Abstract
The National Institute of Standards and Technology has developed reference materials for five human genomes. DNA aliquots are available for purchase and the data, analyses and high-confidence small variant and homozygous reference calls are freely available on the web (www.genomeinabottle.org, last accessed March 12, 2018). These reference materials are useful for evaluating whole genome sequencing methods and can also be used to benchmark targeted sequencing panels, which are commonly used in clinical settings. This paper describes how to use the Genome in a Bottle samples to obtain performance metrics on any germline targeted sequencing panel of interest, as well as the limitations of the reference materials. These materials are useful for understanding the limitations of, and optimizing, targeted sequencing panels and associated bioinformatics pipelines. We present example figures to illustrate ways of accessing the performance metrics of targeted sequencing panels and we include a table of best practices.
Introduction:
In 2015, The National Institute of Standards and Technology (NIST) released the first Genome in a Bottle (GIAB) reference material, RM 8398. To create this reference material, human genomic DNA from a large batch of GM12878 cells was extracted and aliquoted at the Coriell Institute for Medical Research. These homogeneous DNA aliquots were sequenced by multiple unique technologies, each with different capabilities and biases, to obtain a high-confidence “truth set” of small variant and homozygous reference calls1. In 2016, NIST released four additional human genomes as reference materials, a sonfather-mother trio of Ashkenazi Jewish ancestry (RMs 8391 and 8392) and a son in a trio of Chinese ancestry (RM 8393), along with high-confidence calls and regions2,3. All five genomes used for these NIST RMs are also publicly available from the Coriell Institute for Medical Research as cell lines.
Together, these DNA samples and truth sets can be used as reference materials to evaluate assays and analytic pipelines. When the results of a pipeline (“query”) are compared to the truth set, most false positives and false negatives should be errors in the query set. The current high-confidence calls and regions cover about 90 % of the sequence in GRCh37 and GRCh38, but tend to exclude large variants, long tandem repeats, and regions difficult to map with short reads. Ongoing work in GIAB is using new methods to characterize these more challenging variants and regions. The raw data, analyses, and highconfidence calls and regions are freely available online at www.genomeinabottle.org (last accessed March 12, 2018).These genomes and associated data have been widely used in the next-generation sequencing community to obtain performance metrics on whole genome and whole exome sequencing methods4–6. The Global Alliance for Genomics and Health (GA4GH) Benchmarking Team has standardized performance metrics and developed sophisticated variant comparison tools to compare variant calls and output these metrics7.
In addition to their use in evaluating whole genome and whole exome sequencing methods8–10, the GIAB reference materials can also be used with targeted sequencing panels. Next-generation targeted sequencing panels are increasingly being used for clinical purposes due to the higher number of targets that can be covered, relative to Sanger sequencing. Targeted sequencing also has several advantages relative to whole genome sequencing or exome sequencing, including higher coverage for genes of interest at lower cost, and faster analysis time. Targeted sequencing panels have been used clinically for a wide variety of conditions including cystic fibrosis, epilepsy, cardiomyopathies, inherited cancers, disorders of sex development, autoinflammatory diseases, ataxia and retinal disorders11–17 and many others. To ensure the accuracy of these tests, laboratories need well characterized reference materials and associated data sets for test development, validation and quality control.
The recent “Standards and Guidelines for Validating Next-Generation Sequencing Bioinformatics Pipelines” publication recommends the use of reference materials 18. In this work, we describe how the GIAB reference materials can be used to benchmark specific targeted sequencing panels. As an example, we selected germline sequencing panels based on two different library preparation techniques: hybrid capture, which uses oligo probes to capture to regions of interest; and amplicon based, which uses polymerase chain reaction (PCR) to amplify the regions of interest. This work is not intended to be a comprehensive performance assessment of these methods or a comparison between platforms.
Materials and Methods:
DNA Samples
This study used the five genomes contained within three NIST Reference Materials (RMs): RM 8398, RM 8392, and RM 8393 (NIST, Gaithersburg, MD). Each RM contains a 50 μL DNA aliquot at a concentration of approximately 200 ng/μL. RM 8398 contains extracted DNA from a large, homogeneous batch of the GM12878 cell line. RM 8392 contains three separate tubes of DNA extracted from homogeneous large batches of three cell lines (GM24143, GM24149, GM24385) derived from a mother-father-son Ashkenazim Jewish Trio, which is part of the Personal Genome Project (PGP). RM 8393 contains extracted DNA from a cell line (GM24631) derived from a male individual of Chinese descent, who is also part of the PGP.
Library Preparation and Sequencing
Hybrid Capture Library Preparation and Sequencing
Library preparation for the hybrid capture method was performed with the TruSight Rapid Capture kit (catalog #FC-140–1104, Illumina, San Diego, CA) and TruSight Inherited Disease Sequencing Panel (catalog #FC-121–0205, Illumina, San Diego, CA) according to the manufacturer’s protocol.
