Abstract
MacroH2A histone variants suppress tumor progression and act as epigenetic barriers to induced pluripotency. How they impart their influence on chromatin plasticity is not well understood. Here, we analyze how the different domains of macroH2A proteins contribute to chromatin structure and dynamics. By solving the crystal structure of the macrodomain of human macroH2A2 at 1.7 Å, we find that its putative binding pocket exhibits marked structural differences compared with the macroH2A1.1 isoform, rendering macroH2A2 unable to bind ADP‐ribose. Quantitative binding assays show that this specificity is conserved among vertebrate macroH2A isoforms. We further find that macroH2A histones reduce the transient, PARP1‐dependent chromatin relaxation that occurs in living cells upon DNA damage through two distinct mechanisms. First, macroH2A1.1 mediates an isoform‐specific effect through its ability to suppress PARP1 activity. Second, the unstructured linker region exerts an additional repressive effect that is common to all macroH2A proteins. In the absence of DNA damage, the macroH2A linker is also sufficient for rescuing heterochromatin architecture in cells deficient for macroH2A.
Keywords: DNA damage, heterochromatin, histone variants, macroH2A, PARP1
Subject Categories: Chromatin, Epigenetics, Genomics & Functional Genomics; Structural Biology
Introduction
Histone variants can replace replication‐coupled histones in the nucleosome and endow chromatin with unique properties 1. Thus, histone variants are key to epigenetic processes such as differentiation and development. Further, their deregulation is tightly linked with disease, in particular cancer 2. Histone variants differ in sequence, expression timing, and mRNA processing from replication‐coupled histones. Some of the most substantial sequence changes are found in three histone H2A variants known as macroH2As. One event of mutually exclusive exon usage gives rise to the alternative splice protein isoforms macroH2A1.1 and macroH2A1.2, while macroH2A2 is encoded by a second gene 3. All three isoforms are present in mammals and possibly all vertebrates 4. A large number of loss‐of‐function studies have implicated macroH2A proteins in cancer development, differentiation, and somatic cell reprogramming. MacroH2A proteins act as tumor suppressors in a majority of cancer types 5. The reduced expression of macroH2A is associated with poor prognosis in lung cancer 6, while in advanced stage melanoma, the loss of macroH2A expression contributes to metastatic potential 7. During early embryonic development, macroH2A proteins are virtually absent in the mouse zygote 8, but their expression increases as early development proceeds promoting the differentiation of embryonic stem cells 9, 10. Zebrafish embryos lacking macroH2A display multiple developmental defects 11 and macroH2A‐deficient mice are growth‐retarded 12. Moreover, macroH2A histones act as epigenetic barrier to induced pluripotency, no matter whether it is induced by nuclear transfer or using Yamanaka factors 13, 14, 15, 16.
Taken together, these and other studies support the notion that macroH2A proteins promote the maintenance and stability of the epigenome in differentiated cells, acting as barriers to reprogramming and malignant transformation. The molecular basis for this function is not known, but may be related to a major role of macroH2A in regulating chromatin architecture and nuclear organization 17. Knockdown of macroH2A proteins leads to defects in nuclear circularity, disruption of nucleoli, and a global loss of dense heterochromatin. Constitutive heterochromatin repeats marked by trimethylation of lysine 9 on histone H3 (H3K9me3) are disorganized, expanded, and have lost their interaction with the nucleostructural protein lamin B1 in macroH2A‐deficient cells 17, 18. The influence of macroH2A proteins on gene regulation, however, is ambivalent and might be secondary to functions in higher order chromatin structure. In addition to constitutive heterochromatin, macroH2A proteins associate and contribute to Polycomb‐mediated gene repression 11, 19. In vitro, nucleosomes containing macroH2A1 are more stable and become refractory to chromatin remodeling and acetylation 20, 21. However, macroH2A proteins also contribute to gene activation induced by stress and differentiation cues 9, 22, 23. In the context of paracrine senescence, for example, macroH2A‐dependent gene activation occurs upon histone H2B acetylation 24, 25. Further, macroH2A histones reduce transcriptional noise and promote the robustness of gene expression programs 26.
The molecular mechanisms through which the distinct structural elements present in macroH2A histones directly and indirectly regulate gene activity are not well understood. MacroH2A proteins have a tripartite structure consisting of the histone fold, an unstructured linker, and a globular macrodomain 27, 28. Nucleosome core particles containing the macroH2A1 histone fold are highly similar to nucleosomes with the replication‐coupled histone H2A, but place the linker region and macrodomain at an accessible site outside of the nucleosome core 27. Macrodomains are conserved globular folds with an affinity for NAD+‐derived metabolites, including ADP‐ribose and O‐acetyl‐ADP‐ribose 29. These metabolites are generated by NAD+ consuming enzymes that include poly‐ADP‐ribose polymerases (PARPs/ARTDs) and sirtuin deacetylases 3. Interestingly, the splice variant macroH2A1.2 does not show any affinity for O‐acetyl‐ADP‐ribose nor for ADP‐ribose 28. Since the experimentally determined K D value of the macroH2A1.1 macrodomain for ADP‐ribose (~2 μM) is lower than the estimated cellular concentration of O‐acetyl‐ADP‐ribose 30, it is possible that macroH2A1.1 may be ligand‐regulated in physiological contexts. The same may be true upon acute DNA damage, when poly‐ADP‐ribose (PAR) levels are very high following PARP1 activation 31. While some macrodomains exhibit ADP‐ribosyl‐hydrolase activity, this does not appear to be the case for the histone variant macroH2A1.1 32, 33, 34. Since macroH2A1.1 binds ADP‐ribose in a capping mode, it is further able to bind poly‐ and possibly mono‐ADP‐ribosylated proteins 35, 36. The consequence of such a binding is best understood for auto‐modified PARP1. Depending on the cellular context, this can lead to PARP1 recruitment and gene regulation 24 or the inhibition of basal PARP1 activity 23. In muscle cells, the latter globally impacts cellular NAD+ metabolism 37, revealing an important metabolic function for macroH2A1.1.
Here, we have sought to dissect how the distinct structural elements of macroH2A histone impact on chromatin plasticity and architecture. By testing the binding of macroH2A2 macrodomain to ADP‐ribose and solving its crystal structure, we reveal that this histone isoform is unable to recognize ADP‐ribose and inert toward PARP1 signaling. By studying the influence of different macroH2A histone isoforms and their structure elements on chromatin structure, we provide evidence that macroH2A proteins suppress chromatin plasticity through at least two distinct mechanisms: a macroH2A1.1‐specific mechanism that is directly linked to its ability to lower PARP1 signaling and a molecular mechanism shared by all macroH2A histone isoforms that suppresses chromatin reorganization and is mediated by the unstructured linker region connecting the H2A‐like histone fold with the globular macrodomains of macroH2A histones.
Results and Discussion
The macrodomain of macroH2A2 is incompatible with nucleotide binding
In addition to the macroH2A1‐encoding gene H2AFY, vertebrates contain the gene H2AFY2, which encodes the histone isoform macroH2A2 4. MacroH2A2 appears to have a more prominent role than macroH2A1 as a barrier to somatic cell reprogramming or in suppressing melanoma metastasis 7, 15, but its molecular structure and function have received little attention.
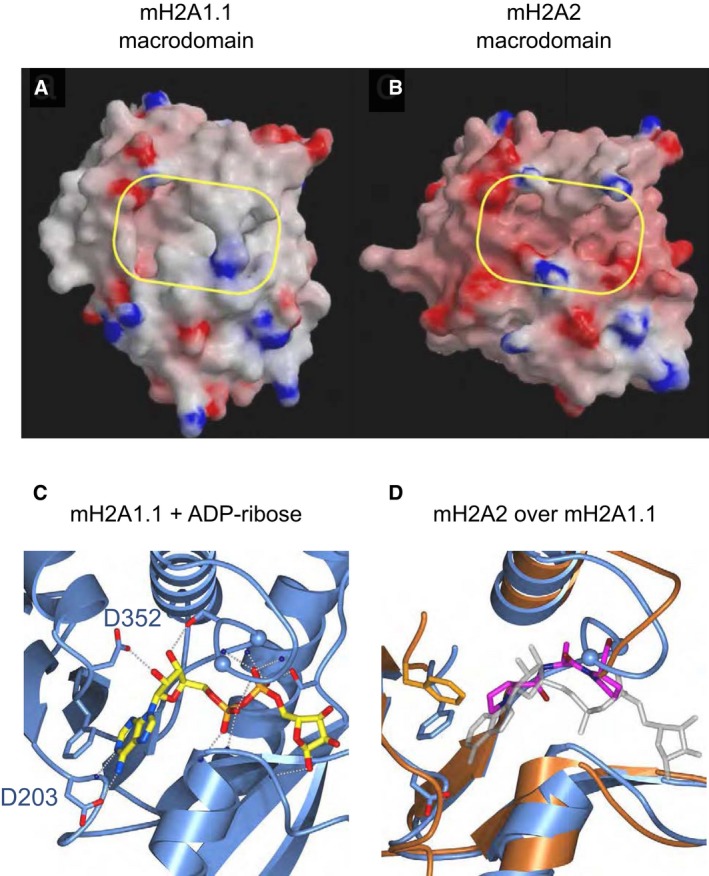
To gain first insight into the potential molecular functions of macroH2A2, we determined the crystal structure of its globular, C‐terminal macrodomain and compared it to the macrodomain of macroH2A1.1. Consistent with the ~65% amino acid sequence identity to macroH2A1.1, the 1.7 Å structure of the macroH2A2 macrodomain reveals a conserved macrodomain fold (Fig 1A and B, for statistical information please see Table EV1). However, macroH2A2 exhibits marked differences in both shape and chemical properties of its major surface pocket when compared to macroH2A1.1. Specifically, in macroH2A2 two prolines insert (Pro 313 and Pro 315) in the P‐loop‐like motif that coordinates the binding of the α‐ and β‐phosphates of the ADP‐ribose ligand in macroH2A1.1 (Fig 1C and D). This distorts the phosphate‐binding loop and is predicted to render the structure incompatible with nucleotide binding (Fig 1D). In contrast, other residues in the binding pocket of macroH2A2 are conserved. This includes Phe 354 that corresponds to Phe 351 in macroH2A.1.1 where it is engaged in an aromatic stacking interaction with the adenine base of the ADP‐ribose moiety (Fig 1C and D).
