Abstract
Cryptochromes are photolyase‐like photoreceptors. Arabidopsis CRY2 (cryptochrome 2) primarily mediates the photoperiodic regulation of floral initiation. CRY2 has been shown to promote FT (FLOWERING LOCUS T) mRNA expression in response to blue light by suppressing the degradation of the CO (CONSTANS) protein and activating CIB1 (CRY2‐interacting bHLH1). Although CIB1 and CO are both transcriptional activators of FT, their relationship is unknown. Here, we show that CIB1 physically interacts with CO and promotes FT transcription in a CO‐dependent manner. CRY2, CIB1, and CO form a protein complex in response to blue light to activate FT transcription, and the complex is regulated by the photoperiod and peaks at dusk along with higher FT expression. We also determined that CRY2 was recruited to the FT chromatin by CIB1 and CO and that all three proteins are bound to the same region within the FT promoter. Therefore, there is crosstalk between the CRY2‐CO and CRY2‐CIBs pathways, and CIB1 and CO act together to regulate FT transcription and flowering.
Keywords: CIB1, CO, cryptochrome, flowering time, FT
Subject Categories: Development & Differentiation, Plant Biology, Transcription
Introduction
Cryptochromes (CRYs) are photolyase‐related blue light receptors that mediate light responses in plants and animals 1, 2, 3. Photoexcited CRYs undergo several biophysical and biochemical changes, including circular electron transfer, phosphorylation, ubiquitination, and conformational changes, to alter gene and protein expression at both transcriptional and posttranslational levels 4, 5. The Arabidopsis genome encodes at least two cryptochromes, cryptochrome 1 (CRY1) and CRY2. The major function of CRY1 is to mediate blue light‐dependent de‐etiolation responses 6, whereas the primary role of Arabidopsis CRY2 is the photoperiodic regulation of floral initiation 7.
CO (CONSTANS) and FT (FLOWERING LOCUS T) are among the most important regulators of floral initiation in response to photoperiod by integrating light and temporal signals 8, 9. CO is a transcriptional regulator that promotes flowering by activating FT mRNA expression 10, 11. FT is a RAF kinase inhibitor protein, which acts as a long‐distance signal by migrating from the leaves through the vascular system to the apical meristem 12, 13. The amount of FT mRNA is very important for the flowering time. The transcriptional activation of FT in response to long‐day (LD) conditions is the limiting step in the photoperiodic induction of flowering in Arabidopsis.
CRY2 has been shown to activate FT mRNA expression in response to blue light by suppressing the degradation of the CO protein 14, 15, 16 and by direct activation of the CIB1 (CRY2‐interacting bHLH1) transcription factor 17, 18, 19. The ability of CRY2 to stabilize the CO protein may be partially explained by the light‐independent formation of the CRY2‐COP1 (CONSTITUTIVE PHOTOMORPHOGENIC 1)‐SPA1 (SUPPRESSOR OF PHYA‐105) complex 20, 21, 22. COP1 is an E3 ubiquitin ligase that targets CO and several other transcription factors for degradation 20, 21, 22, and photoexcited CRY2 interacts with SPA1 to suppress COP1‐dependent degradation of CO. CIB1 is the first blue light‐dependent CRY2‐interacting protein to be described. It promotes flowering in a CRY‐dependent manner by activating the transcription of FT. Several homologs of CIB1 (CIB2, CIB4, and CIB5) function redundantly with CIB1 to activate the FT transcription by forming different heterodimers. All of these homologs positively regulate CRY2‐mediated photoperiodic flowering and FT transcription 17, 18.
Although CIB1 and CO are both transcriptional activators of FT, their relationship is unknown. Here, we demonstrate that CIB1 and CRY2 promote FT transcription and flowering in a CO‐dependent manner, while CO can act independently of CIBs. CIB1 and CO physically interact to induce FT transcription together. CRY2, CIB1, and CO form a protein complex in response to blue light. In addition, CRY2 is recruited to the FT promoter by CIB1 and CO, and they all bind to the same chromatin region; therefore, the CRY2‐CO and CRY2‐CIBs pathways interact to regulate FT transcription and flowering.
Results and Discussion
CIB1 promotes FT transcription and flowering in a CO‐dependent manner
CIB1 is the first blue light‐dependent CRY2‐interacting protein to be described, and it promotes FT transcription and flowering in a CRY2‐dependent manner 17, 18, 19. CO is another transcriptional activator of FT 10, 11, but its relationship with CIB1 in the activation of FT is unknown. When we overexpressed CIB1 in ft‐1, co‐1, co‐2, and gi mutants, our results indicated that the early flowering phenotype of 35S:Myc‐CIB1 was abolished in the ft‐1 and co mutant backgrounds (Figs 1A–C and EV1A–F). These results indicated that the promotion of flowering by CIB1 was dependent on both CO and FT. In addition, CIB1 could not promote FT transcription in the co mutant backgrounds (Figs 1D and EV1G), even though the transcription and protein levels of CIB1 were similar or even higher in the co mutants than in the WT (wild type) background (Figs 1D and E, and EV1G and H). ft‐1 is a recessive mutant with a point mutation at amino acid 171 (G171E) which resulted in non‐functional FT protein 23, 24, CIB1 also promoted the transcription of the mutated FT in ft‐1, though the mutated FT was un‐functional (Fig EV1G and H). CIB1 could not promote flowering in the ft‐1 and co mutant backgrounds not because ft‐1 and co mutants were late flowering, since CIB1 still could promote flowering and FT transcription in the gi (GIGANTEA) mutants which were also late flowering (Appendix Fig S1A–E). Blue light‐dependent CRY2 phosphorylation, degradation, and also the interaction between CRY2 and CIB1 were not affected by GI protein, since CRY2 still gets phosphorylated and degraded under the blue light even in the gi mutant, and CRY2 still could co‐precipitated with CIB1 in the gi mutant (Appendix Fig S2A and B). Interestingly, CIB1 could also promote FT transcription in the cop1‐6 mutant background, and Myc‐CIB1/cop1‐6 showed similar early flowering phenotype as cop1‐6 (Appendix Fig S3). These data indicate that CIB1 promotes flowering and FT transcription in a CO‐dependent manner.
Figure 1. CIB1 promotes FT transcription and flowering in a CO‐dependent manner and CO acts independently of CIBs.
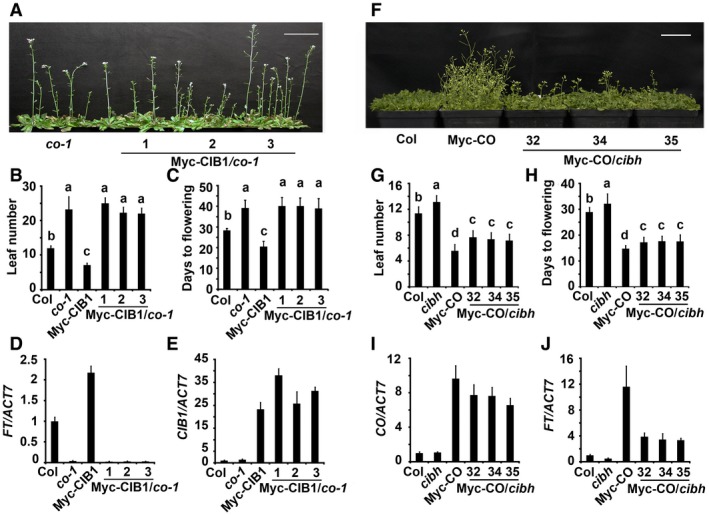
-
A–J(A, F) 52‐day‐old (A), or 25‐day‐old plants (F) of the genotypes indicated grown in LDs. Scale bars: 5 cm. (B, C, G, H) The time to flowering and the number of rosette leaves at the time of flowering of the genotypes shown in (A) (for B and C), (F) (for G and H); the numbers indicate independent transgenic lines. The letters “a” to “c” indicate statistically significant differences between flowering time and leaf numbers of the indicated genotypes. (D, E, I, J) qPCR results showing mRNA expression of CIB1, CO, FT in the genotypes indicated. The numbers indicate independent transgenic lines. The transcript levels of CIB1 or CO or FT in Col were set to 1.0. Samples were collected from 7‐day‐old LD‐grown seedlings at ZT16. In (B, C, G, H), data are presented as mean ± SD, n ≥ 15 per genotypes indicated. P ≤ 0.05 (Tukey's HSD test). For data in (D, E, I, J), error bars indicate standard error within three biological replicates and for each of these two technical replicates were performed.
Figure EV1. CIB1 promotes FT transcription and flowering in a CO‐dependent manner.
