Abstract
Protein S‐palmitoylation is a reversible post‐translational modification that alters the localization, stability, and function of hundreds of proteins in the cell. S‐palmitoylation is essential for the function of both oncogenes (e.g., NRAS and EGFR) and tumor suppressors (e.g., SCRIB, melanocortin 1 receptor). In mammalian cells, the thioesterification of palmitate to internal cysteine residues is catalyzed by 23 Asp‐His‐His‐Cys (DHHC)‐family palmitoyl S‐acyltransferases while the removal of palmitate is catalyzed by serine hydrolases, including acyl‐protein thioesterases (APTs). These enzymes modulate the function of important oncogenes and tumor suppressors and often display altered expression patterns in cancer. Targeting S‐palmitoylation or the enzymes responsible for palmitoylation dynamics may therefore represent a candidate therapeutic strategy for certain cancers.
Keywords: lipid, lipidation, post‐translational modification, S‐palmitoylation, tumor
Subject Categories: Cancer; Post-translational Modifications, Proteolysis & Proteomics; Signal Transduction
Glossary
- APT
acyl‐protein thioesterase
- ATM
ataxia telangiectasia mutated
- ATR
ataxia telangiectasia and Rad3 related
- cAMP
cyclic adenosine monophosphate
- EGFR
epidermal growth factor receptor
- Erf
effect on ras function
- IP3R
inositol 1,4,5‐triphosphate receptor
- MC1R
melanocortin 1 receptor
- PAT
palmitoyl S‐acyltransferase
- PTM
post‐translational modification
- ZDHHC
zinc finger DHHC‐type containing
Introduction
Hallmark cancer cell phenotypes such as sustained proliferation, resistance to cell death, and increased metastasis result from the altered activity of intracellular signaling, metabolic, and gene regulatory networks 1, 2. The function of these networks, both in normal and cancer cells, is regulated by dynamic post‐translational modifications (PTMs) that alter protein localization, stability, and function. Hundreds of distinct PTMs impacting thousands of proteins have been identified in mammalian cells 3, 4. Protein lipidation—the post‐translational addition of a lipid to a protein—comprises one important and diverse class of PTMs 5. Lipidated proteins generally have greater affinity for non‐polar structures such as lipid bilayers, with important consequences for the localization, diffusion, and physical interactions of these proteins within the cell. Indeed, lipidation of key signaling proteins, such as Hedgehog, Wnt, and RAS, is essential for their function, both normally and in cancer 5, 6, 7, 8, 9.
Many protein lipidation events are thought to be essentially irreversible, including N‐myristoylation, S‐farnesylation, O‐palmitoylation, and N‐palmitoylation. By contrast, protein S‐palmitoylation (hereafter simply palmitoylation), which is the thioesterification of a sixteen‐carbon saturated fatty acid (palmitate) to an internal cysteine residue, is a reversible modification. Indeed, proteins can cycle between palmitoylated and de‐palmitoylated forms on timescales that range from seconds to hours 10, 11, 12. Dynamic palmitoylation can impact protein localization, accumulation, secretion, stability, and function by altering membrane affinity 5, 9, 13, 14. Understanding how protein palmitoylation influences the function of individual proteins in normal and cancer cells is an important driver of current research in this area.
Currently, several hundred mammalian proteins, many of which are cancer‐related, have been discovered to be palmitoylated 4, 15. While certain proteins are mono‐palmitoylated, examples of proteins with as many as five or six individual palmitoylated cysteine residues are known 16. Some proteins spontaneously autopalmitoylate 17, 18. However, the majority of protein palmitoylation is catalyzed by the zinc finger DHHC‐type containing (ZDHHC) family of palmitoyl S‐acyltransferases (PATs), comprising 23 distinct proteins in mammals (Table 1) 19. Most ZDHHC proteins are localized to the ER and Golgi apparatus, the primary sites of protein palmitoylation activity within mammalian cells 10, 20, although at least three ZDHHC enzymes (ZDHHC5, ZDHHC20, ZDHHC21) appear primarily localized to the plasma membrane 21. Protein deacylation can be catalyzed by acyl‐protein thioesterases (APTs) 1/2 (APT1/2, LYPLA1/2) belonging to the α/β‐hydrolase family of serine hydrolases 22. Evidence links specific palmitoylated proteins and the function of individual PAT and APT enzymes to carcinogenesis and cancer cell growth, survival, and therapy resistance (Table 1, and see below). Thus, enzymes that modulate protein palmitoylation may be useful targets for anti‐cancer treatments. The nature of protein palmitoylation, the enzymes involved, and the prospects for targeting these enzymes therapeutically are described below.
Table 1.
List of ZDHHC genes, alternative names, associated properties, and links to cancer
| Gene | Alternative names | UniProt ID | Size (aa) | Localization | Cancer links |
|---|---|---|---|---|---|
| ZDHHC1 | Q8WTX9 | 485 | ER | High mRNA expression is a favorable prognostic marker in endometrial, renal, and pancreatic cancers (HPA) | |
| ZDHHC2 | REAM | Q9UIJ5 | 367 | ER, Golgi | In a fusion gene in acute myeloid leukemia; in a region of deletion in hepatocellular carcinoma; high expression is a favorable prognostic marker in gastric cancer; high mRNA expression is a favorable prognostic marker in renal cancer (HPA) |
| ZDHHC3 | GODZ | Q9NYG2 | 327 | Golgi | High expression can be an unfavorable prognostic marker in breast carcinoma; in a region of recurrent loss in cervical cancer; high mRNA expression is a favorable prognostic marker in renal and colorectal cancer, but an unfavorable marker in liver cancer (HPA) |
| ZDHHC4 | Q9NPG8 | 344 | Golgi | High mRNA expression is a favorable prognostic marker in renal cancer, but an unfavorable marker in head and neck, urothelial, and cervical cancer (HPA) | |
| ZDHHC5 | Q9C0B5 | 715 | Plasma membrane | High expression together with mutations in p53 are associated with reduced survival in glioma; high mRNA expression is an unfavorable prognostic marker in pancreatic cancer (HPA) | |
| ZDHHC6 | Q9H6R6 | 413 | ER | – | |
| ZDHHC7 | SERZ‐beta | Q9NXF8 | 308 | Golgi | In a region of deletion in breast carcinoma; deleted in prostate and ovarian cancer; high mRNA expression is an unfavorable prognostic marker in renal and liver cancer (HPA) |
| ZDHHC8 | Q9ULC8 | 765 | Golgi | High mRNA expression is an unfavorable prognostic marker in renal and cervical cancer (HPA) | |
| ZDHHC9 | ZDHHC10, MMSA‐1 | Q9Y397 | 364 | ER, Golgi | Required for palmitoylation of RAS proteins and Nras‐driven leukemias; high expression in a range of cancers including breast and colorectal cancer, and multiple myeloma; high mRNA expression is an unfavorable prognostic marker in cervical, breast, and head and neck cancers (HPA) |
| ZDHHC11 | Q9H8X9 | 412 | ER | High expression in Burkitt lymphoma cells promotes proliferation; frequently deleted in hepatoblastoma; in a region of amplification in non‐small‐cell lung carcinoma and bladder cancers; high mRNA expression of the non‐coding ZDHHC11B transcript is a favorable prognostic marker in lung cancer (HPA) | |
| ZDHHC12 | Q96GR4 | 267 | ER, Golgi | – | |
| ZDHHC13 | HIP14L, HIP3RP | Q8IUH4 | 622 | ER, Golgi | Required for protective function of MC1R against melanoma and to inhibit chemically induced papilloma |
| ZDHHC14 | Q8IZN3 | 488 | ER | Genomic deletions, mutations, and reduced expression in testicular germ cell tumor and prostate cancer; overexpression in scirrhous type gastric cancer and tongue squamous cell carcinoma; activating mutation (gene fusion) in acute biphenotypic leukemia; high mRNA expression is a favorable prognostic marker in pancreatic cancer (HPA) | |
| ZDHHC15 | Q96MV8 | 337 | Golgi | – | |
| ZDHHC16 | Aph2 | Q969W1 | 377 | ER | – |
| ZDHHC17 | HIP14, HIP3, HYPH | Q8IUH5 | 632 | Golgi | High mRNA expression is an unfavorable prognostic marker in renal cancer (HPA) |
| ZDHHC18 | Q9NUE0 | 388 | Golgi | High mRNA expression is an unfavorable prognostic marker in renal cancer, liver cancer, and glioma (HPA) | |
| ZDHHC19 | Q8WVZ1 | 309 | ER | – | |
| ZDHHC20 | Q5W0Z9 | 354 | Plasma membrane | Elevated mRNA expression in ovarian, breast, kidney, colon, and prostate cancer tissue; high mRNA expression is an unfavorable prognostic marker in renal and pancreatic cancer (HPA) | |
| ZDHHC21 | Q8IVQ6 | 265 | Plasma membrane | High mRNA expression is a favorable prognostic marker in urothelial and renal cancer (HPA) | |
| ZDHHC22 | Q8N966 | 263 | ER, Golgi | – | |
| ZDHHC23 | NIDD | Q8IYP9 | 409 | ER | High mRNA expression in B‐precursor acute lymphoblastic leukemia; high mRNA expression is a favorable prognostic marker in renal cancer, but unfavorable in endometrial and thyroid cancer (HPA) |
| ZDHHC24 | Q6UX98 | 284 | ER | High mRNA expression is an unfavorable prognostic marker in glioblastoma (HPA) |
Localization data are from 21 and the UniProt database. Cancer links is defined broadly as any literature link between a ZDHHC‐family member and a particular cancer, including large‐scale and small‐scale experimental studies, animal and cell culture studies, and clinical studies, whether functional or correlative. References: ZDHHC2 70, 129, 130, ZDHHC3 131, 132, 133, ZDHHC5 87, ZDHHC7 47, 134, ZDHHC9 89, 92, 135, 136, ZDHHC11 77, 78, 137, 138, ZDHHC13 49, 73, ZDHHC14 82, 83, 84, 85, ZDHHC20 81, ZDHHC23 139.
HPA, Human Protein Atlas (http://www.proteinatlas.org/).
Protein S‐palmitoylation: basic biochemical mechanisms
Studies of the Saccharomyces cerevisiae integral membrane proteins Erf2 (effect on ras function 2) and Akr1 (ankyrin‐repeat containing 1) were the first to firmly establish the enzymatic basis of protein palmitoylation and provided the first insights into the structure and function of enzymes catalyzing this reaction 23, 24, 25 (Fig 1A). The recent crystal structure of human ZDHHC20 confirmed and extended these earlier biochemical studies 26. ZDHHC20 adopts a teepee‐like structure (wide at the cytoplasmic side, narrow at the membrane‐internal side) with the Asp‐His‐His‐Cys (DHHC) enzyme active site located on the cytosolic linker between transmembrane domains 2 and 3. Here, it is positioned to interact with both palmitoyl‐CoA and substrate proteins at the membrane–cytosol interface. Palmitoyl‐CoA first reacts with the cysteine residue in the DHHC motif itself, forming an acyl‐intermediate and releasing free CoA‐SH (i.e., autopalmitoylation). This intermediate is then transferred from the DHHC motif directly to substrate proteins in the cell.