Figure 1. MacroH2A isoforms show specific structural differences in their macrodomains that translate into functional plasticity.
-
A, BSurface representations of the macrodomain structures for the human macroH2A isoforms macroH2A1.1 (PDB ID 1ZR3) and macroH2A2 (PDB ID 6FY5), respectively, with electrostatic potential included. A yellow‐framed box indicates the location of the major surface pocket accommodating ADP‐ribose in macroH2A1.1.
-
C, DClose‐up view of the nucleotide binding region of ADP‐ribose bound by human macroH2A1.1 (C) overlaid with the corresponding region of macroH2A2 in orange (D). An important structural difference in macroH2A2 that distorts de nucleotide binding loop is highlighted in magenta, and glycines in the nucleotide binding loop are depicted as spheres (G312 and G314). The ADP‐ribose ligand is included in (D) (shaded light gray) to facilitate orientation and comparison. Dashed lines indicate polar interactions.
In addition to structural differences between macroH2A2 and macroH2A1.1 macrodomain modules, the macroH2A histone isoforms also exhibit distinct expression patterns at the protein level. MacroH2A2 is highly expressed in adult brain tissue (Fig EV1A). In the maturing mouse embryo, a strong macroH2A2 mRNA signal can be detected throughout the central nervous system (Fig EV1B). Similarly, macroH2A2 was the macroH2A mRNA transcript predominantly expressed in zebrafish embryos, with high expression in the developing brain and spine 11. Knockdown of macroH2A2 leads to malformation in the mid‐hindbrain boundary 11. Although macroH2A2 knockout mice are viable 12, it is tempting to speculate that the surface pocket of macroH2A2's macrodomain could bind as yet unknown ligands that are involved in either brain development or neuronal plasticity and learning in the adult.
Figure EV1. MacroH2A histones show tissue‐specific expression profiles.
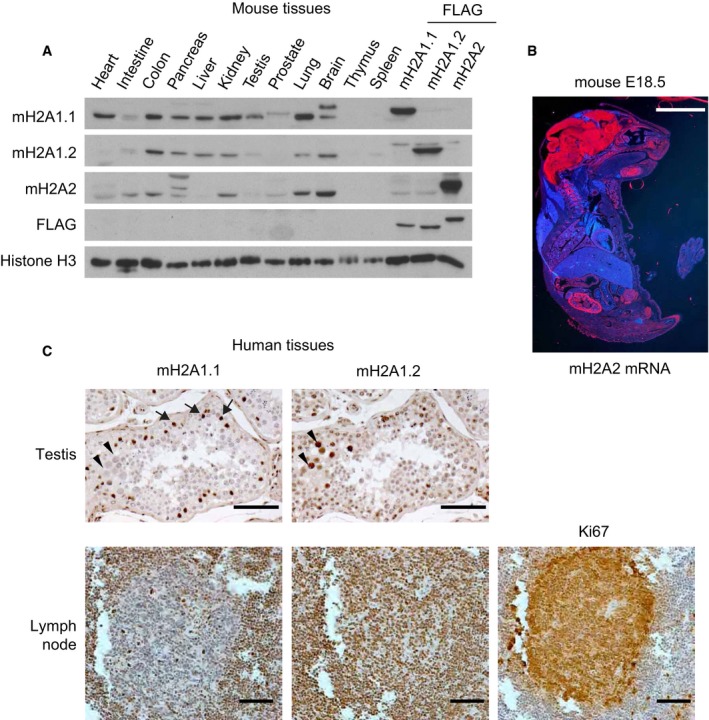
- Western blot showing the expression of macroH2A1.1, macroH2A1.2, and macroH2A2 in different male mouse tissues. Samples of HepG2 cells expressing ectopic FLAG‐tagged forms of the three macroH2A isoforms are included as antibody specificity controls and to aid comparisons. H3 is used as a loading control. A representative blot of three independent analyses is shown.
- In situ mRNA hybridizations of murine macroH2A2 on 20 μm cryosections of E18 mice reveal a high concentration of macroH2A2 mRNA in the entire central nervous system, kidney, and testis. Scale bar is 5 mm.
- Immunohistochemical staining for histone macroH2A1.1 and macroH2A1.2 in paraffin‐embedded human testis sections and lymph nodes. Images reveal a distinct staining consistent with Sertoli cells for macroH2A1.1 (arrows), while macroH2A1.2 is also expressed in proliferating germ cells (arrowheads). In the lymph node, proliferating cells positive for Ki67 express low levels of macroH2A1.1 while they are positively stained for macroH2A1.2. Scale bars are 50 μm.
Source data are available online for this figure.
Analysis of human tissues further strengthens the previous observation that macroH2A1.1 expression inversely correlates with cellular proliferation 6. MacroH2A1.1 is expressed in primary fibroblasts, but absent in most fast proliferating cancer cells 22, 24. Here, we find that macroH2A1.1 levels are particularly high in post‐mitotic Sertoli cells in the testis, for example, while the splice variant macroH2A1.2 is also expressed in highly proliferative lymphocytes forming the inner lymph node (Fig EV1C). Further, during myogenic differentiation, which is associated with cell cycle exit, the splicing of the primary macroH2A1 transcript switches from macroH2A1.2 to the macroH2A1.1 isoform 37.
Taken together, these results suggest that during evolution, macroH2A2 has acquired a binding pocket that appears to be incompatible with nucleotide binding and may have evolved as yet unknown role(‐s) in the mammalian central nervous system.
MacroH2A1, but not the “orphan” macroH2A2, binds ADP‐ribose and inhibits PARP1
Next, we have used isothermal titration calorimetry to measure whether the macroH2A2 macrodomain is capable of binding to monomeric ADP‐ribose. Consistent with our structural analysis, we found that, in contrast to macroH2A1.1, the macrodomain of human macroH2A2 is unable to bind ADP‐ribose (Fig 2A). These highly distinct properties of the macrodomains of the human macroH2A1.1 and macroH2A2 histone isoforms are conserved in the zebrafish (Danio rerio) and medaka (Oryzias latipes) proteins, where macroH2A1.1 binds ADP‐ribose with an equilibrium dissociation constant (K D) of 1.5 μM, identical to the human form (Fig EV2A). This functional conservation is reflected in the high protein sequence conservation of the macrodomains of macroH2A1.1 between human, zebrafish, and medaka (Fig EV2B). In particular, those amino acids directly interacting with the ADP‐ribose molecule are highly conserved (Fig EV2C). The NAD metabolite binding function of macroH2A1.1 thus appears to be a conserved function of this histone isoform across vertebrate evolution. Further ligand analysis reveals that the human macroH2A1.1 macrodomain binds ADP‐ribose with highest affinity compared to other related metabolites and nucleotides, as illustrated by the markedly lower affinity for ADP and a large panel of other tested nucleotides that do not bind this macrodomain module (Table EV2).
Figure 2. Binding of the NAD metabolite ADP‐ribose is specific to macroH2A1.1 and conserved.
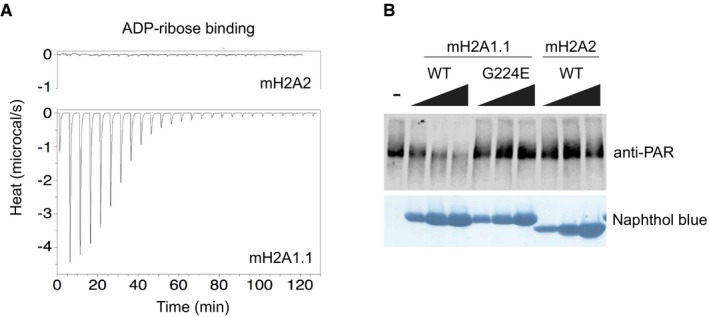
- Isothermal calorimetry (ITC) assays using ADP‐ribose ligand and purified macrodomains of human macroH2A1.1 and macroH2A2.
- In vitro PARP1 activity assay in the presence of increasing concentrations (10, 25, and 50 μM) of purified macrodomains of human macroH2A1.1, macroH2A1.1 G224E mutant, and macroH2A2. PARP1 activity is assessed by its auto‐PARylation detected by an anti‐PAR immunoblot. The naphthol blue staining shows the increasing amounts of purified macrodomain added to each reaction. A representative blot of one of three independent experiments is shown.
Source data are available online for this figure.
Figure EV2. The capacity of macroH2A1.1 to bind ADP‐ribose is conserved in fish.
