Abstract
The generation of big data has enabled systems-level dissections into the mechanisms of cardiovascular pathology. Integration of genetic, proteomic, and pathophysiological variables across platforms and laboratories fosters discoveries through multidisciplinary investigations and minimizes unnecessary redundancy in research efforts. The Mouse Heart Attack Research Tool (mHART) consolidates a large data set of over 10 yr of experiments from a single laboratory for cardiovascular investigators to generate novel hypotheses and identify new predictive markers of progressive left ventricular remodeling after myocardial infarction (MI) in mice. We designed the mHART REDCap database using our own data to integrate cardiovascular community participation. We generated physiological, biochemical, cellular, and proteomic outputs from plasma and left ventricles obtained from post-MI and no-MI (naïve) control groups. We included both male and female mice ranging in age from 3 to 36 mo old. After variable collection, data underwent quality assessment for data curation (e.g., eliminate technical errors, check for completeness, remove duplicates, and define terms). Currently, mHART 1.0 contains >888,000 data points and includes results from >2,100 unique mice. Database performance was tested, and an example is provided to illustrate database utility. This report explains how the first version of the mHART database was established and provides researchers with a standard framework to aid in the integration of their data into our database or in the development of a similar database.
NEW & NOTEWORTHY The Mouse Heart Attack Research Tool combines >888,000 cardiovascular data points from >2,100 mice. We provide this large data set as a REDCap database to generate novel hypotheses and identify new predictive markers of adverse left ventricular remodeling following myocardial infarction in mice and provide examples of use. The Mouse Heart Attack Research Tool is the first database of this size that integrates data sets across platforms that include genomic, proteomic, histological, and physiological data.
Keywords: big data, bioinformatics, cardiovascular disease, myocardial infarction, proteomics
INTRODUCTION
Big data analysis provides a powerful means to facilitate discoveries in science and medicine. Despite efforts by funding agencies and researchers, there remains a significant amount of information not yet captured in digital form (51). Data stored in paper form are not easily accessible or searchable through networks, highlighting a significant source of inefficiency. To capitalize on data already collected and avoid redundancy, researchers need to leverage data-driven strategies to capture the value of existing data and move the research field forward.
A major research focus of the Mississippi Center for Heart Research is to understand the interplay between inflammatory and extracellular matrix (ECM) responses that coordinate infarct scar formation and remodeling of the left ventricle (LV) after myocardial infarction (MI). In addition to physiological variables, we routinely measure biochemical, cellular, genomic, and proteomic outputs. Along with the LV, we collect plasma and individual cell types isolated from the post-MI infarct. We have published on roles of matrix metalloproteinases (MMPs), neutrophils, macrophages, and cardiac fibroblasts as well as the influence of risk factors and comorbidities (e.g., aging, obesity, and chronic inflammation) on LV remodeling alone or after MI (2–6, 9–11, 13, 14, 16, 17, 20, 21, 23, 24, 26–31, 35–37, 39, 41–44, 46–49).
Historically, we have analyzed our results as discrete units, with each output saved in distinct Excel files based on individual projects and individual data types. With the exception of basic regression analysis, variables have not been cross-examined within and across projects. Once a project was published, the information was stored but not further used beyond assessment of reproducibility and variation of new projects in development. To harness our big data, we developed the Mouse Heart Attack Research Tool (mHART) as a means to consolidate results into an easily searchable format amenable to future integration of bioinformatic tools that facilitate data visualization. By consolidating results, we increase study power because of increased sample sizes. Establishing the database is in line with the National Centre for the Replacement, Refinement, and Reduction of Animals in Research recommendations to reduce, refine, or replace animal use in research (34).
The major goals of mHART were to 1) compile our previously collected data into one central database; 2) facilitate broad use of big data by making results discoverable, accessible, and citable; 3) support a data ecosystem that accelerates discovery as part of a digital enterprise; and 4) cross-examine data sets to identify novel patterns, explore new cellular interactions, and identify novel targets that prevent or predict development of heart failure (Fig. 1).
Fig. 1.
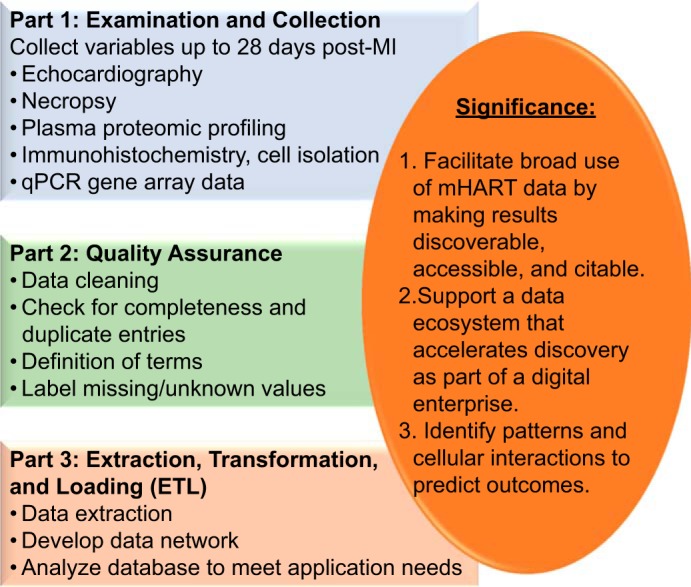
The overarching objective of the Mouse Heart Attack Research Tool (mHART) database. Our goal was to establish a tool that houses data from >10 yr of experiments to generate novel hypotheses and identify new predictive markers of adverse remodeling of the left ventricle following myocardial infarction. qPCR, quantitative PCR.
The development of the mHART database is being conducted over two phases. Phase I (version 1.0) included compiling data from echocardiography, necropsy, immunohistochemistry for macrophage and neutrophil numbers, histological assessment for collagen using picrosirius red staining and for endothelial cells using Griffonia simplicifolia lectin-1 staining, plasma proteomics, and gene expression data (cytokine and ECM arrays). Details on phase I are reported in the present article. Phase II (under development) will integrate bioinformatics tools to facilitate data exploration and interpretation.
The big data revolution relies not only on the exponential growth of data collection but also on improved statistical and computational methods to analyze results paired with bioinformatics tools to harness the data. Creative approaches to visualize data are integral to the process of understanding knowledge of a complex system. Establishing the mHART tool provides a means to improve our ability to obtain insights that otherwise remain obscured. This article explains how this database was established and provides researchers with a standard framework to aid in the integration of their data into our database or in the development of a similar database.
MATERIALS AND METHODS
Data Comprising mHART
The mHART database consists of data from 26 publications with 2 overarching themes. The major focus of the projects incorporated into the mHART tool was to understand the interplay between inflammatory and ECM responses during post-MI cardiac wound repair on its own or superimposed with risk factors of cardiovascular disease (obesity and aging); Table 1 shows the project data collected. All MI surgeries were performed by permanent occlusion of the left coronary artery as previously reported (50). Table 2 shows an example of MI characteristics, including infarct area, necropsy variables, and cardiac physiology measurements for day 0 (naïve, no-MI unoperated control) through day 7 MI time points for C57BL/6J wild-type mice. Our center has published dozens of articles on the roles of MMPs, neutrophils, macrophages, and cardiac fibroblasts as well as the influence of aging, diet, or chronic inflammation on post-MI LV remodeling (2–6, 9–11, 13, 14, 16, 17, 20, 21, 23, 24, 26–31, 35–37, 39, 41–44, 46–49). The two overarching themes are summarized below.