Figure 1. The biochemistry of protein palmitoylation.
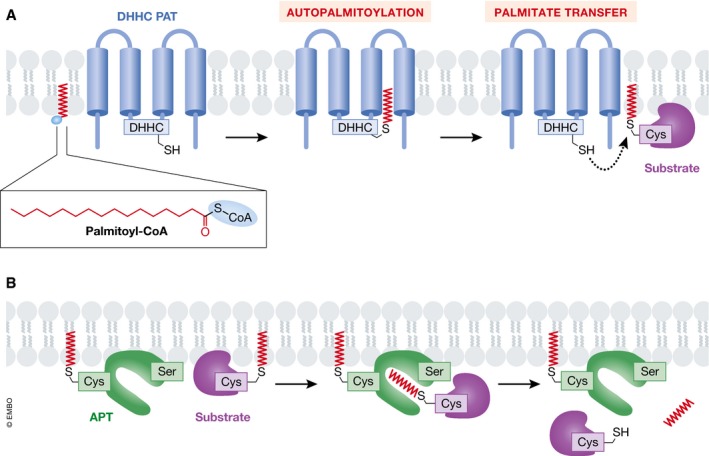
(A) Palmitate (derived from palmitoyl‐CoA) can be thioesterified to substrate proteins by DHHC (Asp‐His‐His‐Cys)‐family protein S‐acyltransferases (PATs). DHHC PATs are integral membrane proteins (blue) with the active site oriented toward the cytosol. These enzymes catalyze palmitoylation on internal cysteine (Cys) residues of substrate proteins (S‐acylation). The DHHC enzyme is first autopalmitoylated on the DHHC cysteine residue with the release of free coenzyme A (CoA), followed by a transfer of the palmitate group to the acceptor cysteine residue of a substrate protein (purple). (B) Acylprotein thioesterase (APT, green) can remove palmitate groups from palmitoylated proteins (purple). APT1/2 are themselves palmitoylated and contain a hydrophobic pocket to accept palmitoylated substrates and position the substrate palmitoylated cysteine near the active site serine (Ser) residue.
Cysteine residues that lie near the catalytic DHHC motif coordinate two structural zinc atoms that are essential for proper enzyme folding and function, but do not play a catalytic role in palmitate transfer 26, 27, 28. Despite overall amino acid similarity, the 23 human ZDHHC enzymes exhibit distinct abilities to autoacylate, implying differences in catalytic efficiency 29, 30. Of note, while often referred to in general as “palmitoylation”, this process can involve not only the addition of palmitate (C16:0), but also other fatty acids with different chain lengths (e.g., C18:0, stearoylation) 31. Indeed, mammalian ZDHHC enzymes display distinct fatty acyl preferences 32, 33, with the ability to preferentially accommodate particular fatty acyl chain lengths specified by amino acids that cap the hydrophobic cavity of the protein 26.
How ZDHHC enzymes select substrate proteins for modification is not entirely clear as there is no consensus palmitoylation sequence. Palmitoylated proteins are often substrates for more than one ZDHHC enzyme, but one particular ZDHHC enzyme often has a stronger effect than others on substrate palmitoylation in the cell (e.g. 29, 34, 35, 36). Chimeric ZDHHC proteins and structure–function studies have defined specific regions of individual ZDHHC enzymes that promote substrate interaction and palmitoylation, indicating that unique substrate‐binding preferences could guide the palmitoylation of certain proteins 5. For other proteins, palmitoylation requires an initial lipidation event (e.g., RAS C‐terminal farnesylation, SRC‐family N‐terminal myristoylation) at an amino acid residue near the to‐be palmitoylated Cys residue(s), which likely helps localize the substrate to ZDHHC‐enriched membranes (e.g., the Golgi) 37, 38, 39. Two ZDHHC enzymes, ZDHHC13 and ZDHHC17, contain unique C‐terminal ankyrin‐repeat domains that can bind certain proteins and enhance their membrane localization, facilitating palmitoylation by other ZDHHC enzymes 30, 40. Efficient palmitoylation by some DHHC enzymes (Erf2/ZDHHC9, ZDHHC6) requires an accessory protein (Erf4/GOLGA7, SELENOK), and speculatively, these accessory proteins could aid in substrate selection 24, 41, 42, 43, 44.
S‐palmitoylation occurs on internal cysteine residues of substrate proteins, and within a given protein, only specific cysteine residues are S‐palmitoylated. Despite progress in computational prediction 45, 46, it remains difficult to determine a priori which specific cysteines within a given protein will be modified, and experimental trial‐and‐error remains essential to define palmitoylated residues (e.g. 47, 48, 49). Due to the location and orientation of the DHHC motif, palmitoylation of integral membrane proteins occurs preferentially on cysteines located within eight angstroms of the membrane–cytosol interface, providing a structural constraint on the palmitoylation potential of certain residues 26, 50. For peripheral membrane proteins, the ZDHHC‐substrate binding conformation appears to be important for effective palmitoylation to occur on specific residues; substrate mutations distal to the sites of palmitoylation can strongly impact the likelihood of modification 47, 49. How substrate conformation or binding influences palmitoylation remains to be worked out in detail.
The enzymatic removal of S‐acyl modifications in mammalian cells is catalyzed by a family of serine hydrolases 51 (Fig 1B). The first enzyme, APT1, was reported to depalmitoylate the alpha subunit of G proteins and HRAS in vitro 52. However, it appears that in cells APT2 is a cytosolic enzyme while APT1 is localized to the mitochondria, which may explain the selective effect of APT2 inhibition on the palmitoylation of cytosolic proteins 53, 54. APT1 and APT2 are not the only thioesterases in the cell and in fact do not impact the palmitoylation of key cancer‐related proteins in the cell 11, 54; ABHD17A, ABHD17B, and ABHD17C are novel depalmitoylation enzymes that regulate the palmitoylation status of NRAS, for example 51, 55, 56. Finally, palmitoyl protein thioesterases 1 and 2 (PPT1/2) localize to and deacylate proteins in the lysosome.
PAT and APT enzymes can exist within complex regulatory networks. ZDHHC5, ZDHHC6, and ZDHHC8 are palmitoylated at cysteine residues outside the catalytic DHHC motif 57, and these modifications are likely essential for normal enzyme localization and function. Recently, ZDHHC16 was shown to physically associate with and palmitoylate ZDHHC6 on Cys328, Cys329, and Cys343 residues found within the ZDHHC6 C‐terminal Src homology 3 (SH3) domain 36 (Fig 2A). Modeling and experimental studies suggest that this palmitoylation “cascade”, from ZDHHC16 to ZDHHC6, enhances the overall stability and activity of ZDHHC6, although different configurations of modifications yield distinct activity and stability profiles. Palmitoylation of ZDHHC6 at these C‐terminal cysteine residues is reversible by APT2, with turnover on Cys328 being especially rapid (Fig 2B). Adding further complexity to this network, APT1/2 and ABDH‐family thioesterases are also themselves palmitoylated, and this modification is necessary for their proper localization and function 57, 58. For example, ABHD17A‐C proteins have a highly conserved cluster of cysteines near the N‐terminus that are required for proper localization to plasma and endosomal membranes and optimal catalytic activity 51. PPT1 is also palmitoylated by ZDHHC3 and ZDHHC7, but in this case, palmitoylation decreases enzyme activity 59. The function of both PAT (e.g., ZDHHC13, ZDHHC3) and APT (e.g., APT1) enzymes can also be regulated by phosphorylation in cancer and other cells, downstream of different signaling pathways 49, 60, 61. Thus, a web of interconnected PTMs regulates both ZDHHC and thioesterase localization and activity, with important implications for the regulation of cancer‐associated substrates.
Figure 2. A highly connected web of palmitoylation events.
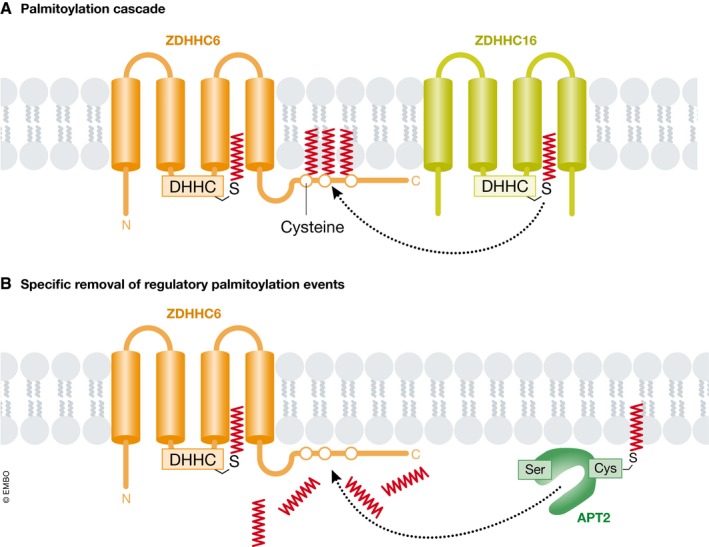
(A) ER‐resident ZDHHC6 can autopalmitoylate, like other DHHC family members. In addition, three cysteine residues within the C‐terminal tail of ZDHHC6 are palmitoylated by a distinct enzyme, ZDHHC16, representing a palmitoylation cascade. (B) The acylprotein thioesterase APT2, which itself requires palmitoylation to function normally, can remove the C‐terminal palmitoyl groups on ZDHHC6.
Protein palmitoylation and cancer: connecting the dots
Several important cancer‐associated proteins are palmitoylated. A classic example is provided by the RAS family of small GTPases 62, 63. In addition to being irreversibly farnesylated at a conserved C‐terminal CaaX sequence, HRAS, NRAS, and the KRAS‐4A isoform (but not the KRAS‐4B isoform) are reversibly S‐palmitoylated at one (NRAS, KRAS‐4A) or two (HRAS) internal cysteine residues. RAS that is both farnesylated and palmitoylated has a 100‐fold greater affinity for membranes than RAS that is only farnesylated 64. Only the dually lipidated (i.e., farnesylated and palmitoylated) forms of HRAS, NRAS, and KRAS‐4A are properly localized to the plasma membrane and capable of transforming cells, with the distinct patterns of lipid modification found between these closely related GTPases dictating trafficking and signaling properties 55, 65, 66, 67, 68. Looking more broadly, intersecting a recent list of 299 validated cancer driver genes 69 with the results of fifteen palmitoylome studies currently available in SwissPalm suggests that 78 (26%) of the encoded proteins may be palmitoylated, highlighting the relevance of this modification to cancer (Fig 3). For most cancer‐relevant palmitoylated proteins, the responsible ZDHHC enzyme(s) is not known.
Figure 3. Many cancer drivers are palmitoylated.