Briefly, DNA was “tagmented,” (a combination of DNA fragmentation and end-polishing, using transposons), adapters and barcodes were added, and then three to eight libraries were pooled for hybridization (varying numbers of libraries were pooled to obtain a broad range of sequencing depths) The library pool was hybridized twice with Inherited Disease Panel Oligos at 58° C. After library preparation, the library was checked on a 2100 Bioanalyzer high sensitivity DNA chip (catalog # 50674626, Agilent, Santa Clara, CA) to assess the quality before sequencing. DNA concentration was measured with the Qubit high sensitivity DNA assay (catalog # Q32851, ThermoFisher, Waltham, MA), diluted to 4 nmol/L, and denatured with 0.2 mol/L NaOH. PhiX DNA (catalog # FC-110–3001, Illumina, San Diego, CA) was spiked in at 5 % volume/volume. The denatured library was then sequenced with a MiSeq Reagent Kit (catalog # MS-102–3003, Illumina, San Diego, CA) for 300 cycles (2×150 bp) on an Illumina MiSeq or Illumina ForenSeq.
Amplicon Library Preparation and Sequencing
For the amplicon sequencing, the Ion AmpliSeq Library Kit 2.0 (catalog # 4475345, ThermoFisher, Waltham, MA) and AmpliSeq Inherited Disease Panel (catalog# 4477686, ThermoFisher, Waltham, MA) were used in accordance with the manufacturer’s protocol. DNA from each genome was amplified in three separate primer pools, then these PCR products were combined for barcoding and library preparation. The concentration of the final library was measured with the Ion Library TaqMan Quantification Kit (catalog #4468802, ThermoFisher, Waltham, MA), then two libraries were adjusted to a concentration of 40 picomol/L and combined before chip loading. 318v2 BC chips (catalog #4488146, ThermoFisher, Waltham, MA) were loaded using the Ion Chef and then sequenced on the Personal Genome Machine using the Ion PGM Hi-Q Chef kit (catalog # A25948, ThermoFisher, Waltham, MA).
Variant Calling
Sequence variants were identified and stored in Variant Call Format (VCF) files using the included commercial software. MiSeq Reporter (BWA Enrichment version 2.5.1.3) was used to generate the VCF files for the hybrid capture targeted sequencing.
Torrent Suite (version 5.0.5) was used to generate VCF files for the amplicon sequencing.
Data Analysis
After generation, the VCF files were compared to the Genome in Bottle High Confidence VCF files using the Global Alliance for Genomics and Health (GA4GH) Benchmarking application on precisionFDA (registration required, http://precision.fda.gov/, last accessed March 12, 2018). The GA4GH Benchmarking Team developed standardized performance metrics for genomic variant calls as well as sophisticated variant comparison tools to robustly compare different representations of the same variant, and a set of standard Browser Extensible Data (BED) files describing difficult genome contexts to stratify performance. The GA4GH Benchmarking application requires a truth VCF file (the GIAB high confidence VCF file), the truth confident regions (the GIAB high confidence BED file), the query VCF file (generated by the included commercial software) and the target regions (the BED file provided by the manufacturer for the targeted sequencing panel). All GIAB files (VCF files, BED files) are available on the web at https://github.com/genome-in-a-bottle (last accessed March 12, 2018). The GA4GH application returns the count of false negatives (FN), false positives (FP) and true positives (TP) in both standardized VCF and comma-separated value (CSV) formats. Performance metrics follow the GA4GH standardized definitions, where genotyping errors are counted both as FP and FN. In addition, the GA4GH application stratifies performance metrics by variant type, size, and genome context to enable understanding strengths and weaknesses of a method. We calculated sensitivity using the formula:
Sensitivity above a specific minimum coverage ‘X’ was calculated by only including TPs and FNs at sites with coverage greater than or equal to ‘X’. Coverage analysis of each locus and common false negatives shared among replicates were determined using Bedtools19. The precisionFDA output VCF was first split into three files: the false negatives, false positives, and true positives. Next, the bedtools “coverage” command was used to determine the coverage at each FN and TP location. The bedtools “multiinter” command was used to identify FN shared between different replicates of the same genome. The number of common FNs are represented using Venn Diagram Plotter20.
Confidence intervals for stratified regions were calculated using Fisher’s Exact test in R. FNs and FPs in the binary alignment map (BAM) and VCF files were visualized using Golden Helix GenomeBrowse version 2.1.221.