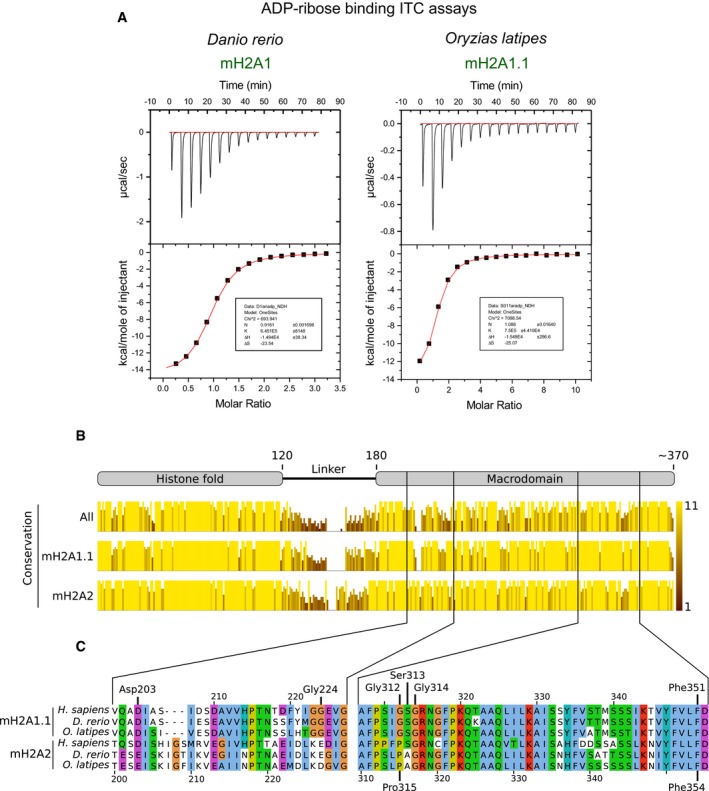
- Isothermal calorimetry demonstrates the capacity of fish macroH2A1.1 macrodomains to bind ADP‐ribose with similar affinity as the human form. Representative blots of more than three independent experiments are shown.
- Conservation scores resulting of aligning macroH2A1.1 and macroH2A2 protein sequences of human, zebrafish, and medaka all together or mH2A1.1 and mH2A2 separately. The conservation score reflects the conservation of physico‐chemical properties in each position of the alignment on a scale that ranges from 0 to 11 and is represented both in the height and color brightness of the bar. The top schematic displays the different domains of macroH2A. The alignment was generated with the online alignment tool PRALINE 59 using the protein sequences with the following accession codes: H. sapiens macroH2A1.1 (NP_613075.1), D. rerio macroH2A1 (NP_001035451.1), O. latipes macroH2A1.1 (XP_011481424.1), human macroH2A2 (NP_061119.1), D. rerio macroH2A2 (NP_001020673.1), and O. latipes macroH2A2 (XP_004076965.2). The alignment and conservation track were edited with Jalview 60.
- Protein sequence alignment of the amino acids that form the binding pocket of the macrodomain of macroH2A1.1 and macroH2A2 from human (H. sapiens), zebrafish (D. rerio), and medaka (O. latipes). Critical amino acids for ADPr binding conserved in macroH2A1.1 are indicated, as well as the amino acids in macroH2A2 cited in the main text. The top amino acid coordinates refer to macroH2A1.1 sequences while the bottom ones refer to macroH2A2 sequences. The alignment was generated as in (B) and colored using the ClustalX color scheme available in Jalview. In summary, amino acids are colored according to their physico‐chemical properties only if that position in the alignment fulfills a conservation criteria for that amino acid type: blue—hydrophobic, red—positive charge, magenta—negative charge, green—polar, pink—cysteine, orange—glycine, yellow—proline, cyan—aromatic, white—no criteria met.
MacroH2A2 proteins from the three species show conserved divergences from macroH2A1.1 such as the presence of a proline that distorts the phosphate‐binding loop (Pro315), a three amino acid insertion in the adenine‐binding site, and a charged aspartate or glutamate at the position corresponding to Gly 224 in macroH2A1.1 (Fig EV2C). Exchange of Gly 224 for glutamate is known to abolish metabolite binding when introduced in macroH2A1.1 28. In contrast to the splice isoform macroH2A1.2, the macroH2A2 macrodomain retains a key aromatic group that in macroH2A1.1 is involved in aromatic stacking with the nucleotide base of the ADP‐ribose ligand 28. These structural features are conserved across vertebrate evolution at the level of primary sequence, suggesting that macroH2A2 has, in principle, retained structural features that would be beneficial for the binding of a nucleobase or related metabolites. This suggests a conserved, but as yet unknown, potential binding function of the macroH2A2 macrodomain. We shall thus refer to macroH2A2 as an “orphan histone variant”.
Our previous structural analysis of the macroH2A1.1 macrodomain in complex with monomeric ADP‐ribose revealed that this macrodomain module engages ADP‐ribose in a manner that would allow macroH2A1.1 to cap oligo‐ADP‐ribose chains as PARP1 synthesizes them on PARP1 targets 35. We hypothesized that such a molecular “capping” function might be essential for the known capacity of macroH2A1.1 to both bind and inhibit auto‐modified PARP1 23, 37. Some controversy exists on whether other macrodomain modules, and in particular the macrodomain of the distinct macroH2A2 histone isoform, can inhibit PARP1 activity 38. To shed light on this issue, we have directly compared the macrodomain modules of macroH2A1.1 and macroH2A2 in an in vitro assay system by monitoring PARP1 enzymatic activity. We find that increasing the amount of the macrodomain of macroH2A1.1 progressively decreases PARP1 activity (Fig 2B). In contrast, PARP1 activity is completely inert to the addition of similar amounts of the macroH2A2 macrodomain or a macroH2A1.1 mutant unable to bind ADP‐ribose. This indicates that the direct, repressive effect of macroH2A histones on PARP1 activity is limited to the histone variant macroH2A1.1 and fully depends on its unique capability of directly recognizing mono‐, oligo‐, and polymeric ADP‐ribose.
MacroH2A histones suppress chromatin dynamics through two distinct mechanisms
The ability of macroH2A.1.1 to lower PARP1 activation in vitro raises the question as to whether macroH2A histones suppress chromatin dynamics. As a model to study this effect, we have utilized the rapid relaxation of chromatin structure that is observed at and near DNA damage sites in vivo. Previous work has shown that this dramatic structural reorganization of chromatin depends on PARP1 activity 39, 40, 41. To study whether and how macroH2A histones impact this process, we have taken advantage of a photo‐activable GFP (PAGFP) fused to canonical H2B 42 and expressed the reporter tool in human U2OS osteosarcoma cells that have been sensitized to DNA damage by pre‐incubation with DNA intercalating Hoechst dye. After simultaneous induction of DNA damage and photo‐activation of the PAGFP‐H2B using a 405 nm laser in the focus of interest, DNA damage‐induced chromatin reorganization can be readily followed and statistically quantitated using live cell imaging (Fig 3A). Importantly, PAGFP‐H2B was shown to remain chromatin bound during this process 42. The rapid expansion of chromatin structure that is seen at and near the DNA damage site is fully suppressed by pre‐treating cells with the PARP inhibitor Olaparib (Fig 3B). Like many other cancer cells 24, human U2OS cells do not express the macroH2A1.1 isoform, while the macroH2A1.2, in particular, and to lesser extent macroH2A2 are endogenously expressed (Fig EV3A). To compare the effect of overexpressing distinct macroH2A isoforms and of canonical H2A on chromatin reorganization upon DNA damage, we transiently expressed mCherry‐tagged histone constructs (Fig EV3B) and induced DNA damage in cells with comparable transgene expression levels that we have selected on the basis of their mCherry fluorescence signal. To test for chromatin incorporation, we have used fluorescence recovery after local photobleaching (FRAP) assays, which is suitable for testing fluorescence‐tagged histones 43. Both H2A and macroH2A histones are readily incorporated into chromatin, as judged by their low mobility (Fig EV3C). When inducing DNA damage in our cell lines, we find that macroH2A1.1 reduced chromatin expansion, while overexpression of canonical H2A did not (Fig 3C). Mutation of a key residue (G224E) in the ADP‐ribose binding pocket of macroH2A1.1 eliminates the binding of ADP‐ribose 28 and abrogates PARP1 binding and inhibition 23, 35, 37.
Figure 3. MacroH2A proteins repress chromatin expansion at DNA damage sites; macroH2A1.1 being the most potent in the reduction.
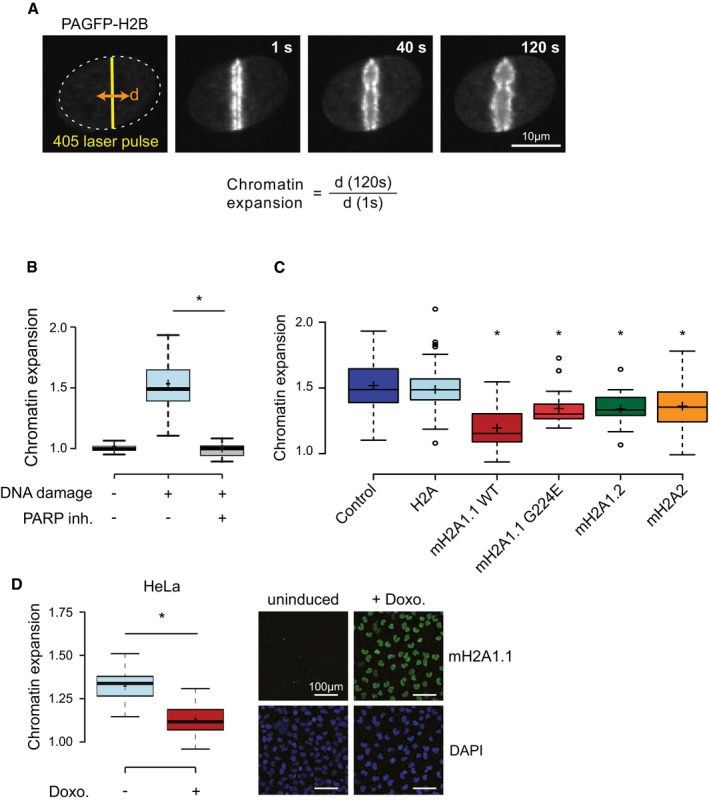
- The diameter (d) of the fluorescence signaling of H2B tagged with photo‐activatable GFP (PAGFP‐H2B) is used to quantify chromatin expansion across 120 s after induction of DNA damage using intercalating DNA dye and local irradiation with a laser.