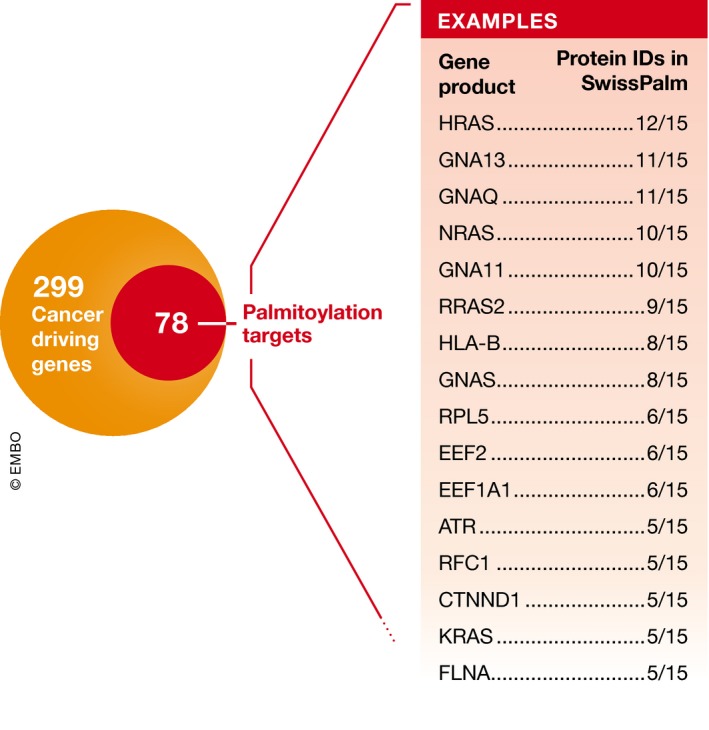
Bailey et al 69 identified a set of 299 cancer drivers, 78 of which are annotated in SwissPalm (Dataset 3) as being palmitoylated in at least one of fifteen proteomics studies. A subset of the most frequently identified proteins are shown in the box, indicating how many times they have been discovered as palmitoylated in high‐throughput studies (15 total).
The role of protein palmitoylation in cancer can also be approached from the direction of the PAT and APT enzymes, many of which display associations with cancer. Here, we focus on ZDHHC enzymes that are linked to cancer as candidate oncoproteins, tumor suppressors, or prognostic markers (Table 1). For example, low ZDHHC2 expression is found in gastric adenocarcinoma and associated with lymph node metastasis 70, and low Zdhhc2 expression is found in highly metastatic murine colorectal adenocarcinoma clones 71, 72. Truncation mutations in Zdhhc13 (Hip14l) increase susceptibility to chemically induced skin cancer in mice, possibly through enhanced NF‐κB signaling 73. Thus, ZDHHC2 and ZDHHC13 could act as tumor suppressors, but links between these enzymes, candidate substrates 74, 75, 76, and tumor suppression are unclear.
A similar situation exists in relation to ZDHHC enzymes that may act as oncoproteins. ZDHHC11 is found on chromosome 5 in a small region of recurrent amplification in non‐small‐cell lung and high‐grade bladder cancers 77, 78. ZDHHC17 (also known as HIP14) is upregulated in several tumor types, including breast and colon, and overexpression of this enzyme can also transform NIH 3T3 cells 79, 80. Overexpression of ZDHHC20 can likewise transform NIH 3T3 cells 81. The specific substrate protein(s) that link increased ZDHHC11, ZDHHC17, or ZDHHC20 expression to cancer are not known.
In some cases, there are conflicting associations between ZDHHC expression and cancer. Many testicular germ cell tumors contain genomic deletions, mutations, or other alterations that reduce ZDHHC14 expression, suggesting that this enzyme could act as a tumor suppressor in this disease 82. Indeed, overexpression of ZDHHC14 can slow xenograft tumor formation and induce apoptosis in cultured HEK 293 cells 82. However, studies of scirrhous type gastric cancer, tongue squamous cell carcinoma, and a patient with acute biphenotypic leukemia indicate that expression and/or activation of ZDHHC14 could be oncogenic in these diseases through enhanced cell migration or reduced terminal differentiation 83, 84, 85. In other cases, functional studies have disclosed a role for a particular ZDHHC enzyme in cancer development without any obvious change in gene or protein expression level. In lung cancer, ZDHHC5 expression levels are not associated with overall survival, but short interfering RNA (siRNA)‐mediated silencing of ZDHHC5 leads to reduced cell proliferation, colony formation, cell migration, and xenograft tumor growth 86. By contrast, in glioma cells, high ZDHHC5 expression is predictive of poorer prognosis, and this overexpression appears to be driven by oncogenic mutations in the tumor suppressor protein p53 87.
These results suggest that individual ZDHHC enzymes can act as either oncoproteins or tumor suppressors in a tissue‐specific and/or cancer type‐specific manner. These opposing roles may be explained by differences in the relative expression of ZDHHC enzymes and one or more key substrates between tissues. The effect of deleting or overexpressing a single ZDHHC gene on cancer phenotypes could also be masked in a given tissue due to functional redundancy. Because of this complexity, how the expression of individual ZDHHC enzymes alters substrate palmitoylation to influence cancer phenotypes is often difficult to establish. To most clearly illustrate the different ways in which protein palmitoylation can impact cancer, we describe below in greater detail several recent examples where enzyme–substrate–phenotype “triads” have been more firmly established.
Regulation of oncogenic RAS signaling from the plasma membrane
Palmitoylation of HRAS, NRAS, and KRAS‐4A at the Golgi ensures the proper delivery of these proteins to the plasma membrane via the secretory system 62. In vivo, NRAS‐driven leukemogenesis is palmitoylation‐dependent: mice transplanted with bone marrow cells expressing oncogenic human NRASG12D die of leukemia‐like diseases within 3 months, while those transplanted with cells expressing an activated but non‐palmitoylatable mutant (NRASG12D,C181S) remain healthy for up to 2 years 88. Mechanistically, inhibition of palmitoylation does not impair GTP loading of NRAS but rather prevents its normal plasma membrane localization and the hyperactivation of downstream signaling effectors such as Akt, Erk1/2, and Ral 88. This clearly demonstrates that oncogenic NRAS signaling requires palmitoylation‐dependent plasma membrane localization. Similarly to S. cerevisiae Ras2, which is palmitoylated by the Erf2‐Erf4 PAT complex, mammalian NRAS and HRAS are palmitoylated by an orthologous Golgi‐localized complex comprising ZDHHC9 and GOLGA7 42, 89 (Fig 4A). Interestingly, mammalian GOLGA7 is itself palmitoylated by an unknown ZDHHC enzyme and this modification is essential for its stability and localization to the Golgi apparatus, as well as for ZDHHC9 function 42, 90. Indeed, GOLGA7 has been identified as a selective dependency in NRAS‐mutant human AML cell lines, highlighting the importance of accessory subunits in ZDHHC function 91. In mice lacking Zdhhc9, NrasG12D‐driven T‐cell acute lymphocytic leukemia and chronic myelomonocytic leukemia are both significantly attenuated but not completely prevented 92. The incomplete phenotypic suppression implies that additional PATs likely palmitoylate Nras in vivo 92.
Figure 4. Examples of protein palmitoylation and cancer.
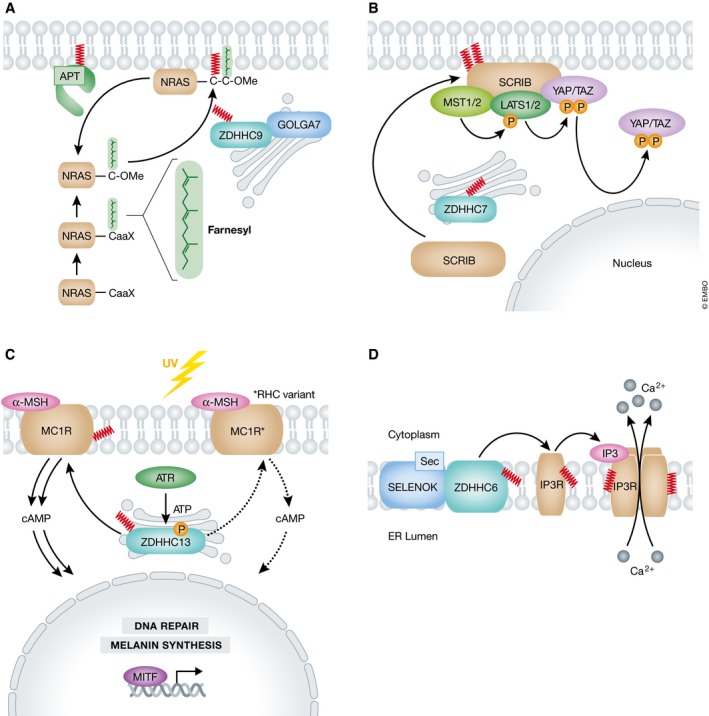
(A) The NRAS GTPase has a conserved C‐terminal CaaX motif that is farnesylated and O‐methylated (OMe), then palmitoylated by the Golgi‐resident ZDHHC9‐GOLGA7 complex. Dually lipidated NRAS is properly anchored in the plasma membrane. (B) SCRIB is localized to the plasma membrane by ZDHHC7‐catalyzed palmitoylation. SCRIB regulates the Hippo kinase cascade, ultimately phosphorylating YAP and TAZ, occluding them from the nucleus. (C) In response to ultraviolet (UV) radiation‐induced DNA damage, ATR phosphorylates ZDHHC13, activating it. ZDHHC13 palmitoylates MC1R, which can then drive cAMP‐dependent MITF transcriptional activation of melanin synthesis and DNA repair to mitigate DNA damage. Certain MC1R alleles, termed RHC variants, are found in cancer‐sensitive populations and have mutations that reduce MC1R palmitoylation. This results in downregulation of the DNA damage response and sensitization of the cells to damage. (D) Inositol‐1,4,5‐triphosphate (IP3) activates the receptor IP3R, a tetrameric Ca2+ channel, to stimulate calcium release. IP3R monomers are palmitoylated by a ZDHHC6‐SELENOK complex, with the selenocysteine (Sec) residue of SELENOK being essential to stabilize the autoacylated ZDHHC6 intermediate.
Regulation of tumor suppressor localization to the plasma membrane
Another example of palmitoylation‐dependent control over protein localization is provided by Scribble (SCRIB). SCRIB is a tumor suppressor critical for regulating epithelial cell polarity 93, 94. Loss of SCRIB inactivates the Hippo kinase cascade, leading to the accumulation of unphosphorylated Yes‐associated protein (YAP) and transcriptional coactivator with PDZ‐binding motif (TAZ) in the nucleus, and increased mitogen‐activated protein kinase (MAPK) and AKT pathway activity 47, 54, 95, 96. To activate the Hippo kinase cascade and repress MAPK and AKT signaling, SCRIB must be localized to the plasma membrane. This localization is dependent upon palmitoylation of Cys4 and Cys10 in the N‐terminus of SCRIB, most likely by ZDHHC7, but with other enzymes potentially contributing to this process 47, 54 (Fig 4B). Overexpression of the transcription factor SNAIL promotes intracellular mislocalization of SCRIB, likely through the coordinate downregulation of multiple ZDHHC enzymes and upregulation of APT2, leading to loss of protein palmitoylation 54. Treatment with the specific APT2 inhibitor ML349 results in stabilization of SCRIB at the plasma membrane by enhancing the palmitoylation of this protein 54. While the effects of this treatment on SCRIB‐dependent tumor cell growth in vivo have not been examined, the expectation is that increased membrane retention would result in decreased tumorigenicity.