Results:
Effect of Average and Locus Coverage on Sensitivity
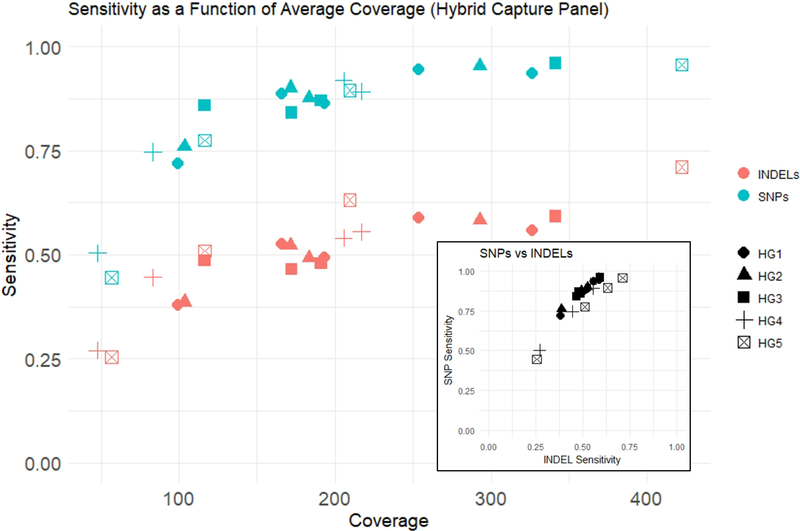
Sensitivity increased with increasing average coverage for both single nucleotide polymorphisms (SNPs) and insertions/deletions (INDELs). With the hybrid capture sequencing, we observed a maximum SNP sensitivity rate of 96 % and a maximum INDEL sensitivity rate of 71 % at the highest mean coverage (422x). For all genomes examined, INDEL sensitivity was lower than SNP sensitivity (Figure 1). Average coverage and sensitivity were similar across replicates for the amplicon-based panel (Supplementary data Figure 1). Depending on the sample, 82 % to 93 % of INDELs in the truth set were 1 to 5 bp in size, so analysis of larger INDELs was limited. For each sample, there were only 7 to 17 INDELs between 6 and 15 bp in size, and 1 to 6 INDELs larger than 15 bp in size.
Figure 1. Effect of Average Coverage on Sensitivity.
As average coverage increases, sensitivity for both SNPs and INDELs increases. SNP sensitivity is higher than INDEL sensitivity. SNP sensitivity and INDEL sensitivity are strongly correlated (inset).
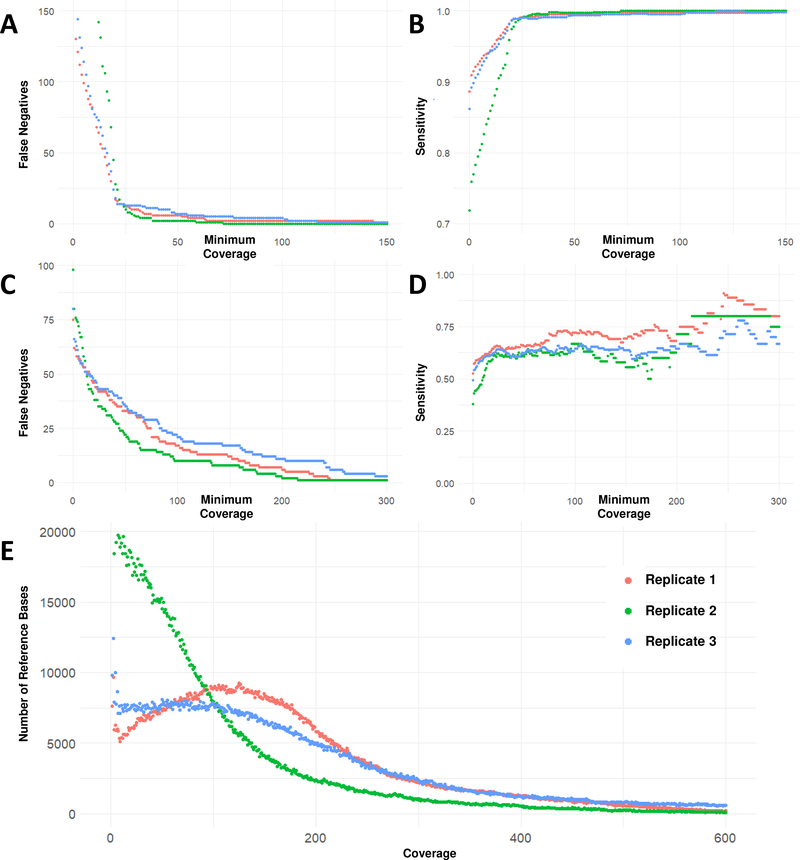
We also analyzed the number of false negatives, true positives and sensitivity within individual datasets when excluding loci below a varying coverage threshold (Figure 2). For SNPs, almost all false negatives occur at locus coverage of 50× or below. For INDELs, many false negatives remain even with high locus coverages (above 100×). We therefore stratified further by genome context to gain insight into the causes for these false negatives.