- Chromatin expansion in absence and presence of intercalating DNA dye (− and + DNA damage) and pre‐treatment of 1 h with 1 μM of Olaparib PARP inhibitor. Boxplots represent single cell measurement of chromatin expansion at 120 s. post‐DNA damage from three biological replicates with n > 30 cells each. The box limits correspond to 25th and 75th percentiles, the bold line indicates median, and cross indicates the average values. The whiskers show the highest and lowest data point within the 1.5× interquantile range of the upper and lower quartile, respectively (*P < 0.05 using unpaired, two‐tailed Student's t‐test).
- Quantified chromatin expansion in cells not transfected (control) and cells transfected with each mCherry‐tagged histone (as indicated). Data are represented as in (B) (n > 30 cells, 3 bio. repl.; *P < 0.05 compared to mCherry‐H2A‐expressing cells using unpaired, two‐tailed Student's t‐test).
- Chromatin expansion in Hela cells containing a doxocycline (doxo.)‐inducible macroH2A1.1 transgene. Chromatin expansion assay was performed 48 h after induction, and expression was controlled by anti‐macroH2A1.1‐immunoblotting. Data are represented as in (B) (n > 30 cells, 3 bio. repl.; *P < 0.05 using unpaired, two‐tailed Student's t‐test).
Source data are available online for this figure.
Figure EV3. Exogenous macroH2A has low mobility in U2OS cells.
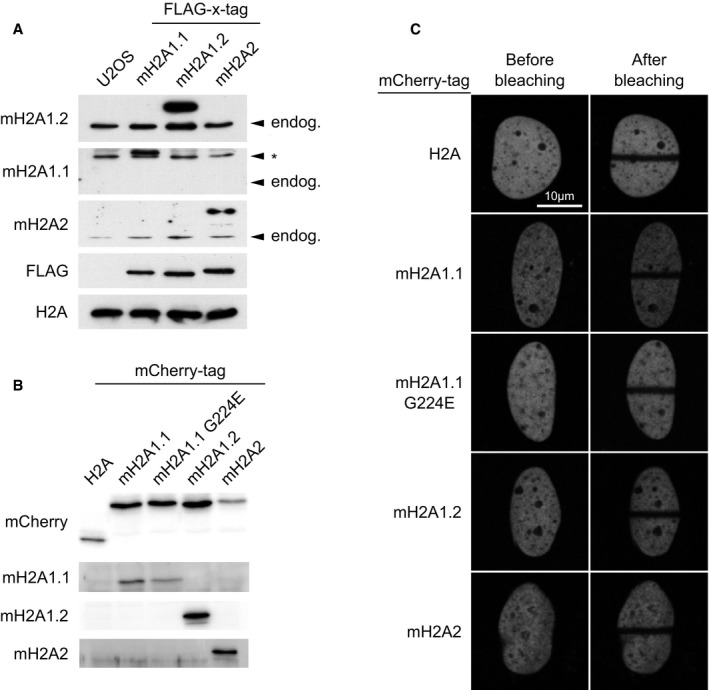
- The comparison of the endogenous levels of all three macroH2A proteins in U2OS cell lysates with cells expressing tagged proteins for reference shows that U2OS cells predominantly express macroH2A1.2, little macroH2A2 while macroH2A1.1 was not detectable. Arrowheads indicate the size of endogenous proteins, asterisk indicates an unspecific band.
- The mCherry‐tagged constructs of H2A and macroH2A proteins used in Fig 4C were transiently expressed in U2OS cells, and migration sizes were examined by immunoblotting.
- Using experimental conditions as in Fig 3A, the fluorescence of mCherry‐tagged H2A and macroH2A proteins is not recovered in the bleached area after 120 s, indicative of stable chromatin incorporation of the exogenously expressed constructs.
Source data are available online for this figure.
In vivo, the G224E mutation reduced but did not abolish the inhibitory effect of macroH2A1.1 on chromatin expansion (Fig 3C). Likewise, macroH2A2 and the splice isoform macroH2A1.2 significantly reduced chromatin expansion to a similar extent as the macroH2A1.1 G224E mutant, albeit clearly less than wild‐type macroH2A1.1. When compared to canonical H2A, macroH2A histones thus all lower chromatin reorganization upon DNA damage, with the strongest suppression seen for the PARP1 inhibiting macroH2A1.1 isoform. To exclude the possibility that the fused fluorescent protein interferes with chromatin dynamics, we have confirmed that the inducible expression of a macroH2A1.1 protein without the fluorescent protein tag also reduced chromatin expansion (Fig 3D).
In summary, we find that macroH2A histones suppress DNA damage‐induced chromatin dynamics, and that this occurs best for macroH2A1.1, which reflects its capacity to inhibit PARP1 activity. Compared to the use of a potent small‐molecule PARP inhibitor, the suppression of chromatin expansion by macroH2A1.1 is partial. Nonetheless, our data indicate that a histone variant is capable of restraining nuclear NAD+ signaling by virtue of its ability of interacting with ADP‐ribosylated PARP1. In HeLa cells, heat shock overcomes the inhibition of PARP1 by macroH2A1.1, and both the prior recruitment of PARP1, as well as its activity is required for the expression of stress response genes 23. In fibroblasts, PARP1 is active and binds and cooperates with macroH2A1.1 in the regulation of genes, causing both increases and decreases in gene expression 24. Taken together, these observations indicate that the relative expression levels of the histone variant macroH2A1.1 may set a threshold for PARP1 activation. Furthermore, the intensity of activating stress signals may determine whether the interaction between macroH2A1.1 and PARP1 results in a global inhibition of PARP1 or the recruitment of the active PARP1 enzyme to regulated genes.
However, the residual inhibitory capacity of the macroH2A1.1 G224E mutant and of macroH2A2 in chromatin expansion indicates that a distinct molecular mechanism also helps to suppress chromatin dynamics.
The linker region of macroH2A histones limits chromatin plasticity in living cells
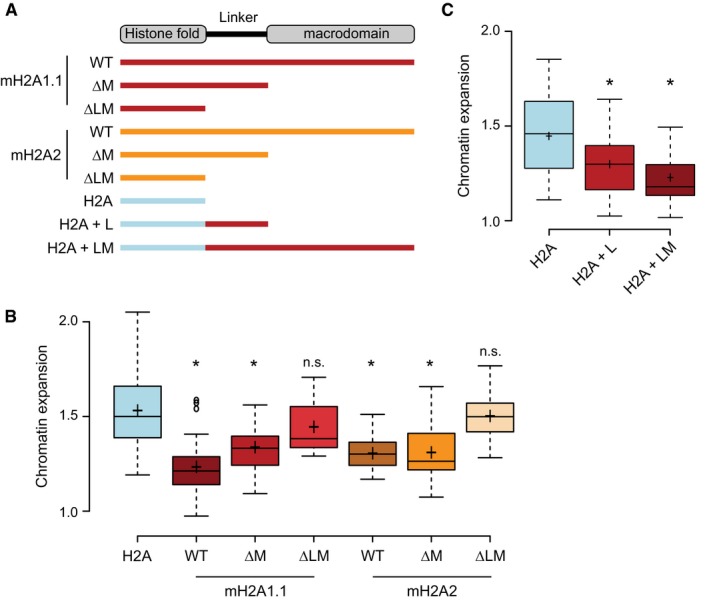
To dissect this second molecular mechanism, we sought to decipher which domains of macroH2A2 suppress chromatin relaxation. As macroH2A proteins have a tripartite structure composed of histone fold, linker, and macrodomain, we generated truncation mutants that eliminate either the macrodomain or the macrodomain and linker of both macroH2A2 and macroH2A1.1 (Fig 4A). We found that both macroH2A1.1 and macroH2A2 were able to reduce chromatin relaxation even in the absence of their respective macrodomains (Fig 4B). However, elimination of the unstructured linker region connecting the H2A‐fold to the macrodomain abolished their ability to reduce chromatin expansion (Fig 4B). Importantly, the linker region of macroH2A1.1 was sufficient to inhibit chromatin relaxation when fused to the canonical, replication‐coupled H2A (Fig 4C). Moreover, additional inhibitory capacity is transferred when also adding the macrodomain of macroH2A1.1 to H2A (Fig 4C). These experiments indicate that the 39‐amino acid long basic linker of macroH2A histones is sufficient to restrain chromatin expansion after acute DNA damage in living human cells.
Figure 4. MacroH2A histones repress chromatin expansion via the basic linker and the macrodomain capable of ADP‐ribose binding.
-
AScheme of macroH2A and H2A variants constructs used in the study: full‐length wild type (WT), constructs with deleted macrodomain (ΔM), constructs with deleted linker and macrodomain (ΔLM), or hybrids consisting of the H2A histone fold fused to the linker of macroH2A1.1 (H2A + L) or the H2A fold fused to the linker and macrodomain of macroH2A1.1 (H2A + LM).
