Abstract
Background
Neurofibromatosis type 1 (NF1) is a dominantly inherited tumor predisposition syndrome that targets the peripheral nervous system. It is caused by mutations of the NF1 gene which serve as a negative regulator of the cellular Ras/MAPK (mitogen-activated protein kinases) signaling pathway. Owing to the complexity in some parts of clinical diagnoses and the need for better understanding of its molecular relationships, a genetic characterization of this disorder will be helpful in the clinical setting.
Methods
In this study, we present a customized targeted gene panel of NF1/KRAS/BRAF/p53 and SPRED1 genes combined with Multiple Ligation-Dependent Probe Amplification analysis for the NF1 mutation screening in a cohort of patients clinically suspected as NF1.
Results
In this study, we identified 73 NF1 mutations and two BRAF novel variants from 100 NF1 patients who were suspected as having NF1. These genetic alterations are heterogeneous and distribute in a complicated way without clustering in either cysteine–serine-rich domain or within the GAP-related domain. We also detected fifteen multi-exon deletions within the NF1 gene by MLPA Analysis.
Conclusions
Our results suggested that a genetic screening using a NGS panel with high coverage of Ras–signaling components combined with Multiple Ligation-Dependent Probe Amplification analysis will enable differential diagnosis of patients with overlapping clinical features.
Keywords: Neurofibromatosis type 1, RASopathies, Targeted NGS, MLPA, Genetic counseling
Background
Neurofibromatosis type 1 (MIM# 162200) is a very common genetic disorder affecting approximately 1 in 3000–4000 individuals worldwide with the penetrance of the mutant gene being close to 100% by 5 years of age [1–4]. Clinically, it is presented with the occurrence of Café-au-lait macules, Lisch nodules, axillary freckling and multiple neurofibromas. Phenotypically, Neurofibromatosis type 1 (NF1) patients have a very heterogeneous condition. Discrete dermal neurofibromas occur in almost all adults with NF1, and the number usually increases with age. If whole-body magnetic resonance imaging (MRI) is used, plexiform neurofibromas are detectable in at least half of NF1 patients. Other complications include learning disabilities, mental retardation, optic gliomas, certain bone abnormalities, CNS tumors, and an increased risk for certain malignancies [5, 6].
NF1 is caused by mutations of the NF1 gene which maps to chromosome 17q11.2. Many evidences have suggested NF1 as a tumor suppressor gene as inactivation of both NF1 alleles would reduce the control of cell proliferation and lead to tumorigenesis [7, 8]. The function of NF1 gene product, neurofibromin, is to stimulate the GTPase activity of the RAS protein and serve as a negative regulator of the cellular Ras/MAPK (mitogen-activated protein kinases) signaling pathway [7, 9–11]. Up to date, more than 1000 pathogenic allelic variants have been identified in the NF1 gene [The Human Gene Mutation Database (HGMD, Institute of Medical Genetics, Cardiff, http://www.hgmd.org/; Leiden Open Variation Database, LOVD: www.lovd.nl/NF1]. Most NF1 mutations are single-base substitutions, insertions, or deletions. Other mutations are single- or multi-exon deletions or duplications and microdeletions encompassing NF1 and its neighboring genes [12–22].
NF1 is a member of RAS-related disorders, which usually show similar clinical features in cutaneous signs, cardiac defects, developmental disabilities and neurocognitive impairment [23–25]. Therefore, molecular diagnosis in NF1 should be of great value to confirm the diagnosis, particularly in the early childhood. However, the procedures for molecular diagnosis of NF1 are usually expensive, time-consuming, and labor-intensive [15–21, 26–28]. The development of next-generation sequencing (NGS) technologies which allows for rapid identification of disease-causing mutations and high-risk alleles has recently been introduced into NF1 diagnosis [29–34]. Owing to the complexity with some aspects of clinical diagnoses and the need for a better understanding of its molecular relationships, an extended genetic characterization of this disorder will be helpful in a clinical setting.
Methods
Patients and sample preparations
One hundred NF1 patients suspected as having NF1 by a clinical evaluation were recruited for this study. From each patient, 10 ml of whole blood samples were collected in EDTA-anticoagulant tube through the Linko Medical Center of the Chang Gung Memorial Hospital. Fifteen patients had a known family history of NF1. Ethical approval for this study was obtained by the institutional review board (102-0226A3) at Chang Gung Memorial Hospital. All participants provided written informed consent. Genomic DNA of each patient was then prepared using the PUREGENE DNA purification kit from GENTRA using standard protein precipitation procedures. The quality of the DNA was estimated using the Nano-Drop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA).
Candidate gene-targeted sequencing
A panel of five NF1-related genes including NF1 (NM_000267, 17q11.2), SPRED1 (NM_152594, 15q14), KRAS (NM_004985, 12p12.1), BRAF (NM_004333, 7q34), and p53 (NM_000546, 17p13.1) was initially created designed to capture, amplify, and sequence specific regions (including exons and splice junctions) of the genome for human cancer screening. The total length was 32.3 kb encompassing 296 amplicons, and the coverage was 507×. Adapter sequences were clonally amplified by emulsion PCR on the high-density array of micro-machined wells. In this study, we took the advantage of this gene panel for the germline mutation analysis of NF1 using the Ion Personal Genome Machine® (PGM™) Sequencer (Life technology).
Sample library preparation
A total of 100 indexed rapid prepared Ion AmpliSeq DNA libraries, starting from 100 ng of gDNA per sample, were prepared according to the manufacturer’s instructions. Template preparation and emulsion PCR and Ion Sphere Particles (ISP) enrichment were performed according to the manufacturer’s instructions. Following the purification and size selection using AMPure beads (Beckman Coulter, Brea, CA, USA), the size distribution of the DNA fragments was analyzed on the Agilent Bioanalyzer using High-Sensitivity DNA chip (Agilent Technologies Inc., Santa Clara, CA) and the quality checking of ion sphere particles for the prepared library was performed using Qubit 2.0 Fluorometer (Life Technologies). Enriched ISPs were prepared for sequencing using the Ion PGM 200 Sequencing Kit v2.0 and were loaded on an Ion 316 chip v2 or Ion 318 chip v2.