Figure 2. False Negatives, Sensitivity and Coverage at each site inside targeted regions.
A. SNP False Negative sites with minimum locus coverage at or below the coverage on the X-axis; there were few SNP false negatives at locus coverages greater than 50x. B. SNP Sensitivity with minimum locus coverage at or below the coverage on the X-axis; SNP sensitivity was above 98% for loci with a coverage of 25x or higher. C. INDEL False Negatives with minimum locus coverage at or below the coverage on the X-axis; there were still a significant number of false negatives at loci with coverages greater than 100x, indicating that read depth is not the only factor affecting INDEL detection. D. INDEL Sensitivity with minimum locus coverage at or below the coverage on the X-axis; even for loci covered at 200x, INDEL sensitivity did not exceed 75 %. E. This histogram shows the distribution of read depths over the total number of reference bases in the manufacturer’s BED file. In replicate 2 (green), the average coverage was lower and more references bases were covered at less than 25x, compared to replicates 1 and 3.
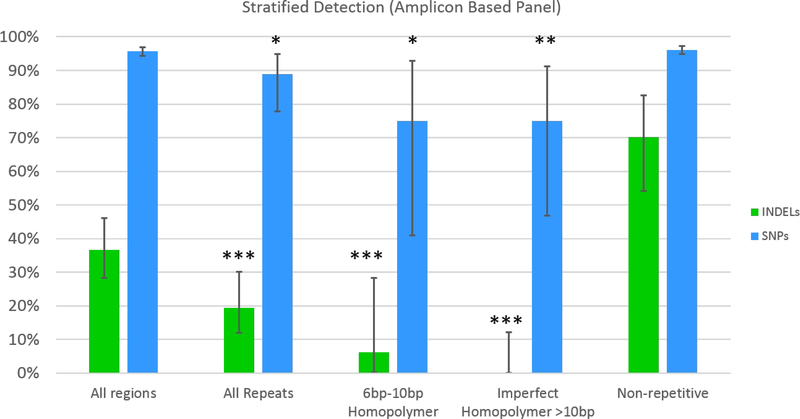
We examined SNP sensitivity and INDEL sensitivity over various stratified regions (Figure 3) in the amplicon based sequencing. Compared to regions with higher complexity, INDEL sensitivity decreases significantly in repetitive regions, imperfect homopolymers >10 bp and perfect homopolymers >10 bp (with no INDEL detection at all in the latter). The overall INDEL detection rate at 273× coverage was 36 %; however, this increased to 70 % in regions that were higher complexity (with no repeats, homopolymers or imperfect homopolymers). SNP sensitivity followed a similar pattern. The overall SNP sensitivity was 96 %, but SNP sensitivity decreased to 75 % in 6 to 10 bp homopolymer regions.
Figure 3. INDEL and SNP Sensitivity Stratified by Region Type.
Overall, the INDEL sensitivity for this replicate of the amplicon based panel assay with 273x coverage was 36% for INDELS and 96% for SNPs. INDEL sensitivity is significantly higher in non-repetitive regions compared to all repetitive regions, 610bp homopolymer regions, and imperfect homopolymer regions (* indicates p-value < 0.05, ** indicates p-value < 0.005, *** indicates p-value < 0.0005.). Vertical black lines indicate 95% confidence intervals. SNP sensitivity was also significantly higher in non-repetitive regions compared to all repetitive regions, 6–10bp homopolymer regions, and imperfect homopolymer regions.