-
B, CQuantified chromatin expansion in cells transfected with indicated histone constructs. Boxplots represent single cell measurement of chromatin expansion at 120 s. post‐DNA damage (n > 30 cells, 3 bio. repl.). The box limits correspond to 25th and 75th percentiles, the bold line indicates median, and cross indicates the average values. The whiskers show the highest and lowest data point within the 1.5× interquantile range of the upper and lower quartile, respectively (*P < 0.05 using unpaired two‐tailed Student's t‐test comparing to H2A; n.s., not significant).
Source data are available online for this figure.
The macroH2A linker region is essential and sufficient for stabilizing heterochromatin architecture
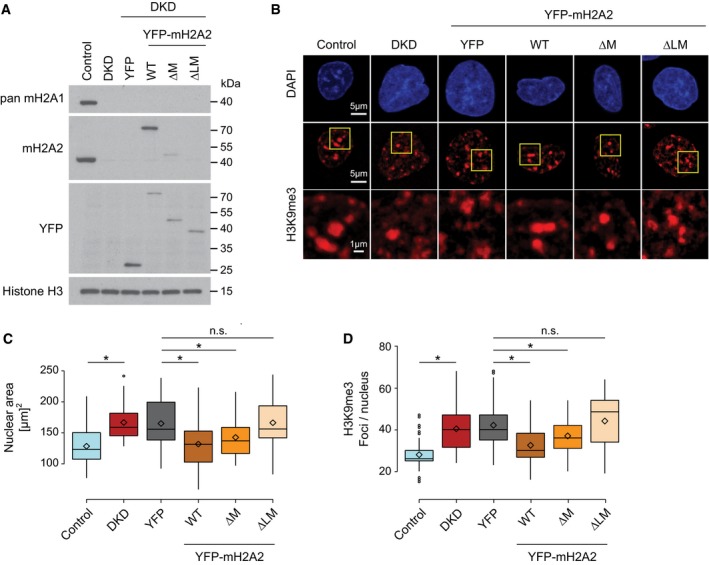
We have previously shown that loss of all macroH2A histones leads to major changes in nuclear organization and chromatin architecture 17. This includes a global increase in nuclear size and a partial dispersion of heterochromatin. Next, we wished to test whether the linker domain of macroH2A proteins contributes to these functions in the absence of acute DNA damage. To this end, we introduced YFP‐tagged versions of wild‐type macroH2A2 and its deletion mutants in hepatoblastoma cells that were depleted of endogenous macroH2A histone isoforms (Fig 5A). We monitored nuclear size using DAPI staining and assessed heterochromatin architecture by counting the number of individual spots marked by H3K9 trimethylation (Fig 5B). We first confirmed that the size of the nucleus, as well as the number of H3K9me3 spots, increased in macroH2A‐deficient cells (Fig 5B–D). Importantly, expression of YFP‐tagged macroH2A2 rescued both nuclear size (Fig 5B and C) and heterochromatin structure (Fig 5B and D). Next, we tested whether a macroH2A2 fragment lacking the macrodomain, but retaining the linker, was able to rescue both phenotypes. We find that the linker region of macroH2A2, but not its macrodomain module, is necessary and sufficient to rescue nuclear size and heterochromatin structure. In conclusion, our results provide evidence that the linker region of macroH2A proteins mediates important in vivo functions, both by regulating nuclear organization and by stabilizing chromatin architecture. This linker domain is intrinsically disordered with very few order‐inducing amino acids in its sequence and a high percentage of basic, positively charged residues 44. Based on these features, the linker bears some resemblance to the C‐terminal tail of the linker histone H1 45. Interestingly, in chicken chromatin, macroH2A1 and histone H1 bind chromatin in a mutually exclusive fashion 46. Pioneering biochemical experiments with the basic linker region of macroH2A1 have shown that macroH2A indeed possesses histone H1‐like properties. In fact, the linker stabilizes DNA at the entry/exit site of the nucleosome 47 and enhances the condensation of nucleosome arrays and fiber–fiber interactions in vitro exclusively in the absence of the macrodomain 44. We hypothesize that the capacity of the linker to generally compact chromatin fibers in vitro could explain its ability to reduce chromatin relaxation upon DNA damage in vivo. The linker residues might also explain how macroH2A1.2 contributes to the compaction of chromatin, as has been observed at a later time point of the double‐strand repair process 48. In the case of H1, the intrinsic disorder of the region and its amino acid composition are essential for its function, rather than the precise sequence 49. It will be exciting to test whether this relatively “loose” sequence requirements holds true also for the histone variants macroH2A1 and macroH2A2, which indeed differ in the exact sequence of their linker regions. In support of the special role of the linker region on chromatin compaction, the plant Arabidopsis thaliana lacks macroH2A, but contains a H2AW histone variant with structural and functional similarities to macroH2A histones 51. H2AW contains an extended C‐terminal tail that resembles part of the linker region of macroH2A as well as the C‐terminus of H1, and cell biological data show that this region mediates heterochromatin condensation in vivo 51.
Figure 5. The linker domain of macroH2A2 is able to maintain heterochromatic structures.
-
AWestern blot showing the expression of macroH2A proteins in control HepG2 cells, knockdown for macroH2A1 and macroH2A2 (DKD) and DKD with ectopic expression of the indicated YFP‐tagged constructs. H3 is used as a loading control.
-
BH3K9me3 immunostaining in the indicated HepG2 cell lines with nuclear counterstaining in DAPI. The indicated areas are zoomed‐in for a better observation of the H3K9me3 signal profile. All images are maximum intensity Z‐projections of confocal stacks.
-
C, DQuantification of the nuclear area and number of H3K9me3 foci per nucleus, respectively (n > 50 cells, 2 biol. repl.). The box limits correspond to 25th and 75th percentiles, the bold line indicates median, and rhombi indicate the average values. The whiskers show the highest and lowest data point within the 1.5× interquantile range of the upper and lower quartile, respectively (*P < 0.05 using unpaired two‐tailed Student's t‐test; n.s., not significant).
Source data are available online for this figure.
Our recent finding macroH2A has a major role in regulating nuclear organization, and higher order heterochromatin architecture has changed how we classically view the function of histone variants 17. Although the linker of macroH2A2 was sufficient to rescue the organization of heterochromatic structures (Fig 5B and D), it is likely that the macrodomain also contributes to the heterochromatin‐promoting function of macroH2A, possibly by mediating additional protein–protein interactions, consistent with macroH2A's established role in promoting the maintenance of the inactive X chromosome 50. An interesting candidate is lamin B1, which interacts with macroH2A at the nuclear periphery 17, 18. Whether this interaction is direct and mediated by the macrodomain remains to be tested.
In conclusion, we have identified two mechanisms through which macroH2A proteins can suppress chromatin plasticity and stabilize nuclear architecture. The inhibition of PARP1 enzymatic activity and the effect of macroH2A on the PARP1‐dependent chromatin functions is specific to the splice isoform macroH2A1.1, while the linker region present in all three macroH2A proteins plays an additional role in promoting or stabilizing a compact chromatin state. These mechanisms provide a molecular rationale for how histone variant macroH2A isoforms may stabilize the epigenome of differentiated cells and a potential unifying explanation for the reported functions of macroH2A. In light of these findings, future work will need to revisit the macroH2A loss‐of‐function phenotypes observed in development, cancer, and somatic cell reprogramming in order to test the individual contributions of the macroH2A linker and the macroH2A1.1‐mediated inhibition of PARP1 activity.
Materials and Methods
Protein expression, protein purification, and binding assays
The macrodomains of human histone variants macroH2A2 (residues 177–372) and macroH2A1.1 (residues 162–352, macroH2A1.1Δ10) were cloned into a modified pET24 (Novagen), providing a tobacco etch virus (TEV) cleavable N‐terminal 6xHis tag. Proteins were expressed in E. coli BL21 (DE3) cells at 37°C for 3 h after induction with 1 mM isopropyl‐β‐D‐thiogalactopyranoside. Cells were harvested by centrifugation, resuspended in lysis buffer (50 mM NaH2PO4, 500 mM NaCl, 1 mM MgCl2, 5 mM β‐mercaptoethanol, 10% [w/v] glycerol), lysed with Emulsiflex‐C5 (Avestin, Canada), and purified by Ni2+ sepharose (GE Lifesciences) affinity chromatography using the batch method. Beads were then washed in lysis buffer containing 1 M NaCl and 40 mM imidazole, followed by step‐elution with lysis buffer supplemented with 500 mM imidazole. Elution fractions were dialyzed against buffer containing 50 mM Tris pH 8.0, 300 mM NaCl, and in the presence of 6xHis‐tagged TEV protease (1:100 molar ratio). The 6xHis tag, TEV protease, and remaining impurities were removed by re‐adsorption of the dialyzed protein to Ni2+ sepharose resin, followed by size exclusion chromatography on a Superdex 75 HR26/60 column (GE Lifesciences), equilibrated in 50 mM Tris pH 8.0, 300 mM NaCl. Monomeric peak fractions were dialyzed against 20 mM Bis‐Tris pH 7.0, 100 mM NaCl, concentrated to 10 mg/ml (macroH2A2) and 20 mg/ml (macroH2A1.1Δ10), respectively, and immediately used for crystallization.