Regulation of G protein‐coupled receptor activity
In melanocytes, ZDHHC13‐dependent palmitoylation of the plasma membrane α‐melanocyte‐stimulating hormone (α‐MSH) receptor, melanocortin‐1 receptor (MC1R), at Cys315 is required for MC1R tumor suppressor function 49. Specifically, only properly palmitoylated MC1R can stimulate sufficient cyclic adenosine monophosphate (cAMP) production to induce a protective transcriptional cascade in response to UV irradiation and consequent DNA damage. Cancer‐sensitizing mutations (e.g., R151C and R160W, found in people with red hair or fair skin, termed red hair color (RHC) variants) reduce Cys315 palmitoylation and attenuate cAMP production (Fig 4C). Whether this attenuation of signaling is due to mislocalization of the RHC‐variant MC1R or due to loss of MC1R activity remains unclear. Regardless, this signaling defect can be rescued by overexpression of ZDHHC13 or treatment with the thioesterase inhibitor palmostatin B, which restores MC1R‐RHC palmitoylation and attenuates the tumorigenic effects of UV irradiation in a mouse BRAFV600E‐driven cancer model 49. Thus, inhibition of depalmitoylation is a potential prophylactic strategy for individuals with certain cancer‐prone mutations affecting palmitoylation of MC1R.
Regulation of ER calcium homeostasis and proliferation
Selenoprotein K (SELENOK) is a cofactor for ZDHHC6 that promotes its stability and activity 43. One important substrate of the ER‐resident ZDHHC6‐SELENOK complex is the inositol 1,4,5‐triphosphate receptor (IP3R, ITPR1) Ca2+ channel 44. Silencing of SELENOK or ZDHHC6 expression reduces palmitoylation of IP3R on at least two cysteine residues (Cys56, Cys849), leading to protein destabilization, decreased IP3‐stimulated Ca2+ release from the ER to the cytosol, loss of Ca2+‐stimulated calcineurin activity, and reduced cell adhesion and migration 44, 97 (Fig 4D). In a mouse model of spontaneous melanoma driven by transgenic expression of glutamate receptor 1, deletion of Selenok reduced tumor formation on both ears and tail to control levels and also attenuated the appearance of metastatic cells within the lymph nodes 97. This particular melanoma model is not lethal, and therefore, it was not possible to assess the effects of SELENOK inactivation on animal survival, but the results are consistent with the possibility that inhibition of ZDHHC6‐SELENOK function could be beneficial in this cancer type. Whether this effect on tumor growth in vivo is mediated through palmitoylation of IP3R remains to be established. One interesting possibility is that this complex might, in addition to IP3R, also regulate the palmitoylation of other ER‐resident proteins involved in Ca2+ handling and cancer metabolism, such as thioredoxin‐related transmembrane protein 1 (TMX1) 98, 99.
Targeting protein palmitoylation for cancer treatment
Inhibition of protein lipidation can be toxic to normal cells and was a main reason for the clinical failure of farnesyltransferase inhibitors (FTIs) developed to inhibit the farnesylation of RAS oncoproteins 100. Notwithstanding this cautionary note, several lines of evidence provide a strong rationale for targeting PAT enzymes as a means to treat cancer. As described above, oncoproteins such as RAS‐family GTPases require palmitoylation to promote tumor formation. Certain ZDHHC enzymes and cofactors (e.g., Zdhhc9 92, Selenok 97) are essential for tumor formation and cancer progression but can be deleted in mice without gross effects on animal development. This suggests that cancer cells may develop non‐oncogene addiction 101 to the function of specific ZDHHC enzymes and cofactors, such that normal tissues are much less sensitive to inhibition of these enzymes. In turn, this should translate into a therapeutic window enabling ZDHHC enzyme inhibition to arrest the growth of or kill cancer cells without adverse effects elsewhere in the body. RAS GTPases also provide an example of a protein that itself is a challenging drug target. For RAS and other potentially undruggable proteins, inhibiting oncoprotein function by blocking a key post‐translational modification is an attractive strategy 102.
Currently, there are no potent and specific ZDHHC inhibitors. Broad‐spectrum protein palmitoylation inhibitors like 2‐bromopalmitate (2‐BP) have been used in pre‐clinical studies to validate the concept that ZDHHC protein inhibition can promote cancer cell death (e.g. 48, 103, 104). However, 2‐BP is not selective for individual ZDHHC enzymes and off‐target acylation of other intracellular proteins makes it unsuitable as a drug lead or candidate therapeutic 105, 106. A screen of 30 million compounds using a scaffold ranking library approach for inhibitors of the yeast ZDHHC9 ortholog Erf2 identified several bis‐cyclic piperazines as candidate inhibitors of this enzyme with low micromolar activity in yeast 107. Whether these lead compounds or derivatives thereof can inhibit mammalian ZDHHC enzymes in cancer cells remains to be determined.
An orthogonal approach to the development of ZDHHC inhibitors could exploit the recent crystal structure of ZDHHC20 to develop inhibitors using structure‐guided methods 26. However, structures of additional ZDHHC enzymes may be necessary to enable inhibitors specific for one enzyme over another to be found. In some cases, the process of discovering specific inhibitors could be facilitated by targeting unique domains. For example, the ankyrin‐repeat domains unique to ZDHHC13 and ZDHHC17 recognize and bind consensus short linear motifs in normally unstructured regions of substrate proteins 30, 40. A small molecule could conceivably inhibit these protein–protein interactions, which as noted above are necessary for substrate palmitoylation by a different set of ZDHHC enzymes.
Many proteins can be palmitoylated by more than one ZDHHC enzyme 29, 34, 35. This functional redundancy suggests that specific inhibitors of individual ZDHHC enzymes may not be as clinically useful as molecules that inhibit multiple family members, akin to the polypharmacology that is central to the efficacy of many kinase inhibitors used in cancer therapy 108. One potential solution to this problem is to search for pan‐ZDHHC inhibitors that can block the function of several enzymes required for the palmitoylation of a single target. For example, membrane preparations from Madin–Darby canine kidney (MDCK) cells were used as a source of PAT activity in a miniaturized screening assay looking for specific inhibitors of NRAS palmitoylation 109. It remains to be seen whether this approach will yield any candidate inhibitors, but this approach is an exciting illustration of how technical challenges in this area are being overcome to develop useful screening assays. A complementary ZDHHC‐agnostic approach is to identify compounds that directly prevent substrate palmitoylation via irreversible covalent modification of individual cysteine residues. For example, cell‐based screening and structure–activity relationship analysis have yielded a series of compounds that covalently modify the stimulator of interferon genes (STING) protein in mouse and human cells at one specific cysteine residue (Cys91), blocking the normal function of this protein in promoting inflammation 110. Identifying covalent modifiers for specific palmitoylated cysteine residues on cancer‐associated proteins would eliminate the problem of functional redundancy between individual ZDHHC enzymes.
Rather than inhibiting protein palmitoylation, in some cases it may be more effective to prevent depalmitoylation. APTs in general are highly druggable targets 55, 63, and as described above, inhibition of APT enzymes can suppress tumor formation by promoting the proper localization of SCRIB and enhancing the activity of MC1R 47, 49, 54. This paradigm could be extended to other situations where reduced PAT expression or substrate palmitoylation leads to loss of a tumor‐suppressive effect. For example, inhibition of ZDHHC7 function lowers the palmitoylation and membrane localization of the FAS death receptor, reducing the sensitivity of colorectal cancer cells to FAS ligand‐induced cell death 111. Inhibition of the appropriate thioesterase could enhance FAS receptor cell surface expression and the receipt of a lethal tumor‐suppressive signal. However, the potential toxicity associated with long‐term inhibition of acylprotein thioesterases is an important consideration that would need to be addressed.
Beyond inhibition of APT1/2 to regulate tumor suppressor function, inhibition of lysosomal PPT1‐mediated depalmitoylation may lead to a more direct toxic effect in cancer cells. Dual inhibition of PPT1 and eukaryotic translation elongation factor 1 alpha 1 (EEF1A1)‐dependent protein synthesis by the natural product didemnin B can trigger apoptosis in certain “exceptional responder” cell lines 112. Here, apoptosis is caused by loss of the rapidly degraded pro‐survival protein MCL1 and inhibition of PPT1, which may inhibit lysosomal acidification. Intriguingly, dimeric quinacrine compounds (e.g., DQ661) were recently described to inhibit PPT1 function and cause the concurrent inhibition of both mTOR signaling and autophagy, leading to protein synthesis inhibition, lysosomal membrane permeabilization, and apoptosis induction 113. DQ661 inhibits the mTOR pathway by preventing RAGC, MTOR, and other key molecules from associating with the lysosomal surface. How this molecule blocks the adaptive catabolic process of autophagy, which is normally active in response to mTOR inhibition, is unclear, but could be as simple as an increase in lysosomal pH, as observed with didemnin B. The ability of DQ661 to simultaneously inhibit mTOR and autophagy is remarkable and may underlie the promising activity of this compound in models of melanoma, pancreatic, and colon cancers 113. Identification of PPT1 substrates will contribute to a better understanding of the mechanism(s) by which DQ661 or didemnin B might cause disruption of lysosome function, which could occur through the inhibition of PPT1 and/or the accumulation of excess palmitoylated proteins.
How PPT1 inhibition can trigger cell death selectively in cancer cells is unclear. One possibility is that cancer cells have a higher number of palmitoylated proteins, or greater flux through the PPT1/lysosomal degradation pathway, rendering cells hypersensitive to any accumulation of palmitoylated proteins upon PPT1 inhibition. This would be analogous to how certain cancer cells are killed by the accumulation of reactive oxygen species (ROS) due to an already high pre‐existing ROS load 114. Interestingly, the small molecule caspase‐independent lethal 56 (CIL56) triggers cancer cell death through an unknown pathway that is inhibited by deletion of ZDHHC5 115, 116. One possibility is that like didemnin B and DQ661, CIL56 inhibits PPT1 or a related thioesterase, resulting in ZDHHC5‐driven accumulation of palmitoylated proteins to toxic levels. Further characterization of compounds that exhibit lethal mechanisms tied to specific ZDHHC enzymes could help unravel the function of these proteins and suggest new ways to target palmitoylation pathways.
Protein palmitoylation as a modulator of anti‐cancer treatment efficacy
Above, we described how directly targeting PAT or APT/PPT enzymes could be beneficial for cancer prevention or treatment. Below, we describe how targeting protein palmitoylation could enhance the therapeutic effects of existing anti‐cancer agents.