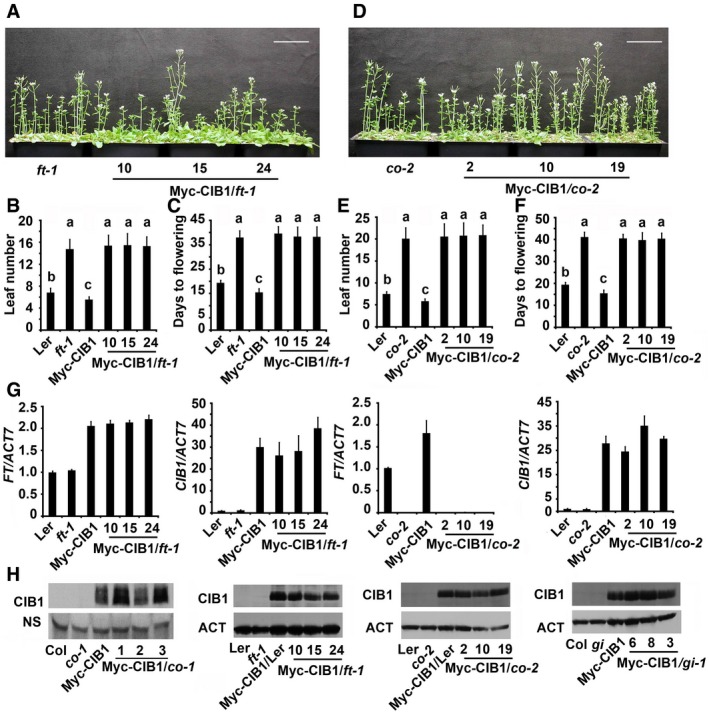
-
A–F(A, D) 48‐day‐old (A) or 52‐day‐old (D) of the genotypes indicated grown in LDs. Scale bars: 5 cm. (B, C, E, F) The time to flowering and the number of rosette leaves at the time of flowering of the genotypes shown in (A) (for B and C), (D) (for E and F); the numbers indicate independent transgenic lines. Data are presented as mean ± SD, n ≥ 15. The letters “a” to “c” indicate statistically significant differences between flowering time and leaf numbers of the indicated genotypes, as determined by Tukey's HSD test (P ≤ 0.05).
-
GqPCR results showing mRNA expression of CIB1 or FT in the genotypes indicated. The transcript levels in Col are set to 1.0. Error bars represent SE of three biological replicates. Samples were collected from 7‐day‐old seedlings of the genotypes indicated at ZT16.
-
HImmunoblots showing Myc‐CIB1 protein levels in the genotypes indicated.
CO can act independently of CIBs
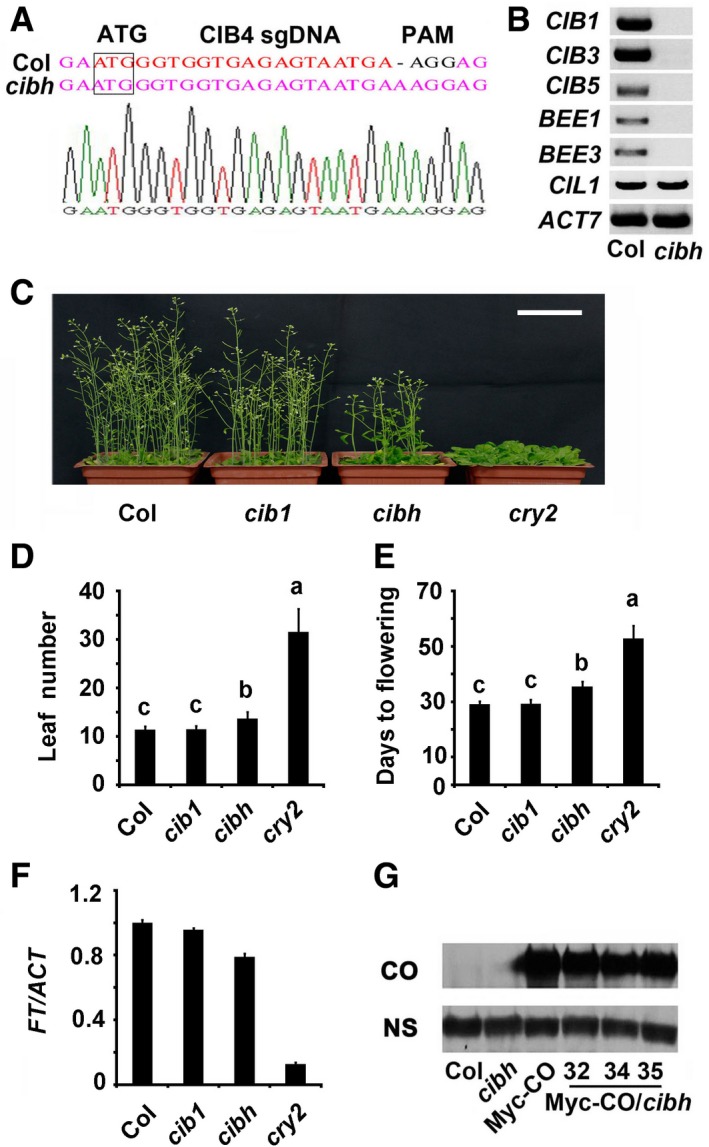
There are about 170 bHLH proteins in Arabidopsis thaliana. CIB1 belongs to the 17‐member bHLH subfamily 18; of this family, CIB4, CIB5, CIL1, and HBI1 are also involved in flowering 17, 18, 25, suggesting a functional redundancy among these genes. Indeed, although cib1 cib5 double and cib1 cib2 cib5 triple mutants showed statistically significant delays in flowering under the photoperiodic inductive condition, but not under the LD (long‐day) condition 17, 18, the transgenic lines expressing the dominant repressor version of CIB1 (CIB1‐EAR) showed a very late flowering phenotype under LD (Appendix Fig S4) 18. When CIB1‐EAR was expressed in a co mutant background, it consistently showed a late flowering phenotype and decreased FT expression similar to the co single mutant (Appendix Fig S4). We generated a cib4 c mutant using the CRISPR/Cas9 system 26 and then made the cib1 cib3 cib5 bee1 bee3 cib4 c hextuple (cibh) mutant (Fig EV2A and B). The cibh mutants exhibited significantly late flowering under LD conditions though not as late as cry2 mutant (Fig EV2C–E), whereas the expression of FT was lower in cibh mutants than in the WT plants (Fig EV2F). Interestingly, the cibh mutant partially suppressed the early flowering phenotype and elevated FT expression in 35S:Myc‐CO plants, even though the transcription and the protein levels of CO were similar in the cibh and WT plants (Figs 1F–J and EV2G). gi mutants could not suppress the early flowering phenotype and elevated FT expression in 35S:Myc‐CO plants though gi mutants exhibit more severe late flowering phenotype than cibh (Appendix Fig S1F–J), the expression pattern of CO was not affected by GI in our condition (Appendix Fig S2C), indicating that CO can act independently of CIBs, and CIBs may regulate CO activity. Taken together, these results demonstrate that CO can act independently of CIBs while cibh may attenuate CO activity in the activation of FT transcription.
Figure EV2. cib1 cib3 cib5 bee1 bee3 cib4 c hextuple (cibh) mutant exhibits late flowering phenotype.
-
ADNA sequence of CIB4 in the WT and the cibh mutants.
-
BRT–PCR results showing mRNA expression of CIBs in the WT and cibh mutants.
-
C42‐day‐old plants of the genotypes indicated grown in LDs. Scale bar: 5 cm.
-
D, EThe time to flowering and the number of rosette leaves at the time of flowering of the genotypes indicated. Data are presented as mean ± SD, n ≥ 15. The letters “a” to “c” indicate statistically significant differences between leaf numbers of the indicated genotypes, as determined by Tukey's HSD test (P ≤ 0.01).
-
FqPCR results showing mRNA expression of FT in the genotypes indicated. The transcript levels in Col are set to 1.0. Error bars represent SE of three biological replicates. Samples were collected from 7‐day‐old seedlings of the genotypes indicated at ZT16.
-
GImmunoblots showing Myc‐CO protein levels in the genotypes indicated.
CIB1 and CO are co‐expressed
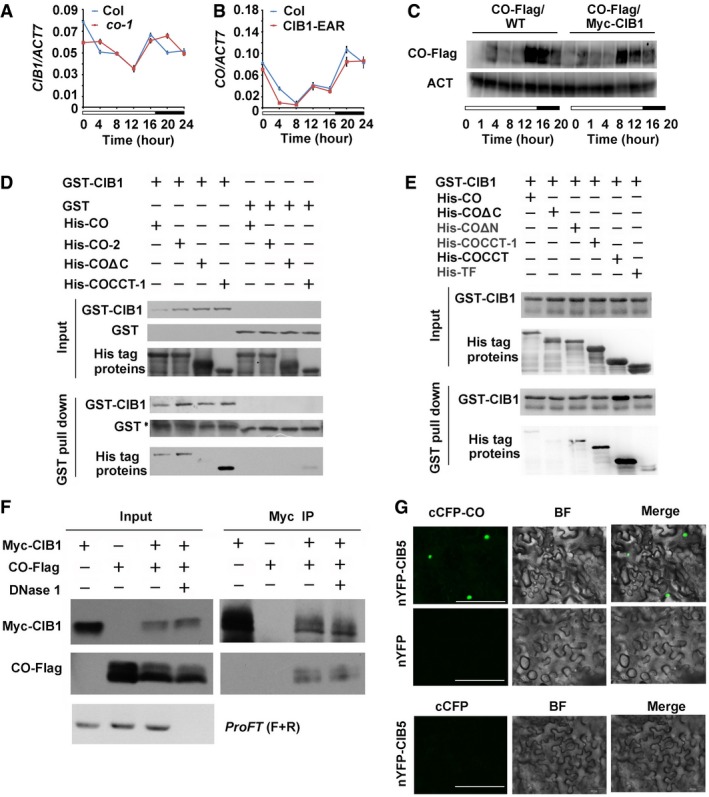
CIB1 promotes flowering and FT transcription in a CO‐dependent manner, and CO can act independently of CIBs. To figure out the underlying mechanism, we first checked whether they affect each other's expression. Our quantitative PCR results indicated that the transcription of CIB1 in WT and the co‐1 mutant were similar, and the transcription of CO in WT and CIB1‐EAR transgenic lines were also similar (Fig EV3A and B). CIB1 and CO did not affect one another's transcription. Furthermore, using transgenic lines that overexpress CO and CIB1 in different reciprocal mutant background with similar expression level, we found CIB1 and CO did not affect one another's protein stability. The protein level of CO in WT, cibh, and Myc‐CIB1 overexpression lines or CIB1 in WT and the co‐1 mutant were similar (Figs 2A–D and EV3C). Interestingly, CIB1 and CO were both found to be regulated by photoperiod and are co‐expressed; their proteins both peaked in abundance early in the morning and again at dusk in LDs with FT expression (Fig 2A–D).
Figure EV3. CIB1 and CO are Co‐expressed and CIB1 interacts with CO .
-
AqPCR results showing the mRNA expression of CIB1 in the WT and the co‐1. Error bars represent SD of three technical replicates of qPCR. The experiment was performed at least three times with similar results.
-
BqPCR results showing the mRNA expression of CO in the WT and CIB1‐EAR transgenic lines. Error bars represent SD of three technical replicates of qPCR. The experiment was performed at least three times with similar results.
-
CImmunoblots showing CO protein levels in transgenic lines expressing 35S:CO‐Flag and 35S:CO‐Flag/35S:Myc‐CIB1 over the course of 20 h under LDs. Samples were separated on 10% SDS–PAGE, blotted, probed with the anti‐Flag antibody, stripped and re‐probed with the anti‐actin antibody (ACT).
-
D, EIn vitro pull‐down assays showing the interaction between CIB1 and CO, or CIB1 and COΔN, or CIB1 and COCCT‐1, or CIB1 and COCCT and the lack of interaction between CIB1 and COΔC. His‐TF tagged CO, CO‐2, COΔN, COΔC, COCCT‐1, or COCCT was mixed with GST‐CIB1 purified from Escherichia coli, and TF antibody was used for the in vitro pull‐down assay. The products were analyzed by immunoblots probed with the anti‐GST (CIB1) or the anti‐TF antibody (CO, CO‐2, COΔN, COΔC, COCCT‐1, COCCT). *Indicates non‐specific bands.
-
FCo‐IP assays showing in planta interactions between CO and CIB1 with or without FT DNA. Protein extracts from transgenic lines co‐expressing Myc‐CIB1 and CO‐Flag were treated with or without DNase I at 22°C for 15 min before used in the co‐IP assay. ProFT (F+R): 1,000 bp FT promoter was amplified using PCR to verify the digestion of the DNase I.
-
GBiFC assays of the in vivo protein interaction. Epidermal cells of Nicotiana benthamiana leaf were co‐transformated with cCFP‐CO and nYFP‐CIB5, with cCFP‐CO and nYFP. BF, bright field. Merge, overlay of the YFP and bright field images. Scale bars: 50 μm.
Figure 2. CIB1 and CO are Co‐expressed.
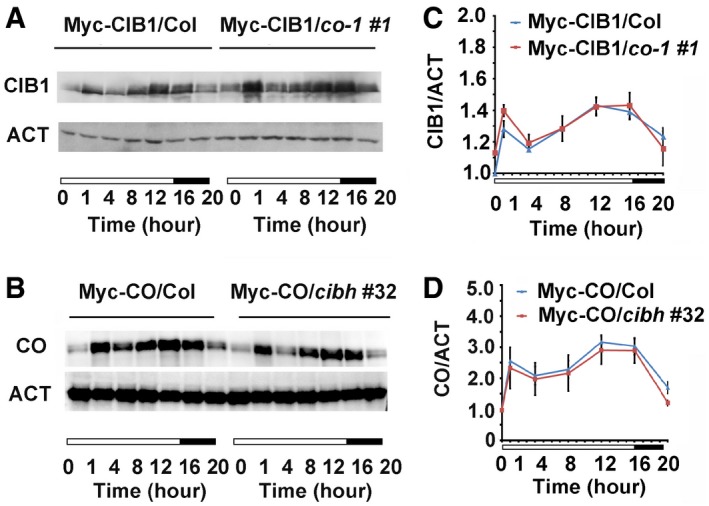
-
A, BImmunoblots showing CIB1 (A) and CO (B) protein levels in WT and co‐1 backgrounds (A) or WT and cibh backgrounds (B) over the course of 20 h under LDs. Samples were separated on 10% SDS–PAGE, blotted, probed with the anti‐Myc antibody, stripped and re‐probed with the anti‐actin antibody (ACT).
-
C, DThe relative levels of CIB1 (A) and CO proteins (B) in WT and co‐1 backgrounds (A) or WT and cibh backgrounds (B) over the course of 20 h under LDs were calculated as described (see Materials and Methods). Error bars represent the standard deviation of three biological replicates of the Western blots.
CIB1 and CO interact directly
Because CIB1 depends on CO in the induction of FT transcription and CIB1 and CO are co‐expressed, we examined the interaction between CO and CIB1. CO and CIB1 were co‐localized in the nuclei (Fig 3A); therefore, we examined their interaction in plant cells using the bimolecular fluorescence complementation (BiFC) assay. Strong fluorescence was detected in the nuclei of cells co‐transformed with cCFP‐CO and nYFP‐CIB1, but no fluorescence was detected in cells transformed separately with either cCFP‐CO or nYFP‐CIB1 (Fig 3B). Bimolecular luminescence complementation (BiLC) assays confirmed that CO interacted directly with CIB1 (Fig 3C). Next, we investigated the CO‐CIB1 interaction using co‐IP; our results indicated that CO co‐immunoprecipitated with CIB1 (Fig 3D). Furthermore, the in vitro pull‐down assay indicated that the C‐terminus of CO interacted with the C‐terminus of CIB1 (Figs 3E and F, and EV3D and E, and Appendix Fig S5), whereas CRY2 interacted with the N‐terminus of CIB1 17. co‐2 is a recessive mutant with a point mutation at amino acid 59 (R59H) on B‐BOX domain of CO protein. Consistent with that CCT domain interacted with the C‐terminus of CIB1, the in vitro pull‐down assays showed that CO‐2 mutant protein could still interact with CIB1 (Fig EV3D). CO and CIB1 had been previously shown to bind to a similar region in the FT promoter 17, 18, 27. To investigate whether the CO‐CIB1 interaction could be detected in planta just because they both bind to the FT promoter and the DNA pull them together, we did a co‐IP experiment with DNase treatment. Our results indicated that CO co‐precipitated with CIB1 even after DNase treatment, and they interacted with each other without FT promoter (Fig EV3F). In addition to CIB1, CIB5 was shown to interact with CO by the BiFC assay (Fig EV3G), indicating that multiple CIBs interact with CO.
Figure 3. CIB1 and CO interact directly.
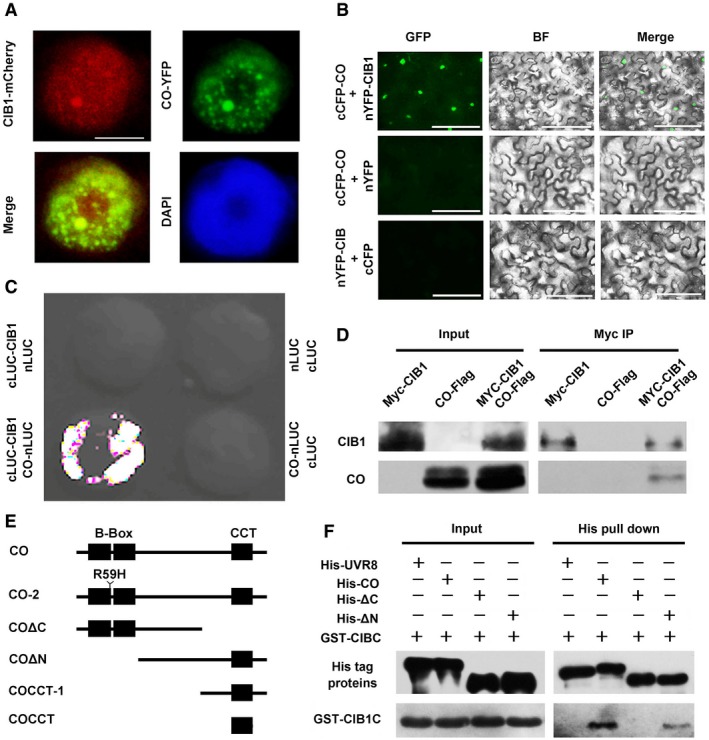
- Fluorescent microscopy images showing that CIB1 co‐localizes with CO in nuclear speckles. Scale bars: 2 μm.
- BiFC assays to show in vivo protein interactions. Epidermal cells of Nicotiana benthamiana leaves were co‐transformed with cCFP‐CO and nYFP‐CIB1, with cCFP‐CO and nYFP, or with cCFP and nYFP‐CIB1. BF, bright field. Merge, overlay of the YFP and bright field images. Scale bars: 50 μm.
- Split‐LUC assay showing that CIB1 interacts with CO. Leaf epidermal cells of N. benthamiana were co‐transformed with CO‐nLUC and cLUC‐CIB1 or cLUC or nLUC with cLUC‐CIB or cLUC.
- Co‐IP assays of samples prepared from 10‐day‐old 35S:Myc‐CIB1, 35S:CO‐Flag, or 35S:Myc‐CIB1/35S:CO‐Flag seedlings grown in LDs.
- Schematic representation of CO used in this work was showing. CO contains two B‐Box domains and a CCT domain.
- In vitro pull‐down assays showing the interaction between CIBC and CO, or CIBC and COΔN and the lack of interaction between CIB1C and COΔC. His‐TF tagged CO, COΔN, COΔC, or His‐TF tagged UVR8 was mixed with GST‐CIB1C purified from Escherichia coli, and His antibody was used for the in vitro pull‐down assay. The products were analyzed by immunoblots probed with the anti‐CIB1C antibody or the anti‐His antibody (UVR8, CO, COΔC, COΔN).
CIB1 and CO act together to induce FT transcription
To investigate the biological significance of the CIB1 and CO interaction, we checked the flowering phenotype of transgenic lines that co‐overexpressed CO and CIB1, they flowered earlier and exhibited dramatically higher FT transcription than lines that only overexpressed CO (Figs 4A–E and EV4). Together with the finding that CIB1 promoted flowering and FT transcription in a CO‐dependent manner, this result indicated that CO and CIB1 might act together to induce FT transcription. A transient dual‐LUC assay was used to further study whether CO and CIB1 act together to induce FT transcription. The co‐expression of CIB1 and CO promoted the expression of the FT promoter:LUC to a much higher level compared with the expression of CIB1 or CO alone (Fig 4F and G). Consistent with the functional dependency of CIB1 on CO in the induction of FT transcription, the activities of CIB1, CO, and CIB1 plus CO were all decreased on the FT promoter lacking the CO binding site (ProFTm2; Fig 4F and G). The 1,034 bp FT promoter fragment contains 5 E‐box motifs, which are putative binding sites for CIB1 (Fig 4F and Appendix Fig S6A). The mutation of each E‐box motif individually had no effect on CIB1‐induced FT transcription, while the mutation of E‐box 1 and E‐box 2 (ProFTE1E2m) together or of all five E‐box motifs (ProFTm1) significantly reduced CIB1‐mediated activation of FT transcription. The mutation of all five E‐box motifs (ProFTm1) also slightly reduced CO‐mediated activation of FT transcription (Fig 4G and Appendix Fig S6B). CIB1 and CO could not promote the transcription of the FT promoter lacking both the CO binding site and all E‐box motifs (proFTm3; Fig 4G). These results indicate that CO and CIB1 bind to the DNA of the FT promoter together to activate its transcription, and CO and CIB1 act together to induce FT transcription.
Figure 4. CO and CIB1 promote flowering and FT transcription together.
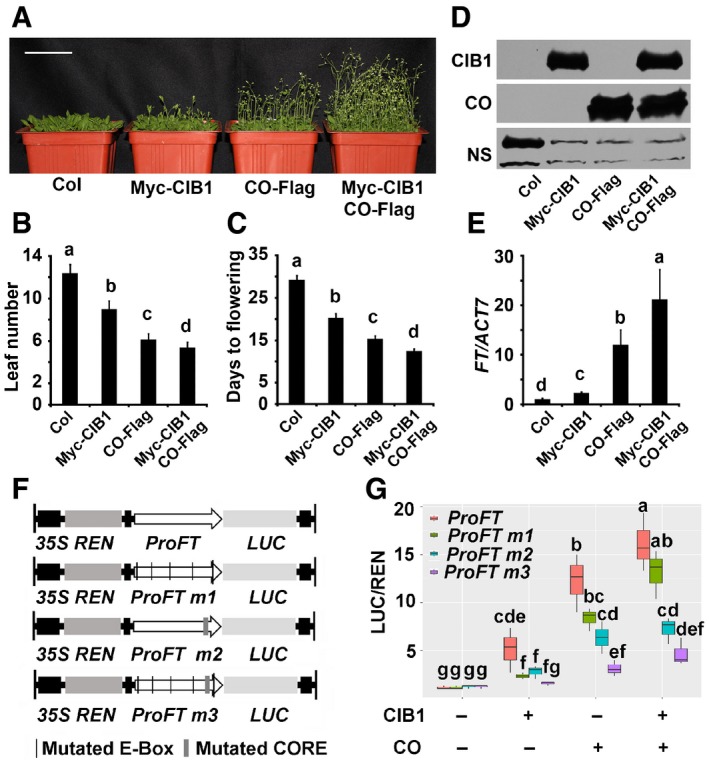
-
A28‐day‐old plants of the genotypes indicated grown in LDs. Scale bars: 5 cm.
-
B, CThe time to flowering and the number of rosette leaves at the time of flowering of the genotypes shown in (A). The letters “a” to “c” indicate statistically significant differences between flowering time and leaf numbers of the indicated genotypes.
-
DImmunoblots showing Myc‐CIB1 and CO‐Flag protein levels in the genotypes indicated. Samples were separated on 10% SDS–PAGE, blotted, probed with the anti‐Myc antibody, stripped and re‐probed with the anti‐Flag antibody.
-
EqPCR results showing mRNA expression of FT in the genotypes indicated. The transcript levels in Col are set to 1.0. Samples were collected from 7‐day‐old seedlings of the genotypes indicated at ZT16.
-
FStructure of the FT promoter‐driven dual‐Luc reporter gene. 35S promoter, FT promoter (−1,034 to −1 bp) (proPFT), FT promoter with mutated E‐boxes (ProFTm1), FT promoter without CO binding site (ProFTm2), FT promoter without CO binding site plus with mutated E‐boxes (ProFTm3). REN luciferase (REN) and firefly luciferase (LUC) are indicated.
-
GBox plots (the minimum, first quartile, median, third quartile, and maximum are shown, with data > 1.5 interquartile ranges denoted with circles) for relative reporter activity (LUC/REN) in plants expressing different reporters and effectors. The relative LUC activities normalized to the REN activity are shown. The LUC/REN in plants expressing ProFT alone is set to 1.0.
Figure EV4. CIB1 and CO act together to promote FT transcription.
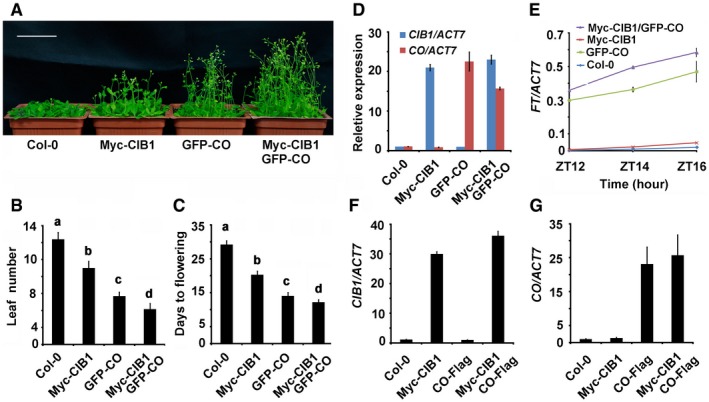
-
A28‐day‐old of the genotypes indicated grown in LDs. Scale bar: 5 cm.
-
B, CThe time to flowering and the number of rosette leaves at the time of flowering of the genotypes indicated. SDs (n > 15) are indicated. The letters “a” to “d” indicate statistically significant differences between leaf numbers of the indicated genotypes, as determined by Tukey's HSD test (P ≤ 0.01).
-
DqPCR results showing mRNA expression of CIB1 and CO in the genotypes indicated. The transcript levels of CIB1 or CO in Col are set to 1.0. Samples were collected from 7‐day‐old seedlings of the genotypes indicated at ZT16. Error bars represent SD of three technical replicates.
-
EqPCR results showing mRNA expression of FT in the genotypes indicated. Samples were collected from 7‐day‐old seedlings of the genotypes indicated at ZT12, ZT14, and ZT16. Error bars represent standard deviation of three technical replicates.
-
F, GqPCR results showing mRNA expression of CIB1 and CO in the genotypes indicated. The transcript levels of CIB1 or CO in Col are set to 1.0. Samples were collected from 7‐day‐old seedlings of the genotypes indicated at ZT16. Error bars represent SE of three biological replicates.
CRY2 activates FT mRNA expression in response to blue light by suppressing the degradation of the CO protein 14, 15, 16 and by direct activation of the CIB1 17, 18, 19. CIB1 and CO acted together to regulate FT transcription, and CIB1 promoted flowering in a CO‐dependent manner, then CRY2 might also promote flowering in a CO‐dependent manner. Overexpression of the constitutively active CRY2 form CRY2W374A leads to early flowering phenotype in both long‐day and short‐day conditions 28. The early flowering phenotype of 35S:CRY2 W374A was abolished in co‐1 mutant background (Appendix Fig S7A–C), and the higher level of FT transcription detected in 35S:CRY2 W374A plants was not observed in the co‐1 mutant background (Appendix Fig S7D and E). Furthermore, cry2 co and cry2 ft double mutants exhibited late flowering that was similar to the late flowering phenotype observed in co and ft mutants (Appendix Fig S7F and G). These results indicate that CRY2 promotes flowering and the transcription of FT in a CO‐dependent manner.
CIB1 and CO act together to mediate CRY2‐dependent regulation of FT transcription
CIB1 physically interacts with both CRY2 and CO; the three proteins might be in the same protein complex. We observed that CRY2, CIB1, and CO co‐localized in nuclear speckles of tobacco (Fig 5A), in agreement with the previous observation that CRY2‐GFP formed distinct nuclear bodies in response to blue light 29. More importantly, endogenous CRY2 could co‐immunoprecipitate both CIB1 and CO in samples that were irradiated with white or blue light but not in samples irradiated with red light, indicating that CRY2, CIB1, and CO form a complex in vivo in a blue light‐dependent manner (Figs 5B and EV5A). CIB1 and CO were both found to be regulated by photoperiod and they are co‐expressed, whereas CRY2 was not (Figs 5C, and EV5B and C), and endogenous CRY2 could co‐immunoprecipitate more CIB1 and CO when more CIB1 and CO proteins were present at dusk along with higher FT expression (Fig 5C). CO and CIB1 had been previously shown to bind to a similar region in the FT promoter 17, 18, 27. CRY2 might also bind to the same sites as part of a complex with CIB1‐CO. We therefore tested this possibility using ChIP. The ChIP‐qPCR results indicated that CRY2, CIB1, and CO bound to the same chromatin region of FT (Fig 5D and E), suggesting that CIB1 and CO recruit CRY2 to the FT chromatin and that the three proteins form a complex to promote FT transcription. Our ChIP results also showed that CIB1 bound to the FT promoter in co‐1, gi‐1, and cry2 mutant backgrounds, but the enrichment of CIB1 on the FT promoter was significantly lower in cry2 mutant than in the WT, indicating that CRY2 might affect the binding of CIB1 to FT (Appendix Fig S8A–C). CO also bound to the FT promoter in cry2 mutant background, indicating that the binding of CO to FT was not affected by CRY2 (Appendix Fig S8C). Interestingly, CRY2 bound to the FT promoter in co‐9 mutant and cibh mutant (Appendix Fig S8D), given that the late flowering of cibh was not as late as the flowering of cry2, suggesting that more CIB1 homologs are involved in flowering, other CIB1 homologs might mediated the binding of CRY2 to the FT promoter. Consistent with that CRY2, CIB1 and CO bound to the same chromatin region of FT, the co‐expression of CRY2, CIB1, and CO promoted the expression of the proFT:LUC to a much higher level compared with the expression of CO alone or CO and CIB1 together (Fig 5F). These results indicate that CIB1 and CO act together to mediate CRY2‐dependent regulation of FT transcription.
Figure 5. CRY2 forms a complex with CO and CIB1 in response to blue light to activate FT transcription.
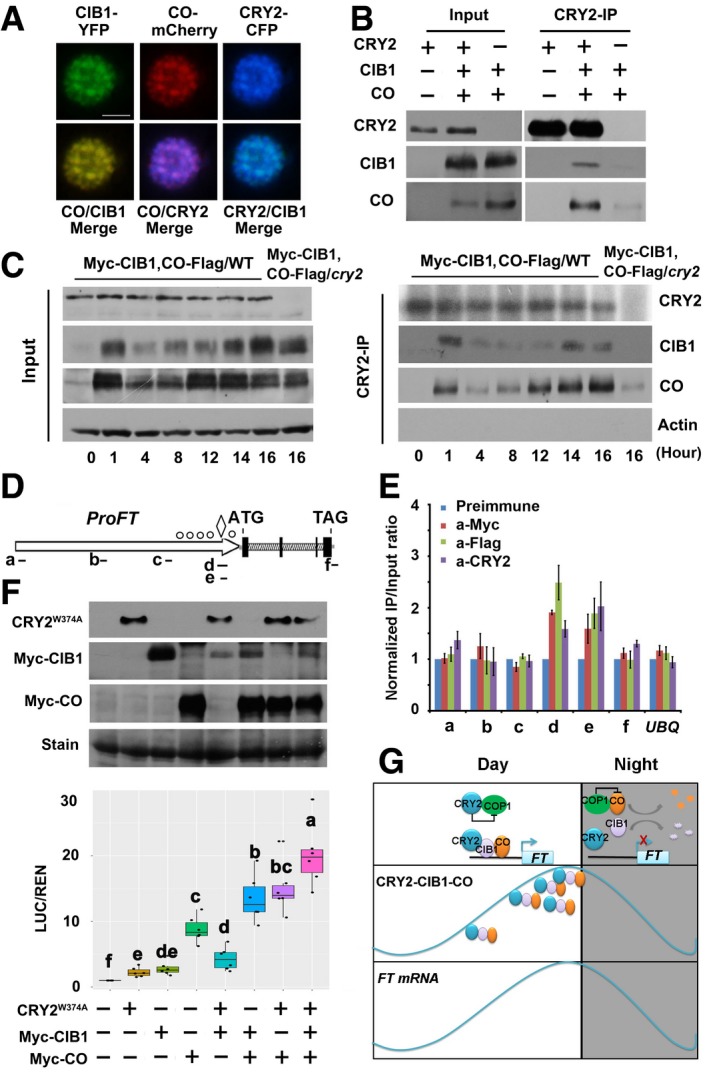
- Fluorescent microscopy images showing that CRY2, CIB1, and CO co‐localized to the nucleus. Scale bars: 2 μm.
- Co‐IP assays showing that CRY2, CIB1, and CO form a protein complex. Samples were prepared from 10‐day‐old WT or 35S:Myc‐CIB1/35S:CO‐Flag or 35S:Myc‐CIB1/35S:CO‐Flag/cry2‐1 seedlings grown in LDs (16L/8D), and samples were collected at ZT16.
- Co‐IP assays of samples prepared from 10‐day‐old 35S:Myc‐CIB1/35S:CO‐Flag 35S:Myc‐CIB1/35S:CO‐Flag/cry2‐1 seedlings grown in LDs (16L/8D). Samples were collected at the times indicated over the course of 1 day.
- Diagram depicting the putative promoter and genomic region of FT. The diamond symbol indicates the position of CORE. The white circles indicate positions of E‐boxes (CANNTG) located at proximal promoter of FT.
- Representative results of the ChIP‐qPCR assays. Chromatin fragments (˜500 bp) were prepared from 10‐day‐old 35S:Myc‐CIB1/35S:CO‐Flag seedlings, immunoprecipitated by the anti‐CRY2, anti‐Flag (CO‐Flag), anti‐Myc (Myc‐CIB1) antibody, or anti‐preimmune IgG, and the precipitated DNA was analyzed by qPCR using the primer pairs indicated. The IP/input ratio of anti‐preimmune is set to 1.0.
- Box plots (the minimum, first quartile, median, third quartile, and maximum are shown, with data > 1.5 interquartile ranges denoted with circles) for relative reporter activity (LUC/REN) in plants expressing different reporters and effectors. The LUC/REN in plants expressing ProFT alone is set to 1.0. Immunoblots were done to show GFP‐CRY2W374A, Myc‐CIB1, and Myc‐CO protein levels in the transient Dual‐LUC assay. Samples were separated on 10% SDS–PAGE, blotted, probed with the anti‐Myc antibody and with the anti‐GFP antibody.
- Model representing the action of CIB1 and CO on FT expression under long‐day condition.
Figure EV5. CRY2, CIB1, and CO could be found in the same protein complex.
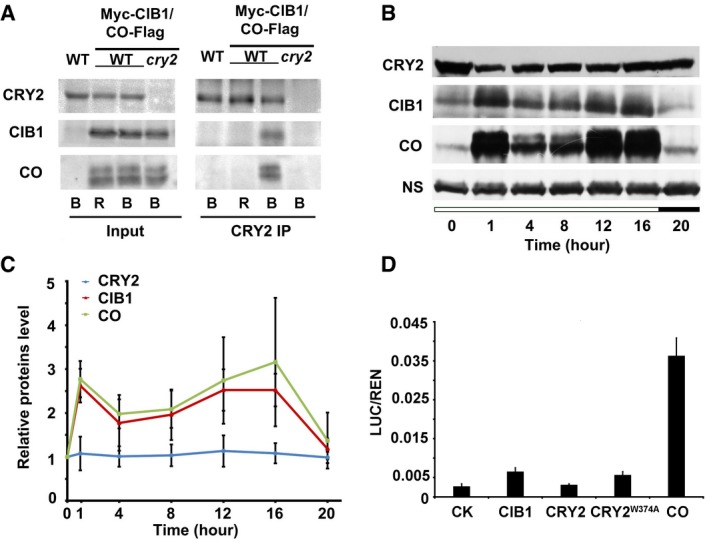
- Co‐IP assays showing that blue light stimulates formation of the CRY2‐CIB1‐CO complex. 10‐day‐old LD‐grown seedlings that co‐expressed CO‐Flag and Myc‐CIB1 together were placed in the dark for 12 h and then were treated with 50 μM MG132 for 2 h before exposure to red light (R 10 μmol m−2 s−1) or blue light (B 40 μmol m−2 s−1) for 60 min.
- Immunoblots showing how CRY2, CIB1, and CO protein levels vary over the course of 1 day. 35S:Myc‐CIB1/35S:CO‐Flag double transgenic lines were used. The results indicate that CRY2 protein levels remain constant over a day, whereas CIB1 and CO both fluctuate in the same way over the day and peak at dusk.
- The relative levels of CRY2, CO, and CIB1 proteins in 35S:Myc‐CIB1/35S:CO‐Flag double transgenic lines over the course of 20 h under LDs. The protein levels were calculated as described (see Materials and Methods). Error bars represent the SD of three biological replicates of the Western blots.
- Relative reporter activity (LUC/REN) in plant with different effectors expression. Tobacco leaves were transfected with the FT promoter (ProFT) reporter alone or with either CRY2, CRY2W374A, CIB1, or CO. Error bars represent the SD of three biological repeats.
Flowering time is regulated by very complex regulatory networks to ensure reproductive success. The photoperiodic pathway mainly perceives external light signals from the environment to optimize seed production in specific environments. CO is the main component of the photoperiodic pathway and the main transcriptional activator of FT 8, 9, 10, 11. CIB1 is another transcriptional activator of FT, a key component of blue light signaling; however, the relationship between CIB1 and CO is unknown. Here, we showed that CIB1 promoted FT transcription and flowering in a CO‐dependent manner, and they interacted physically and act together to induce FT transcription. Furthermore, CRY2 displayed a higher co‐immunoprecipitation efficiency for CIB1 and CO in particular when their protein abundance peaked at dusk on one hand and overlapped with the peak in expression of FT on the other hand. Moreover, the observed physical interaction between CIB1 and CO strongly argues for a cooperation of the CIBs‐CO complex with CRY2 to regulate FT transcription.
CO binds to the proximal region of the FT promoter by recognizing two CO responsive elements (CORE) that are required for FT activation 30. One CORE is located at −221 to −210 bp, whereas the other one is located at −161 to −151 bp upstream of the FT transcription start site. CIB1 and its homologs (CIB2, CIB4, and CIB5) function redundantly to activate the transcription of FT by forming different heterodimers that bind to the E‐boxes of the FT promoter. There are two E‐box elements around the CORE, one located at −23 to −18 bp and the other one located at −325 to −320 bp. The DNA‐binding sites of the CIBs and CO are close to one another. CIBs and CO all bind to the FT promoter directly, and they physically interact with one another to promote each other's transcriptional activity. TARGET OF EAT1 (TOE1) can bind to the FT promoter near the CO binding site, and it interacts with CO to inhibit CO activity 31. Nuclear factor Y (NF‐Y) binds to a CCAAT element located 5.3 kb upstream of the transcriptional start site of FT 32, and certain NF‐Y subunit paralogs are required for CO activation of FT 33. NF‐Y can also interact with CO directly to regulate the transcription of SOC1 34. It has also been reported that ASYMMETRIC LEAVES 1 (AS1) physically interacts with CO to promote FT transcription and binds directly to the FT promoter 35. Multiple transcription factors or DNA‐binding proteins could interact with CO on the FT promoter to regulate its transcription.
Our ChIP data indicate that CRY2 associates with the FT promoter together with CIB1 and CO; therefore, the CIB1‐CO transcription factors recruit CRY2 to the FT promoter, and CRY2 supports the transcriptional activity of CO‐CIB1. Thus, we propose that CRY2, CIB1, and CO form a complex on the DNA so that changes in the external environment can rapidly lead to phenotypic changes. We also find that CRY2 affects the binding of CIB1 to FT, since the enrichment of CIB1 on the FT promoter was significantly lower in the cry2 mutant than in the WT. It is therefore possible that CRY2 supports or stabilizes the binding of CIB1 to FT promoter. Along with Pedmale et al, we showed that the transcription factor PIF4 (PHYTOCHROME INTERACTING FACTOR 4) could recruit CRY1/CRY2 to the promoters of PIF4 targets 36, 37. CRYs are likely localized to DNA indirectly via interactions with other transcription factors, similar to PhyB, which lacks a DNA‐binding domain, is actively recruited to chromatin at PIF TF binding sites 38. In animals, CRY interacts with CLOCK‐BMAL1 at the promoter and inhibits CLOCK‐BMAL1‐dependent transcription 39, 40; therefore, in both plants and animals, CRYs interact with transcription factors on the DNA to regulate transcription. The molecular mechanisms underlying CRY2‐mediated transcriptional inhibition and activation will need to be further investigated.
In summary, we found that CIB1 promoted FT transcription and flowering in a CO‐dependent manner. CIB1 physically interacts with both CRY2 and CO, the three proteins could be found in the same protein complex, and the complex is regulated by both blue light and photoperiod. CIB1 and CO recruit CRY2 to the FT promoter, forming a protein complex to promote FT transcription (Fig 5G). Therefore, there is crosstalk between the CRY2‐CO and CRY2‐CIBs pathways, and CIB1 and CO act together to regulate FT transcription and flowering.
Materials and Methods
Plant materials
Plants were grown on soil or Murashige and Skoog (MS) media containing 1% sucrose in plant incubators (Percival Scientific) or phytotron at 22°C under full‐spectrum white fluorescent light with a fluence rate of 90 μmol m−2 s−1 in long‐day (16 h light/8 h dark) conditions. For the light treatment, red or blue light was provided by red LEDs or blue LEDs, respectively, with light intensities of 40 μmol m−2 s−1.
For the transient assay, tobacco plants (Nicotiana benthamiana) were grown in phytotron at 22°C in long‐day condition. The tobacco leaves were ready for infiltration 4–5 weeks after germination.
To analyze the flowering time, the number of rosette leaves were counted when inflorescences were seen. The days to flowering were calculated as how many days needed from the seed germination to the inflorescences be seen. All flowering experiments were repeated at least twice independently, and similar results were obtained. Western blot or qPCR or both were done to verify the overexpression of the transgenes.
The cry2‐1, co‐1, co‐2, co‐9, ft‐1, ft‐10, gi‐1, cib1 cib2 cib5, bee1, bee3, and CRY2W374A mutants and transgenic lines were described previously 7, 8, 18, 23, 28, 41, 42, 43, 44, 45, 46, 47, 48. The T‐DNA insertion mutants cib3 (AT3G07340 SALK_091124) were obtained from ABRC. The cry2 co‐9, cry2 ft‐10, and cib1 cib3 cib5 bee 1 bee3 cib4 c mutants were prepared by carrying out genetic crosses and their identities were verified by genotyping and RT–PCR. Transgenic Arabidopsis that overexpress 35S:Myc‐CIB1 in the Col, Ler, co‐1, co‐2, ft‐1, and gi‐1 backgrounds, 35S:MycCIB1EAR in the co‐2 background, 35S:CO‐Flag, 35S:GFP‐CO in Col background, 35S:Myc‐CO expressed in the cib1 cib2 cib5 bee1 bee3 cib4 c backgrounds, 35S:HA‐CO expressed in gi‐1 backgrounds, 35S:CRY2 W374A in the co‐1 mutant background were prepared by the floral dip method 49. Transgenic Arabidopsis that overexpress 35S:Myc‐CIB1 and 35S:CO‐Flag together or 35S:Myc‐CIB1 and 35S:GFP‐CO together was prepared by carrying out genetic crosses between transgenic Arabidopsis that overexpress 35S:Myc‐CIB1 and transgenic Arabidopsis that overexpress 35S:CO‐Flag or 35S:GFP‐CO. For every transformation, more than 10 independent transgenic lines were generated. All the independent transgenic lines overexpress the same construct showed the same phenotype.
The binary plasmid encoding 35S:CO‐Flag was pCambia 1306; the coding sequence of CO was inserted between BamHI and SalI restriction sites. To construct Myc‐CO and Myc‐CIB1, the coding sequences of CIB1 and CO were fused to the vector pEGAD by inserting the sequences between the EcoRI and XhoI restriction sites. 35S:HA‐CO was prepared by gateway method and was cloned into pEarly‐201. 35S:CO‐YFP, 35S:CO‐mCherry, 35S:CIB1‐YFP, 35S:CRY2‐CFP, and 35S:CIB1‐mCherry were cloned into pCambia 1301 with modification by conventional restriction digestion and ligation method. The vectors pCold‐TF (Takara) and pGEX4T‐1 were used in the pull‐down assays. BiFC and BiLC vectors were prepared using the GATEWAY method; the vectors backbone was described as before 50. All of the prepared constructs were verified by DNA sequencing.
Immunoblots
Our attempts to prepare anti‐CO and anti‐CIB1 antibodies were unsuccessful (they were able to recognize CO and CIB1 expressed in Escherichia coli but not in plant extracts, and they were used in the in vitro pull‐down assays). Therefore, all analyses of CO and CIB1 proteins were carried out using transgenic plants expressing the Myc‐, Flag‐, or GFP‐tagged CO and CIB1. Immunoblots were carried out as described previously 51. Anti‐Myc antibody (Millipore, 05‐724, diluted 1:4,000), anti‐GFP antibody (Abiocode, M0802‐3a, diluted 1:4,000), anti‐Flag antibody (Sigma, F3165, diluted 1:3,000), anti‐actin antibody (Abmart, M20009, diluted 1:3,000), anti‐His antibody (MBL, D291‐3, diluted 1:4,000), and anti‐GST antibody (Abmart, M20007, diluted 1:5,000) were used.
CIB1 or CO signals were quantified by digitizing the band signal (ImageJ), subtracting the background signal, normalizing against the internal control (CRY1, ACT, or NS), and the relative level calculated against the value of CIB1, CO, or CRY2 in the WT background at the ZT0. The relative level of CIB1, CO, or CRY2 (Figs 2C and D, and EV5C) is calculated by the formula [(CIB1ZT − bZT)/(CZT − bZT)]/[CIB1ZT0W − bZT0W)/(CZT0W − bZT0W)], or [(COZT − bZT)/(CZT − bZT)]/[COZT0W − bZT0W)/(CZT0W − bZT0W)]in which CIB1ZT or COZT represents the signal intensity of CIB1 or CO from a sample collected at different ZT time (ZT0 to ZT20), bZT represents the background signal of the CIB1ZT or COZT lane in the immunoblot, CZT represents signal intensity of the loading control (CRY1 or ACT or NS), bZT represents the background signal of the respective CZT lane in the immunoblot, and CIB1ZT0W, COZT0W, CZT0W, bZT0W represent CIB1, CO, loading control, or background signals from sample in the WT background collected at ZT0.
Co‐localization, BiLC, and BiFC assays
The co‐localization assay was carried out as described previously with the following modifications 17, 52: CO was fused to YFP or mCherry, CIB1 was fused to GFP or mCherry, and CRY2 was fused to CFP. These constructs were then transformed into Agrobacterium strain GV3101 containing the pSoup‐P19 plasmid that encodes the suppressor of gene silencing 53. Overnight cultures of Agrobacteria were collected by centrifugation, re‐suspended in MES buffer (10 mM MES pH 5.8, 10 mM MgCl2, 200 μM acetosyringone) to an OD600 of 0.8, mixed, and incubated at room temperature for 2 h prior to infiltration. The Agrobacteria suspension was drawn into a 1‐ml syringe (without the needle) and carefully press‐infiltrated by hand into healthy leaves of 3‐week‐old N. benthamiana plants. The plants were left under continuous white light for 3 days after the infiltration. For the BiLC assay, 1 mM luciferin was infiltrated before LUC activity was monitored after 3 days. The LUC signal was photographed with a cool CCD camera (Andor DW936N‐BV).
Co‐immunoprecipitation (co‐IP) experiments
The co‐immunoprecipitation (co‐IP) procedure was described previously 17. Samples were ground in liquid nitrogen, homogenized in Binding Buffer [20 mM HEPES (pH 7.5), 40 mM KCl, 1 mM EDTA, 0.5% Triton X‐100, 1 mM PMSF, 1 mg ml−1 Complete Protease Inhibitor Cocktail Tablets (Roche), 50 μg ml−1 MG132], and centrifuged at 14,000 g for 15 min. One millilitre of supernatant was mixed with 25 μl of anti‐CRY2‐IgG‐coupled protein A‐Sepharose or anti‐Myc affinity gel (Sigma E6654) incubated at 4°C for 20 min. Three microlitre of anti‐CRY2 antiserum was incubated with 20 μl of protein A‐Sepharose beads in 100 μl of Binding Buffer at 4°C for 2 h and used shortly after its preparation. The beads were washed (20 s each) three times with Washing Buffer [20 mM HEPES (pH 7.5), 40 mM KCl, 1 mM EDTA, 0.1% Triton X‐100]. The bound proteins were then eluted from the affinity beads with 4X SDS–PAGE sample buffer (40% Glycerol, 0.4% Bromophenol Blue, 1% SDS, 400 mM Tris–HCl pH6.8, 1% DTT) and analyzed by immunoblotting.
In vitro pull‐down assays
The in vitro pull‐down protein–protein interaction assay was described previously with modifications 18. The coding sequences of CIB1 and CIB1C (residues 172–335) were fused to the C‐terminus of GST in the vector pGEX4T (GE Healthcare) by inserting the sequences between the EcoRI and XhoI restriction sites. The coding sequences of CO, CO‐2, COΔC (residues 1–253), COΔN (residues 106–374), CO‐CCT (residues 208–374), and UVR8 (UV RESISTANCE LOCUS 8) were fused to the C‐terminus of the His tag in the vector pCold‐TF (Takara) by inserting the sequences between the EcoRI and SalI restriction sites. The fusion proteins were expressed in E. coli (strain BL21), purified and bound to Glutathione Sepharose 4B or Ni‐NTA beads according to the manufacturer's instructions (Amersham).
Transient transcription dual‐luciferase (dual‐LUC) assays
Transient transcription dual‐LUC assays in N. benthamiana plants were carried out as described previously 17, 18. To generate the constructs needed, 1,034 bp FT promoter was cloned into pBlueScript SK to generate ProFT‐SK construct. The ProFTm1‐SK ProFTm2‐SK and ProFTm3‐SK constructs carrying mutated E‐box element (CANNTG to AAAAAA) or deleted CORE element were generated by overlapping PCR reaction from the ProFT‐KS construct. After verifying by DNA sequencing, the ProFT, ProFTm1, ProFTm2, and ProFTm3 fragments were inserted to pGreen II 0800 to generate the final constructs for dual‐LUC assay. All the primers used for dual‐LUC assay were listed in Appendix Table S1. The LUC activity of the plant extracts was analyzed with a luminometer (GloMax 20/20, Promega) using LUC detection reagents according to the manufacturer's instructions (Promega).
mRNA expression analyses
Total RNA was isolated using the RNAiso Plus (Takara). cDNA was synthesized from 500 ng of total RNA using the PrimeScript RT Reagent Kit with gDNA Eraser (Takara). SYBR Premix Ex Taq (Takara) was used in the qPCR reactions, and the reactions were carried out using the Mx3000 System (Stratagene). Transcript levels for each sample were normalized to the level of ACTIN7 mRNA expression (AT5G09810, Appendix Table S1). Biological replicates represent three independent experiments involving about 30 seedlings per experiment. Three technical replicates were done for each experiment.
Chromatin immunoprecipitation (ChIP) assays
ChIP was performed as described previously 17. Two‐week‐old Arabidopsis seedlings grown on soil were harvested at ZT14 and were treated with 1% formaldehyde under vacuum for 16 min. The cross‐linking was stopped by adding glycine to a final concentration of 0.125 M. The seedlings were rinsed with water, frozen in liquid nitrogen, and ground to a fine powder. Nuclei were extracted with Extraction Buffer 1 [10 mM Tris–HCl pH8.0, 0.4 M sucrose, 10 mM MgCl2 0.1 mM PMSF, Complete Protease Inhibitor Cocktail Tablets (Roche)] and washed with Extraction Buffer 2 [10 mM Tris–HCl pH8.0, 0.25 M sucrose, 10 mM MgCl2, 1% Triton X‐100, 0.1 mM PMSF, Complete Protease Inhibitor Cocktail Tablets (Roche)]. Nuclei were lysed in Nuclei Lysis Buffer [50 mM Tris–HCl pH 8.0, 10 mM EDTA, 1% SDS, 0.1 mM PMSF, Complete Protease Inhibitor Cocktail Tablets (Roche)] and sonicated to shear the DNA into fragments that were approximately 0.5–1 kb. The chromatin solution was diluted 10‐fold with ChIP Dilution Buffer [16.7 mM Tris–HCl pH 8.0, 167 mM NaCl, 1.1% Triton X‐100, 1.2 mM EDTA, 0.1 mM PMSF, Complete Protease Inhibitor Cocktail Tablets (Roche)]. Immobilized protein G (Pierce) was used for the immunoprecipitation. Anti‐Myc antibody was prebound to the beads by incubating them together for 2 h in ChIP Dilution Buffer. Immunocomplexes were washed with Low Salt Buffer (20 mM Tris–HCl pH 8.0, 150 mM NaCl, 0.2% SDS, 0.5% Triton X‐100, 2 mM EDTA), High Salt Buffer (20 mM Tris–HCl pH 8.0, 500 mM NaCl, 0.2% SDS, 0.5% Triton X‐100, 2 mM EDTA), LiCl Wash Buffer (20 mM Tris–HCl pH 8.0, 0.25 M LiCl, 1% NP‐40, 1% sodium deoxycholate, 1 mM EDTA,), and TE buffer (10 mM Tris–HCl pH 8.0, 1 mM EDTA) and then eluted with Elution Buffer (50 mM Tris–HCl pH 8.0, 10 mM EDTA, 1% SDS). The cross‐linking of the samples was reversed by incubating at 65°C overnight and then by treatment with Proteinase K to remove the proteins. DNA was purified by using the FastDNA Kit (MP Biomedicals) and eluted with 50 μl of TE buffer. One microlitre aliquots were used for qPCR. The primer pairs that were used to amplify the genomic sequence are listed in Appendix Table S1.
Author contributions
YL and HL conceived the project. YL performed most of the experiments, XL performed the pull‐down assays, DM aided in the performance of the ChIP assays, ZC helped to prepare constructs, J‐WW provided materials, YL and HL analyzed the data, and HL and YL wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Review Process File
Acknowledgements
We thank Drs. Zhiyong Wang, Roger P. Hellens, and Jiankang Zhu for materials and technical assistance. We thank Dr. Candace Webb for copyediting the manuscript. This work is supported in part by the National Natural Science Foundation of China (31730009, 31721001, 31670282, 31670307), the National Key Research and Development Program of China (2017YFA 0503800), and the Strategic Priority Research Program “Molecular Mechanism of Plant Growth and Development” (XDPB04).
EMBO Reports (2018) 19: e45762
References
- 1. Cashmore AR (2003) Cryptochromes: enabling plants and animals to determine circadian time. Cell 114: 537–543 [PubMed] [Google Scholar]
- 2. Sancar A (2003) Structure and function of DNA photolyase and cryptochrome blue‐light photoreceptors. Chem Rev 103: 2203–2237 [DOI] [PubMed] [Google Scholar]
- 3. Lin C, Shalitin D (2003) Cryptochrome structure and signal transduction. Annu Rev Plant Biol 54: 469–496 [DOI] [PubMed] [Google Scholar]
- 4. Liu B, Liu H, Zhong D, Lin C (2010) Searching for a photocycle of the cryptochrome photoreceptors. Curr Opin Plant Biol 13: 578–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liu H, Liu B, Zhao C, Pepper M, Lin C (2011) The action mechanisms of plant cryptochromes. Trends Plant Sci 16: 684–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ahmad M, Cashmore AR (1993) HY4 gene of A. thaliana encodes a protein with characteristics of a blue‐light photoreceptor. Nature 366: 162–166 [DOI] [PubMed] [Google Scholar]
- 7. Guo H, Yang H, Mockler TC, Lin C (1998) Regulation of flowering time by Arabidopsis photoreceptors. Science 279: 1360–1363 [DOI] [PubMed] [Google Scholar]
- 8. Putterill J, Robson F, Lee K, Simon R, Coupland G (1995) The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80: 847–857 [DOI] [PubMed] [Google Scholar]
- 9. Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T (1999) A pair of related genes with antagonistic roles in mediating flowering signals. Science 286: 1960–1962 [DOI] [PubMed] [Google Scholar]
- 10. Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz‐Sommer Z, Yanofsky MF, Coupland G (2000) Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis . Science 288: 1613–1616 [DOI] [PubMed] [Google Scholar]
- 11. Onouchi H, Igeno MI, Perilleux C, Graves K, Coupland G (2000) Mutagenesis of plants overexpressing CONSTANS demonstrates novel interactions among Arabidopsis flowering‐time genes. Plant Cell 12: 885–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C et al (2007) FT protein movement contributes to long‐distance signaling in floral induction of Arabidopsis . Science 316: 1030–1033 [DOI] [PubMed] [Google Scholar]
- 13. Lifschitz E, Eviatar T, Rozman A, Shalit A, Goldshmidt A, Amsellem Z, Alvarez JP, Eshed Y (2006) The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc Natl Acad Sci USA 103: 6398–6403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G (2004) Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303: 1003–1006 [DOI] [PubMed] [Google Scholar]
- 15. Yanovsky MJ, Kay SA (2002) Molecular basis of seasonal time measurement in Arabidopsis . Nature 419: 308–312 [DOI] [PubMed] [Google Scholar]
- 16. Liu LJ, Zhang YC, Li QH, Sang Y, Mao J, Lian HL, Wang L, Yang HQ (2008) COP1‐mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis . Plant Cell 20: 292–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu H, Yu X, Li K, Klejnot J, Yang H, Lisiero D, Lin C (2008) Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis . Science 322: 1535–1539 [DOI] [PubMed] [Google Scholar]
- 18. Liu Y, Li X, Li K, Liu H, Lin C (2013) Multiple bHLH proteins form heterodimers to mediate CRY2‐dependent regulation of flowering‐time in Arabidopsis . PLoS Genet 9: e1003861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu H, Wang Q, Liu Y, Zhao X, Imaizumi T, Somers DE, Tobin EM, Lin C (2013) Arabidopsis CRY2 and ZTL mediate blue‐light regulation of the transcription factor CIB1 by distinct mechanisms. Proc Natl Acad Sci USA 110: 17582–17587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang H, Ma LG, Li JM, Zhao HY, Deng XW (2001) Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science 294: 154–158 [DOI] [PubMed] [Google Scholar]
- 21. Jang S, Marchal V, Panigrahi KC, Wenkel S, Soppe W, Deng XW, Valverde F, Coupland G (2008) Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J 27: 1277–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zuo Z, Liu H, Liu B, Liu X, Lin C (2011) Blue light‐dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis . Curr Biol 21: 841–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Koornneef M, Hanhart CJ, van der Veen JH (1991) A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana . Mol Gen Genet 229: 57–66 [DOI] [PubMed] [Google Scholar]
- 24. Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D (1999) Activation tagging of the floral inducer FT. Science 286: 1962–1965 [DOI] [PubMed] [Google Scholar]
- 25. Bai MY, Fan M, Oh E, Wang ZY (2012) A triple helix‐loop‐helix/basic helix‐loop‐helix cascade controls cell elongation downstream of multiple hormonal and environmental signaling pathways in Arabidopsis . Plant Cell 24: 4917–4929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Feng Z, Mao Y, Xu N, Zhang B, Wei P, Yang DL, Wang Z, Zhang Z, Zheng R, Yang L et al (2014) Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas‐induced gene modifications in Arabidopsis . Proc Natl Acad Sci USA 111: 4632–4637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Song YH, Smith RW, To BJ, Millar AJ, Imaizumi T (2012) FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 336: 1045–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li X, Wang Q, Yu X, Liu H, Yang H, Zhao C, Liu X, Tan C, Klejnot J, Zhong D et al (2011) Arabidopsis cryptochrome 2 (CRY2) functions by the photoactivation mechanism distinct from the tryptophan (trp) triad‐dependent photoreduction. Proc Natl Acad Sci USA 108: 20844–20849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yu X, Sayegh R, Maymon M, Warpeha K, Klejnot J, Yang H, Huang J, Lee J, Kaufman L, Lin C (2009) Formation of nuclear bodies of Arabidopsis CRY2 in response to blue light is associated with its blue light‐dependent degradation. Plant Cell 21: 118–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tiwari SB, Shen Y, Chang HC, Hou Y, Harris A, Ma SF, McPartland M, Hymus GJ, Adam L, Marion C et al (2010) The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis‐element. New Phytol 187: 57–66 [DOI] [PubMed] [Google Scholar]
- 31. Zhang B, Wang L, Zeng L, Zhang C, Ma H (2015) Arabidopsis TOE proteins convey a photoperiodic signal to antagonize CONSTANS and regulate flowering time. Genes Dev 29: 975–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cao S, Kumimoto RW, Gnesutta N, Calogero AM, Mantovani R, Holt BF III (2014) A distal CCAAT/NUCLEAR FACTOR Y complex promotes chromatin looping at the FLOWERING LOCUS T promoter and regulates the timing of flowering in Arabidopsis . Plant Cell 26: 1009–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kumimoto RW, Zhang Y, Siefers N, Holt BF III (2010) NF‐YC3, NF‐YC4 and NF‐YC9 are required for CONSTANS‐mediated, photoperiod‐dependent flowering in Arabidopsis thaliana . Plant J 63: 379–391 [DOI] [PubMed] [Google Scholar]
- 34. Hou X, Zhou J, Liu C, Liu L, Shen L, Yu H (2014) Nuclear factor Y‐mediated H3K27me3 demethylation of the SOC1 locus orchestrates flowering responses of Arabidopsis . Nat Commun 5: 4601 [DOI] [PubMed] [Google Scholar]
- 35. Song YH, Lee I, Lee SY, Imaizumi T, Hong JC (2012) CONSTANS and ASYMMETRIC LEAVES 1 complex is involved in the induction of FLOWERING LOCUS T in photoperiodic flowering in Arabidopsis . Plant J 69: 332–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pedmale UV, Huang SS, Zander M, Cole BJ, Hetzel J, Ljung K, Reis PA, Sridevi P, Nito K, Nery JR et al (2016) Cryptochromes interact directly with PIFs to control plant growth in limiting blue light. Cell 164: 233–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ma D, Li X, Guo Y, Chu J, Fang S, Yan C, Noel JP, Liu H (2016) Cryptochrome 1 interacts with PIF4 to regulate high temperature‐mediated hypocotyl elongation in response to blue light. Proc Natl Acad Sci USA 113: 224–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jung JH, Domijan M, Klose C, Biswas S, Ezer D, Gao M, Khattak AK, Box MS, Charoensawan V, Cortijo S et al (2016) Phytochromes function as thermosensors in Arabidopsis . Science 354: 886–889 [DOI] [PubMed] [Google Scholar]
- 39. Ye R, Selby CP, Chiou YY, Ozkan‐Dagliyan I, Gaddameedhi S, Sancar A (2014) Dual modes of CLOCK:BMAL1 inhibition mediated by Cryptochrome and Period proteins in the mammalian circadian clock. Genes Dev 28: 1989–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Griffin EA Jr, Staknis D, Weitz CJ (1999) Light‐independent role of CRY1 and CRY2 in the mammalian circadian clock. Science 286: 768–771 [DOI] [PubMed] [Google Scholar]
- 41. Mockler TC, Yu X, Shalitin D, Parikh D, Michael TP, Liou J, Huang J, Smith Z, Alonso JM, Ecker JR et al (2004) Regulation of flowering time in Arabidopsis by K homology domain proteins. Proc Natl Acad Sci USA 101: 12759–12764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Balasubramanian S, Sureshkumar S, Lempe J, Weigel D (2006) Potent induction of Arabidopsis thaliana flowering by elevated growth temperature. PLoS Genet 2: e106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yoo SK, Chung KS, Kim J, Lee JH, Hong SM, Yoo SJ, Yoo SY, Lee JS, Ahn JH (2005) CONSTANS activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 through FLOWERING LOCUS T to promote flowering in Arabidopsis . Plant Physiol 139: 770–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fowler S, Lee K, Onouchi H, Samach A, Richardson K, Morris B, Coupland G, Putterill J (1999) GIGANTEA: a circadian clock‐controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane‐spanning domains. EMBO J 18: 4679–4688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R et al (2003) Genome‐wide insertional mutagenesis of Arabidopsis thaliana . Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- 46. Schonrock N, Bouveret R, Leroy O, Borghi L, Kohler C, Gruissem W, Hennig L (2006) Polycomb‐group proteins repress the floral activator AGL19 in the FLC‐independent vernalization pathway. Genes Dev 20: 1667–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Somers D, Sharrock R, Tepperman J, Quail P (1991) The hy3 long hypocotyl mutant of Arabidopsis is deficient in phytochrome B. Plant Cell 3: 1263–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Friedrichsen DM, Nemhauser J, Muramitsu T, Maloof JN, Alonso J, Ecker JR, Furuya M, Chory J (2002) Three redundant brassinosteroid early response genes encode putative bHLH transcription factors required for normal growth. Genetics 162: 1445–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Weigel D, Ahn JH, Blazquez MA, Borevitz JO, Christensen SK, Fankhauser C, Ferrandiz C, Kardailsky I, Malancharuvil EJ, Neff MM et al (2000) Activation tagging in Arabidopsis . Plant Physiol 122: 1003–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou JM (2008) Firefly luciferase complementation imaging assay for protein‐protein interactions in plants. Plant Physiol 146: 368–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yu X, Shalitin D, Liu X, Maymon M, Klejnot J, Yang H, Lopez J, Zhao X, Bendehakkalu KT, Lin C (2007) Derepression of the NC80 motif is critical for the photoactivation of Arabidopsis CRY2. Proc Natl Acad Sci USA 104: 7289–7294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bai MY, Zhang LY, Gampala SS, Zhu SW, Song WY, Chong K, Wang ZY (2007) Functions of OsBZR1 and 14‐3‐3 proteins in brassinosteroid signaling in rice. Proc Natl Acad Sci USA 104: 13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Review Process File