Table 1.
Project data summary
| Project Description | Echocardiogram | Necropsy | Plasma | Histology | Gene Array |
|---|---|---|---|---|---|
| Myocardial infarction in wild-type animals or genetic mutants with or without inhibitor treatments (3, 6, 9, 13, 14, 16, 23, 24, 29, 30, 32, 35, 37, 40, 47–49) | 922 | 470 | 285 | 332 | 312 |
| Myocardial infarction superimposed on cardiovascular disease risk factors (aging, obesity, and chronic inflammation) (2, 5, 12–14, 21, 26, 27, 39, 43, 45) | 753 | 642 | 590 | 51 | 302 |
Values are numbers of individual data sets.
Table 2.
MI characteristics, including infarct area, necropsy variables, and cardiac physiology measurements, for wild-type mice at ages of 3–14 mo at day 0 (no MI) and days 1, 3, 5, and 7 post-MI
| Day 0 (No MI) | Day 1 MI | Day 3 MI | Day 5 MI | Day 7 MI | |
|---|---|---|---|---|---|
| n | 193 | 82 | 72 | 92 | 150 |
| Infarct area, % | NA | 58 ± 1* | 56 ± 1* | 48 ± 1*†‡ | 46 ± 1*†‡ |
| Body wt, mg | 25 ± 0 | 23 ± 0* | 23 ± 0* | 23 ± 0* | 23 ± 0* |
| Left ventricular mass, mg | 78 ± 0 | 83 ± 0* | 107 ± 1*† | 98 ± 0*†‡ | 100 ± 0*†‡§ |
| Right ventricular mass, mg | 17 ± 0 | 16 ± 0 | 18 ± 0 | 18 ± 0 | 23 ± 0*†‡§ |
| Lung wet weight, mg | 95 ± 0 | 137 ± 1* | 161 ± 1*† | 158 ± 1*†‡ | 207 ± 1*†‡§ |
| Heart rate, beats/min | 462 ± 0 | 461 ± 1 | 462 ± 1 | 466 ± 1 | 479 ± 0 |
| Fractional shortening, % | 35 ± 0 | 8 ± 0* | 8 ± 0* | 7 ± 0*†‡ | 6 ± 0*†‡§ |
| Ejection fraction, % | 65 ± 0 | 16 ± 0* | 16 ± 0* | 15 ± 0*†‡ | 13 ± 0*†‡§ |
Data are means ± SE; n, number of data elements. MI, myocardial infarction; NA, not applicable.
P < 0.05 vs. day 0;
P < 0.05 vs. day 1;
P < 0.05 vs. day 3;
P < 0.05 vs. day 5.
MI in wild-type C57BL/6J or genetic mutants with or without inhibitor treatments.
Our team leads efforts to understand how MMPs serve as upstream signaling molecules to coordinate remodeling. While many groups measure MMPs as output signals, we are one of a few groups that investigate the downstream effects after MMP activation. To date, we have published results evaluating the roles of MMP-7, MMP-9, MMP-12, and MMP-28 during remodeling of the LV post-MI (2, 7–9, 15, 16, 23, 24, 27, 29, 32, 35, 38, 45, 47–49). We used genetic mutants and pharmaceutical reagents to evaluate whether or not individual MMPs and their downstream targets are involved in MI remodeling. Our studies elucidate MMP roles, particularly with regard to inflammatory and ECM signaling pathways.
MI superimposed on cardiovascular disease risk factors (aging, obesity, or chronic inflammation).
Aging, obesity, and chronic inflammation are major risk factors and comorbidities for cardiovascular disease. We have extensively evaluated these particular risk factors as well as during the MI response (2, 5, 6, 13, 14, 21, 26, 27, 33, 38, 39, 43, 45). The results from our aging studies assign MMPs as mediators of the inflammaging profile by directly and indirectly modifying leukocyte polarization. Using a mouse model for obesity, we have shown reduced post-MI survival but paradoxical improved cardiac function in the survivors, an effect mediated by decreased inflammation, ECM accumulation, and neovascularization in the infarct region (13). These results indicate a dual role of obesity in the post-MI response. Additionally, in a chronic inflammatory state, MMP-9 exacerbated post-MI inflammation by accelerating macrophage-mediated inflammation (5).
Database Workflow
The database was constructed using three main steps (Table 3). The first step was data collection, the second step was quality assessment, and the third step was extraction, transformation, and loading of the data.
Table 3.
Schematic of the workflow to generate the Mouse Heart Attack Research Tool database
| Step | Potential Pitfalls | Solutions |
|---|---|---|
| 1. Data collection | • Interobserver variability • Missing values • Technical advancement |
• Standard operating procedures • Data collection templates |
| 2. Quality assessment | • Data inconsistency • Data inaccuracy |
• Quality assurance checks • Data curation |
| 3. Extraction, transformation, and loading | • Coding errors • Formatting errors • Sharing/open access |
• Validation requirement • Provide restricted access |
Data Collection
Data were collected from projects starting in 2007. We obtained data from seven different mouse genotypes, from both male and female mice, ranging from 3 to 36 mo (Fig. 2, A–C). All genotypes were on the C57BL6/J background strain. We collected temporal and spatial results from day 0 unoperated, no-MI, control mice to day 28 post-MI (Fig. 2D). Each mouse received a unique identification number when entered into the colony, which allowed examination of variables across platforms. All mice were obtained from the Jackson Laboratory or from in-house breeding colonies, and all animal use was approved by the local institutional animal care and use committee. Data were collected in a systematic and uniform manner, with investigators blinded to groups during acquisition and analysis. Below, we discuss three potential pitfalls one may encounter during data collection and how we handled these issues.
Fig. 2.