For the zebrafish macrodomain construct, we carried out sequence alignments between fish and human macroH2A histones and chose domain boundaries identical to those of human macroH2A constructs. PCR fragments were cloned into a GST‐fusion plasmid and the proteins expressed in E. coli at 18°C, and purified using glutathione‐coupled sepharose beads and lysis/wash buffer containing 0.5 M NaCl (50 mM Tris, pH 7.9, 0.1 mM MgCl2, 1 mM EDTA, and 1 mM DTT). Purified macrodomains were cleaved from GST using TEV protease and dialyzed into ITC buffer 28. Binding of nucleotides to all macrodomain constructs was assessed using isothermal titration calorimetry (ITC), as previously reported.
Crystallization and data collection
MacroH2A2 crystallized in sitting drop setups using 12.5% polyethylene glycol 1,000, 0.2 M Na(CH3COO), 0.1 M MES (pH 6.5) as crystallization buffer. Ethylene glycol was added to a final concentration of 15% (v/v), and crystals of up to 200 × 40 × 40 μm were flash‐frozen in liquid nitrogen. A dataset of 1.65 Å resolution has been recorded at beam line PX01, Swiss Light Source, Villigen, Switzerland.
Crystals of an engineered form of macroH2A1.1 omitting the last ten C‐terminal residues developed in 20% (w/v) polyethylene glycol 3,350, 0.2 M KCl in sitting drops using micro‐seeding protocols. Plate‐like crystals of up to 500 × 300 × 30 μm were transferred in reservoir solution supplemented with 20% (v/v) glycerol and directly frozen in the cryo‐stream at ESRF beam line ID29. Data processing and scaling were carried out with XDS 52.
Structure solution, refinement, and accessibility
The molecular replacement method as implemented in PHASER 53 was used to solve the structure of macroH2A in all crystal forms using the previously determined human macrodomain as search model (28, PDB ID 1ZR3). The structures were completed in alternating cycles of manual model correction in COOT 22 and restrained TLS‐refinement in autoBUSTER (Global Phasing Limited). The relatively high free R‐factor observed with monoclinic macroH2A2 macrodomain crystals is likely due to one of two macrodomains in the asymmetric unit being partly disordered. Inspection of the refined model with MolProbity revealed excellent stereochemistry. Structural visualization was done with POVSCRIPT and POVRAY (http://www.povray.org).
Antibodies
The following antibodies have been used in this study: anti‐macroH2A1.1 6; anti‐macroH2A1.2 (6 used for immunohistochemistry or Cell Signaling 4827S used for Western blotting in Fig 5); anti‐pan‐mH2A1 and anti‐mH2A2 17; anti‐GFP (SantaCruz, sc‐9996); anti‐histone H3 (AbCam, ab‐1791); anti‐H3K9me3 (Abcam, ab‐8898); anti‐H2A (ab15653), anti‐K67 (DakoCytomation, monoclonal mouse clone MIB‐1; anti‐FLAG (clone M2, Sigma‐Aldrich); and secondary anti‐mouse and anti‐rabbit conjugated to HRP (DakoCytomation or Bio‐Rad).
Plasmids
We generated mammalian expression constructs of human full‐length macroH2A.1.1, macroH2A1.2, and macroH2A2 by cloning the PCR amplified cDNA representative of each macroH2A histone, into pmCherry‐C3. For WB analysis, full‐length macroH2A isoforms, fused to tandem FLAG and HA tag on N terminus, were cloned in the pTREtight vector as described 35. Single residue mutants or whole domain deletions (via stop‐codon introduction) were generated using QuikChange site‐directed mutagenesis (Stratagene). MacroH2A2 constructs were cloned into pBabe plasmids with an N‐terminal YFP fusion tag. The histone H2B‐PAGFP was described before 54. The histone H2A cDNA was provided by Sandra Hake and subcloned into pmCherry‐C1. All constructs were sequence‐verified.
Immunodetection
For Western blotting, proteins were separated via SDS–PAGE system, transferred to nitrocellulose membrane (Protran Whatman). After 1 h blocking in 5% milk in TBST (10 mM Tris–HCl, pH 7.6, 150 mM NaCl, 0.2% Tween‐20) at RT, proteins were incubated with primary antibodies diluted 1:1,000 in 1% milk in TBST overnight at 4°C. The secondary antibodies were used in a dilution 1:10,000 ratio for 30 min at RT. The membrane was developed with Immobilon Western Chemiluminescent HRP Substrate (Millipore, WBKLS0050) and Fuji medical X‐ray films (Super RX). Fresh tissues from healthy 6 weeks old male NOD mice were kindly provided by Marta Vives, Arce Garcia‐Jaraquemada, and Josep Manyé. Organs were cleaned with PBS, frozen in liquid nitrogen, and powdered using a liquid nitrogen‐cooled mortar. In order to obtain total protein extracts, 0.1 g of powdered tissue was resuspended in 500 μl Laemmli's buffer, sonicated with an ultrasonic homogenizer (Omni Ruptor 4000), and then boiled at 95°C for 10 min for Western blot analysis.
Immunohistochemical stainings were performed essentially as described before 55 in successive slides of tissue microarrays (Superbiochips, Seoul, Korea; BioChain, Hayward, CA). For immunofluorescence, cells were seeded on poly‐L‐lysine coated glass slides (Thermo Scientific) and processed as described previously 17.
In situ mRNA hybridization
Tissues were fixed in 10% formalin. Hybridization with 35S‐labeled antisense RNA probes was carried out on 20 μm cryosections, as described previously 56. Hybridization signals were detected by autoradiography using Kodak NTB‐2 liquid emulsion. Exposure time varied from 2 to 4 weeks depending on signal strength, as determined by previous exposure to Kodak BioMax MR films. Tissue sections were counterstained with 0.001% bisbenzimide in PBS. Pictures of signal and counter stain were overlaid using Adobe Photoshop software.
Cell culture and gene transduction
U2OS (ATCC, HTB‐96) and HepG2 cells (ATCC, HB‐8065) were cultured in Dulbecco's modified Eagle medium containing 4.5 g/l glucose and supplemented with 10% fetal bovine serum, 2 mM glutamine, 50 U/ml penicillin, and 50 mg/ml streptomycin at 37°C in 5% CO2. U2OS cells were transiently transfected with XtremeGENE HP DNA transfection reagent (Roche). The generation and culture of HeLa Kyoto cells with an inducible macroH2A.1.1 transgene have been described 35. For induction, 1 μg/ml doxycycline (Clontech) was added 48 h before analysis. HepG2 cells stably depleted for all macroH2A proteins (DKD) have been described previously 17 and periodically reselected with 150 μg/ml hygromycin and bleomycin 30 μg/ml. DKD cell lines re‐expressing rescue constructs were generated by transduction with retroviral vectors that was performed as described previously 57 and selected with 2 μg/ml puromycin. HepG2 cells were periodically tested for mycoplasma contamination.
Imaging of DNA damage induced by laser micro‐irradiation
U2OS cells were plated on Lab‐Tek chambered coverglass (Nunc). For the induction and imaging of DNA damage, cells were pre‐sensitized by addition of 0.3 μg/ml Hoechst 33285 into the culture media for 1 h. When indicated cells were treated with 1 μM Olaparib (Selleckchem) for 1 h. Immediately before imaging, the growth medium was replaced by Leibovit's L‐15 medium, supplemented with 10% fetal bovine serum, 2 mM glutamine, 50 U/ml penicillin, and 50 mg/ml streptomycin. Laser micro‐irradiation experiments were performed on a Zeiss AxioObserver Z1 confocal spinning‐disk microscope equipped with an AxioCam HRm CCD camera (Zeiss) or a sCMOS ORCA Flash 4.0 camera (Hamamatsu) through a Zeiss Plan/Apo 63 Å~/1.4 water‐immersion objective lens. Laser micro‐irradiation and PAGFP activation were performed simultaneously using 405 nm laser set to 100 μW for 350 ms; subsequently, the images were taken every 4 s, using 488 nm laser, for 3 min.
FRAP assay
FRAP experiments were performed with an inverted Leica SP8X WLL microscope equipped with a WLL2 laser (470–670 nm) and acusto‐optical beam splitter. Live cells were recorded at 37°C in supplemented Leibovit's L15 medium. Images were acquired with a 40 × 1.3 objective at 600 Hz. Cells were bleached for 10 frames using four different wavelengths (585, 577, 569 and 561 nm).
Image analysis
Changes in chromatin expansion were analyzed automatically using a custom‐made routine written in MatLab (MathWorks) as previously described in detail 41. HepG2 H3K9me3 immunofluorescence images were loaded and analyzed in Fiji, a distribution of ImageJ 58, using custom macro scripts that are available from the authors upon request. Briefly, image stacks were converted to maximum intensity z‐projections and the DAPI signal was automatically thresholded with built‐in methods and used to define and measure individual nuclei with particle analyzer. The H3K9me3 channel signal was cropped for each individual nucleus and analyzed by subtracting the background with a rolling‐ball algorithm, applying a median filter and automatically thresholding the signal. Artifacts were removed after thresholding with noise removal and binary processing functions of the software. H3K9me3 foci were automatically identified and counted with particle analyzer. Single cell measurements were grouped and plotted as box plots using a web‐tool developed by Tyers and Rappsilber laboratories (http://boxplot.tyerslab.com) and R.
Statistics
In all box plots, the horizontal lines (whiskers) represent the maximum and minimum values, the box signifies the upper (75th) and lower quartiles (25th), the median is represented by a short line within the box, and the mean is represented by a square or cross within the box. Unless stated otherwise, P‐values were calculated using a two‐tailed Student's t‐test assuming unequal variances.