Data analysis
We used IT platform-specific pipeline software Torrent Suite, version 4.2, with the plug-in “variant caller” program (Life Technologies) to perform reference genome alignment, base calling, and filtering of poor signal reads. The Integrative Genome Viewer (IGV) (http://software.broadinstitute.org/software/igv/) was used for visualizing the status of each read alignment. The selected variants were classified as deleterious mutation by mutation type if they were identified as nonsynonymous, frameshift, or stopgain at the exonic region. ACMG Standards and Guidelines for the interpretation of sequence variants were followed in this study [35]. In an appropriate reference population, the pathogenic variant should have a frequency of much less than 1%. We removed all the common variants (Minor Allele Frequency, MAF > 1%) reported in the following public databases: 1000 Genomes Project (http://www.1000genomes.org/), dbSNP database and ClinVar database (https://www.ncbi.nlm.nih.gov/snp/; https://www.ncbi.nlm.nih.gov/clinvar/). Variants with amino acid changes were further examined for whether the changes were potentially damaging alterations using Sorting Tolerant From Intolerant (SIFT) and Polymorphism Phenotyping v2 (PolyPhen2) softwares, which can predict the possible impact of an amino acid substitution on the structure and function of a protein. The nomenclature of novel variants followed the rules of the Human Genome Variation Society (http://www.hgvs.org/mutnomen/). The genetic variants in the Human Gene Mutation Database (HGMD, Institute of Medical Genetics, Cardiff, http://www.hgmd.org/) and Leiden Open Variation Database (LOVD: www.lovd.nl/NF1) were also considered as references.
PCR amplification and sanger sequencing verification
We performed Sanger validation for all putatively pathogenic SNVs and indels variants on each detected patient (and their family members, if available) by PCR amplification and sequenced with Applied Biosystems 3730 Genetic Analyzer. PCR amplification was performed under standard conditions with 30 PCR cycles and 55°–60 °C annealing. PCR products were sequenced using the Big Dye Terminator cycle sequencing kit (Life Technologies) according to the manufacturer’s cycling conditions and analyzed on an Applied Biosystems 3730xl Automated Sequencer Genetic Analyzer (Life Technologies). Sequence alignments and analysis were further performed using the Autoassembler computer program (Life Technologies).
Multiplex ligation-dependent probe amplification (MLPA) analysis
We used SALSA P081/P082 NF1 MLPA kit (MRC Holland, Amsterdam, The Netherlands) to confirm and identify single and multiple exon deletions/duplications according to the manufacturer’s protocol. Each samples containing 100 ng of genomic DNA was used for overnight hybridization with the probemixes. After ligation and amplification were performed with FAM-labeled primers, the PCR products were analyzed on a Genetic Analyzer 3730 capillary electrophoresis system and interpreted using Genotyper version 2.0 (Applied Biosystems, CA, USA). In this study, we used the Coffalyser program (version 3.5) for peak area normalization and gene dosage calculation.
Results
Genetic alterations identified from a targeted NGS gene panel screening
A total of 100 individuals from 95 families who were clinically suspected as carrying NF1 were referred for this genetic testing. A brief summary of the clinical data collected for each patient is given in Table 1. Fifteen patients (15%) had a family history of NF1 in this cohort. Café-au-lait spots and Lisch nodules in the iris were observed in 93 and 19 patients, respectively. Cutaneous neurofibromas, plexiform neurofibromas and malignant peripheral nerve sheath tumors were identified in 32, 13, and 2, patients, respectively. Five patients had optic gliomas and two patients had brain tumors. Among these individuals, we have identified seventy-three NF1 mutations (Table 2) and two BRAF novel variants from a targeted NGS gene panel of NF1/KRAS/BRAF/p53 and SPRED1 analyses. SPRED1 genetic mutations were not detected in this study. Variants with amino acid changes were further examined to check if the changes were potentially damaging alterations using Sorting Intolerant from Tolerant (SIFT) algorithm and Polymorphism Phenotyping v2 (PolyPhen2) software, which can predict the possible impact of an amino acid substitution on the structure and function of a protein.
Table 1.
Clinical features of 100 Taiwanese NF1 patients
| Clinical features | Patientsa (%) |
|---|---|
| Café-au-lait spots | 93 (93%) |
| Lisch nodules in the Iris | 19 (19%) |
| Cutaneous neurofibroma | 32 (32%) |
| Plexiform neurofibroma | 13 (13%) |
| Malignant peripheral nerve sheath tumor | 2 (2%) |
| Optic glioma | 5 (5%) |
| Brain tumor | 2 (2%) |
| Scoliosis | 10 (10%) |
| Heart defects | 8 (8%) |
| Learning disability | 4 (4%) |
| Craniofacial disability | 9 (9%) |
| Family history | 15 (15%) |
a11 patients are under 12 years old; male: female = 53:47
Table 2.
NF1 Mutational profile of the 100 NF1 blood samples tested in NGS study
| Patient | Codinga | Amino Acid Change | Variant Effect | NM_000267.3 | SIFT | Polyphen2 |
|---|---|---|---|---|---|---|
| Wu p001 | c.492_495 del AACT/Het | p.Val166fs | Frameshift Deletion | Exon 5 | ||
| Wu p002 | c.5844C > A | p.Tyr1948Ter | nonsense | Exon 40 | ||
| Wu p003 | c.1466A > G, c.1400C > T, c.1448A > G, c.1513A > G | p.Tyr489Cys | missense | Exon 13 | Tolerated | Benign |
| Wu p004 | c.6855C > A | p.Tyr2285Ter | nonsense | Exon 46 | Tolerated | |
| Wu p006 | c.2982_2982delT | p.Leu995fs | Frameshift Deletion | Exon 22 | ||
| Wu p007 | c.1105C > T | p.Gln369Ter | nonsense | Exon 10 | Tolerated | |
| Wu p009 | c.7862_7862delC | p.Thr2621fs | Frameshift Deletion | Exon 54 | ||
| Wu p010 | c.5902C > T | p.Arg1968Ter | nonsense | Exon 40 | Tolerated | |
| Wup011 | c.7152_7157del TAACTT | p.2384_2386del | Deletion | Exon 49 | ||