Activating mutations and amplifications of epidermal growth factor receptor (EGFR) are observed in non‐small‐cell lung, breast, and other cancers, leading to the development of targeted inhibitors such as gefitinib 117, 118. In non‐small‐cell lung carcinoma cells, palmitoylation of EGFR on Cys797 by ZDHHC1, ZDHHC2, and/or ZDHHC21 is essential for the stability, membrane localization, dimerization, and activation of this receptor 104. Inhibition of EGFR palmitoylation using 2‐BP therefore enhances the lethal potency of gefitinib 104. In a similar vein, but through a distinct mechanism, ZDHHC20‐dependent palmitoylation of EGFR at Cys1025 within the C‐terminal tail physically anchors this region of the receptor to the plasma membrane, preventing phosphorylation and activation of downstream ERK and AKT signaling pathways 48. An EGFR mutant that cannot be palmitoylated at Cys1025 has hyperactive EGFR pathway activity and exhibits a heightened dependency on (i.e., addiction to) EGFR signaling, such that the lethal potency of gefitinib is increased 48. However, this mechanism is cell type specific, and is modulated by other mutations (e.g., KRAS), which means it may not be universal 103. Though they describe different mechanisms, these reports show that combining an EGFR‐specific kinase inhibitor with ZDHHC inhibitors could result in a useful anti‐cancer synergy. Inhibition of fatty acid synthase (FASN), which causes a reduction in de novo palmitate synthesis and EGFR palmitoylation, could provide an alternative approach to blocking EGFR activity 119. While inhibition of de novo palmitate synthesis is likely to have additional effects on the cell beyond those attributable to protein palmitoylation (e.g., de novo phospholipid synthesis and re‐acylation), the availability of existing drugs targeting FASN (e.g., orlistat, Ref. 119) is one advantageous aspect of this approach.
In addition to drug sensitivity, protein palmitoylation impacts sensitivity to radiation therapy. In response to radiation, cells will arrest proliferation to repair the damage. Those cells unable to arrest the cell cycle are more likely to undergo apoptosis due to mitotic catastrophe. HEK 293 cells exhibit increased sensitivity to x‐ray irradiation when ZDHHC8 is silenced, similar to what is observed when genes encoding the DNA damage response kinases ATM or ATR or the cell cycle regulatory protein p21CIP1/WAF1 are silenced 120. Further analysis in mesothelioma cell lines pinpointed a specific defect in G2/M cell arrest and the formation of micronuclei following irradiation 120, 121. In a mouse xenograft model, injection of siRNAs targeting ZDHHC8 increased the lethal potency of X‐ray irradiation toward sarcomatoid mesothelioma cells 121. The specific target(s) of palmitoylation that are involved in regulating cell cycle progression following irradiation remain unclear. These results suggest that selective inhibition of ZDHHC8 could be useful in combination with irradiation for the treatment of certain cancers.
Concluding remarks
The full complement of mammalian PAT enzymes is now well‐established, the biochemical mechanism of reversible protein palmitoylation is increasingly well‐understood, and our knowledge of the palmitoylome has increased substantially in recent years 9, 122, 123, 124. Looking ahead, one immediate goal is to link this information together to better understand how different ZDHHC and APT enzymes regulate specific substrates to impact cancer, especially in vivo. Amazingly, even for well‐established palmitoylated oncoproteins such as KRAS‐4A, the identity of the ZDHHC palmitoyltransferase(s) responsible for this modification in vivo is not clear. Another question, given that high ZDHHC expression is a negative prognostic marker in several cancers (Table 1), is to understand biochemically how high ZDHHC enzyme expression promotes tumorigenesis. Experimental and modeling studies indicate that proteins are rarely fully palmitoylated at all possible sites and that ZDHHC overexpression can enhance substrate palmitoylation 36, 50. Thus, one possibility is that ZDHHC overexpression could produce populations of more fully palmitoylated substrate proteins that exhibit enhanced (oncogenic) activity.
Even in the absence of ZDHHC overexpression, it is conceivable that alterations in global lipid metabolism could directly influence the breadth or stoichiometry of protein palmitoylation in a given cell. For example, in cancer cells, boosting FASN expression or activity could result in increased de novo palmitate synthesis and palmitoyl‐CoA levels, thereby enhancing the modification of key oncoproteins such as EGFR 119, 125. This would be akin to how enhanced glycolysis in cancer can lead to changes in the production of acetyl‐CoA, a substrate for histone modification affecting gene expression 126. A similar effect could possibly even be achieved by changes in the dietary uptake of fatty acids: Stearoylation of the transferrin receptor in Drosophila melanogaster is sensitive to dietary stearate consumption 31. While speculative, diets high in saturated fats could conceivably promote cancer by enhancing the palmitoylation of certain oncoproteins. Interestingly, the glucose transporter GLUT4 is a target for ZDHHC7‐mediated palmitoylation in adipose cells 127, and endoplasmic reticulum Ca2+ levels and cancer cell metabolism are regulated by the palmitoylation of ER‐/mitochondrial‐associated membrane‐resident TMX1 98. Thus, enhanced or inhibited palmitoylation of metabolic enzymes could directly reinforce or maintain the altered metabolic state of cancer cells.
Finally, much work remains to understanding how ZDHHC and APT/PPT enzyme expression and activity are integrated with and regulated by oncogenic pathways. In NIH 3T3 cells, overexpression of NRAS or KRAS can increase the levels of ZDHHC17 (HIP14) 79. Likewise, growth factors (e.g., EGF, FGF) can prevent the proteolytic destruction of plasma membrane‐localized ZDHHC5 128. Thus, oncogenic signaling pathways likely alter protein palmitoylation dynamics by affecting ZDHHC enzyme stability. Less directly, overexpression of transcription factors such as SNAIL can reduce the expression of several ZDHHC enzymes while increasing expression of APT2, resulting in coordinate net depalmitoylation of many proteins 54. APT enzymes are also targets that can be engaged by oncogenic networks to promote disease progression. In melanoma cells, Wnt5a‐driven phosphorylation of APT1 reduces its dimerization and increases its depalmitoylation activity, leading to enhanced metastasis 60. Tumor suppressor pathways also engage ZDHHC enzymes. In melanoma cells, ZDHHC13 activity is positively regulated by ATR‐dependent phosphorylation at Ser8, which promotes association between ZDHHC13 and its substrate MC1R, increasing palmitoylation of the latter and generating a protective anti‐tumor response 49. How phosphorylation promotes this association is not known, but hints that in some cases ZDHHC substrate selection and activity could both hinge on phosphorylation‐dependent structural modifications. Adding further potential complexity, ATR itself may be palmitoylated (Fig 3) suggesting the potential for reciprocal regulation of ATR localization or activity in some contexts by ZDHHC function. Understanding these functional relationships and other important questions provides many avenues for future research in this field (see also Box 1).
Conflict of interest
The authors declare that they have no conflict of interest.
Box 1: In need of answers.
What is the full complement of palmitoylated proteins and how does this vary between normal and cancer cells? Can we link the palmitoylation of specific proteins to the activity of one or more ZDHHC enzymes in each case? How do proteins with altered palmitoylation contribute to hallmark cancer phenotypes such as growth, proliferation, metastasis, or evasion of cell death?
How can the same ZDHHC enzyme act as an oncogene in one cancer and a tumor suppressor in another? What cell‐type or context‐specific differences account for these distinct functions? This ties in with the question of how individual ZDHHC enzymes select substrates at the biochemical level.
Can we identify specific inhibitors for individual ZDHHC‐family enzymes? Will such inhibitors offer a means to selectively inhibit the function of important and difficult‐to‐drug oncoproteins such as NRAS?
Does the palmitoylation state of individual cysteine residues within a given protein vary over time in normal and cancer cells? Are there interactions at individual cysteine residues between different types of post‐translational modifications, including disulfide formation, glutathionylation, and oxidation.
How are protein palmitoylation cascades integrated within the broader signaling and metabolic networks of the cell? ZDHHC and APT function appears to be regulated by phosphorylation and ubiquitination—how prevalent are post‐translational modifications and how do they impact enzyme function? How do the common cancer‐induced alterations in cellular lipid metabolism impact the import, de novo synthesis, compartmentalization, and degradation of palmitate or other free fatty acids used as substrates for palmitoylation?
Acknowledgements
We thank Leslie Magtanong for comments on the manuscript. This work was supported by T32 GM007276 to P‐J.K. and a Damon Runyon‐Rachleff Innovation award to S.J.D.
EMBO Reports (2018) 19: e46666
See the Glossary for abbreviations used in this article.