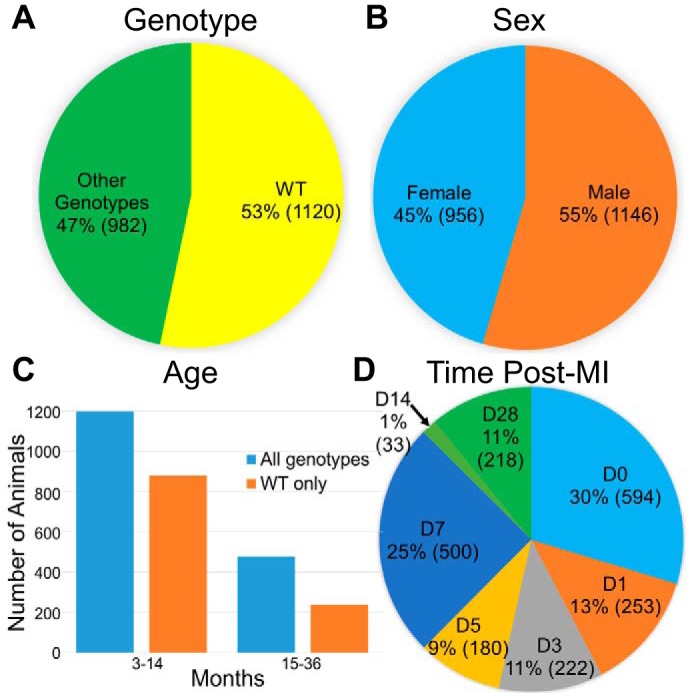
Summary of data included in Mouse Heart Attack Research Tool (mHART) 1.0. The mHART 1.0 contains results from both wild-type (WT) and genetically modified mice (A), male and female mice (B), 3- to 36-mo-old mice (C), and mice evaluated at days 0–28 after myocardial infarction (MI; D). D, day.
Potential pitfall 1: interobserver variability.
During database curation, technical aspects such as site-to-site variability in data acquisition, interpretation, and analysis must be standardized. Data were only included in mHART after being reviewed for inconsistencies in acquisition, interpretation, and analysis.
Although our laboratory is a one-site facility, multiple investigators within our team worked on different projects. One way our laboratory prevented investigator-to-investigator variability was by developing standard operating procedures (SOPs) that were reviewed annually by all members of the team to prevent deviations in protocol (50). The success of our SOPs is evident by the reproducibility of experiments within our own center and by the fact that other laboratories from around the country ask to use our SOPs within their laboratory and yield results that are similar to ours. All SOPs are readily available and can be obtained by contacting the corresponding author. Data collected after 2007 were selected as the starting point because SOPs were routinely in use by that time for all techniques to ensure uniformity in acquisition and analysis.
A further source of variability was a lack of consistency in language across users, making it difficult to interpret data across projects. For example, echocardiography variables were labeled differently (e.g., interventricular septum wall thickness and septal wall thickness) in the Excel worksheets used by various team members. This was primarily because of the evolution from one ultrasound instrument to another across time and the development of abbreviations to shorten the width of Excel spreadsheets. Small differences in terminology can make queries of the database difficult and inefficient. Because each project was performed by a separate individual and therefore potentially had its own layout and terminology nuances, it was important to convert all into a single template. To remove variation in terminology, standard names or abbreviations were given to each data point.
An additional source for potential variability was differences in data analysis. For example, gene array data from each project were inspected to confirm gene data were normalized to the same housekeeping gene. Hypoxanthine guanine phosphoribosyl transferase 1 (Hprt1) was chosen because, of five reference genes [β-glucuronidase (GusB), heat shock protein 90ab1 (Hsp90ab1), β-actin (Actb), and and glyceraldehyde 3-phosphate dehydrogenase (Gapdh)] Gapdh], Hprt1 is the only gene that shows no significant change in expression after MI (24). Thus, projects using other genes (e.g., Gapdh or all five reference genes) were reanalyzed for consistency.
Potential pitfall 2: missing values.
Missing data (data that should have been collected but were not) are an unfortunate complication when data are compiled and consolidated among a variety of projects. If not organized from the start and continuously monitored, data have the potential to be incomplete, noisy, and inconsistent. When preprocessing data sets, it is imperative to fill in missing values when possible and correct inconsistencies to improve statistical power, remove bias in parameter estimations, and increase sample representativeness. Missing data make results difficult to interpret (18). If data points are truly missing, accounting for why values are missing is important for others to assess data quality. The best way to control for missing data is to establish uniform data collection practices across studies. To ensure all data parameters were collected for each project to the fullest extent possible, we developed data templates that users filled out as they acquired and analyzed results. This ensured that data parameters such as age, birthdate, and sex were collected for each animal. We also duplicated information provided across data templates when possible as a backup plan to increase the chance to obtain any missing information.
Potential pitfall 3: technological advancement.
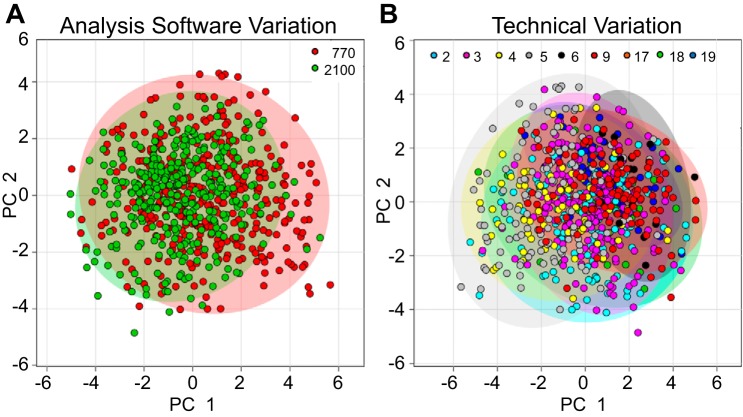
With time comes advancement of technology; one example of this is technological development in echocardiography ultrasound instruments. In 2012, we transitioned from the Vevo 770 (Fujifilm VisualSonics) to the Vevo 2100 ultrasound instrument. To evaluate whether images were consistent across instrument platforms, principal component analysis (PCA) was performed. PCA is an unsupervised learning method, similar to clustering, that compares two different data sets and locates the directions that best explain the variance in a data set. For example, if one instrument consistently overestimated cardiac volumes, PCA would identify this pattern. Comparing the components identified by PCA allowed us to determine if echocardiography analysis software contributed variability within the data. Based on strong overlap between the 95% confidence ellipses around the clusters, there was no significant variation between image analysis software platforms (Fig. 3A).
Fig. 3.
Principal component analysis of echocardiography data showed low analysis software or technical variation. A: we tested technical variation using VisualSonics Vevo 770 vs. Vevo 2100. B: laboratory members were assigned a unique identifying number (colored numbers at the top of each image) to assess interperson variation. Principal component analysis showed low variation between analysis software and reader, as indicated by the 95% confidence ellipses around the clusters.
Quality Assessment
Data curation and quality assurance checks are necessary to determine reliability and validity. All data underwent at least two quality assessments to certify precision and accuracy, one quality assessment at the initial time of collection and analysis and another quality assessment at the time of inclusion in the database. By ensuring data infrastructure was stable and easily accessible, we were able to derive new data-driven hypotheses. Two potential pitfalls one may encounter during quality assessment are data inconsistency and data inaccuracy.
Potential pitfall 1: data inconsistency.
Data first underwent internal checks for consistency of measurements within groups, and any potential outliers were reanalyzed by two or more investigators in a blinded manner. By using more than one investigator for the reanalysis, we could certify that the data generated were accurate and minimized error potential. This assured all data generated could be combined and analyzed across projects. If quality could not be confirmed, data were not included; a total of 52 data points were removed for this reason. All 52 data points were echocardiography images of insufficient acquisition quality.