| Wu p014 | c.3113 + 1 G > A | . | splicing | Exon 23 | ||
| Wu p015 | c.4700C > G | p.Ser1567Ter | nonsense | Exon 36 | Tolerated | |
| Wu p017 | c.487G > T | p.Glu163Ter | nonsense | Exon 5 | ||
| Wu p018 | c.6970C > T | p.Gln2324Ter | nonsense | Exon 47 | Damaging | |
| c.8386A > C | p.Lys2796Gln | missense | Exon 58 | Damaging | Possibly damaging | |
| c.8520 + 125 del C (Intron) | Frameshift Deletion | Exon 58 | ||||
| Wu p019 | c.575G > A | p.Arg192Gln | missense | Exon 5 | Tolerated | Benign |
| c.1422_1422delC | p.Lys476fs | Frameshift Deletion | Exon 13 | |||
| Wu p021 | c.1080_1083delAAGT | p.Lys362fs | Frameshift Deletion | Exon 10 | ||
| Wu p022 | c.1062G > C | p.Lys354Asn | missense | Exon 9 | Tolerated | Possibly damaging |
| Wu p023 | c.1062G > C | p.Lys354Asn | missense | Exon 9 | Tolerated | Possibly damaging |
| Wu p024 | c.1658A > C | p.His553Pro | missense | Exon 15 | D | Possibly damaging |
| Wu p025 | c.4316 T > A | p.Leu1439Ter | nonsense | Exon 32 | Tolerated | |
| Wu p027 | c.1754_1757delTAAC | p.Thr586fs | Frameshift Deletion | Exon 16 | ||
| Wu p030 | c.5665G > T | p.Glu1889Ter | nonsense | Exon 39 | Tolerated | |
| Wu p032 | c.2266C > T | p.Gln756Ter | nonsense | Exon 19 | Damaging | |
| Wu p033 | c.7348C > T | p.Arg2450Ter | nonsense | Exon 50 | Tolerated | |
| Wu p034 | c.910C > T | p.Arg304Ter | nonsense | Exon 9 | Tolerated | |
| Wu p035 | c.5580_5581insA | p.Asn1861fs | Frameshift Insertion | Exon 38 | ||
| Wu p038 | c.1246C > T | p.Arg416Ter | nonsense | Exon 11 | Tolerated | |
| Wu p039 | c.492_495 del AACT/He | p.Val166fs | Frameshift Deletion | Exon 5 | ||
| Wu p041 | c.910C > T | p.Arg304Ter | nonsense | Exon 9 | Tolerated | |
| Wu p043 | c. 3796 G > T | p.Glu1266Ter | nonsense | Exon28 | Tolerated | |
| Wu p044 | c.86_87delAC | p.29_29del | frameshift deletion | Exon2 | ||
| Wu p045 | c.6618_6618 delA | p.Thr2206fs | frameshift deletion | Exon43 | ||
| Wu p047 | c. 6818 A > C | p.Lys2273 Thr | missense | Exon46 | Tolerated | Possibly damaging |
| Wu p048 | c. 910 C > T | p.Arg304Ter | nonsense | Exon9 | Tolerated | |
| Wu p050 | c.2212dupT | p.Phe738fs | frameshift insertion | Exon18 | ||
| Wu p051 | c. 5170 C > T | p.Gln1724Ter | nonsense | Exon37 | Tolerated | |
| Wu p052 | c. 1224 T > A | p.Tyr408Ter | nonsense | Exon11 | Tolerated | |
| Wu p053 | c.7266_7267del AC | p.2422_2423del | frameshift deletion | Exon49 | ||
| Wu p054 | c. 574 C > T | p.Arg192Ter | nonsense | Exon5 | Tolerated | |
| Wu p055 | c. 574 C > T | p.Arg192Ter | nonsense | Exon5 | Tolerated | |
| Wu p058 | c. 3040 A > T | p.K1014Ter | nonsense | Exon23 | ||
| Wu p059 | c.288 + 1G > T | . | splicing | Exon3 | ||
| Wu p060 | c.4509dupT | p.Asn1503fs | frameshift insertion | Exon34 | ||
| Wu p064 | c. 479 G > T | p.Arg160Met | missense | Exon4 | Damaging | Possibly damaging |
| Wu p066 | c.1592delA | p.Gln531fs | frameshift deletion | Exon14 | ||
| Wu p067 | c.8070dupC | p.Tyr2690fs | frameshift insertion | Exon56 | ||
| Wu p068 | c.288 + 1G > T | . | splicing | Exon3 | ||
| Wu p070 | c.4990_4992AAA (GTT) | . | nonframeshift substitution | Exon37 | ||
| Wu p071 | c. 3826 C > T | p.Arg1276Ter | nonsense | Exon28 | Tolerated | |
| Wu p073 | c.2340_2346delACATGCA | p.780_782del | frameshift deletion | Exon20 | ||
| Wu p074 | c. 4107 C > A | p.Tyr1369Ter | nonsense | Exon30 | Tolerated | |
| Wu p075 | c. 5651 T > G | p.Phe1884Cys | missense | Exon39 | Damaging | Damaging |
| Wu p076 | c. 3888 T > G | p.Tyr1296Ter | nonsense | Exon29 | Tolerated | |
| Wu p077 | c. 3484 A > G | p.Met1162Val | missense | Exon26 | Tolerated | Benign |
| Wu p077 | c. 7189 G > A | p.Gly2397Arg | missense | Exon49 | Damaging | Damaging |
| Wu p080 | c. 1933 A > G | p.Met645Val | missense | Exon17 | Tolerated | Benign |
| Wu p081 | c.1754_1757del | p.Leu585fs | frameshift deletion | Exon16 | ||
| Wu p083 | c.2953dupC | p.Gly984fs | frameshift insertion | Exon22 | ||
| Wu p086 | c.6855C > A | p.Tyr2285Ter | nonsense | Exon46 | Tolerated | |
| Wu p087 | c. 4940 A > C | p.His1647Pro | missense | Exon37 | Tolerated | Damaging |
| Wu p088 | c.1754_1757del | p.Leu585fs | frameshift deletion | Exon16 | ||
| Wu p089 | c. 1466 A > G | p.Tyr489Cys | missense | Exon13 | Tolerated | Damaging |
| Wu p090 | c. 376 G > T | p.Glu126Ter | nonsense | Exon4 | Damaging | |
| Wu p092 | c. 3827 G > A | p.Arg1276Gln | missense | Exon28 | Damaging | |
| Wu p094 | c. 3796 G > T | p.Glu1266Ter | nonsense | Exon28 | Tolerated | Damaging |
| Wu p095 | c.1693dupG | p.Asp564fs | frameshift insertion | Exon15 | ||
| Wu p098 | c.1754_1757del | p.Leu585fs | frameshift deletion | Exon16 | ||
| Wu p100 | c. 1318 C > T | p.Arg440Ter | nonsense | Exon12 | Tolerated |
abold lettering indicated as novel variants
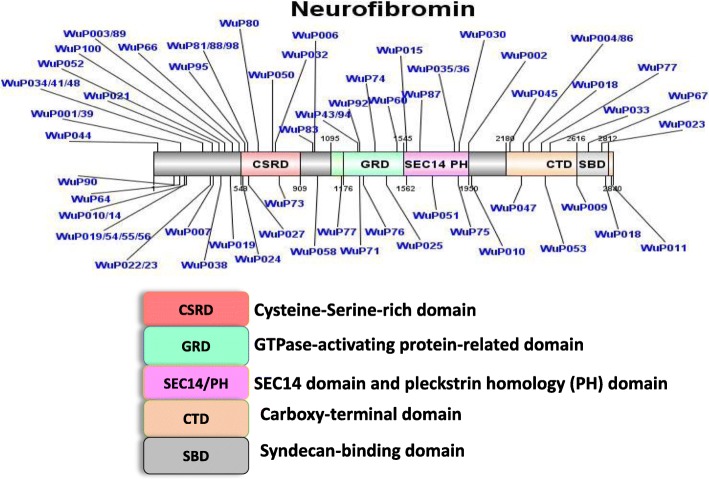
Genetic alterations in the NF1 gene were detected as frameshift, nonsense, splice, missense mutations, and frame deletions or duplications from the first NGS panel screening (Fig. 1). These variants distributed along the NF1 gene without any clustering hotspot domain. Intragenic NF1 point mutations were found in 46 patients, 28 nonsense and 18 missense mutations. Small insertions and/or deletions were identified in 24 patients and most of them with frameshift consequences. Splice alterations were detected only in three patients. Four patients (Wu p003, Wu p018, Wu p019, and Wu p077) possessed more than one NF1 variant. Two patients with BRAF variants (c.74C > T in Exon1: p.Pro25Leu; c.G316A in Exon 3: p.Gly106Arg) were identified from this NGS screening. Both these patients also carried NF1 mutations (Fig. 2). On comparing with the Human Gene Mutation Database (HGMD, Institute of Medical Genetics, Cardiff, http://www.hgmd.org/), and Leiden Open Variation Database (LOVD: www.lovd.nl/NF1), we found that 48 variants of NF1 gene and two of BRAF gene are supposed to be novel (presented in bold in Table 2 and Table 3). All these novel mutations in this study were tested in 100 normal alleles.