Consistency of False Negatives between Replicates
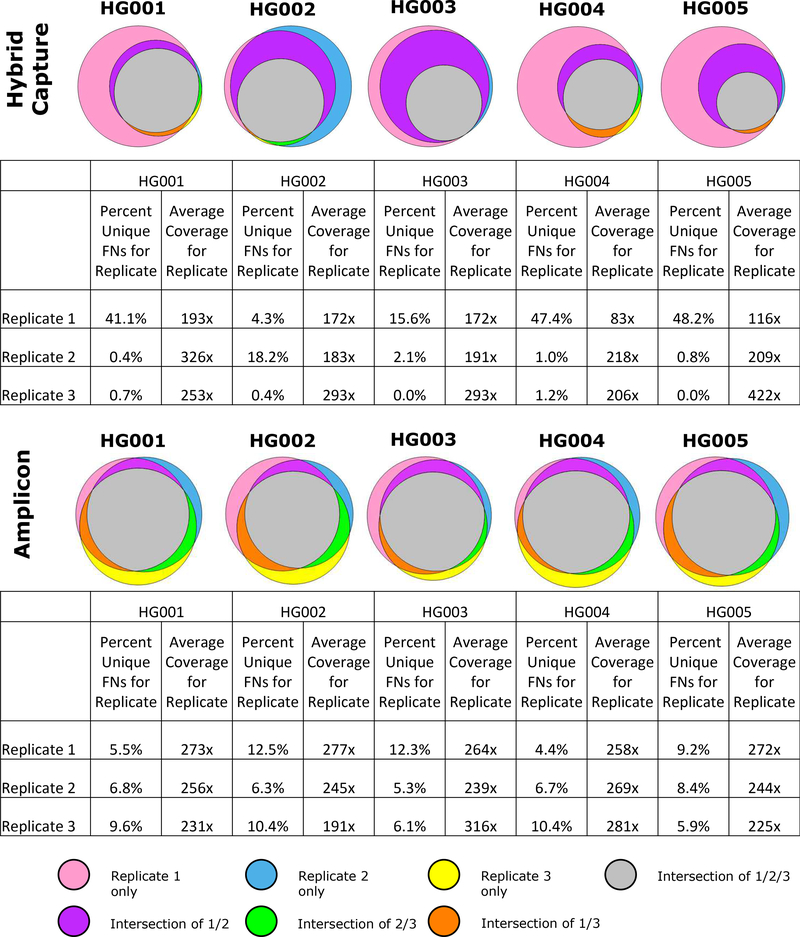
For both amplicon and hybrid capture panels, the locations of false negative calls were similar between replicates (Figure 4). The number of total false negatives varies more in the hybrid capture assay because there was more variability in average coverage; however, almost all false negatives contained within the higher coverage replicates also occur in the lower coverage replicates. For amplicon based sequencing, the average coverage was very similar between replicates, and approximately 40 % of false negatives are shared by all replicates.
Figure 4. False Negatives shared across replicates.
Venn diagrams show the overlap of false negatives between replicates. In addition, we include a table that shows the average coverage for each replicate and the percentage of false negatives that were unique to that replicate. For both amplicon and hybrid capture panels, false negatives appear to be non-random; a high number of the same false negatives appear in multiple replicates, with lower coverage replicates having most of the same false negatives as higher coverage replicates, plus additional false negatives.
Causes of False Positives
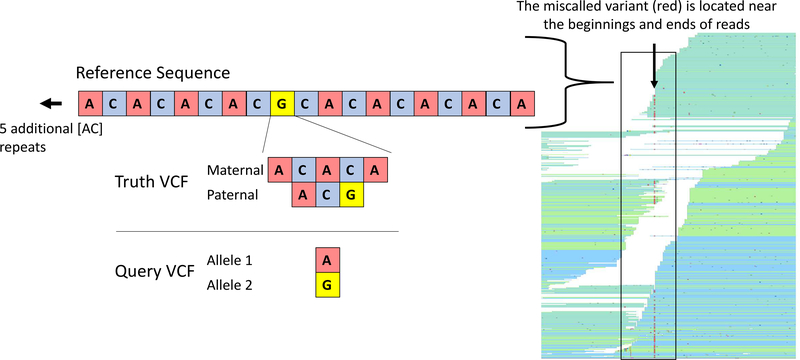
False Positives were less common than false negatives and tended to occur near actual variants, in repetitive regions, and near the ends and beginnings of reads. In the example shown (Figure 5), the true variant is a complex, compound heterozygous mutation. The GIAB high confidence VCF shows that for the Ashkenazi son, there is a 2 base pair insertion on the paternal allele and a 4 base pair insertion followed by a G to A SNP on the maternal allele. The variant caller incorrectly called this as location as simply having a heterozygous [G/A] SNP.
Figure 5. False positive (and false negative) call near a true variant.
False positives were most likely to occur in repetitive regions, near the ends of reads and near true variants. In the example shown here, there was a 4 base pair insertion on the maternal allele, followed by a G to A SNP, and a 2 base pair insertion on the paternal allele. The variant caller incorrectly only identified a G to A SNP. The region had 10 [AC] repeats preceding the variant and 5 [CA] repeats after the variant. The read pileup on the right is shown to indicate the location of the miscalled variant, which is near the beginnings and ends of the reads. The miscalled variant is due to misaligned reads that do not encompass the entire repeat and its flanking sequences.
Discussion:
Targeted sequencing panels are increasingly used in clinical settings because they offer higher coverage depth at a lower cost, relative to whole genome and whole exome sequencing. There are two main types of targeted sequencing panels: probe capture-based and amplicon based. Selection of efficient probes or primers, careful library preparation and appropriate bioinformatic pipelines all have impacts on panel sensitivity22. Potential pathogenic variants are typically confirmed using Sanger sequencing23 which can identify Next-Generation Sequencing (NGS) false positives. We have shown that these NGS false positives often occur near true variants, which may often be identified by follow-up Sanger sequencing if the false positive was flagged for follow-up. There is some debate about whether Sanger sequencing is necessary when specific conditions are met by the NGS sequencing24. Minimizing false negatives is also important, and it is critical both to ensure sufficient coverage at every locus and assess whether the pipeline can detect more difficult variants even at high coverage.