Data availability
The protein structure data from this publication have been deposited to the protein data bank database [https://www.rcsb.org] and assigned the identifier 6FY5.
Author contributions
AGL, MB, GT, and MK conceived and designed the study; MK, IKM, and CB performed and analyzed live cell imaging assays; DC designed, performed, and analyzed experiments; MH conducted crystallography and macrodomain biochemistry; KS and MT supervised and provided valuable input; IG performed in vitro PARP1 inhibition assays; AG‐T conducted ITC experiments; JS conducted mRNA expression analysis; TP conducted ISH experiments; DC, IKM, CB, and RS performed Western blots; SH provided reagents and Matlab scripts for data analysis. AGL, DC, MB, and MK wrote the manuscript.
Conflict of interest
AGL is on the Scientific Advisory Board of VolitionRx, Inc. The remaining authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Table EV1
Table EV2
Source Data for Expanded View
Review Process File
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Acknowledgements
We thank the staff at the Microscopy Rennes Imaging Center (BIOSIT, University Rennes 1), beam line ID29 of the ESRF (Grenoble, France), and beam line PX01 of the SLS (Villigen, Switzerland) for technical assistance; Sandra Hake (University Giessen, Germany) for the H2A‐GFP construct; Marta Vives, Arce Jaraquemada, and Josep Manyé (IGTP, Spain) for samples; Vladimir Rybin (EMBL) for help with ITC experiments; Miriam Bortfeld, Bianca Nijmeijer, Anna Hegele, and Julia Preisser for technical help; as well as the members of the Buschbeck, Huet, and Timinszky labs for helpful discussions. This project was supported by the Ludwig‐Maximilians‐Universität München (to AGL); the Deutsche Forschungsgemeinschaft SFB 646 and SFB 1064 collaborative research centers (to AGL); the Deutsche Forschungsgemeinschaft CIPSM and SyNergy research excellence clusters (to AGL); the Bavarian BioSysNet program (to AGL); the ERA‐NET Neuron project Food4Thought funded by the Bundesministerium für Bildung und Forschung (to AGL); the Marie Skłodowska Curie Training network “ChroMe” H2020‐MSCA‐ITN‐2015‐675610 (to MB and AGL), MINECO BFU2015‐66559‐P (to MB), and the MECD fellowship FPU14/06542 (to DC). Work in the Buschbeck lab is further supported by MINECO‐ISCIII (PIE16/00011), the Deutsche José Carreras Leukämie Stiftung, and AGAUR (2017‐SGR‐305). Research at the IJC is supported by the “La Caixa” Foundation, the Fundació Internacional Josep Carreras, Celgene Spain, and the CERCA Programme/Generalitat de Catalunya.
EMBO Reports (2018) 19: e44445
Contributor Information
Marcus Buschbeck, Email: mbuschbeck@carrerasresearch.org.
Andreas G Ladurner, Email: andreas.ladurner@bmc.med.lmu.de.
References
- 1. Biterge B, Schneider R (2014) Histone variants: key players of chromatin. Cell Tissue Res 356: 457–466 [DOI] [PubMed] [Google Scholar]
- 2. Buschbeck M, Hake SB (2017) Variants of core histones and their roles in cell fate decisions, development and cancer. Nat Rev Mol Cell Biol 18: 299–314 [DOI] [PubMed] [Google Scholar]
- 3. Posavec M, Timinszky G, Buschbeck M (2013) Macro domains as metabolite sensors on chromatin. Cell Mol Life Sci 70: 1509–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rivera‐Casas C, Gonzalez‐Romero R, Cheema MS, Ausió J, Eirin‐López JM (2016) The characterization of macroH2A beyond vertebrates supports an ancestral origin and conserved role for histone variants in chromatin. Epigenetics 11: 415–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Corujo D, Buschbeck M (2018) Post‐translational modifications of H2A histone variants and their role in cancer. Cancers 10: 59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sporn JC, Kustatscher G, Hothorn T, Collado M, Serrano M, Muley T, Schnabel P, Ladurner AG (2009) Histone macroH2A isoforms predict the risk of lung cancer recurrence. Oncogene 28: 3423–3428 [DOI] [PubMed] [Google Scholar]
- 7. Kapoor A, Goldberg MS, Cumberland LK, Ratnakumar K, Segura MF, Emanuel PO, Menendez S, Vardabasso C, LeRoy G, Vidal CI et al (2010) The histone variant macroH2A suppresses melanoma progression through regulation of CDK8. Nature 468: 1105–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nashun B, Yukawa M, Liu H, Akiyama T, Aoki F (2010) Changes in the nuclear deposition of histone H2A variants during pre‐implantation development in mice. Development 137: 3785–3794 [DOI] [PubMed] [Google Scholar]
- 9. Creppe C, Janich P, Cantarino N, Noguera M, Valero V, Musulen E, Douet J, Posavec M, Martin‐Caballero J, Sumoy L et al (2012) MacroH2A1 regulates the balance between self‐renewal and differentiation commitment in embryonic and adult stem cells. Mol Cell Biol 32: 1442–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Barrero MJ, Sese B, Marti M, Izpisua Belmonte JC (2013) Macro histone variants are critical for the differentiation of human pluripotent cells. J Biol Chem 288: 16110–16116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Buschbeck M, Uribesalgo I, Wibowo I, Rué P, Martin D, Gutiérrez A, Morey L, Guigó R, López‐Schier H, Di Croce L (2009) The histone variant macroH2A is an epigenetic regulator of key developmental genes. Nat Struct Mol Biol 16: 1074–1079 [DOI] [PubMed] [Google Scholar]
- 12. Pehrson JR, Changolkar LN, Costanzi C, Leu NA (2014) Mice without macroH2A histone variants. Mol Cell Biol 34: 4523–4533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pasque V, Gillich A, Garrett N, Gurdon JB (2011) Histone variant macroH2A confers resistance to nuclear reprogramming. EMBO J 30: 2373–2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pasque V, Radzisheuskaya A, Gillich A, Halley‐Stott RP, Panamarova M, Zernicka‐Goetz M, Surani MA, Silva JCR (2013) Histone variant macroH2A marks embryonic differentiation in vivo and acts as an epigenetic barrier to induced pluripotency. J Cell Sci 125: 6094–6104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gaspar‐Maia A, Qadeer ZA, Hasson D, Ratnakumar K, Leu NA, LeRoy G, Liu S, Costanzi C, Valle‐Garcia D, Schaniel C et al (2013) MacroH2A histone variants act as a barrier upon reprogramming towards pluripotency. Nat Commun 4: 1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Barrero MJ, Sesé B, Kuebler B, Bilic J, Boue S, Martí M, Belmonte JCI (2013) Macrohistone variants preserve cell identity by preventing the gain of H3K4me2 during reprogramming to pluripotency. Cell Rep 3: 1005–1011 [DOI] [PubMed] [Google Scholar]
- 17. Douet J, Corujo D, Malinverni R, Renauld J, Sansoni V, Posavec Marjanović M, Cantariño N, Valero V, Mongelard F, Bouvet P et al (2017) MacroH2A histone variants maintain nuclear organization and heterochromatin architecture. J Cell Sci 130: 1570–1582 [DOI] [PubMed] [Google Scholar]
- 18. Fu Y, Lv P, Yan G, Fan H, Cheng L, Zhang F, Dang Y, Wu H, Wen B (2015) MacroH2A1 associates with nuclear lamina and maintains chromatin architecture in mouse liver cells. Sci Rep 5: 17186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gamble M, Kraus WL (2010) Multiple facets of the unique histone variant macroH2A: from genomics to cell biology. Cell Cycle 9: 2568–2574 [DOI] [PubMed] [Google Scholar]
- 20. Angelov D, Molla A, Perche P‐Y, Hans F, Côté J, Khochbin S, Bouvet P, Dimitrov S (2003) The histone variant macroH2A interferes with transcription factor binding and SWI/SNF nucleosome remodeling. Mol Cell 11: 1033–1041 [DOI] [PubMed] [Google Scholar]
- 21. Doyen CM, An W, Angelov D, Bondarenko V, Mietton F, Studitsky VM, Hamiche A, Roeder RG, Bouvet P, Dimitrov S (2006) Mechanism of polymerase II transcription repression by the histone variant macroH2A. Mol Cell Biol 26: 1156–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gamble MJ, Frizzell KM, Yang C, Krishnakumar R, Kraus WL (2010) The histone variant macroH2A1 marks repressed autosomal chromatin, but protects a subset of its target genes from silencing. Genes Dev 24: 21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ouararhni K, Hadj‐Slimane R, Ait‐Si‐Ali S, Robin P, Mietton F, Harel‐Bellan A, Dimitrov S, Hamiche A (2006) The histone variant mH2A1.1 interferes with transcription by down‐regulating PARP‐1 enzymatic activity. Genes Dev 20: 3324–3336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen H, Ruiz PD, Novikov L, Casill AD, Park JW, Gamble MJ (2014) MacroH2A1.1 and PARP‐1 cooperate to regulate transcription by promoting CBP‐mediated H2B acetylation. Nat Struct Mol Biol 21: 981–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen H, Ruiz PD, McKimpson WM, Novikov L, Kitsis RN, Gamble MJ (2015) MacroH2A1 and ATM play opposing roles in paracrine senescence and the senescence‐ associated secretory phenotype. Mol Cell 59: 719–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lavigne MD, Vatsellas G, Polyzos A, Mantouvalou E, Sianidis G, Maraziotis I, Agelopoulos M, Thanos D (2015) Composite macroH2A/NRF‐1 nucleosomes suppress noise and generate robustness in gene expression. Cell Rep 11: 1090–1101 [DOI] [PubMed] [Google Scholar]