Of the results acquired, echocardiography data had the greatest potential for issues with quality resulting from potential observer-related differences in both acquiring and analyzing images. At the same time, echocardiography is of crucial importance to assess cardiac physiology to determine LV size, shape, and function. Echocardiography data within the mHART database were read by nine laboratory members who underwent quarterly calibration and were each assigned a unique identifying laboratory member number. During calibration assessments, each member imaged the same mouse and read images of all other members to assess group variation. Based on coefficient of variation calculations, any members who were not within range (<10% for day 0 images) were retrained. We performed PCA on echocardiography data within mHART to evaluate whether images were consistent among readers. There was no significant variation between readers as shown by the strong overlap between confidence ellipses around the clusters (Fig. 3B).
Potential pitfall 2: data inaccuracy.
The data also underwent multiple steps of quality control. If imperfections were found, data underwent an additional round of curation steps to ensure quality control and data validation for consistency. One way data underwent curation was ensuring all gene array data were set to the same threshold value. Data sets that did not use the same threshold were reanalyzed to ensure data were comparable across projects. In addition, poor image acquisition can affect the accuracy of echocardiography data. During M-mode image acquisition, if the LV was not centered within the frame or the cursor was not perpendicular to the anterior and posterior walls the echocardiography data for this animal were excluded as this would result in overestimated LV volumes. Only four animals were excluded, illustrating the strength of the training protocol and the vigilance in continual performance evaluation and calibration.
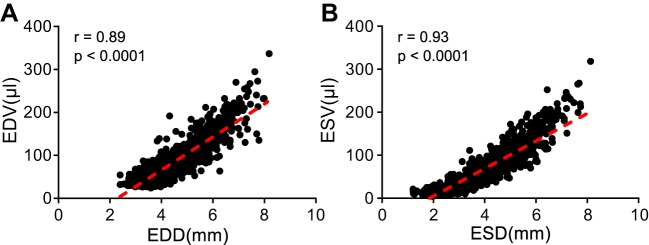
The generally accepted practice for parasternal long-axis B-mode image acquisition is to line up the apex and aorta horizontally. However, even when the apex and aorta are lined up, a significant variation in image quality can exist because of inconsistency in the tilt angle of the probe or inclusion of the mitral annulus in the image. To address this complication and attain anatomically correct images, the LV should be centered in the frame during parasternal long-axis B-mode image acquisition. Proper short-axis views require that the M-mode cursor cuts the LV anterior and posterior wall perpendicularly. Linear regression analysis showed an excellent fit for both diastolic (r = 0.89) and systolic (r = 0.93; Fig. 4) measurements, indicating measurements taken at the short-axis M-mode were congruent with those taken at the long-axis B-mode. By ensuring all images acquired closely adhere to this standard, we were able to incorporate strong cardiac physiology data within our database.
Fig. 4.
Short- and long-axis measurements were consistent in the same mice. Accuracy of each measurement was determined by comparing long-axis volumes with short-axis dimensions. Linear regression analysis showed good correlation for both (A) diastolic (r = 0.89, P < 0.0001) and (B) systolic (r = 0.93, P < 0.0001) measurements, indicating values determined by long axis were in line with changes observed in short axis. EDD, end-diastolic dimensions; EDV, end-diastolic volume; ESD, end-systolic dimensions; ESV, end-systolic volume.
Extraction, Transformation, and Loading
After examining data sources and formats, data sets were uploaded into REDCap (https://projectREDCap.org/). A REDCap database was created to work as a user-friendly interface on which different instruments or data libraries could readily be added to accommodate the different data types collected. A REDCap database is also amenable to future revisions to incorporate new types of data. The data were organized to facilitate sorting results, for example, by experiment, laboratory member, project, echocardiogram acquisition day, genotype, sex, or age. This setup resulted in an accessible and searchable database; furthermore, security was ensured by setting user authorization and login requirements. One potential pitfall during extraction, transformation, and loading is coding or formatting errors.
Potential pitfall 1: coding/formatting error.
The format for the REDCap database is already established, which was a strength since no coding was required on our part. To upload data into REDCap, a data dictionary must be generated. The data dictionary is a description of all the field (variable) names, field types, and formatting (i.e., decimal place). REDCap has a tool called Online Designer that will generate the data dictionary, which minimizes errors. In the Online Designer, you can choose to have a validation requirement. This serves as a check system to ensure the data uploaded are in the correct format. Once the data dictionary is generated, you can upload the data into REDCap as a csv file. A strength of using the REDCap program is that any changes made to the database are tracked. This allows one to backtrack and catch errors that may have been generated during the upload. We followed this process for data upload.
Potential pitfall 2: sharing/open access.
A drawback of using the REDCap format for the database is that, because REDCap was developed to contain human data, sharing the database in open format is not allowed. The database owner/administrator can, however, provide access to outside users. In addition, the database is easily downloaded as an Excel csv file, or as SAS or STATA statistical software files, which provide easy mechanisms to share data from the database. Future data-sharing tools and integration of bioinformatics software are currently being developed. Access to the REDCap format of the database will be controlled to ensure all data within mHART passes quality check points.
RESULTS
Assembly of a Cardiac Remodeling Phenotype
We compiled >888,000 data points for >2,100 unique mouse identification numbers, including information on >100 autopsies on nonsurviving mice. The database went into production on February 8, 2018, and is available for others to use. In principle, integrating an immense amount of data into the mHART 1.0 tool yielded the potential for overly complex analyses. We avoided this problem by using efficient database queries. We tested the strength of the data by evaluating the overlap among analyses. For example, 59% of the data within mHART have at least two groups of variable measurements (echocardiography, gene analysis, or plasma proteomics) measured for the same mouse (Fig. 5). This allows the user to develop a complete report that encompasses the multidimensionality of the data.
Fig. 5.
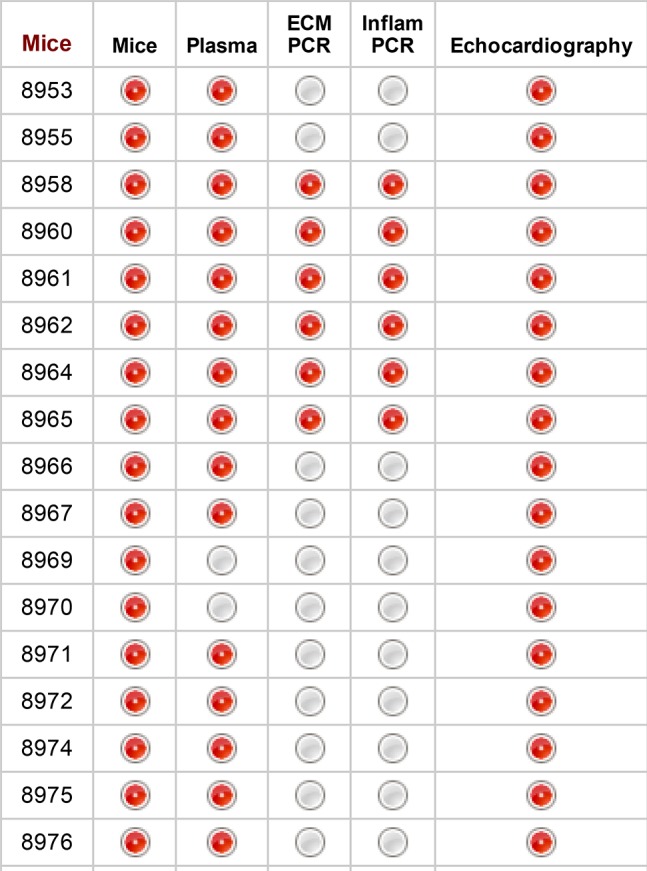
Data within the database show strong overlap. Image generated from REDCap illustrating strong overlap among variables acquired from the same mouse. Red circles indicate data have been collected for the instrument. ECM, extracellular matrix.