Fig. 1.
Details of the 73 NF1 genetic variations identified by NGS targeted gene sequencing. The position of genetic variations detected in the NF1 gene from each patient is shown and their relationship to a possible defect of NF1 gene was also included. Known functional domains of Neurofibromin: CSRD > cysteine–serine-rich domain; GRD > GTPase-activating protein-related domain; SEC14/PH > SEC14 domain and pleckstrin homology (PH) domain; CTD > Carboxy-terminal domain; SBD > Syndecan-binding domain
Fig. 2.
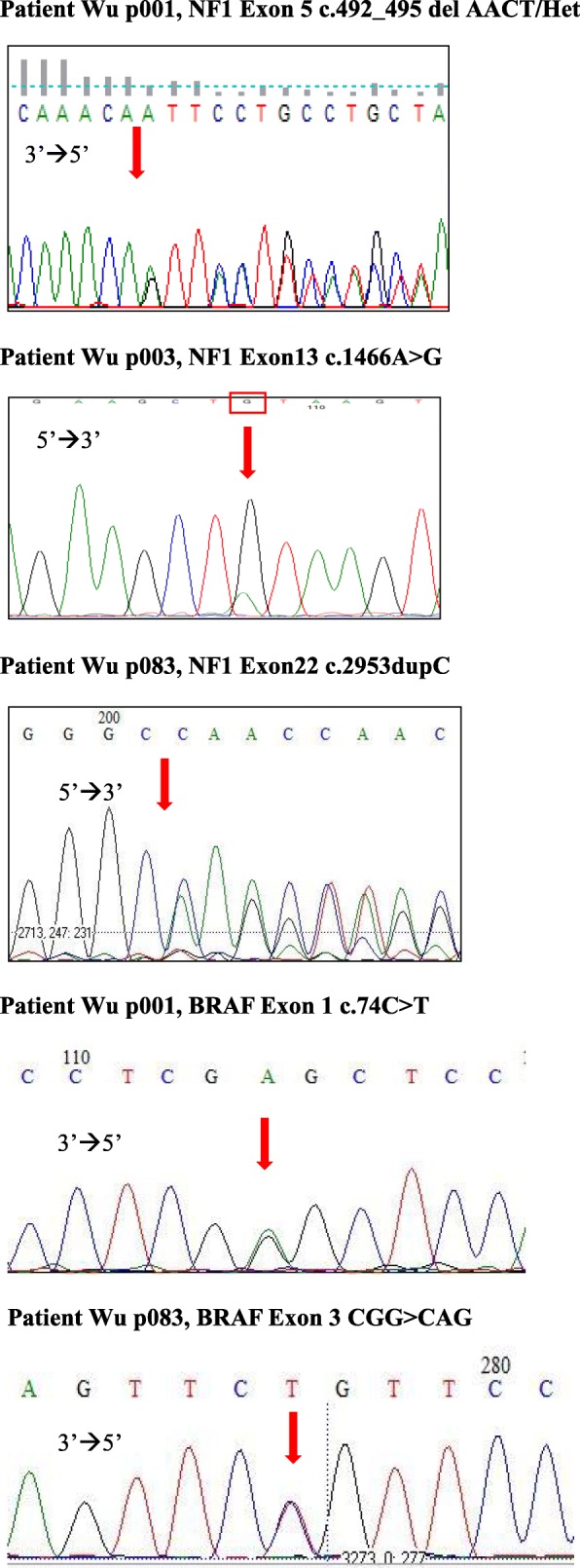
Some represented results of Sanger sequencing at the mutation site with blood sample
Table 3.
NF1 multi-exon deletions or duplications
| NAME | MLPA | Clinical features | Tumor type |
|---|---|---|---|
| Wu p008 | 3’ UTR del/He | café-au-lait spot | Multiple cutaneous tumor |
| Wu p013 | Exon 10 ~ 58 del/He | café-au-lait spot | whole body |
| Wu p016 | Exon 1 ~ 58 del/He | café-au-lait spot, skin nodules | Two nodules of tumor involving the dermis and composed of spindle cells with wavy elongated nuclei |
| Wu p020 | Exon 28~ 39 del/He | café-au-lait spot | Neurofibroma |
| Wu p029 | Exon 4C~ 6 (no Exon 5) | café-au-lait spot, skin nodules | Right facial plexiform neurofibroma |
| Wu p031 | Exon 1B~ 49 | café-au-lait spot, skin nodules | multiple nodules over face and bilateral forearms |
| Wu p037 | Exon1~ 58 del/He | café-au-lait spot/List Nodules in the Iris | multiple nodules over face |
| Wu p061 | Exon37~ 51 del/He | café-au-lait spot | NF1 with optic nerve glioma |
| Wu p062 | Exon2~ 8 del/He | café-au-lait spot/List Nodules in the Iris | right thigh subcutaneous layer soft tissue nodule |
| Wu p065 | Exon 28–29 del/He | café-au-lait spot | lower limb plexiform NF |
| Wu p069 | Exon1~ 58 del/He | café-au-lait spot/List Nodules in the Iris | Neurofibroma over back |
| Wu p082 | Exon2~ 5 del/He | café-au-lait spot | plexiform neurofibroma over buttock |
| Wu p085 | Exon2~ 5 del/He | café-au-lait spot | left optic nerve glioma & liposarcoma |
| Wu p093 | Exon 1B ~ 4B | skin nodules/List Nodules in the Iris | Plexiform Neurofibroma |
| Wu p099 | Exon 4C~ 6 (no Exon 5) | café-au-lait spot, skin nodules | skin and soft tissue on right face, plexiform neurofibroma |
Spectrum of NF1 mutations identified by MLPA analysis
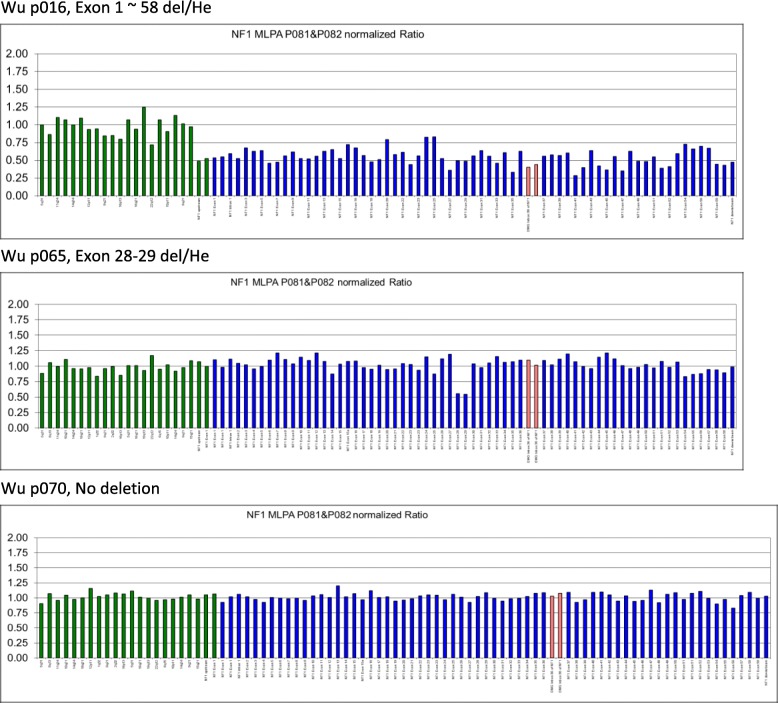
For patients who showed no detected mutations by our NGS panel screening, we then analyzed possible exons deletion/duplication within the NF1 gene using multiplex ligation-dependent probe amplification (MLPA) approach. Whole NF1 gene deletions were found in three patients and fifteen multi-exon deletions within the NF1 gene were obtained in this cohort of NF1 patients. Most of these exon deletions were only seen once in this study (Fig. 3, Table 3).
Fig. 3.
Examples of multi-exon deletions detected by multiplex ligation-dependent probe amplification. In this study, we used the Coffalyser program (version 3.5) for peak area normalization and gene dosage calculation. Two copies of the genome have a relative peak area value of approximately 1.0. A reduction in the peak area value to < 0.7 indicates the occurrence of a deletion
Clinical features of NF1 patients with concurrence of NF1-BRAF mutations
Patient (Wu p001) was diagnosed as NF1 at the age of seven years by the presence of left craniofacial plexiform neurofibromas, infiltrative at the left temporal scalp, nodular subcutaneous tissue of the cheek, masticator space and probably the parotid space. He had multiple café-au-lait spots but no Lisch nodules. His brain magnetic resonance image (MRI) showed multiple unknown bright objects at pons, bilateral cerebellar hemisphere, globus pallidus and right thalamus. His left temporal and zygomatic bone showed progressive enlargement. His father (Wu p039) was the first patient with NF1 in this family and was diagnosed as having NF1 because of the presence of Lisch nodules, skeletal dysplasia, hundreds of café-au-lait spots and cutaneous nodular neurofibromas all over the body. This father and son are both intellectually normal. They both share the same genetic alterations on NF1 (c.492_495 del AACT/p.Val166fs) and BRAF (c.74C > T/p.Pro25Leu) gene (Fig.2). Another patient (Wu p083) was diagnosed as having NF1 at age of five years. He had multiple café-au-lait spots and Lisch nodules with soft tissue mass over the right back. He also had unspecified heart anomaly and T-spine scoliosis. His brain MRI showed white matter hyperintensity suggesting spongiform change at the left globus pallidus, dorsal pons, and bilateral cerebellar hemisphere (dentate nuclei) but no definite evidence of optic gliomas. He was detected as having NF1 (c.2953dupC/p.Gly984fs) and BRAF (c.316 G > A/p.Gly106Arg) genetic variants in the first NGS screening (Fig. 2, Table 2). These three patients presented no data for their definite atrial septal defect, ventricular septal defect and patent ductus arteriosus. In addition, none of these patients show the typical features of NF1–Noonan syndrome, Noonan syndrome or CFC syndrome.
Discussion
We here assessed a DNA-based approach combining targeted gene panel screening with MLPA analysis in a cohort of clinically suspected NF1 patients. On targeted gene panel screening, we identified 73 NF1 mutations and two BRAF variants (c.74C > T: p.Pro25Leu; c.316 G > A: p.Gly106Arg) in a total of 100 NF1 patients from 95 families diagnosed as having NF1 on the basis of the clinical criteria. These mutations are heterogeneous and distribute without clustering in either cysteine–serine-rich domain or within the GAP-related domain. For patients in whom mutations were not detected by our NGS panel screening, we detected fifteen multi-exon deletions within the NF1 gene by Multiplex Ligation-Dependent Probe Amplification (MLPA) analysis (~ 15% of detected NF1 alterations). A multi-step mutation detection protocol has been used for over 95% of pathogenic NF1 mutations in different laboratories [15–21, 26–28]. The NF1 mutations were detected in our study was in 92.6% (88/95) of the subjects when five patients who did not completely met the clinical diagnostic criteria were excluded. Our analysis and this study may have missed the genetic variants residing in the promotor and intronic untranscribed non-coding regions or those involved in large genomic rearrangements or epigenetic mechanisms. We anticipate that whole-genome analysis may provide further insights for the information related to this issue.
NF1 is a progressive disorder complicated by the variability of disease expression. Beyond the primary concern of cutaneous/dermal neurofibromas, pigmented lesions, and the occasional limb abnormalities, the majority of NF1 patients do not fulfill the NIH criteria. Only ~ 30% of NF1 patients develop clinically detectable plexiform neurofibromas, and many features of NF1 only display café-au-lait spots and mild symptoms or no major disease complications in their early life [5, 36, 37]. Although neurofibromatosis type 1 is the most common syndrome seen in children with multiple café-au-lait spots, other syndromes associated with one or more café-au-lait spots include McCune-Albright syndrome, Legius syndrome, Noonan syndrome and other neuro-cardio-facio-cutaneous syndromes [38]. It also shares some features including reduced growth, facial dysmorphia, cardiac defects, skeletal and ectodermal anomalies, variable cognitive deficits, and susceptibility to certain malignancies with a group of clinically distinct developmental disorders [23–25]. Neurofibromatosis type I, Noonan syndrome, LEOPARD syndrome, and cardiofaciocutaneous syndromes were usually grouped as “neuro-cardio-facio-cutaneous” (NCFC) syndromes but are now called as “RaSopathies”. All these disorders involve a common Ras–Raf–signaling pathway [39–41]. To our knowledge, germline KRAS mutations occasionally occur in Noonan (2–4%) [42–46] and CFC syndromes (< 2%) [43–45, 47, 49]. Germline BRAF mutations can cause CFC syndrome (approximately 50–75%) [44, 47–50], Noonan syndrome [47, 50], and LEOPARD syndrome type 3 (< 2%) [50, 51]. However, these individuals usually are not associated with neurofibromas (Table 4).
Table 4.
BRAF mutations in patients with RASopathies
| Patient | Germline mutation | Clinical Phenotypes | Tumor type |
|---|---|---|---|
| Wu p001 (this study) |
NF1 Exon 5, c.492_495 del AACT/p.Val166fs |
Café-au-lait spots, Cutaneous neurofibroma, left zygoms progressive enlargement | plexiform neurofibroma |
| BRAF Exon 1, c.74C > T/p.Pro25Leu | |||
| Wu p083 (this study) | NF1 Exon22, c.2953dupC/p.Gly984fs | Café-au-lait spots, unspecified cardiac anomaly, Lisch Nodules in the Iris, T-spine scoliosis | paraspinal plexiform neurofibroma |
| BRAF Exon 3, c. 316 G > A/p.Gly106Arg | |||
| Noonan syndrome (NS) | BRAF (T241 M; T241R; W531C; L597 V) | Short stature, dysmorphic facial features, mild-to-moderate cognitive deficits, skeletal anomalies, and hypotonia | |
| Cardio-facio-cutaneous syndrome (CFCS) | BRAF (L245F; A246P; T241P; Q257R; G469E; etc) | Dysmorphic facies, cardiac defects, and skin and skeletal anomalies | |
| Leopard syndrome Type 3 | BRAF (T241P; L245F) | Craniofacial anomalies, short and webbed neck, cardiac conduction defects, Multiple pigmented skin lesions and showed growth retardation, delayed puberty, and delayed bone age. | undetected |
*bold lettering indicated as novel variants
Phenotypic variation could result from different expression patterns of mutated genes, as well as from different mechanisms that disturb RAS signaling through specific interactions with effector and regulatory proteins for different mutants. Variability could also result from the feedback mechanisms that can affect upstream molecules (like RAS) but not downstream molecules [40]. Therefore, a NGS panel with high coverage of Ras–signaling components should be very useful in clinical diagnosis. However, we cannot yet explain how the concurrence of NF1 and BRAF variants contributes to NF1 in these patients.
Conclusion
Differential diagnosis of NF1-like patients is still challenging owing to its clinical complexity. A genetic screening using a NGS panel in high coverage of Ras–signaling components combined with Multiple Ligation-Dependent Probe Amplification analysis should enable us to get the molecular control of these clinically overlapping disorders. We believe that the availability of whole-genome analysis will provide an opportunity for the genetic diagnosis of NF1 and will bring more insights for the development of NF1.
Acknowledgements
We are grateful to all the participating patients and their families who made these studies possible. The authors also thank the DNA Sequencing Core Laboratory at the Chang Gung Memorial Hospital, Linko.
Funding
Dr. Wu-Chou received funding by grants from the National Science Council, Taiwan (NSC94–2320-B-182A-020) and the Chang Gung Research Foundation (CMRPG 33139, CMRPG 331393, CMRPG 3D0391–3).
Availability of data and materials
The datasets generated in the current study are available from the corresponding author on request.
Abbreviations
- CSRD
Cysteine–serine-rich domain
- CTD
Carboxy-terminal domain
- GAP-related domain
GTPase activating protein-related domain
- GRD
GTPase-activating protein-related domain
- HGMD
Human Gene Mutation Database
- IGV
Integrative Genome Viewer
- indels variants
insertions/deletions variants
- ISP
Ion Sphere Particles
- LOVD
Leiden Open Variation Database
- MAF
Minor Allele Frequency
- MLPA
Multiple Ligation-Dependent Probe Amplification
- MRI
Magnetic Resonance Image
- NCFC syndromes
Neuro-Cardio-Facio-Cutaneous syndromes
- NF1
Neurofibromatosis type 1
- NGS
Next-Generation Sequencing
- PCR
Polymerase Chain Reaction
- PH
pleckstrin homology domain
- PolyPhen2
Polymorphism Phenotyping v2
- Ras/MAPK signaling pathway
Ras/Mitogen-Activated Protein Kinases signaling pathway
- SBD
Syndecan-binding domain
- SEC14/PH
SEC14 domain and Pleckstrin Homology domain
- SIFT
Sorting Intolerant from Tolerant
- SNVs
Single Nucleotide Variants
Authors’ contributions
YHWC conceived and designed the study, carried out the lab data analysis, interpreted the results and drafted the manuscript. The study subjects were assessed by the pediatric physician JLL, neurological physician THY, and plastic surgeons CHL, CCY, KTC, and YRC at the Chang Gung Craniofacial Center. YTL contributed to participant recruitment, acquisition of samples and experimental data. ZCH and HWC performed molecular genetic experiments. All authors have approved the final manuscript for submission.
Ethics approval and consent to participate
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. Ethics approval was obtained by the institutional review board (102-0226A3) at the Chang Gung Memorial Hospital. Informed consent was individually obtained from all participants included in the study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yah-Huei Wu-Chou, Phone: +886-3-3281200, Email: yhwc8876@gmail.com.
Tzu-Chao Hung, Email: s9315144@yahoo.com.tw.
Yin-Ting Lin, Email: ingjylin@gmail.com.
Hsing-Wen Cheng, Email: aimee10221@gmail.com.
Ju-Li Lin, Email: lin001@cgmh.org.tw.
Chih-Hung Lin, Email: bossbusiness1505@gmail.com.
Chung-Chih Yu, Email: yu@drccyu.com.
Kuo-Ting Chen, Email: philip.ktchen@gmail.com.
Tu-Hsueh Yeh, Email: neuroyeh@gmail.com.
Yu-Ray Chen, Phone: 886-3-3281200, Email: uraychen@cgmh.org.tw, Email: uraychen@adm.cgmh.org.tw.
References
- 1.Ferner RE, Huson SM, Thomas N, Moss C, Willshaw H, Evans DG, et al. Guidelines for the diagnosis and management of individuals with neurofibromatosis 1. J Med Genet. 2007;44:81e8. doi: 10.1136/jmg.2006.045906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyd KP, Korf BR, Theos A. Neurofibromatosis type 1. J Am Acad Dermatol. 2009;61:1–14. doi: 10.1016/j.jaad.2008.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Williams VC, Lucas J, Babcock MA, Gutmann DH, Korf B, Maria BL. Neurofibromatosis type 1 revisited. Pediatrics. 2009;123:124–133. doi: 10.1542/peds.2007-3204. [DOI] [PubMed] [Google Scholar]
- 4.Ferner RE. The neurofibromatoses. Pract Neurol. 2010;10:82–93. doi: 10.1136/jnnp.2010.206532. [DOI] [PubMed] [Google Scholar]
- 5.DeBella K, Szudek J, Friedman JM. Use of the national institutes of health criteria for diagnosis of neurofibromatosis 1 in children. Pediatrics. 2000;105:608–614. doi: 10.1542/peds.105.3.608. [DOI] [PubMed] [Google Scholar]
- 6.Jett K, Friedman JM. Clinical and genetic aspects of neurofibromatosis. Genet Med. 2010;12:1–11. doi: 10.1097/GIM.0b013e3181bf15e3. [DOI] [PubMed] [Google Scholar]
- 7.Cichowski K, Jacks T. NF1 tumor suppressor gene function: narrowing the gap. Cell. 2001;104:593–604. doi: 10.1016/S0092-8674(01)00245-8. [DOI] [PubMed] [Google Scholar]
- 8.Gottfried ON, Viskochil D, Couldwell WT. Neurofibromatosis type 1 and tumorigenesis: molecular mechanisms and therapeutic implications. Neurosurg Focus. 2010;28:1–9. doi: 10.3171/2009.11.FOCUS09221. [DOI] [PubMed] [Google Scholar]
- 9.Xu GF, O'Connell P, Viskochil D, Cawthon R, Robertson M, Culver M, et al. The neurofibromatosis type 1 gene encodes a protein related to GAP. Cell. 1990;62:599–608. doi: 10.1016/0092-8674(90)90024-9. [DOI] [PubMed] [Google Scholar]
- 10.Basu TN, Gutmann DH, Fletcher JA, Glover TW, Collins FS, Downward J. Aberrant regulation of ras proteins in malignant tumour cells from type 1 neurofibromatosis patients. Nature. 1992;356:713–715. doi: 10.1038/356713a0. [DOI] [PubMed] [Google Scholar]
- 11.Viskochil D. Genetics of neurofibromatosis 1 and the NF1 gene. J Child Neurol. 2002;17:562–570. doi: 10.1177/088307380201700804. [DOI] [PubMed] [Google Scholar]
- 12.Cawthon RM, Weiss R, Xu GF, Viskochil D, Culver M, Stevens J, et al. A major segment of the neurofibromatosis type 1 gene: cDNA sequence, genomic structure, and point mutations. Cell. 1990;62:193–201. doi: 10.1016/0092-8674(90)90253-B. [DOI] [PubMed] [Google Scholar]
- 13.Kluwe L, Siebert R, Gesk S, Friedrich RE, Tinschert S, Kehrer-Sawatzki H, et al. Screening 500 unselected neurofibromatosis type 1 patients for deletions of the NF1 gene. Hum Mutat. 2004;23:111–116. doi: 10.1002/humu.10299. [DOI] [PubMed] [Google Scholar]
- 14.Griffiths S, Thompson P, Frayling I, Upadhyaya M. Molecular diagnosis of neurofibromatosis type 1: 2 years’ experience. Familial Cancer. 2007;6:21–34. doi: 10.1007/s10689-006-9001-3. [DOI] [PubMed] [Google Scholar]
- 15.Wimmer K, Yao S, Claes K, Kehrer-Sawatzki H, Tinschert S, De Raedt T, et al. Spectrum of single- and multiexon NF1 copy number changes in a cohort of 1,100 unselected NF1 patients. Genes Chromosomes Cancer. 2006;45:265–276. doi: 10.1002/gcc.20289. [DOI] [PubMed] [Google Scholar]
- 16.De Luca A, Bottillo I, Dasdia MC, Morella A, Lanari V, Bernardini L, et al. Deletions of NF1 gene and exons detected by multiplex ligation-dependent probe amplification. J Med Genet. 2007;44:800–808. doi: 10.1136/jmg.2007.053785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Messiaen LM, Wimmer K. NF1 mutational spectrum. Neurofibromatoses. In: Kaufmann D, editor. Monographs in Human Genetics. Basel: Karger; 2008. pp. 63–77. [Google Scholar]
- 18.Pasmant E, Sabbagh A, Spurlock G, Laurendeau I, Grillo E, Hamel MJ, et al. NF1 microdeletions in neurofibromatosis type 1: from genotype to phenotype. Hum Mutat. 2010;31:E1506–E1518. doi: 10.1002/humu.21271. [DOI] [PubMed] [Google Scholar]
- 19.Sabbagh A, Pasmant E, Imbard A, Luscan A, Soares M, Blanché H, et al. NF1 molecular characterization and neurofibromatosis type I genotype-phenotype correlation: the French experience. Hum Mutat. 2013;34:1510–1518. doi: 10.1002/humu.22392. [DOI] [PubMed] [Google Scholar]
- 20.van Minkelen R, van Bever Y, Kromosoeto JN, Withagen-Hermans CJ, Nieuwlaat A, Halley DJ, et al. A clinical and genetic overview of 18 years neurofibromatosis type 1 molecular diagnostics in the Netherlands. Clin Genet. 2014;85:318–327. doi: 10.1111/cge.12187. [DOI] [PubMed] [Google Scholar]
- 21.Imbard A, Pasmant E, Sabbagh A, Luscan A, Soares M, Goussard P, et al. NF1 single and multi-exons copy number variations in neurofibromatosis type 1. J Hum Genet. 2015;60:221–224. doi: 10.1038/jhg.2015.6. [DOI] [PubMed] [Google Scholar]
- 22.Bianchessi D, Morosini S, Saletti V, Ibba MC, Natacci F. Esposito S, et al. 126 novel mutations in Italian patients with neurofibromatosis type 1. Mol Genet Genomic Med. 2015;3:513–525. doi: 10.1002/mgg3.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aoki Y, Niihori T, Narumi Y, Kure S, Matsubara Y. The RAS/MAPK syndromes: novel roles of the RAS pathway in human genetic disorders. Hum Mutat. 2008;29:992–1006. doi: 10.1002/humu.20748. [DOI] [PubMed] [Google Scholar]