References
- 1. Hanahan D, Weinberg RA (2000) The hallmarks of cancer. Cell 100: 57–70 [DOI] [PubMed] [Google Scholar]
- 2. Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144: 646–674 [DOI] [PubMed] [Google Scholar]
- 3. Lothrop AP, Torres MP, Fuchs SM (2013) Deciphering post‐translational modification codes. FEBS Lett 587: 1247–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Blanc M, David F, Abrami L, Migliozzi D, Armand F, Bürgi J, van der Goot FG (2015) SwissPalm: protein palmitoylation database. F1000Res 4: 261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jiang H, Zhang X, Chen X, Aramsangtienchai P, Tong Z, Lin H (2018) Protein lipidation: occurrence, mechanisms, biological functions, and enabling technologies. Chem Rev 118: 919–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen B, Sun Y, Niu J, Jarugumilli GK, Wu X (2018) Protein lipidation in cell signaling and diseases: function, regulation, and therapeutic opportunities. Cell Chem Biol 25: 817–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Milligan G, Parenti M, Magee AI (1995) The dynamic role of palmitoylation in signal transduction. Trends Biochem Sci 20: 181–187 [DOI] [PubMed] [Google Scholar]
- 8. Burnaevskiy N, Peng T, Reddick LE, Hang HC, Alto NM (2015) Myristoylome profiling reveals a concerted mechanism of ARF GTPase deacylation by the bacterial protease IpaJ. Mol Cell 58: 110–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lanyon‐Hogg T, Faronato M, Serwa RA, Tate EW (2017) Dynamic protein acylation: new substrates, mechanisms, and drug targets. Trends Biochem Sci 42: 566–581 [DOI] [PubMed] [Google Scholar]
- 10. Rocks O, Gerauer M, Vartak N, Koch S, Huang Z‐P, Pechlivanis M, Kuhlmann J, Brunsveld L, Chandra A, Ellinger B et al (2010) The palmitoylation machinery is a spatially organizing system for peripheral membrane proteins. Cell 141: 458–471 [DOI] [PubMed] [Google Scholar]
- 11. Won SJ, Martin BR (2018) Temporal profiling establishes a dynamic S‐palmitoylation cycle. ACS Chem Biol 13: 1560–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Martin BR, Wang C, Adibekian A, Tully SE, Cravatt BF (2011) Global profiling of dynamic protein palmitoylation. Nat Methods 9: 84–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dunphy JT, Linder ME (1998) Signalling functions of protein palmitoylation. Biochim Biophys Acta 1436: 245–261 [DOI] [PubMed] [Google Scholar]
- 14. Percherancier Y, Planchenault T, Valenzuela‐Fernandez A, Virelizier JL, Arenzana‐Seisdedos F, Bachelerie F (2001) Palmitoylation‐dependent control of degradation, life span, and membrane expression of the CCR5 receptor. J Biol Chem 276: 31936–31944 [DOI] [PubMed] [Google Scholar]
- 15. Sanders SS, Martin DDO, Butland SL, Lavallée‐Adam M, Calzolari D, Kay C, Yates JR, Hayden MR (2015) Curation of the mammalian palmitoylome indicates a pivotal role for palmitoylation in diseases and disorders of the nervous system and cancers. PLoS Comput Biol 11: e1004405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang J, Huang Y, Chen J, Zhu H, Whiteheart SW (2018) Dynamic cycling of t‐SNARE acylation regulates platelet exocytosis. J Biol Chem 293: 3593–3606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chan P, Han X, Zheng B, DeRan M, Yu J, Jarugumilli GK, Deng H, Pan D, Luo X, Wu X (2016) Autopalmitoylation of TEAD proteins regulates transcriptional output of the Hippo pathway. Nat Chem Biol 12: 282–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Noland CL, Gierke S, Schnier PD, Murray J, Sandoval WN, Sagolla M, Dey A, Hannoush RN, Fairbrother WJ, Cunningham CN (2016) Palmitoylation of TEAD transcription factors is required for their stability and function in Hippo pathway signaling. Structure 24: 179–186 [DOI] [PubMed] [Google Scholar]
- 19. Linder ME, Deschenes RJ (2007) Palmitoylation: policing protein stability and traffic. Nat Rev Mol Cell Biol 8: 74–84 [DOI] [PubMed] [Google Scholar]
- 20. Gorleku OA, Barns A‐M, Prescott GR, Greaves J, Chamberlain LH (2011) Endoplasmic reticulum localization of DHHC palmitoyltransferases mediated by lysine‐based sorting signals. J Biol Chem 286: 39573–39584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ohno Y, Kihara A, Sano T, Igarashi Y (2006) Intracellular localization and tissue‐specific distribution of human and yeast DHHC cysteine‐rich domain‐containing proteins. Biochim Biophys Acta 1761: 474–483 [DOI] [PubMed] [Google Scholar]
- 22. Long JZ, Cravatt BF (2011) The metabolic serine hydrolases and their functions in mammalian physiology and disease. Chem Rev 111: 6022–6063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bartels DJ, Mitchell DA, Dong X, Deschenes RJ (1999) Erf2, a novel gene product that affects the localization and palmitoylation of Ras2 in Saccharomyces cerevisiae . Mol Cell Biol 19: 6775–6787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lobo S, Greentree WK, Linder ME, Deschenes RJ (2002) Identification of a Ras palmitoyltransferase in Saccharomyces cerevisiae . J Biol Chem 277: 41268–41273 [DOI] [PubMed] [Google Scholar]
- 25. Roth AF, Feng Y, Chen L, Davis NG (2002) The yeast DHHC cysteine‐rich domain protein Akr1p is a palmitoyl transferase. J Cell Biol 159: 23–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rana MS, Kumar P, Lee C‐J, Verardi R, Rajashankar KR, Banerjee A (2018) Fatty acyl recognition and transfer by an integral membrane S‐acyltransferase. Science 359: 6326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gottlieb CD, Zhang S, Linder ME (2015) The cysteine‐rich domain of the DHHC3 palmitoyltransferase is palmitoylated and contains tightly bound zinc. J Biol Chem 290: 29259–29269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. González Montoro A, Quiroga R, Valdez Taubas J (2013) Zinc co‐ordination by the DHHC cysteine‐rich domain of the palmitoyltransferase Swf1. Biochem J 454: 427–435 [DOI] [PubMed] [Google Scholar]
- 29. Ohno Y, Kashio A, Ogata R, Ishitomi A, Yamazaki Y, Kihara A (2012) Analysis of substrate specificity of human DHHC protein acyltransferases using a yeast expression system. Mol Biol Cell 23: 4543–4551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lemonidis K, Gorleku OA, Sanchez‐Perez MC, Grefen C, Chamberlain LH (2014) The Golgi S‐acylation machinery comprises zDHHC enzymes with major differences in substrate affinity and S‐acylation activity. Mol Biol Cell 25: 3870–3883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Senyilmaz D, Virtue S, Xu X, Tan CY, Griffin JL, Miller AK, Vidal‐Puig A, Teleman AA (2015) Regulation of mitochondrial morphology and function by stearoylation of TFR1. Nature 525: 124–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jennings BC, Linder ME (2012) DHHC protein S‐acyltransferases use similar ping‐pong kinetic mechanisms but display different acyl‐CoA specificities. J Biol Chem 287: 7236–7245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Greaves J, Munro KR, Davidson SC, Riviere M, Wojno J, Smith TK, Tomkinson NCO, Chamberlain LH (2017) Molecular basis of fatty acid selectivity in the zDHHC family of S‐acyltransferases revealed by click chemistry. Proc Natl Acad Sci USA 114: E1365–E1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tian L, McClafferty H, Jeffries O, Shipston MJ (2010) Multiple palmitoyltransferases are required for palmitoylation‐dependent regulation of large conductance calcium‐ and voltage‐activated potassium channels. J Biol Chem 285: 23954–23962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lemonidis K, Salaun C, Kouskou M, Diez‐Ardanuy C, Chamberlain LH, Greaves J (2017) Substrate selectivity in the zDHHC family of S‐acyltransferases. Biochem Soc Trans 45: 751–758 [DOI] [PubMed] [Google Scholar]
- 36. Abrami L, Dallavilla T, Sandoz PA, Demir M, Kunz B, Savoglidis G, Hatzimanikatis V, van der Goot FG (2017) Identification and dynamics of the human ZDHHC16‐ZDHHC6 palmitoylation cascade. Elife 6: 309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Castrec B, Dian C, Ciccone S, Ebert CL, Bienvenut WV, Le Caer J‐P, Steyaert J‐M, Giglione C, Meinnel T (2018) Structural and genomic decoding of human and plant myristoylomes reveals a definitive recognition pattern. Nat Chem Biol 14: 671–679 [DOI] [PubMed] [Google Scholar]
- 38. Wolven A, Okamura H, Rosenblatt Y, Resh MD (1997) Palmitoylation of p59fyn is reversible and sufficient for plasma membrane association. Mol Biol Cell 8: 1159–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. van't Hof W, Resh MD (1997) Rapid plasma membrane anchoring of newly synthesized p59fyn: selective requirement for NH2‐terminal myristoylation and palmitoylation at cysteine‐3. J Cell Biol 136: 1023–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lemonidis K, Sanchez‐Perez MC, Chamberlain LH (2015) Identification of a novel sequence motif recognized by the ankyrin repeat domain of zDHHC17/13 S‐acyltransferases. J Biol Chem 290: 21939–21950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mitchell DA, Hamel LD, Ishizuka K, Mitchell G, Schaefer LM, Deschenes RJ (2012) The Erf4 subunit of the yeast Ras palmitoyl acyltransferase is required for stability of the Acyl‐Erf2 intermediate and palmitoyl transfer to a Ras2 substrate. J Biol Chem 287: 34337–34348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Swarthout JT, Lobo S, Farh L, Croke MR, Greentree WK, Deschenes RJ, Linder ME (2005) DHHC9 and GCP16 constitute a human protein fatty acyltransferase with specificity for H‐ and N‐Ras. J Biol Chem 280: 31141–31148 [DOI] [PubMed] [Google Scholar]
- 43. Fredericks GJ, Hoffmann FW, Hondal RJ, Rozovsky S, Urschitz J, Hoffmann PR (2017) Selenoprotein K increases efficiency of DHHC6 catalyzed protein palmitoylation by stabilizing the acyl‐DHHC6 intermediate. Antioxidants (Basel) 7: 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fredericks GJ, Hoffmann FW, Rose AH, Osterheld HJ, Hess FM, Mercier F, Hoffmann PR (2014) Stable expression and function of the inositol 1,4,5‐triphosphate receptor requires palmitoylation by a DHHC6/selenoprotein K complex. Proc Natl Acad Sci USA 111: 16478–16483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xie Y, Zheng Y, Li H, Luo X, He Z, Cao S, Shi Y, Zhao Q, Xue Y, Zuo Z et al (2016) GPS‐Lipid: a robust tool for the prediction of multiple lipid modification sites. Sci Rep 6: 28249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shi S‐P, Sun X‐Y, Qiu J‐D, Suo S‐B, Chen X, Huang S‐Y, Liang R‐P (2013) The prediction of palmitoylation site locations using a multiple feature extraction method. J Mol Graph Model 40: 125–130 [DOI] [PubMed] [Google Scholar]