- 27. Chakravarthy S, Gundimella SKY, Caron C, Perche P‐Y, Pehrson JR, Khochbin S, Luger K (2005) Structural characterization of the histone variant macroH2A. Mol Cell Biol 25: 7616–7624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kustatscher G, Hothorn M, Pugieux C, Scheffzek K, Ladurner AG (2005) Splicing regulates NAD metabolite binding to histone macroH2A. Nat Struct Mol Biol 12: 624–625 [DOI] [PubMed] [Google Scholar]
- 29. Karras GI, Kustatscher G, Buhecha HR, Allen MD, Pugieux C, Sait F, Bycroft M, Ladurner AG (2005) The macro domain is an ADP‐ribose binding module. EMBO J 24: 1911–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee S, Tong L, Denu JM (2008) Quantification of endogenous sirtuin metabolite O‐acetyl‐ADP‐ribose. Anal Biochem 383: 174–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Haince J‐F, McDonald D, Rodrigue A, Déry U, Masson J‐Y, Hendzel MJ, Poirier GG (2008) PARP1‐dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J Biol Chem 283: 1197–1208 [DOI] [PubMed] [Google Scholar]
- 32. Jankevicius G, Hassler M, Golia B, Rybin V, Zacharias M, Timinszky G, Ladurner AG (2013) A family of macrodomain proteins reverses cellular mono‐ADP‐ribosylation. Nat Struct Mol Biol 20: 508–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sharifi R, Morra R, Appel CD, Tallis M, Chioza B, Jankevicius G, Simpson MA, Matic I, Ozkan E, Golia B et al (2013) Deficiency of terminal ADP‐ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease. EMBO J 32: 1225–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rosenthal F, Feijs KLH, Frugier E, Bonalli M, Forst AH, Imhof R, Winkler HC, Fischer D, Caflisch A, Hassa PO et al (2013) Macrodomain‐containing proteins are new mono‐ADP‐ribosylhydrolases. Nat Struct Mol Biol 20: 502–507 [DOI] [PubMed] [Google Scholar]
- 35. Timinszky G, Till S, Hassa PO, Hothorn M, Kustatscher G, Nijmeijer B, Colombelli J, Altmeyer M, Stelzer EHK, Scheffzek K et al (2009) A macrodomain‐containing histone rearranges chromatin upon sensing PARP1 activation. Nat Struct Mol Biol 16: 923–929 [DOI] [PubMed] [Google Scholar]
- 36. Mehrotra PV, Ahel D, Ryan DP, Weston R, Wiechens N, Kraehenbuehl R, Owen‐Hughes T, Ahel I (2011) DNA repair factor APLF is a histone chaperone. Mol Cell 41: 46–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Posavec Marjanović M, Hurtado‐Bagès S, Lassi M, Valero V, Malinverni R, Delage H, Navarro M, Corujo D, Guberovic I, Douet J et al (2017) MacroH2A1.1 regulates mitochondrial respiration by limiting nuclear NAD+ consumption. Nat Struct Mol Biol 24: 902–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nusinow DA, Hernández‐Muñoz I, Fazzio TG, Shah GM, Kraus WL, Panning B (2007) Poly(ADP‐ribose) polymerase 1 is inhibited by a histone H2A variant, MacroH2A, and contributes to silencing of the inactive X chromosome. J Biol Chem 282: 12851–12859 [DOI] [PubMed] [Google Scholar]
- 39. Strickfaden H, McDonald D, Kruhlak MJ, Haince J‐F, Th'ng JPH, Rouleau M, Ishibashi T, Corry GN, Ausió J, Underhill DA et al (2016) Poly(ADP‐ribosyl)ation‐dependent transient chromatin decondensation and histone displacement following laser microirradiation. J Biol Chem 291: 1789–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Luijsterburg MS, de Krijger I, Wiegant WW, Shah RG, Smeenk G, de Groot AJL, Pines A, Vertegaal ACO, Jacobs JJL, Shah GM et al (2016) PARP1 links CHD2‐mediated chromatin expansion and H3.3 deposition to DNA repair by non‐ homologous end‐joining. Mol Cell 61: 547–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sellou H, Lebeaupin T, Chapuis C, Smith R, Hegele A, Singh HR, Kozlowski M, Bultmann S, Ladurner AG, Timinszky G et al (2016) The poly(ADP‐ribose)‐dependent chromatin remodeler Alc1 induces local chromatin relaxation upon DNA damage. Mol Biol Cell 27: 3791–3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kruhlak MJ, Celeste A, Dellaire G, Fernandez‐Capetillo O, Müller WG, McNally JG, Bazett‐Jones DP, Nussenzweig A (2006) Changes in chromatin structure and mobility in living cells at sites of DNA double‐strand breaks. J Cell Biol 172: 823–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bönisch C, Schneider K, Pünzeler S, Wiedemann SM, Bielmeier C, Bocola M, Eberl HC, Kuegel W, Neumann J, Kremmer E et al (2012) H2A.Z.2.2 is an alternatively spliced histone H2A.Z variant that causes severe nucleosome destabilization. Nucleic Acids Res 40: 5951–5964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Muthurajan UM, McBryant SJ, Lu X, Hansen JC, Luger K (2011) The linker region of MacroH2A promotes self‐association of nucleosomal arrays. J Biol Chem 286: 23852–23864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Caterino TL, Fang H, Hayes JJ (2011) Nucleosome linker DNA contacts and induces specific folding of the intrinsically disordered H1 carboxyl‐terminal domain. Mol Cell Biol 31: 2341–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Abbott DW, Chadwick BP, Thambirajah AA, Ausió J (2005) Beyond the Xi: macroH2A chromatin distribution and post‐translational modification in an avian system. J Biol Chem 280: 16437–16445 [DOI] [PubMed] [Google Scholar]
- 47. Chakravarthy S, Patel A, Bowman GD (2012) The basic linker of macroH2A stabilizes DNA at the entry/exit site of the nucleosome. Nucleic Acids Res 40: 8285–8295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Khurana S, Kruhlak MJ, Kim J, Tran AD, Liu J, Nyswaner K, Shi L, Jailwala P, Sung M‐H, Hakim O et al (2014) A macrohistone variant links dynamic chromatin compaction to BRCA1‐dependent genome maintenance. Cell Rep 8: 1049–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lu X, Hamkalo B, Parseghian MH, Hansen JC (2009) Chromatin condensing functions of the linker histone C‐terminal domain are mediated by specific amino acid composition and intrinsic protein disorder †. Biochemistry 48: 164–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hernández‐Muñoz I, Lund AH, van der Stoop P, Boutsma E, Muijrers I, Verhoeven E, Nusinow DA, Panning B, Marahrens Y, van Lohuizen M (2005) Stable X chromosome inactivation involves the PRC1 Polycomb complex and requires histone MACROH2A1 and the CULLIN3/SPOP ubiquitin E3 ligase. Proc Natl Acad Sci USA 102: 7635–7640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yelagandula R, Stroud H, Holec S, Zhou K, Feng S, Zhong X, Muthurajan UM, Nie X, Kawashima T, Groth M et al (2014) The histone variant H2A.W defines heterochromatin and promotes chromatin condensation in Arabidopsis . Cell 158: 98–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kabsch W (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr 26: 795–800 [Google Scholar]
- 53. McCoy AJ, Grosse‐Kunstleve RW, Storoni LC, Read RJ (2005) Likelihood‐enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr 61: 458–464 [DOI] [PubMed] [Google Scholar]
- 54. Beaudouin J, Mora‐Bermúdez F, Klee T, Daigle N, Ellenberg J (2006) Dissecting the contribution of diffusion and interactions to the mobility of nuclear proteins. Biophys J 90: 1878–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cantarino N, Musulen E, Valero V, Peinado MA, Perucho M, Moreno V, Forcales SV, Douet J, Buschbeck M (2016) Downregulation of the deiminase PADI2 is an early event in colorectal carcinogenesis and indicates poor prognosis. Mol Cancer Res 14: 841–848 [DOI] [PubMed] [Google Scholar]
- 56. Treier M, Gleiberman AS, O'Connell SM, Szeto DP, McMahon JA, McMahon AP, Rosenfeld MG (1998) Multistep signaling requirements for pituitary organogenesis in vivo . Genes Dev 12: 1691–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cantariño N, Fernández‐Figueras MT, Valero V, Musulen E, Malinverni R, Granada I, Goldie SJ, Martín‐Caballero J, Douet J, Forcales SV et al (2016) A cellular model reflecting the phenotypic heterogeneity of mutant HRAS driven squamous cell carcinoma. Int J Cancer 139: 1106–1116 [DOI] [PubMed] [Google Scholar]
- 58. Schindelin J, Arganda‐Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B et al (2012) Fiji: an open‐source platform for biological‐image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bawono P, Heringa J (2013) PRALINE: a versatile multiple sequence alignment toolkit In Methods in molecular biology, Russell D. (ed.), pp 245–262. Totowa, NJ: Humana Press; [DOI] [PubMed] [Google Scholar]
- 60. Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ (2009) Jalview version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded View Figures PDF
Table EV1
Table EV2
Source Data for Expanded View
Review Process File
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Data Availability Statement
The protein structure data from this publication have been deposited to the protein data bank database [https://www.rcsb.org] and assigned the identifier 6FY5.