- 24.Tidyman E, Rauen KA. The RASopathies: developmental syndromes of Ras/MAPK pathway dysregulation. Curr Opin Genet Dev. 2009;19:230–236. doi: 10.1016/j.gde.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rauen KA. The RASopathies. Annu Rev Genomics Hum Genet. 2013;14:355–369. doi: 10.1146/annurev-genom-091212-153523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fahsold R, Hoffmeyer S, Mischung C, Gille C, Ehlers C, Kücükceylan N, et al. Minor lesion mutational spectrum of the entire NF1 gene does not explain its high mutability but points to a functional domain upstream of the GAP-related domain. Am J Hum Genet. 2000;66:790–818. doi: 10.1086/302809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valero MC, Martín Y, Hernández-Imaz E, Marina Hernández A, Meleán G, Valero AM, et al. A highly sensitive genetic protocol to detect NF1 mutations. J Mol Diagn. 2011;13:113–122. doi: 10.1016/j.jmoldx.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ko JM, Sohn YB, Jeong SY, Kim HJ, Messiaen LM. Mutation spectrum of NF1 and clinical characteristics in 78 Korean patients with neurofibromatosis type 1. Pediatr Neurol. 2013;48:447–453. doi: 10.1016/j.pediatrneurol.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 29.Uusitalo E, Hammais A, Palonen E, Brandt A, Mäkelä VV, Kallionpää R, et al. Neurofibromatosis type 1 gene mutation analysis using sequence capture and high-throughput sequencing. Acta Derm Venereol. 2014;94:663–666. doi: 10.2340/00015555-1843. [DOI] [PubMed] [Google Scholar]
- 30.Balla B, Árvai K, Horváth P, Tobiás B, Takács I, Nagy Z, et al. Fast and robust next-generation sequencing technique using ion torrent personal genome machine for the screening of neurofibromatosis type 1 (NF1) gene. J Mol Neurosci. 2014;53:204–210. doi: 10.1007/s12031-014-0286-7. [DOI] [PubMed] [Google Scholar]
- 31.Maruoka R, Takenouchi T, Torii C, Shimizu A, Misu K, Higasa K, et al. The use of next-generation sequencing in molecular diagnosis of neurofibromatosis type 1: a validation study. Genet Test Mol Biomarkers. 2014;18:722–735. doi: 10.1089/gtmb.2014.0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pasmant E, Parfait B, Luscan A, Goussard P, Briand-Suleau A, Laurendeau I, et al. Neurofibromatosis type 1 molecular diagnosis: what can NGS do for you when you have a large gene with loss of function mutations? Eur J Hum Genet. 2015;23:596–601. doi: 10.1038/ejhg.2014.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang J, Tong H, Fu X, Zhang Y, Liu J, Cheng R, et al. Molecular characterization of NF1 and neurofibromatosis type 1 genotype-phenotype correlations in a Chinese population. Sci Rep 2015; 5: 11291. [DOI] [PMC free article] [PubMed]
- 34.Cunha KS, Oliveira NS, Fausto AK, de Souza CC, Gros A, Bandres T, et al. Hybridization capture-based next-generation sequencing to evaluate coding sequence and deep Intronic mutations in the NF1 gene. Genes (Basel) 2016;7:133. doi: 10.3390/genes7120133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. ACMG laboratory quality assurance committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gutmann DH. Eliminating barriers to personalized medicine. Neurology. 2014;83:463–471. doi: 10.1212/WNL.0000000000000652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tadini G, Milani D, Menni F, Pezzani L, Sabatini C, Esposito S. Is it time to change the neurofibromatosis 1 diagnostic criteria? Eur J Intern Med. 2014;25:506–510. doi: 10.1016/j.ejim.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 38.Shah KN. The diagnostic and clinical significance of café-au-lait macules. Pediatr Clin N Am. 2010;57(5):1131–1153. doi: 10.1016/j.pcl.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 39.Bentires-Alj M, Kontaridis MI, Neel BG. Stops along the RAS pathway in human genetic disease. Nat Med. 2006;12:283–285. doi: 10.1038/nm0306-283. [DOI] [PubMed] [Google Scholar]
- 40.Denayer E, de Ravel T., Legius E. Clinical and molecular aspects of RAS related disorders. Journal of Medical Genetics. 2008;45(11):695–703. doi: 10.1136/jmg.2007.055772. [DOI] [PubMed] [Google Scholar]
- 41.Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer. 2007;7:295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- 42.Carta C, Pantaleoni F, Bocchinfuso G, Stella L, Vasta I, Sarkozy A, et al. Germline missense mutations affecting KRAS isoform B are associated with a severe Noonan syndrome phenotype. Am J Hum Genet. 2006;79:129–135. doi: 10.1086/504394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schubbert S, Zenker M, Rowe SL, Böll S, Klein C, Bollag G, et al. Germline KRAS mutations cause Noonan syndrome. Nat Genet. 2006;38:331–336. doi: 10.1038/ng1748. [DOI] [PubMed] [Google Scholar]
- 44.Nava C, Hanna N, Michot C, Pereira S, Pouvreau N, Niihori T, et al. Cardiofacio-cutaneous and Noonan syndromes due to mutations in RAS/MAPK signaling pathway: genotype/phenotype relationships and overlap with Costello syndrome. J Med Genet. 2007;44:763–771. doi: 10.1136/jmg.2007.050450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zenker M, Lehmann K, Schulz AL, Barth H, Hansmann D, Koenig R, et al. Expansion of the genotypic and phenotypic spectrum in patients with KRAS germline mutations. J Med Genet. 2007;44:131–135. doi: 10.1136/jmg.2006.046300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lo FS, Lin JL, Kuo MT, Chiu PC, Shu SG, