 |
| Axon guidance | 120 |
| TGF-β signaling pathway | 81 |
| Basal cell carcinoma | 55 |
| Regulation of actin cytoskeleton | 191 |
| Colorectal cancer | 61 |
| Adherens junction | 71 |
| Chronic myeloid leukemia | 71 |
| ECM-receptor interaction | 81 |
| Endocytosis | 186 |
| Pyrimidine metabolism | 97 |
| Non-small cell lung cancer | 57 |
| Hedgehog signaling pathway | 54 |
| Neurotrophin signaling pathway | 116 |
| Glioma | 63 |
| Endometrial cancer | 52 |
| VEGF signaling pathway | 71 |
| ErbB signaling pathway | 80 |
| Small cell lung cancer | 80 |
| Lysosome | 110 |
| Metabolic pathways | 964 |
| Calcium signaling pathway | 159 |
| Purine metabolism | 150 |
| Ubiquitin mediated proteolysis | 128 |
| Insulin signaling pathway | 128 |
| Notch signaling pathway | 45 |
| Protein processing in endoplasmic reticulum | 156 |
| RNA polymerase | 32 |
| Hepatitis C | 116 |
| Renal cell carcinoma | 61 |
| Aminoacyl-tRNA biosynthesis | 42 |
| B cell receptor signaling pathway | 69 |
| Thyroid cancer | 29 |
| Melanoma | 65 |
| Oocyte meiosis | 103 |
| Adipocytokine signaling pathway | 64 |
| Melanogenesis | 94 |
| Vascular smooth muscle contraction | 115 |
| Selenocompound metabolism | 26 |
| p53 signaling pathway | 63 |
Discussion
DNA methylation is the main epigenetic modification and regulator of gene expression in humans. Aberrant changes in genomic methylation patterns have been observed in many cancer cell lines; these are regarded as the major type of molecular aberration in malignancies (7,8). Previous studies have evaluated aberrant DNA methylation in HCC by analyzing tumor tissues and adjacent non-tumor tissues (14,21–23). Their main aims were to identify novel potential biomarkers for the diagnosis of HCC or to study the associations between methylation and cirrhosis-associated HCC; the study design and enrolment criteria differed when selecting various patients for study. In addition, variations in ethnicity and the effects of the cutting edge of adjacent non-tumor tissues may lead to differing results observed across the different studies. Although previous studies have indicated a few novel DNA methylation markers that are associated with HCC, specific DNA methylation patterns associated with the progression of HCC and alterations in methylation between HCC and normal liver cells have yet to be identified. In the present study, Illumina Infinium HumanMethylation 450K BeadChip was used to identify global DNA methylation profiles in Huh7 and L02 cells.
In the present study, a total of 102,254 differentially methylated CpG sites were detected across the whole-genome of Huh7 and L02 cells; more hypermethylated CpG sites (62,702; 61.3%) were observed than hypomethylated (39,552; 38.7%) CpG sites. The results indicated that within Huh7 cells, aberrant DNA methylation was a very common event and that hypermethylation of CpG sites occurred more frequently than hypomethylation. In addition, stringent criteria were employed to select the significantly differentially methylated CpG sites, genes and DMRs. Finally, 5,285 (66.5%) significantly hypermethylated and 2,659 (33.5%) hypomethylated CpG sites were identified. It has been reported that, in many types of diseases including cancers, aberrant DNA methylation is a common event, particularly aberrant DNA methylation of CpG islands or within promoter regions, which are associated with tumor suppressor gene inactivation or oncogene activation (16). The results of the present study indicated that within a CpG island, a greater number of significantly hypermethylated CpG sites (1,544) were observed than significantly hypomethylated CpG sites (1,201). This result is consistent with previous HCC genome-wide methylation studies (14,24–27). Yates et al (18) and Dudziec et al (19) demonstrated that aberrant DNA methylation occurs in CpG islands, but can also be detected in the regions adjacent to CpG islands, CpG shores and CpG shelves (15), and may lead to tumorigenesis (20,28,29). The present study also demonstrated these points; significantly hypermethylated CpG sites in the CpG shores regions were more abundant than significantly hypomethylated CpG sites (1,137 to 632). In addition, in the CpG shelf regions, the significantly hypermethylated CpG sites were more frequent than significantly hypomethylated CpG sites (655 to 133).
DMRs are stretches of DNA in the genome. Varied DNA methylation patterns are seen between different organisms, and adjacent sites or a group of sites in proximity to each other tend to have different methylation patterns between different diseases (30). DMRs are associated with many diseases including several types of cancer (31). There are also many types of DMRs: Tissue-specific DMRs, cDMRs, rDMRs, imprinting-specific DMRs and aging-specific DMRs (20). In the present study, there were 390 differentially hypermethylated CpG sites located within DMRs, 233 (59.7%) were in island regions, 119 (30.5%) were in shore regions, and 15 (3.8%) were in shelf regions. In addition, there were 208 differentially hypomethylated CpG sites located within DMRs, 87 (41.8%) were in CpG island regions, 89 (42.8%) were in shore regions and 6 (2.9%) were in shelf regions. These results indicated that within HCC cells, aberrant DNA methylation may occur within CpG shore regions, which can also cause DNA transcriptional silencing and inactivation of gene function. Hepatocarcinogenesis was also associated with genomic instability and inactivation of gene function; the results of the present study concerning DMRs suggests that aberrant methylation within these sites may be an important epigenetic mechanism associated with hepatocarcinogenesis. These results may provide more information regarding the associations between HCC and aberrant DNA methylation.
Furthermore, the present study listed the top 20 significantly hyper- and hypo-methylated CpG sites, and genes in DMRs within Huh7 cells compared with L02 cells. The top 20 significantly hypermethylated genes, which were high-ranking with notable differences in the absolute value of β-difference, included the following: ERBB4, paired box 6, splat like transcription factor 4, protocadherin (PCDH)-8, NK2 homeobox 6, B-cell lymphoma (BCL)-3, glypican 6, family with sequence similarity 196 member A, dedicator of cytokinesis 1, Meis homeobox 2, Zic family member 4, transcription factor 21, zinc finger and BTB domain containing 32, leucine rich repeat and fibronectin type III domain containing 2, BCL2, PCDH9, DPY30 domain-containing protein 2, zinc finger CCHC-type containing 24, brain-derived neurotrophic factor, cg00057722 and cg08640046. The functional role of these genes in HCC requires further study. The top 20 significant differentially hyper- and hypo-methylated genes from GO enrichment were also listed. These genes, which were located within DMRs, were mainly associated with ‘cell differentiation development’, ‘transcription factor activity’, ‘sequence-specific DNA binding’, ‘cellular development process’ and ‘cell junction’.
Additionally, through GO enrichment analysis, the present study revealed that aberrant DNA methylation in HCC was associated with cell differentiation and proliferation, and through KEGG pathway analysis, 43 signaling pathways associated with HCC were identified, including pathways in cancer, MAPK signaling, Wnt signaling, VEGF signaling and p53 signaling pathways. Previous studies have demonstrated that aberrant DNA hypermethylation can downregulate the expression of cell cycle inhibitors, p16INK4A, p53 and factors involved in TGF-β/mothers against decapentaplegic signaling (32,33). Thus far, researchers have revealed that the inactivation of Wnt pathway-associated antagonists is linked to the aberrant DNA hypermethylation of some genes (34,35). Activation of the ERB receptor and MAPK signaling pathways, as well as the regulation of epigenetic proteins that were previously demonstrated to promote cancer growth and metastasis, have been reported to be possible candidate targets for anticancer treatment in multiple types of cancer, including HCC (32,36).
In addition, HCC cells may escape or become tolerant to chemotherapy via various mechanisms, therefore, identifying novel drugs is very important for the future therapy of HCC. The application of inhibitors of DNA methylated drugs in the treatment of cancer has gradually attracted the attention of researchers (37), including 5-azacytidine (5-aza-C), decitabine (5-aza-2′-deoxycytidine, 5-aza-dC), 1-β-D-arabinofuranosyl-5-azacytosine, dihydro-5-azacytosine (38), SGI-110 (previously known as S110), a dinucleotide of 5-aza-2′-deoxycytidine and deoxyguanosine, containing 5-azaCdR moiety, which has been revealed to be very effective in inhibiting DNA methylation, though its stability and cytotoxicity are comparable to that of decitabine (39), and a non-nucleoside DNA methyltransferase inhibitor, SGI-1027 (40,41). To the best of our knowledge, there have been only a few studies investigating the effects of demethylation agents on HCC in vitro.
In conclusion, the present study detected genome-wide DNA methylation patterns occurring in Huh7 cells, and identified numerous differentially hypo- and hypermethylated CpG sites, genes, DMRs and signaling pathways associated with HCC. Additionally, the diversity in methylation within Huh7 cells was also observed. The results of the present study may provide important information regarding the molecular mechanisms underlying methylation in Huh7 cells, which may be useful in future research into the underlying mechanisms associated with HCC. In addition, HCC cells may escape or develop tolerance to chemotherapy via various mechanisms, therefore, identifying novel drugs is very important for future therapies of HCC. The application of inhibitors of DNA methylation for the treatment of cancer has gradually attracted more attention within the field (37), and there have been a few studies investigating the effects of demethylation agents in HCC in vitro. The results of the present study may provide a useful basis for future research into effective HCC therapies.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- PLC
primary liver cancer
- HCC
hepatocellular carcinoma
- DMR
differentially methylated regions
- GO
Gene Ontology
Funding
The present study was supported by The Science and Technology project of Shenyang (grant no. F13-212-9-00).
Availability of data and materials
The analyzed datasets generated during this study are available from the corresponding author on reasonable request.
Authors' contributions
JZ conceived and designed the study. NS, CZ, YS, BZ and BC performed the experiments. NS and AJ analyzed the data. NS wrote the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Zuo TT, Zheng RS, Zhang SW, Zeng HM, Chen WQ. Incidence and mortality of liver cancer in China in 2011. Chin J Cancer. 2015;34:56. doi: 10.1186/s40880-015-0056-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 4.Tanaka M, Katayama F, Kato H, Tanaka H, Wang J, Qiao YL, Inoue M. Hepatitis B and C virus infection and hepatocellular carcinoma in China: A review of epidemiology and control measures. J Epidemiol. 2011;21:401–416. doi: 10.2188/jea.JE20100190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Panayiotidis MI. Cancer epigenetics as biomarkers of clinical significance. Cancer Lett. 2014;342:168–169. doi: 10.1016/j.canlet.2013.10.027. [DOI] [PubMed] [Google Scholar]
- 6.Udali S, Guarini P, Moruzzi S, Ruzzenente A, Tammen SA, Guglielmi A, Conci S, Pattini P, Olivieri O, Corrocher R, et al. Global DNA methylation and hydroxymethylation differ in hepatocellular carcinoma and cholangiocarcinoma and relate to survival rate. Hepatology. 2015;62:496–504. doi: 10.1002/hep.27823. [DOI] [PubMed] [Google Scholar]
- 7.Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 8.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 9.Jain S, Xie L, Boldbaatar B, Lin SY, Hamilton JP, Meltzer SJ, Chen SH, Hu CT, Block TM, Song W, Su YH. Differential methylation of the promoter and first exon of the RASSF1A gene in hepatocarcinogenesis. Hepatol Res. 2015;45:110–123. doi: 10.1111/hepr.12449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hinrichsen I, Kemp M, Peveling-Oberhag J, Passmann S, Plotz G, Zeuzem S, Brieger A. Promoter methylation of MLH1, PMS2, MSH2 and p16 is a phenomenon of advanced-stage HCCs. PLoS One. 2014;9:e84453. doi: 10.1371/journal.pone.0084453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu B, Nie Y, Liu X, Feng S, Yang Z, Wang Z, Zheng Q, Luo X. Quantitative analysis of APC promoter methylation in hepatocellular carcinoma and its prognostic implications. Oncol Lett. 2014;7:1683–1688. doi: 10.3892/ol.2014.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jain S, Chen S, Chang KC, Lin YJ, Hu CT, Boldbaatar B, Hamilton JP, Lin SY, Chang TT, Chen SH, et al. Impact of the location of CpG methylation within the GSTP1 gene on its specificity as a DNA marker for hepatocellular carcinoma. PLoS One. 2012;7:e35789. doi: 10.1371/journal.pone.0035789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qu Z, Jiang Y, Li H, Yu DC, Ding YT. Detecting abnormal methylation of tumor suppressor genes GSTP1, P16, RIZ1, and RASSF1A in hepatocellular carcinoma and its clinical significance. Oncol Lett. 2015;10:2553–2558. doi: 10.3892/ol.2015.3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen J, Wang S, Zhang YJ, Wu HC, Kibriya MG, Jasmine F, Ahsan H, Wu DP, Siegel AB, Remotti H, Santella RM. Exploring genome-wide DNA methylation profiles altered in hepatocellular carcinoma using Infinium HumanMethylation 450 BeadChips. Epigenetics. 2013;8:34–43. doi: 10.4161/epi.23062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bibikova M, Barnes B, Tsan C, Ho V, Klotzle B, Le JM, Delano D, Zhang L, Schroth GP, Gunderson KL, et al. High density DNA methylation array with single CpG site resolution. Genomics. 2011;98:288–295. doi: 10.1016/j.ygeno.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 16.Esteller M. Epigenetic gene silencing in cancer: The DNA hypermethylome. Hum Mol Genet. 2007;16:R50–R59. doi: 10.1093/hmg/ddm018. [DOI] [PubMed] [Google Scholar]
- 17.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 18.Yates DR, Rehman I, Meuth M, Cross SS, Hamdy FC, Catto JW. Methylational urinalysis: A prospective study of bladder cancer patients and age stratified benign controls. Oncogene. 2006;25:1984–1988. doi: 10.1038/sj.onc.1209209. [DOI] [PubMed] [Google Scholar]
- 19.Dudziec E, Miah S, Choudhry HM, Owen HC, Blizard S, Glover M, Hamdy FC, Catto JW. Hypermethylation of CpG islands and shores around specific microRNAs and mirtrons is associated with the phenotype and presence of bladder cancer. Clin Cancer Res. 2011;17:1287–1296. doi: 10.1158/1078-0432.CCR-10-2017. [DOI] [PubMed] [Google Scholar]
- 20.Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishida N, Kudo M, Nagasaka T, Ikai I, Goel A. Characteristic patterns of altered DNA methylation predict emergence of human hepatocellular carcinoma. Hepatology. 2012;56:994–1003. doi: 10.1002/hep.25706. [DOI] [PubMed] [Google Scholar]
- 22.Shen J, Wang S, Zhang YJ, Kappil M, Wu HC, Kibriya MG, Wang Q, Jasmine F, Ahsan H, Lee PH, et al. Genome-wide DNA methylation profiles in hepatocellular carcinoma. Hepatology. 2012;55:1799–1808. doi: 10.1002/hep.25569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Udali S, Guarini P, Ruzzenente A, Ferrarini A, Guglielmi A, Lotto V, Tononi P, Pattini P, Moruzzi S, Campagnaro T, et al. DNA methylation and gene expression profiles show novel regulatory pathways in hepatocellular carcinoma. Clin Epigenetics. 2015;7:43. doi: 10.1186/s13148-015-0077-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao W, Kondo Y, Shen L, Shimizu Y, Sano T, Yamao K, Natsume A, Goto Y, Ito M, Murakami H, et al. Variable DNA methylation patterns associated with progression of disease in hepatocellular carcinomas. Carcinogenesis. 2008;29:1901–1910. doi: 10.1093/carcin/bgn170. [DOI] [PubMed] [Google Scholar]
- 25.Shin SH, Kim BH, Jang JJ, Suh KS, Kang GH. Identification of novel methylation markers in hepatocellular carcinoma using a methylation array. J Korean Med Sci. 2010;25:1152–1159. doi: 10.3346/jkms.2010.25.8.1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ammerpohl O, Pratschke J, Schafmayer C, et al. Distinct DNA methylation patterns in cirrhotic liver and hepatocellular carcinoma. Int J Cancer. 2012;130:1319–1328. doi: 10.1002/ijc.26136. [DOI] [PubMed] [Google Scholar]
- 27.Kohles N, Nagel D, Jüngst D, Durner J, Stieber P, Holdenrieder S. Prognostic relevance of oncological serum biomarkers in liver cancer patients undergoing transarterial chemoembolization therapy. Tumour Biol. 2012;33:33–40. doi: 10.1007/s13277-012-0504-2. [DOI] [PubMed] [Google Scholar]
- 28.Doi A, Park IH, Wen B, Murakami P, Aryee MJ, Irizarry R, Herb B, Ladd-Acosta C, Rho J, Loewer S, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet. 2009;41:1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ogoshi K, Hashimoto S, Nakatani Y, Qu W, Oshima K, Tokunaga K, Sugano S, Hattori M, Morishita S, Matsushima K. Genome-wide profiling of DNA methylation in human cancer cells. Genomics. 2011;98:280–287. doi: 10.1016/j.ygeno.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 30.Rakyan VK, Down TA, Balding DJ, Beck S. Epigenome-wide association studies for common human diseases. Nat Rev Genet. 2011;12:529–541. doi: 10.1038/nrg3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schübeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 32.Calvisi DF, Pascale RM, Feo F. Dissection of signal transduction pathways as a tool for the development of targeted therapies of hepatocellular carcinoma. Rev Recent Clin Trials. 2007;2:217–236. doi: 10.2174/157488707781662715. [DOI] [PubMed] [Google Scholar]
- 33.Kuo KK, Jian SF, Li YJ, Wan SW, Weng CC, Fang K, Wu DC, Cheng KH. Epigenetic inactivation of transforming growth factor-beta1 target gene HEYL, a novel tumor suppressor, is involved in the P53-induced apoptotic pathway in hepatocellular carcinoma. Hepatol Res. 2015;45:782–793. doi: 10.1111/hepr.12414. [DOI] [PubMed] [Google Scholar]
- 34.Umer M, Qureshi SA, Hashmi ZY, Raza A, Ahmad J, Rahman M, Iqbal M. Promoter hypermethylation of Wnt pathway inhibitors in hepatitis C virus-induced multistep hepatocarcinogenesis. Virol J. 2014;11:117. doi: 10.1186/1743-422X-11-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ding SL, Yang ZW, Wang J, Zhang XL, Chen XM, Lu FM. Integrative analysis of aberrant Wnt signaling in hepatitis B virus-related hepatocellular carcinoma. World J Gastroenterol. 2015;21:6317–6328. doi: 10.3748/wjg.v21.i20.6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stefanska B, Cheishvili D, Suderman M, Arakelian A, Huang J, Hallett M, Han ZG, Al-Mahtab M, Akbar SM, Khan WA, et al. Genome-wide study of hypomethylated and induced genes in patients with liver cancer unravels novel anticancer targets. Clin Cancer Res. 2014;20:3118–3132. doi: 10.1158/1078-0432.CCR-13-0283. [DOI] [PubMed] [Google Scholar]
- 37.Yan W, Herman JG, Guo M. Epigenome-based personalized medicine in human cancer. Epigenomics. 2016;8:119–133. doi: 10.2217/epi.15.84. [DOI] [PubMed] [Google Scholar]
- 38.Ghoshal K, Bai S. DNA methyltransferases as targets for cancer therapy. Drugs Today (Barc) 2007;43:395–422. doi: 10.1358/dot.2007.43.6.1062666. [DOI] [PubMed] [Google Scholar]
- 39.Yoo CB, Jeong S, Egger G, Liang G, Phiasivongsa P, Tang C, Redkar S, Jones PA. Delivery of 5-aza-2′-deoxycytidine to cells using oligodeoxynucleotides. Cancer Res. 2007;67:6400–6408. doi: 10.1158/0008-5472.CAN-07-0251. [DOI] [PubMed] [Google Scholar]
- 40.Datta J, Ghoshal K, Denny WA, Gamage SA, Brooke DG, Phiasivongsa P, Redkar S, Jacob ST. A new class of quinoline-based DNA hypomethylating agents reactivates tumor suppressor genes by blocking DNA methyltransferase 1 activity and inducing its degradation. Cancer Res. 2009;69:4277–4285. doi: 10.1158/0008-5472.CAN-08-3669. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Gros C, Fleury L, Nahoum V, Faux C, Valente S, Labella D, Cantagrel F, Rilova E, Bouhlel MA, David-Cordonnier MH, et al. New insights on the mechanism of quinoline-based DNA Methyltransferase inhibitors. J Biol Chem. 2015;290:6293–6302. doi: 10.1074/jbc.M114.594671. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed datasets generated during this study are available from the corresponding author on reasonable request.