- 47. Chen B, Zheng B, DeRan M, Jarugumilli GK, Fu J, Brooks YS, Wu X (2016) ZDHHC7‐mediated S‐palmitoylation of Scribble regulates cell polarity. Nat Chem Biol 12: 686–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Runkle KB, Kharbanda A, Stypulkowski E, Cao X‐J, Wang W, Garcia BA, Witze ES (2016) Inhibition of DHHC20‐mediated EGFR palmitoylation creates a dependence on EGFR signaling. Mol Cell 62: 385–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chen S, Zhu B, Yin C, Liu W, Han C, Chen B, Liu T, Li X, Chen X, Li C et al (2017) Palmitoylation‐dependent activation of MC1R prevents melanomagenesis. Nature 549: 399–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rodenburg RNP, Snijder J, van de Waterbeemd M, Schouten A, Granneman J, Heck AJR, Gros P (2017) Stochastic palmitoylation of accessible cysteines in membrane proteins revealed by native mass spectrometry. Nat Commun 8: 1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lin DTS, Conibear E (2015) ABHD17 proteins are novel protein depalmitoylases that regulate N‐Ras palmitate turnover and subcellular localization. Elife 4: e11306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Duncan JA, Gilman AG (1998) A cytoplasmic acyl‐protein thioesterase that removes palmitate from G protein alpha subunits and p21(RAS). J Biol Chem 273: 15830–15837 [DOI] [PubMed] [Google Scholar]
- 53. Kathayat RS, Cao Y, Elvira PD, Sandoz PA, Zaballa M‐E, Springer MZ, Drake LE, Macleod KF, van der Goot FG, Dickinson BC (2018) Active and dynamic mitochondrial S‐depalmitoylation revealed by targeted fluorescent probes. Nat Commun 9: 334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hernandez JL, Davda D, Cheung See Kit M, Majmudar JD, Won SJ, Gang M, Pasupuleti SC, Choi AI, Bartkowiak CM, Martin BR (2017) APT2 inhibition restores scribble localization and S‐palmitoylation in snail‐transformed cells. Cell Chem Biol 24: 87–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lin DTS, Davis NG, Conibear E (2017) Targeting the Ras palmitoylation/depalmitoylation cycle in cancer. Biochem Soc Trans 45: 913–921 [DOI] [PubMed] [Google Scholar]
- 56. Yokoi N, Fukata Y, Sekiya A, Murakami T, Kobayashi K, Fukata M (2016) Identification of PSD‐95 depalmitoylating enzymes. J Neurosci 36: 6431–6444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yang W, Di Vizio D, Kirchner M, Steen H, Freeman MR (2010) Proteome scale characterization of human S‐acylated proteins in lipid raft‐enriched and non‐raft membranes. Mol Cell Proteomics 9: 54–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kong E, Peng S, Chandra G, Sarkar C, Zhang Z, Bagh MB, Mukherjee AB (2013) Dynamic palmitoylation links cytosol‐membrane shuttling of acyl‐protein thioesterase‐1 and acyl‐protein thioesterase‐2 with that of proto‐oncogene H‐ras product and growth‐associated protein‐43. J Biol Chem 288: 9112–9125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Segal‐Salto M, Sapir T, Reiner O (2016) Reversible cysteine acylation regulates the activity of human palmitoyl‐protein thioesterase 1 (PPT1). PLoS One 11: e0146466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sadeghi RS, Kulej K, Kathayat RS, Garcia BA, Dickinson BC, Brady DC, Witze ES (2018) Wnt5a signaling induced phosphorylation increases APT1 activity and promotes melanoma metastatic behavior. Elife 7: R19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lievens PM‐J, Kuznetsova T, Kochlamazashvili G, Cesca F, Gorinski N, Galil DA, Cherkas V, Ronkina N, Lafera J, Gaestel M et al (2016) ZDHHC3 tyrosine phosphorylation regulates neural cell adhesion molecule palmitoylation. Mol Cell Biol 36: 2208–2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Schmick M, Kraemer A, Bastiaens PIH (2015) Ras moves to stay in place. Trends Cell Biol 25: 190–197 [DOI] [PubMed] [Google Scholar]
- 63. Dekker FJ, Rocks O, Vartak N, Menninger S, Hedberg C, Balamurugan R, Wetzel S, Renner S, Gerauer M, Schölermann B et al (2010) Small‐molecule inhibition of APT1 affects Ras localization and signaling. Nat Chem Biol 6: 449–456 [DOI] [PubMed] [Google Scholar]
- 64. Schroeder H, Leventis R, Rex S, Schelhaas M, Nägele E, Waldmann H, Silvius JR (1997) S‐Acylation and plasma membrane targeting of the farnesylated carboxyl‐terminal peptide of N‐ras in mammalian fibroblasts. Biochemistry 36: 13102–13109 [DOI] [PubMed] [Google Scholar]
- 65. Chen ZQ, Ulsh LS, DuBois G, Shih TY (1985) Posttranslational processing of p21 ras proteins involves palmitylation of the C‐terminal tetrapeptide containing cysteine‐186. J Virol 56: 607–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hancock JF, Magee AI, Childs JE, Marshall CJ (1989) All ras proteins are polyisoprenylated but only some are palmitoylated. Cell 57: 1167–1177 [DOI] [PubMed] [Google Scholar]
- 67. Jackson JH, Cochrane CG, Bourne JR, Solski PA, Buss JE, Der CJ (1990) Farnesol modification of Kirsten‐ras exon 4B protein is essential for transformation. Proc Natl Acad Sci USA 87: 3042–3046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lynch SJ, Snitkin H, Gumper I, Philips MR, Sabatini D, Pellicer A (2015) The differential palmitoylation states of N‐Ras and H‐Ras determine their distinct Golgi subcompartment localizations. J Cell Physiol 230: 610–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bailey MH, Tokheim C, Porta‐Pardo E, Sengupta S, Bertrand D, Weerasinghe A, Colaprico A, Wendl MC, Kim J, Reardon B et al (2018) Comprehensive characterization of cancer driver genes and mutations. Cell 173: 371–385.e18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yan S‐M, Tang J‐J, Huang C‐Y, Xi S‐Y, Huang M‐Y, Liang J‐Z, Jiang Y‐X, Li Y‐H, Zhou Z‐W, Ernberg I et al (2013) Reduced expression of ZDHHC2 is associated with lymph node metastasis and poor prognosis in gastric adenocarcinoma. PLoS One 8: e56366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Tsuruo T, Yamori T, Naganuma K, Tsukagoshi S, Sakurai Y (1983) Characterization of metastatic clones derived from a metastatic variant of mouse colon adenocarcinoma 26. Cancer Res 43: 5437–5442 [PubMed] [Google Scholar]
- 72. Oyama T, Miyoshi Y, Koyama K, Nakagawa H, Yamori T, Ito T, Matsuda H, Arakawa H, Nakamura Y (2000) Isolation of a novel gene on 8p21.3‐22 whose expression is reduced significantly in human colorectal cancers with liver metastasis. Genes Chromosom Cancer 29: 9–15 [DOI] [PubMed] [Google Scholar]
- 73. Perez CJ, Mecklenburg L, Jaubert J, Martinez‐Santamaria L, Iritani BM, Espejo A, Napoli E, Song G, Del Río M, DiGiovanni J et al (2015) Increased susceptibility to skin carcinogenesis associated with a spontaneous mouse mutation in the Palmitoyl Transferase Zdhhc13 gene. J Invest Dermatol 135: 3133–3143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sharma C, Yang XH, Hemler ME (2008) DHHC2 affects palmitoylation, stability, and functions of tetraspanins CD9 and CD151. Mol Biol Cell 19: 3415–3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Planey SL, Keay SK, Zhang C‐O, Zacharias DA (2009) Palmitoylation of cytoskeleton associated protein 4 by DHHC2 regulates antiproliferative factor‐mediated signaling. Mol Biol Cell 20: 1454–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Shen L‐F, Chen Y‐J, Liu K‐M, Haddad ANS, Song I‐W, Roan H‐Y, Chen L‐Y, Yen JJY, Chen Y‐J, Wu J‐Y et al (2017) Role of S‐palmitoylation by ZDHHC13 in mitochondrial function and metabolism in liver. Sci Rep 7: 2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kang JU, Koo SH, Kwon KC, Park JW, Kim JM (2008) Gain at chromosomal region 5p15.33, containing TERT, is the most frequent genetic event in early stages of non‐small cell lung cancer. Cancer Genet Cytogenet 182: 1–11 [DOI] [PubMed] [Google Scholar]
- 78. Yamamoto Y, Chochi Y, Matsuyama H, Eguchi S, Kawauchi S, Furuya T, Oga A, Kang JJ, Naito K, Sasaki K (2007) Gain of 5p15.33 is associated with progression of bladder cancer. Oncology 72: 132–138 [DOI] [PubMed] [Google Scholar]
- 79. Ducker CE, Stettler EM, French KJ, Upson JJ, Smith CD (2004) Huntingtin interacting protein 14 is an oncogenic human protein: palmitoyl acyltransferase. Oncogene 23: 9230–9237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ducker CE, Griffel LK, Smith RA, Keller SN, Zhuang Y, Xia Z, Diller JD, Smith CD (2006) Discovery and characterization of inhibitors of human palmitoyl acyltransferases. Mol Cancer Ther 5: 1647–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Draper JM, Smith CD (2010) DHHC20: a human palmitoyl acyltransferase that causes cellular transformation. Mol Membr Biol 27: 123–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Yeste‐Velasco M, Mao X, Grose R, Kudahetti SC, Lin D, Marzec J, Vasiljević N, Chaplin T, Xue L, Xu M et al (2014) Identification of ZDHHC14 as a novel human tumour suppressor gene. J Pathol 232: 566–577 [DOI] [PubMed] [Google Scholar]
- 83. Yu L, Reader JC, Chen C, Zhao XF, Ha JS, Lee C, York T, Gojo I, Baer MR, Ning Y (2011) Activation of a novel palmitoyltransferase ZDHHC14 in acute biphenotypic leukemia and subsets of acute myeloid leukemia. Leukemia 25: 367–371 [DOI] [PubMed] [Google Scholar]
- 84. Oo HZ, Sentani K, Sakamoto N, Anami K, Naito Y, Uraoka N, Oshima T, Yanagihara K, Oue N, Yasui W (2014) Overexpression of ZDHHC14 promotes migration and invasion of scirrhous type gastric cancer. Oncol Rep 32: 403–410 [DOI] [PubMed] [Google Scholar]
- 85. Onda T, Yamamoto N, Kuroiwa T, Katakura A, Takano N, Shibahara T (2010) Aberrant expression of the ZDHHC14 gene in squamous cell carcinoma of the human tongue. Asian J Oral Maxillofac Surg 22: 187–192 [Google Scholar]
- 86. Tian H, Lu J‐Y, Shao C, Huffman KE, Carstens RM, Larsen JE, Girard L, Liu H, Rodriguez‐Canales J, Frenkel EP et al (2015) Systematic siRNA screen unmasks NSCLC growth dependence by palmitoyltransferase DHHC5. Mol Cancer Res 13: 784–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Chen X, Ma H, Wang Z, Zhang S, Yang H, Fang Z (2017) EZH2 palmitoylation mediated by ZDHHC5 in p53‐mutant glioma drives malignant development and progression. Cancer Res 77: 4998–5010 [DOI] [PubMed] [Google Scholar]
- 88. Cuiffo B, Ren R (2010) Palmitoylation of oncogenic NRAS is essential for leukemogenesis. Blood 115: 3598–3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Young E, Zheng Z‐Y, Wilkins AD, Jeong H‐T, Li M, Lichtarge O, Chang EC (2014) Regulation of Ras localization and cell transformation by evolutionarily conserved palmitoyltransferases. Mol Cell Biol 34: 374–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Ohta E, Misumi Y, Sohda M, Fujiwara T, Yano A, Ikehara Y (2003) Identification and characterization of GCP16, a novel acylated Golgi protein that interacts with GCP170. J Biol Chem 278: 51957–51967 [DOI] [PubMed] [Google Scholar]
- 91. Wang T, Yu H, Hughes NW, Liu B, Kendirli A, Klein K, Chen WW, Lander ES, Sabatini DM (2017) Gene essentiality profiling reveals gene networks and synthetic lethal interactions with oncogenic ras. Cell 168: 890–903.e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Liu P, Jiao B, Zhang R, Zhao H, Zhang C, Wu M, Li D, Zhao X, Qiu Q, Li J et al (2016) Palmitoylacyltransferase Zdhhc9 inactivation mitigates leukemogenic potential of oncogenic Nras. Leukemia 30: 1225–1228 [DOI] [PubMed] [Google Scholar]
- 93. Pearson HB, Perez‐Mancera PA, Dow LE, Ryan A, Tennstedt P, Bogani D, Elsum I, Greenfield A, Tuveson DA, Simon R et al (2011) SCRIB expression is deregulated in human prostate cancer, and its deficiency in mice promotes prostate neoplasia. J Clin Invest 121: 4257–4267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Zhan L, Rosenberg A, Bergami KC, Yu M, Xuan Z, Jaffe AB, Allred C, Muthuswamy SK (2008) Deregulation of scribble promotes mammary tumorigenesis and reveals a role for cell polarity in carcinoma. Cell 135: 865–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Cordenonsi M, Zanconato F, Azzolin L, Forcato M, Rosato A, Frasson C, Inui M, Montagner M, Parenti AR, Poletti A et al (2011) The Hippo transducer TAZ confers cancer stem cell‐related traits on breast cancer cells. Cell 147: 759–772 [DOI] [PubMed] [Google Scholar]
- 96. Johnson R, Halder G (2014) The two faces of Hippo: targeting the Hippo pathway for regenerative medicine and cancer treatment. Nat Rev Drug Discov 13: 63–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Marciel MP, Khadka VS, Deng Y, Kilicaslan P, Pham A, Bertino P, Lee K, Chen S, Glibetic N, Hoffmann FW et al (2018) Selenoprotein K deficiency inhibits melanoma by reducing calcium flux required for tumor growth and metastasis. Oncotarget 9: 13407–13422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Raturi A, Gutiérrez T, Ortiz‐Sandoval C, Ruangkittisakul A, Herrera‐Cruz MS, Rockley JP, Gesson K, Ourdev D, Lou P‐H, Lucchinetti E et al (2016) TMX1 determines cancer cell metabolism as a thiol‐based modulator of ER‐mitochondria Ca2+ flux. J Cell Biol 214: 433–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Lynes EM, Bui M, Yap MC, Benson MD, Schneider B, Ellgaard L, Berthiaume LG, Simmen T (2012) Palmitoylated TMX and calnexin target to the mitochondria‐associated membrane. EMBO J 31: 457–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Berndt N, Hamilton AD, Sebti SM (2011) Targeting protein prenylation for cancer therapy. Nat Rev Cancer 11: 775–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Luo J, Solimini NL, Elledge SJ (2009) Principles of cancer therapy: oncogene and non‐oncogene addiction. Cell 136: 823–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Vasan N, Boyer JL, Herbst RS (2014) A RAS renaissance: emerging targeted therapies for KRAS‐mutated non‐small cell lung cancer. Clin Cancer Res 20: 3921–3930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Kharbanda A, Runkle K, Wang W, Witze ES (2017) Induced sensitivity to EGFR inhibitors is mediated by palmitoylated cysteine 1025 of EGFR and requires oncogenic Kras. Biochem Biophys Res Commun 493: 213–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Bollu LR, Katreddy RR, Blessing AM, Pham N, Zheng B, Wu X, Weihua Z (2015) Intracellular activation of EGFR by fatty acid synthase dependent palmitoylation. Oncotarget 6: 34992–35003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Zheng B, DeRan M, Li X, Liao X, Fukata M, Wu X (2013) 2‐Bromopalmitate analogues as activity‐based probes to explore palmitoyl acyltransferases. J Am Chem Soc 135: 7082–7085 [DOI] [PubMed] [Google Scholar]
- 106. Davda D, El Azzouny MA, Tom CTMB, Hernandez JL, Majmudar JD, Kennedy RT, Martin BR (2013) Profiling targets of the irreversible palmitoylation inhibitor 2‐bromopalmitate. ACS Chem Biol 8: 1912–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Hamel LD, Lenhart BJ, Mitchell DA, Santos RG, Giulianotti MA, Deschenes RJ (2016) Identification of protein palmitoylation inhibitors from a scaffold ranking library. Comb Chem High Throughput Screen 19: 262–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Klaeger S, Heinzlmeir S, Wilhelm M, Polzer H, Vick B, Koenig P‐A, Reinecke M, Ruprecht B, Petzoldt S, Meng C et al (2017) The target landscape of clinical kinase drugs. Science 358: eaan4368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Ganesan L, Shieh P, Bertozzi CR, Levental I (2017) Click‐chemistry based high throughput screening platform for modulators of ras palmitoylation. Sci Rep 7: 41147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Haag SM, Gulen MF, Reymond L, Gibelin A, Abrami L, Decout A, Heymann M, Goot FGVD, Turcatti G, Behrendt R et al (2018) Targeting STING with covalent small‐molecule inhibitors. Nature 559: 269–273 [DOI] [PubMed] [Google Scholar]
- 111. Rossin A, Durivault J, Chakhtoura‐Feghali T, Lounnas N, Gagnoux‐Palacios L, Hueber A‐O (2015) Fas palmitoylation by the palmitoyl acyltransferase DHHC7 regulates Fas stability. Cell Death Differ 22: 643–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Potts MB, McMillan EA, Rosales TI, Kim HS, Ou Y‐H, Toombs JE, Brekken RA, Minden MD, MacMillan JB, White MA (2015) Mode of action and pharmacogenomic biomarkers for exceptional responders to didemnin B. Nat Chem Biol 11: 401–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Rebecca VW, Nicastri MC, McLaughlin N, Fennelly C, McAfee Q, Ronghe A, Nofal M, Lim C‐Y, Witze E, Chude CI et al (2017) A unified approach to targeting the lysosome's degradative and growth signaling roles. Cancer Discov 7: 1266–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Gorrini C, Harris IS, Mak TW (2013) Modulation of oxidative stress as an anticancer strategy. Nat Rev Drug Discov 12: 931–947 [DOI] [PubMed] [Google Scholar]
- 115. Dixon SJ, Winter GE, Musavi LS, Lee ED, Snijder B, Rebsamen M, Superti‐Furga G, Stockwell BR (2015) Human haploid cell genetics reveals roles for lipid metabolism genes in nonapoptotic cell death. ACS Chem Biol 10: 1604–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Shimada K, Skouta R, Kaplan A, Yang WS, Hayano M, Dixon SJ, Brown LM, Valenzuela CA, Wolpaw AJ, Stockwell BR (2016) Global survey of cell death mechanisms reveals metabolic regulation of ferroptosis. Nat Chem Biol 12: 497–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Herbst RS, Fukuoka M, Baselga J (2004) Gefitinib–a novel targeted approach to treating cancer. Nat Rev Cancer 4: 956–965 [DOI] [PubMed] [Google Scholar]
- 118. Sharma MR, Schilsky RL (2011) Role of randomized phase III trials in an era of effective targeted therapies. Nat Rev Clin Oncol 9: 208–214 [DOI] [PubMed] [Google Scholar]
- 119. Ali A, Levantini E, Teo JT, Goggi J, Clohessy JG, Wu CS, Chen L, Yang H, Krishnan I, Kocher O et al (2018) Fatty acid synthase mediates EGFR palmitoylation in EGFR mutated non‐small cell lung cancer. EMBO Mol Med 10: e8313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Sudo H, Tsuji AB, Sugyo A, Imai T, Saga T, Harada Y‐N (2007) A loss of function screen identifies nine new radiation susceptibility genes. Biochem Biophys Res Commun 364: 695–701 [DOI] [PubMed] [Google Scholar]
- 121. Sudo H, Tsuji AB, Sugyo A, Ogawa Y, Sagara M, Saga T (2012) ZDHHC8 knockdown enhances radiosensitivity and suppresses tumor growth in a mesothelioma mouse model. Cancer Sci 103: 203–209 [DOI] [PubMed] [Google Scholar]
- 122. Gao X, Hannoush RN (2018) A decade of click chemistry in protein palmitoylation: impact on discovery and new biology. Cell Chem Biol 25: 236–246 [DOI] [PubMed] [Google Scholar]
- 123. Li W, Li W, Zou L, Ji S, Li C, Liu K, Zhang G, Sun Q, Xiao F, Chen D (2017) Membrane targeting of inhibitory Smads through palmitoylation controls TGF‐β/BMP signaling. Proc Natl Acad Sci USA 114: 13206–13211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Zhang X, Zhang Y, Fang C, Zhang L, Yang P, Wang C, Lu H (2018) Ultradeep palmitoylomics enabled by dithiodipyridine‐functionalized magnetic nanoparticles. Anal Chem 90: 6161–6168 [DOI] [PubMed] [Google Scholar]
- 125. Menendez JA, Lupu R (2007) Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer 7: 763–777 [DOI] [PubMed] [Google Scholar]
- 126. Lee JV, Carrer A, Shah S, Snyder NW, Wei S, Venneti S, Worth AJ, Yuan Z‐F, Lim H‐W, Liu S et al (2014) Akt‐dependent metabolic reprogramming regulates tumor cell histone acetylation. Cell Metab 20: 306–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Du K, Murakami S, Sun Y, Kilpatrick CL, Luscher B (2017) DHHC7 palmitoylates glucose transporter 4 (Glut4) and regulates Glut4 membrane translocation. J Biol Chem 292: 2979–2991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Li Y, Martin BR, Cravatt BF, Hofmann SL (2012) DHHC5 protein palmitoylates flotillin‐2 and is rapidly degraded on induction of neuronal differentiation in cultured cells. J Biol Chem 287: 523–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Wang L, Sun Y, Sun Y, Meng L, Xu X (2018) First case of AML with rare chromosome translocations: a case report of twins. BMC Cancer 18: 458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Peng C, Zhang Z, Wu J, Lv Z, Tang J, Xie H, Zhou L, Zheng S (2014) A critical role for ZDHHC2 in metastasis and recurrence in human hepatocellular carcinoma. Biomed Res Int 2014: 832712–832719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Sharma C, Wang H‐X, Li Q, Knoblich K, Reisenbichler ES, Richardson AL, Hemler ME (2017) Protein acyltransferase DHHC3 regulates breast tumor growth, oxidative stress, and senescence. Cancer Res 77: 6880–6890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Kinney N, Varghese RT, Anandakrishnan R, Garner HRS (2017) ZDHHC3 as a risk and mortality marker for breast cancer in African American women. Cancer Inform 16: 1176935117746644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Choi Y‐W, Bae SM, Kim Y‐W, Lee HN, Kim YW, Park TC, Ro DY, Shin JC, Shin SJ, Seo J‐S et al (2007) Gene expression profiles in squamous cell cervical carcinoma using array‐based comparative genomic hybridization analysis. Int J Gynecol Cancer 17: 687–696 [DOI] [PubMed] [Google Scholar]
- 134. Hungermann D, Schmidt H, Natrajan R, Tidow N, Poos K, Reis‐Filho JS, Brandt B, Buerger H, Korsching E (2011) Influence of whole arm loss of chromosome 16q on gene expression patterns in oestrogen receptor‐positive, invasive breast cancer. J Pathol 224: 517–528 [DOI] [PubMed] [Google Scholar]
- 135. Mansilla F, Birkenkamp‐Demtroder K, Kruhøffer M, Sørensen FB, Andersen CL, Laiho P, Aaltonen LA, Verspaget HW, Orntoft TF (2007) Differential expression of DHHC9 in microsatellite stable and instable human colorectal cancer subgroups. Br J Cancer 96: 1896–1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Meng S, Lu C, Zhang W, Shen W, Wei Y, Su D, Zhou F (2016) MMSA‐1 expression pattern in multiple myeloma and its clinical significance. Clin Exp Med 16: 599–609 [DOI] [PubMed] [Google Scholar]
- 137. Dzikiewicz‐Krawczyk A, Kok K, Slezak‐Prochazka I, Robertus J‐L, Bruining J, Tayari MM, Rutgers B, de Jong D, Koerts J, Seitz A et al (2017) ZDHHC11 and ZDHHC11B are critical novel components of the oncogenic MYC‐miR‐150‐MYB network in Burkitt lymphoma. Leukemia 31: 1470–1473 [DOI] [PubMed] [Google Scholar]
- 138. Wu J‐F, Lee C‐H, Chen H‐L, Ni Y‐H, Hsu H‐Y, Sheu J‐C, Tsuei D‐J, Chang M‐H (2013) Copy‐number variations in hepatoblastoma associate with unique clinical features. Hepatol Int 7: 208–214 [DOI] [PubMed] [Google Scholar]
- 139. Edwards H, Rubenstein M, Dombkowski AA, Caldwell JT, Chu R, Xavier AC, Thummel R, Neely M, Matherly LH, Ge Y et al (2016) Gene signature of high white blood cell count in B‐precursor acute lymphoblastic leukemia. PLoS One 11: e0161539 [DOI] [PMC free article] [PubMed] [Google Scholar]