We have shown how one can use the GIAB benchmark genomes to evaluate a targeted sequencing panel of interest. We performed multiple sequencing replicates, with five different genomes, on both hybrid capture and amplicon based sequencing panels. This allowed us to compare the results of the targeted sequencing panels to the GIAB high confidence calls, using freely available bioinformatic data and tools. We examined overall sensitivity, site specific sensitivity, false negatives, and false positives. The results were similar for both types of panels.
In the targeted sequencing panels we tested, we found that average coverage is the main determinant of sensitivity, with the individual genome having no noticeable effect. Replicates with similar coverage have mostly the same false negatives, with lower coverage replicates having additional false negatives.
For the targeted sequencing panels examined in this study, low coverage regions are not random – they are likely caused by either inefficient PCR primers in amplicon sequencing or inefficient capture probes in hybrid capture sequencing.
On a per site level, in the assays tested, we observed that most SNP false negatives were caused by low coverage; this demonstrates the usefulness of evaluating the effect of coverage on the false negative rate. If one excludes all regions with low coverage, SNP sensitivity is very high. For instance, if only loci with coverage greater than 50x are considered, the SNP sensitivity is above 99 % for all genome replicates we examined. In contrast, only about half of all INDEL false negatives appeared to be caused by low coverage. We therefore used the GA4GH Benchmarking tool’s stratification functionality, which showed that INDEL false negatives with high coverage mostly occurred in repetitive regions. Although there were few false positives in the targeted sequencing panels examined here, similar analyses and figures could be generated for false positives when more false positives occur.
For the targeted NGS panels we examined, false positives do not occur randomly, but instead are most likely to occur at or around complex variants, in repetitive regions and near the beginnings and ends of reads. In contrast to whole genome sequencing, targeted sequencing has reads that begin and end near the same location; the start and stop points are either centered around the capture probe, or occur at the ends of the PCR primer regions. For this reason, although a region within a targeted sequencing panel and region from whole genome sequencing may be sequenced at the same coverage, it is more likely that reads in the targeted panel will have more non-random start and end points. When these start and end points occur in repetitive regions, it can be difficult for the variant caller to properly align the read and make the correct call. This could potentially be eliminated with multiple primer sets and capture probes. These observations were true of the vendor-supplied pipelines used, but variance in performance between pipelines is expected. We show example figures derived from comparing the targeted panel calls to the GIAB benchmark calls that can help to highlight whether these factors are important for any pipeline.
One limitation of our current work is that the Genome in a Bottle high confidence calls are biased towards the relatively simple calls. The high confidence regions include a relatively small number of larger INDELs, especially in coding regions, and no structural variant or copy number variation calls. A panel may perform well over the GIAB high confidence regions and still perform poorly on more difficult variants and difficult regions of the genome. Ideally, one should test a large number of variants of different types, sizes, and sequence contexts; this is usually possible for whole genome sequencing with only small number of benchmark genomes, but this small number of genomes is unlikely to contain enough variants for targeted sequencing tests. This is particularly important because some clinical tests are enriched for more difficult variants.25
The available GIAB genomes and bioinformatics data are a resource for benchmarking the performance of targeted clinical gene sequencing panels. These performance benchmarks can then be used to inform practical recommendations for the use of particular targeted sequencing panels; e.g., necessary target coverage levels and the identification of regions where variant calls can be made with sufficient confidence. Finally, benchmark observations can suggest principles that could be used in the design of probes or primers for targeted sequencing panels, such as the need to avoid placing read boundaries in repetitive regions or the importance of knowing the limitations of the test in these regions. Table 1 outlines our recommendations for best practices.
Table 1:
Best practices for using reference materials to assess performance of targeted assays
| Manual curation | Manually curate false positives and false negatives to help understand their source (e.g., they are located near true variants, in repetitive regions or at the edges of reads) |
| Identify low coverage | Determine how many false negatives are associated with low coverage regions |
| Stratify | Stratify false negatives and false positives according to variant type and genome context (e.g., homopolymers, tandem repeats, difficult to map regions) |
| Confidence intervals | Calculate confidence intervals for performance metrics for variants of different types in different genome contexts, since some variant types and genome contexts may have limited numbers of examples in targeted regions |
| Use additional samples | The GIAB samples are not intended to be used as the only validation method for clinical tests, because there are a limited number of variants in the targeted regions of most clinical assays, the variants in the GIAB samples are likely not representative of the variants of interest clinically, etc. |
| Use high-confidence bed file |
The GIAB samples are useful for benchmarking, but comparisons should generally only be made within the high confidence bed file |
| Most difficult regions are outside the bed file |
The high confidence regions are not yet comprehensive, so they exclude the most difficult regions and variants. |
Supplementary Material
Effect of Average Coverage on Sensitivity for Amplicon Data. Average coverage and sensitivity were similar for all replicates with the amplicon-based targeted sequencing panels, with almost all replicates between 220x and 280x coverage. SNP sensitivity was higher than INDEL sensitivity.
Acknowledgments:
Certain commercial equipment, instruments and materials are identified in order to specify experimental procedures as completely as possible. In no case does such identification imply a recommendation or it imply that any of the materials, instruments or equipment identified are necessarily the best available for the purpose. Information presented does not necessarily represent the official position of the National Institute of Standards and Technology.
All work presented has been reviewed and approved by the National Institute of Standards and Technology Human Subjects Protections Office.
Institution/Address where research was performed: National Institute of Standards and Technology, 100 Bureau Dr, Gaithersburg, MD 20899
References
- 1.Zook JM, McDaniel J, Parikh H, Heaton H, Irvine SA, Truty R, Mclean CY, Vega FMD La, Salit M. Reproducible integration of multiple sequencing datasets to form high-confidence SNP, indel, and reference calls for five human genome reference materials, 2018. 10.1101/281006 [DOI]
- 2.Zook JM, Chapman B, Wang J, Mittelman D, Hofmann O, Hide W, Salit M. Integrating human sequence data sets provides a resource of benchmark SNP and indel genotype calls. Nat Biotechnol, 2014, 32:246–51 [DOI] [PubMed] [Google Scholar]
- 3.Zook JM, Catoe D, McDaniel J, Vang L, Spies N, Sidow A, Weng Z, Liu Y, Mason CE, Alexander N, Henaff E, McIntyre ABR, Chandramohan D, Chen F, Jaeger E, Moshrefi A, Pham K, Stedman W, Liang T, Saghbini M, Dzakula Z, Hastie A, Cao H, Deikus G, Schadt E, Sebra R, Bashir A, Truty RM, Chang CC, Gulbahce N, Zhao K, Ghosh S, Hyland F, Fu Y, Chaisson M, et al. Extensive sequencing of seven human genomes to characterize benchmark reference materials. Sci Data, 2016, 3:160025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laurie S, Fernandez-Callejo M, Marco-Sola S, Trotta JR, Camps J, Chacón A, Espinosa A, Gut M, Gut I, Heath S, Beltran S. From Wet-Lab to Variations: Concordance and Speed of Bioinformatics Pipelines for Whole Genome and Whole Exome Sequencing. Hum Mutat, 2016, 37:1263–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cornish A, Guda C. A Comparison of Variant Calling Pipelines Using Genome in a Bottle as a Reference. Biomed Res Int, 2015, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cleary JG, Braithwaite R, Gaastra K, Hilbush BS, Inglis S, Irvine SA, Jackson A, Littin R, Rathod M, Ware D, Zook JM, Trigg L, De La Vega FMM. Comparing Variant Call Files for Performance Benchmarking of Next-Generation Sequencing Variant Calling Pipelines, 2015. 10.1101/023754 [DOI]
- 7.Krusche P, Trigg L, Boutros PC, Mason CE, De La Vega FM, Moore BL, Gonzalez-Porta M, Eberle MA, Tezak Z, Lababidi S, Truty R, Asimenos G, Funke B, Fleharty M, Salit M, Zook JM. Best Practices for Benchmarking Germline Small Variant Calls in Human Genomes, 2018. 10.1101/270157 [DOI] [PMC free article] [PubMed]
- 8.Hwang S, Kim E, Lee I, Marcotte EM. Systematic comparison of variant calling pipelines using gold standard personal exome variants. Sci Rep, 2015, 5:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meynert AM, Ansari M, FitzPatrick DR, Taylor MS. Variant detection sensitivity and biases in whole genome and exome sequencing. BMC Bioinformatics, 2014, 15:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Linderman MDh, Brandt T, Edelmann L, Jabado O, Kasai Y, Kornreich R, Mahajan M, Shah H, Kasarskis A, Schadt EE. Analytical validation of whole exome and whole genome sequencing for clinical applications. BMC Med Genomics, 2014, 7:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eggers S, Sadedin S, van den Bergen JA, Robevska G, Ohnesorg T, Hewitt J, Lambeth L, Bouty A, Knarston IM, Tan TY, Cameron F, Werther G, Hutson J, O’Connell M, Grover SR, Heloury Y, Zacharin M, Bergman P, Kimber C, Brown J, Webb N, Hunter MF, Srinivasan S, Titmuss A, Verge CF, Mowat D, Smith G, Smith J, Ewans L, Shalhoub C, Crock P, Cowell C, Leong GM, Ono M, Lafferty AR, et al. Disorders of sex development: insights