The mHART database is user friendly and incorporates tools, such as a comparison tool that allows the user to readily compare two samples within the database. Because the REDCap platform was used for data development, querying the database is quick and easy. Average search time is 1–2 s, and the time to download a specific data set takes <2 min. There is also an option to view the data set before download to ensure the user wants all of the data selected.
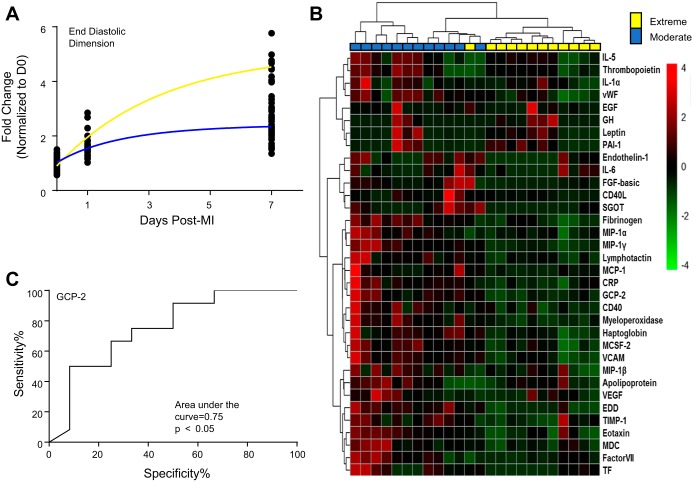
To assess the analytical performance of the underlying database, we analyzed the echocardiogram and plasma data sets. Clustering of the echocardiography variables identified two distinct patterns in the day 7 post-MI end-diastolic dimensions that indicated moderate and extreme LV dilation responses (Fig. 6A). This was not because of sex or other physiological attributes such as heart rate or infarct size. Analysis of 52 plasma analytes from these same mice identified potential drivers that correlated with dilation outcome (Fig. 6B). Of the 52 analytes assessed in the panel, 34 analytes were significantly different between dilation groups (all P < 0.05). Many of the proteins evaluated, including interleukin-6, monocyte chemotactic protein-1, and tissue inhibitor of metalloproteinase-1, are known influencers of LV remodeling (16, 23, 35). Receiver operating characteristic analysis of the 34 analytes identified granulocyte chemotactic protein-2 as being the strongest marker for increased dilation, since it had the largest area under the curve value of the 34 analytes (Fig. 6C). Overexpression of granulocyte chemotactic protein-2 is beneficial post-MI, since it stimulates angiogenesis (19). This could explain why lower granulocyte chemotactic protein-2 plasma protein concentration correlated with the extreme dilation phenotype. Our example illustrates the strength and the advantage of the mHART database in identifying potential biomarkers or targets.
Fig. 6.
An example that demonstrates the use of the Mouse Heart Attack Research Tool (mHART). A: by querying the mHART database, we identified two distinct left ventricle (LV) dilation response groups, based on their end-diastolic dimensions. Blue line, moderate response; yellow line, extreme response. B: analysis of 52 plasma proteins identified potential drivers that correlate with dilation outcome. n = 12 mice/group. C: receiver operating characteristic (ROC) analysis of the 34 analytes identified granulocyte chemotactic protein (GCP)-2 as having the largest area under the curve for increased dilation. D0, day 0; MI, myocardial infarction.
DISCUSSION
Our goal was to establish a database of past results to generate novel hypotheses and identify new predictive markers of adverse remodeling of the LV post-MI. The consolidation of this large data set into one format was not previously available. We designed the mHART database using our own data, with plans to incorporate data from additional laboratories using the same data quality (data inclusion and exclusion criteria) and check points described in materials and methods. In addition to the results currently uploaded, we have data on immunoblotting from targeted analyses, glycoproteomic profiles, and gene evaluations from macrophages and fibroblasts isolated from post-MI LV that characterize cell polarization phenotypes (32, 37, 47, 48). The next iteration of this tool will incorporate those results following the same protocol as well as additional studies published in the last 18 mo that have all been performed under consistent SOPs (9, 24, 26, 30, 39, 40). Now that we have data upload templates developed, future import will be routine.
The National Institute of Aging and the National Heart, Lung, and Blood Institute have recently released new guidelines to implement rigor and transparency in research. Included in these guidelines was the principle of data and material sharing. While data sets for omic studies are usually deposited in public repositories and are often a requirement for manuscript acceptance, other types of data sets (especially physiological variables) are rarely shared. In a recent guidelines article on cardiac physiology published by the American Journal of Physiology: Heart and Circulatory Physiology, the majority of publications did not provide sufficient details on echocardiography methods (25). Similar guidelines have recently been published to cover antibody use and ischemia and infarction animal models (1, 22). The mHART database pioneers systematic data sharing for cardiovascular studies and helps to promote rigor and transparency in cardiovascular research. Querying the database will provide a way to develop novel hypotheses, identify new predictive markers of adverse LV remodeling, and move translational research forward. In addition, the mHART database will provide a means to reduce, refine, and replace animal use.
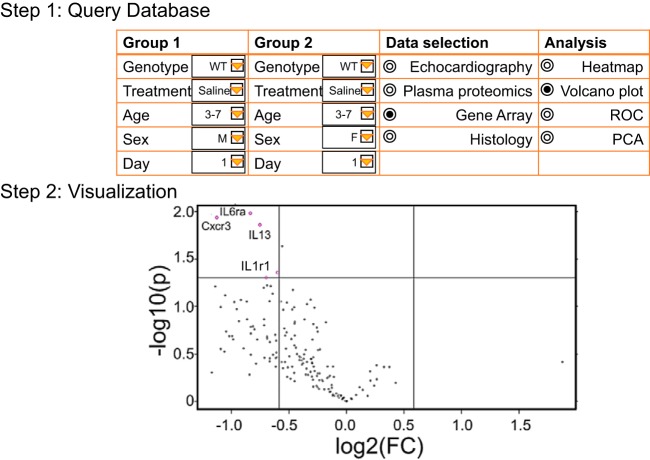
To encourage research community engagement, our database is currently available for external investigators who request to have access to the data present in mHART. Any investigator wishing access to the database is encouraged to contact the corresponding author for access. Future integration of bioinformatics visualization tools (Fig. 7) will be part of the next phase. We invite others who have results from their own post-MI studies in mice that measured remodeling variables to contact us about merging their data sets within our database. This will provide assessment of interlaboratory evaluations and further strengthen this resource. In conclusion, we have assembled the mHART 1.0 database to combine results that span many dimensions into one collective research tool.
Fig. 7.
Our future goal for the Mouse Heart Attack Research Tool (mHART) 1.0 is to integrate bioinformatics software for easier data visualization. Using the mHART database, users will be able to select their groups, data sets, and analysis. This will allow the user to visualize data and will facilitate in developing new hypotheses. WT, wild type; ROC, receiver operating characteristic; PCA, principal component analysis; FC, fold change.
GRANTS
This work was supported by American Heart Association Grant 14SDG18860050 to L. E. de Castro Brás and 15SDG22930009 to Y. Ma; by National Institutes of Health Grants AT-006704 and HL-132989 to G. V. Halade, HL-075360, HL-129823, and GM-114833 to M. L. Lindsey, and HL-051971, GM-104357, and GM-115428; and by Biomedical Laboratory Research and Development Service of the Veterans Affairs Office of Research and Development Award 5I01BX000505 to M. L. Lindsey and IK2BX003922 to K. Y. DeLeon-Pennell.
DISCLAIMERS
The views expressed in this manuscript are those of the authors and do not necessarily represent the views of the National Institutes of Health, the United States Department of Health and Human Services, the American Heart Association, or the United States Department of Veterans Affairs.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
K.Y.D.-P., R.P.I., Y.M., A.Y., R.Z., Y.A.C., P.C., A.K., C.C., E.R.F., G.V.H., L.E.d.C.B., and M.L.L. performed experiments; K.Y.D.-P., R.P.I., Y.M., A.Y., R.Z., Y.A.C., P.C., A.K., C.C., E.R.F., G.V.H., L.E.d.C.B., and M.L.L. analyzed data; K.Y.D.-P., R.P.I., Y.M., A.Y., R.Z., Y.A.C., G.V.H., L.E.d.C.B., and M.L.L. interpreted results of experiments; K.Y.D.-P. and M.L.L. prepared figures; K.Y.D.-P. and M.L.L. drafted manuscript; K.Y.D.-P., R.P.I., Y.M., A.Y., R.Z., Y.A.C., P.C., A.K., C.C., E.R.F., G.V.H., L.E.d.C.B., and M.L.L. edited and revised manuscript; K.Y.D.-P., R.P.I., Y.M., A.Y., R.Z., Y.A.C., P.C., A.K., C.C., E.R.F., G.V.H., L.E.d.C.B., and M.L.L. approved final version of manuscript; M.L.L. conceived and designed research.
ACKNOWLEDGMENTS
We thank De’Aries Shannon and Jeffrey Henry for help in gathering data for the mHART database.
REFERENCES
- 1.Brooks HL, Lindsey ML. Guidelines for authors and reviewers on antibody use in physiology studies. Am J Physiol Heart Circ Physiol 314: H724–H732, 2018. doi: 10.1152/ajpheart.00512.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiao YA, Ramirez TA, Zamilpa R, Okoronkwo SM, Dai Q, Zhang J, Jin YF, Lindsey ML. Matrix metalloproteinase-9 deletion attenuates myocardial fibrosis and diastolic dysfunction in ageing mice. Cardiovasc Res 96: 444–455, 2012. doi: 10.1093/cvr/cvs275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Castro Brás LE, Cates CA, DeLeon-Pennell KY, Ma Y, Iyer RP, Halade GV, Yabluchanskiy A, Fields GB, Weintraub ST, Lindsey ML. Citrate synthase is a novel in vivo matrix metalloproteinase-9 substrate that regulates mitochondrial function in the postmyocardial infarction left ventricle. Antioxid Redox Signal 21: 1974–1985, 2014. doi: 10.1089/ars.2013.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deleon-Pennell KY, Altara R, Yabluchanskiy A, Modesti A, Lindsey ML. The circular relationship between matrix metalloproteinase-9 and inflammation following myocardial infarction. IUBMB Life 67: 611–618, 2015. doi: 10.1002/iub.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deleon-Pennell KY, Bras LE, Lindsey ML. Circulating lipopolysaccharide resets cardiac homeostasis in mice through a matrix metalloproteinase-9 dependent mechanism. Physiol Rep 1: e00079, 2013. doi: 10.1002/phy2.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeLeon-Pennell KY, de Castro Brás LE, Iyer RP, Bratton DR, Jin YF, Ripplinger CM, Lindsey ML. P. gingivalis lipopolysaccharide intensifies inflammation post-myocardial infarction through matrix metalloproteinase-9. J Mol Cell Cardiol 76: 218–226, 2014. doi: 10.1016/j.yjmcc.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeLeon-Pennell KY, Iyer RP, Ero OK, Cates CA, Flynn ER, Cannon PL, Jung M, Shannon D, Garrett MR, Buchanan W, Hall ME, Ma Y, Lindsey ML. Periodontal-induced chronic inflammation triggers macrophage secretion of Ccl12 to inhibit fibroblast-mediated cardiac wound healing. JCI Insight 2: 94207, 2017. doi: 10.1172/jci.insight.94207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeLeon-Pennell KY, Meschiari CA, Jung M, Lindsey ML. Matrix metalloproteinases in myocardial infarction and heart failure. Prog Mol Biol Transl Sci 147: 75–100, 2017. doi: 10.1016/bs.pmbts.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeLeon-Pennell KY, Tian Y, Zhang B, Cates CA, Iyer RP, Cannon P, Shah P, Aiyetan P, Halade GV, Ma Y, Flynn E, Zhang Z, Jin YF, Zhang H, Lindsey ML. cd36 is a matrix metalloproteinase-9 substrate that stimulates neutrophil apoptosis and removal during cardiac remodeling. Circ Cardiovasc Genet 9: 14–25, 2016. doi: 10.1161/CIRCGENETICS.115.001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeLeon KY, de Castro Brás LE, Lange RA, Lindsey ML. Extracellular matrix proteomics in cardiac ischemia/reperfusion: the search is on. Circulation 125: 746–748, 2012. doi: 10.1161/CIRCULATIONAHA.111.086835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeLeon KY, Yabluchankiy A, Winniford MD, Lange RA, Chilton RJ, Lindsey ML. Modifying matrix remodeling to prevent heart failure. In: The Pathogenesis of Congestive Heart Failure, edited by Li RK, Weisel RD. New York: Elsevier, 2014. [Google Scholar]
- 12.Ducharme A, Frantz S, Aikawa M, Rabkin E, Lindsey M, Rohde LE, Schoen FJ, Kelly RA, Werb Z, Libby P, Lee RT. Targeted deletion of matrix metalloproteinase-9 attenuates left ventricular enlargement and collagen accumulation after experimental myocardial infarction. J Clin Invest 106: 55–62, 2000. doi: 10.1172/JCI8768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Halade GV, Ma Y, Ramirez TA, Zhang J, Dai Q, Hensler JG, Lopez EF, Ghasemi O, Jin YF, Lindsey ML. Reduced BDNF attenuates inflammation and angiogenesis to improve survival and cardiac function following myocardial infarction in mice. Am J Physiol Heart Circ Physiol 305: H1830–H1842, 2013. doi: 10.1152/ajpheart.00224.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heaberlin JR, Ma Y, Zhang J, Ahuja SS, Lindsey ML, Halade GV. Obese and diabetic KKAy mice show increased mortality but improved cardiac function following myocardial infarction. Cardiovasc Pathol 22: 481–487, 2013. doi: 10.1016/j.carpath.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iyer RP, de Castro Brás LE, Patterson NL, Bhowmick M, Flynn ER, Asher M, Cannon PL, Deleon-Pennell KY, Fields GB, Lindsey ML. Early matrix metalloproteinase-9 inhibition post-myocardial infarction worsens cardiac dysfunction by delaying inflammation resolution. J Mol Cell Cardiol 100: 109–117, 2016. doi: 10.1016/j.yjmcc.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iyer RP, Patterson NL, Zouein FA, Ma Y, Dive V, de Castro Brás LE, Lindsey ML. Early matrix metalloproteinase-12 inhibition worsens post-myocardial infarction cardiac dysfunction by delaying inflammation resolution. Int J Cardiol 185: 198–208, 2015. doi: 10.1016/j.ijcard.2015.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jourdan-Lesaux C, Zhang J, Lindsey ML. Extracellular matrix roles during cardiac repair. Life Sci 87: 391–400, 2010. doi: 10.1016/j.lfs.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang H. The prevention and handling of the missing data. Korean J Anesthesiol 64: 402–406, 2013. doi: 10.4097/kjae.2013.64.5.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim SW, Lee DW, Yu LH, Zhang HZ, Kim CE, Kim JM, Park TH, Cha KS, Seo SY, Roh MS, Lee KC, Jung JS, Kim MH. Mesenchymal stem cells overexpressing GCP-2 improve heart function through enhanced angiogenic properties in a myocardial infarction model. Cardiovasc Res 95: 495–506, 2012. doi: 10.1093/cvr/cvs224. [DOI] [PubMed] [Google Scholar]
- 20.Lambert JM, Lopez EF, Lindsey ML. Macrophage roles following myocardial infarction. Int J Cardiol 130: 147–158, 2008. doi: 10.1016/j.ijcard.2008.04.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin J, Lopez EF, Jin Y, Van Remmen H, Bauch T, Han HC, Lindsey ML. Age-related cardiac muscle sarcopenia: Combining experimental and mathematical modeling to identify mechanisms. Exp Gerontol 43: 296–306, 2008. doi: 10.1016/j.exger.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindsey ML, Bolli R, Canty JM Jr, Du XJ, Frangogiannis NG, Frantz S, Gourdie RG, Holmes JW, Jones SP, Kloner RA, Lefer DJ, Liao R, Murphy E, Ping P, Przyklenk K, Recchia FA, Schwartz Longacre L, Ripplinger CM, Van Eyk JE, Heusch G. Guidelines for experimental models of myocardial ischemia and infarction. Am J Physiol Heart Circ Physiol 314: H812–H838, 2018. doi: 10.1152/ajpheart.00335.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindsey ML, Escobar GP, Mukherjee R, Goshorn DK, Sheats NJ, Bruce JA, Mains IM, Hendrick JK, Hewett KW, Gourdie RG, Matrisian LM, Spinale FG. Matrix metalloproteinase-7 affects connexin-43 levels, electrical conduction, and survival after myocardial infarction. Circulation 113: 2919–2928, 2006. doi: 10.1161/CIRCULATIONAHA.106.612960. [DOI] [PubMed] [Google Scholar]
- 24.Lindsey ML, Iyer RP, Zamilpa R, Yabluchanskiy A, DeLeon-Pennell KY, Hall ME, Kaplan A, Zouein FA, Bratton D, Flynn ER, Cannon PL, Tian Y, Jin YF, Lange RA, Tokmina-Roszyk D, Fields GB, de Castro Brás LE. A novel collagen matricryptin reduces left ventricular dilation post-myocardial infarction by promoting scar formation and angiogenesis. J Am Coll Cardiol 66: 1364–1374, 2015. doi: 10.1016/j.jacc.2015.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindsey ML, Kassiri Z, Virag JAI, de Castro Brás LE, Scherrer-Crosbie M. Guidelines for measuring cardiac physiology in mice. Am J Physiol Heart Circ Physiol 314: H733–H752, 2018. doi: 10.1152/ajpheart.00339.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma Y, Chiao YA, Clark R, Flynn ER, Yabluchanskiy A, Ghasemi O, Zouein F, Lindsey ML, Jin YF. Deriving a cardiac ageing signature to reveal MMP-9-dependent inflammatory signalling in senescence. Cardiovasc Res 106: 421–431, 2015. doi: 10.1093/cvr/cvv128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma Y, Chiao YA, Zhang J, Manicone AM, Jin YF, Lindsey ML. Matrix metalloproteinase-28 deletion amplifies inflammatory and extracellular matrix responses to cardiac aging. Microsc Microanal 18: 81–90, 2012. doi: 10.1017/S1431927611012220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma Y, de Castro Brás LE, Toba H, Iyer RP, Hall ME, Winniford MD, Lange RA, Tyagi SC, Lindsey ML. Myofibroblasts and the extracellular matrix network in post-myocardial infarction cardiac remodeling. Pflugers Arch 466: 1113–1127, 2014. doi: 10.1007/s00424-014-1463-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma Y, Halade GV, Zhang J, Ramirez TA, Levin D, Voorhees A, Jin YF, Han HC, Manicone AM, Lindsey ML. Matrix metalloproteinase-28 deletion exacerbates cardiac dysfunction and rupture after myocardial infarction in mice by inhibiting M2 macrophage activation. Circ Res 112: 675–688, 2013. doi: 10.1161/CIRCRESAHA.111.300502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma Y, Yabluchanskiy A, Iyer RP, Cannon PL, Flynn ER, Jung M, Henry J, Cates CA, Deleon-Pennell KY, Lindsey ML. Temporal neutrophil polarization following myocardial infarction. Cardiovasc Res 110: 51–61, 2016. doi: 10.1093/cvr/cvw024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma Y, Yabluchanskiy A, Lindsey ML. Neutrophil roles in left ventricular remodeling following myocardial infarction. Fibrogenesis Tissue Repair 6: 11, 2013. doi: 10.1186/1755-1536-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCurdy SM, Dai Q, Zhang J, Zamilpa R, Ramirez TA, Dayah T, Nguyen N, Jin YF, Bradshaw AD, Lindsey ML. SPARC mediates early extracellular matrix remodeling following myocardial infarction. Am J Physiol Heart Circ Physiol 301: H497–H505, 2011. doi: 10.1152/ajpheart.01070.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meschiari CA, Jung M, Iyer RP, Yabluchanskiy A, Toba H, Garrett MR, Lindsey ML. Macrophage overexpression of matrix metalloproteinase-9 in aged mice improves diastolic physiology and cardiac wound healing after myocardial infarction. Am J Physiol Heart Circ Physiol 314: H224–H235, 2018. doi: 10.1152/ajpheart.00453.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prescott MJ, Lidster K. Improving quality of science through better animal welfare: the NC3Rs strategy. Lab Anim (NY) 46: 152–156, 2017. doi: 10.1038/laban.1217. [DOI] [PubMed] [Google Scholar]
- 35.Ramirez TA, Iyer RP, Ghasemi O, Lopez EF, Levin DB, Zhang J, Zamilpa R, Chou YM, Jin YF, Lindsey ML. Aliskiren and valsartan mediate left ventricular remodeling post-myocardial infarction in mice through MMP-9 effects. J Mol Cell Cardiol 72: 326–335, 2014. doi: 10.1016/j.yjmcc.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Squires CE, Escobar GP, Payne JF, Leonardi RA, Goshorn DK, Sheats NJ, Mains IM, Mingoia JT, Flack EC, Lindsey ML. Altered fibroblast function following myocardial infarction. J Mol Cell Cardiol 39: 699–707, 2005. doi: 10.1016/j.yjmcc.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 37.Tian Y, Koganti T, Yao Z, Cannon P, Shah P, Pietrovito L, Modesti A, Aiyetan P, DeLeon-Pennell K, Ma Y, Halade GV, Hicks C, Zhang H, Lindsey ML. Cardiac extracellular proteome profiling and membrane topology analysis using glycoproteomics. Proteomics Clin Appl 8: 595–602, 2014. doi: 10.1002/prca.201400009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toba H, Cannon PL, Yabluchanskiy A, Iyer RP, D’Armiento J, Lindsey ML. Transgenic overexpression of macrophage matrix metalloproteinase-9 exacerbates age-related cardiac hypertrophy, vessel rarefaction, inflammation, and fibrosis. Am J Physiol Heart Circ Physiol 312: H375–H383, 2017. doi: 10.1152/ajpheart.00633.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toba H, de Castro Brás LE, Baicu CF, Zile MR, Lindsey ML, Bradshaw AD. Increased ADAMTS1 mediates SPARC-dependent collagen deposition in the aging myocardium. Am J Physiol Endocrinol Metab 310: E1027–E1035, 2016. doi: 10.1152/ajpendo.00040.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Voorhees AP, DeLeon-Pennell KY, Ma Y, Halade GV, Yabluchanskiy A, Iyer RP, Flynn E, Cates CA, Lindsey ML, Han HC. Building a better infarct: Modulation of collagen cross-linking to increase infarct stiffness and reduce left ventricular dilation post-myocardial infarction. J Mol Cell Cardiol 85: 229–239, 2015. doi: 10.1016/j.yjmcc.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Y, Han HC, Yang JY, Lindsey ML, Jin Y. A conceptual cellular interaction model of left ventricular remodelling post-MI: dynamic network with exit-entry competition strategy. BMC Syst Biol 4, Suppl 1: S5, 2010. doi: 10.1186/1752-0509-4-S1-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yabluchanskiy A, Li Y, Chilton RJ, Lindsey ML. Matrix metalloproteinases: drug targets for myocardial infarction. Curr Drug Targets 14: 276–286, 2013. [PMC free article] [PubMed] [Google Scholar]
- 43.Yabluchanskiy A, Ma Y, Chiao YA, Lopez EF, Voorhees AP, Toba H, Hall ME, Han HC, Lindsey ML, Jin YF. Cardiac aging is initiated by matrix metalloproteinase-9-mediated endothelial dysfunction. Am J Physiol Heart Circ Physiol 306: H1398–H1407, 2014. doi: 10.1152/ajpheart.00090.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yabluchanskiy A, Ma Y, DeLeon-Pennell KY, Altara R, Halade GV, Voorhees AP, Nguyen NT, Jin YF, Winniford MD, Hall ME, Han HC, Lindsey ML. Myocardial infarction superimposed on aging: MMP-9 deletion promotes M2 macrophage polarization. J Gerontol A Biol Sci Med Sci 71: 475–483, 2016. doi: 10.1093/gerona/glv034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yabluchanskiy A, Ma Y, DeLeon-Pennell KY, Jin YF, Lindsey ML. Matrix metalloproteinase-9 deletion shifts macrophage polarization towards M2 phenotype in aged left ventricles post-myocardial infarction. Cardiovasc Res 103: S6, 2014. doi: 10.1093/cvr/cvu079.3. [DOI] [Google Scholar]
- 46.Yabluchanskiy A, Ma Y, Iyer RP, Hall ME, Lindsey ML. Matrix metalloproteinase-9: Many shades of function in cardiovascular disease. Physiology (Bethesda) 28: 391–403, 2013. doi: 10.1152/physiol.00029.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zamilpa R, Ibarra J, de Castro Brás LE, Ramirez TA, Nguyen N, Halade GV, Zhang J, Dai Q, Dayah T, Chiao YA, Lowell W, Ahuja SS, D’Armiento J, Jin YF, Lindsey ML. Transgenic overexpression of matrix metalloproteinase-9 in macrophages attenuates the inflammatory response and improves left ventricular function post-myocardial infarction. J Mol Cell Cardiol 53: 599–608, 2012. doi: 10.1016/j.yjmcc.2012.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zamilpa R, Kanakia R, Cigarroa J IV, Dai Q, Escobar GP, Martinez H, Jimenez F, Ahuja SS, Lindsey ML. CC chemokine receptor 5 deletion impairs macrophage activation and induces adverse remodeling following myocardial infarction. Am J Physiol Heart Circ Physiol 300: H1418–H1426, 2011. doi: 10.1152/ajpheart.01002.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zamilpa R, Lopez EF, Chiao YA, Dai Q, Escobar GP, Hakala K, Weintraub ST, Lindsey ML. Proteomic analysis identifies in vivo candidate matrix metalloproteinase-9 substrates in the left ventricle post-myocardial infarction. Proteomics 10: 2214–2223, 2010. doi: 10.1002/pmic.200900587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zamilpa R, Zhang J, Chiao YA, de Castro Brás LE, Halade GV, Ma Y, Hacker SO, Lindsey ML. Cardiac wound healing post-myocardial infarction: a novel method to target extracellular matrix remodeling in the left ventricle. Methods Mol Biol 1037: 313–324, 2013. doi: 10.1007/978-1-62703-505-7_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zong NC, Li H, Li H, Lam MP, Jimenez RC, Kim CS, Deng N, Kim AK, Choi JH, Zelaya I, Liem D, Meyer D, Odeberg J, Fang C, Lu HJ, Xu T, Weiss J, Duan H, Uhlen M, Yates JR III, Apweiler R, Ge J, Hermjakob H, Ping P. Integration of cardiac proteome biology and medicine by a specialized knowledgebase. Circ Res 113: 1043–1053, 2013. doi: 10.1161/CIRCRESAHA.113.301151. [DOI] [PMC free article] [PubMed] [Google Scholar]