from targeted gene sequencing of a large international patient cohort. Genome Biol, 2016, 17:243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X, Zein WM, D’Souza L, Roberson C, Wetherby K, He H, Villarta A, Turriff A, Johnson KR, Fann YC. Applying next generation sequencing with microdroplet PCR to determine the disease-causing mutations in retinal dystrophies. BMC Ophthalmol, 2017, 17:157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Omoyinmi E, Standing A, Keylock A, Price-Kuehne F, Melo Gomes S, Rowczenio D, Nanthapisal S, Cullup T, Nyanhete R, Ashton E, Murphy C, Clarke M, Ahlfors H, Jenkins L, Gilmour K, Eleftheriou D, Lachmann HJ, Hawkins PN, Klein N, Brogan PA. Clinical impact of a targeted next-generation sequencing gene panel for autoinflammation and vasculitis. PLoS One, 2017, 12:1–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iqbal Z, Rydning SL, Wedding IM, Koht J, Pihlstrøm L, Rengmark AH, Henriksen SP, Tallaksen CME, Toft M. Targeted high throughput sequencing in hereditary ataxia and spastic paraplegia. PLoS One, 2017, 12:1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Celestino-Soper PBS, Gao H, Lynnes TC, Lin H, Liu Y, Spoonamore KG, Chen P- S, Vatta M. Validation and Utilization of a Clinical Next-Generation Sequencing Panel for Selected Cardiovascular Disorders. Front Cardiovasc Med, 2017, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lucarelli M, Porcaro L, Biffignandi A, Costantino L, Giannone V, Alberti L, Bruno SM, Corbetta C, Torresani E, Colombo C, Seia M. A New Targeted CFTR Mutation Panel Based on Next-Generation Sequencing Technology. J Mol Diagnostics, 2017, 19:788–800 [DOI] [PubMed] [Google Scholar]
- 17.LaDuca H, Pesaran T, Elliott AM, Speare V, Dolinsky JS, Gau CL, Chao E. Utilization of multigene panels in hereditary cancer predisposition testing. Next Gener Seq Cancer Res Vol 2 From Basepairs to Bedsides, 2015, 16:459–82 [Google Scholar]
- 18.Roy S, Coldren C, Karunamurthy A, Kip NS, Klee EW, Lincoln SE, Leon A, Pullambhatla M, Temple-Smolkin RL, Voelkerding K V., Wang C, Carter AB. Standards and Guidelines for Validating Next-Generation Sequencing Bioinformatics Pipelines. J Mol Diagnostics, 2017, 0:1–24 [DOI] [PubMed] [Google Scholar]
- 19.Quinlan AR. BEDTools: The Swiss-Army Tool for Genome Feature Analysis. Curr. Protoc. Bioinforma, vol. 2014, Hoboken, NJ, USA, John Wiley & Sons, Inc., 2014, p. 1112.1–11. 12.34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pacific Northwest National Laboratory. Venn Diagram Plotter, 2017
- 21.Golden Helix Inc. Golden Helix GenomeBrowse visualization tool (Version 2.1.2), n.d.
- 22.Jennings LJ, Arcila ME, Corless C, Kamel-Reid S, Lubin IM, Pfeifer J, Temple-Smolkin RL, Voelkerding K V, Nikiforova MN. Guidelines for Validation of Next-Generation Sequencing–Based Oncology Panels. J Mol Diagnostics, 2017, 19:341–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rehm HL, Bale SJ, Bayrak-Toydemir P, Berg JS, Brown KK, Deignan JL, Friez MJ, Funke BH, Hegde MR, Lyon E. ACMG clinical laboratory standards for next-generation sequencing. Genet Med, 2013, 15:733–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mu W, Lu H, Chen J, Li S, Elliott AM. Sanger Confirmation Is Required to Achieve Optimal Sensitivity and Specificity in Next-Generation Sequencing Panel Testing. J Mol Diagnostics, 2016, 18:923–32 [DOI] [PubMed] [Google Scholar]
- 25.Lincoln SE, Zook JM, Chowdhury S, Mahamdallie S, Fellowes A, Klee EW, Truty R, Huang C, Tomson FL, Cleveland MH, Vallone PM, Ding Y, Seal S, Desilva W, Garlick RK, Salit M, Rahman N, Lincoln SE. An interlaboratory study of complex variant detection, 2017. 10.1101/218529 [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Effect of Average Coverage on Sensitivity for Amplicon Data. Average coverage and sensitivity were similar for all replicates with the amplicon-based targeted sequencing panels, with almost all replicates between 220x and 280x coverage. SNP sensitivity was higher than INDEL sensitivity.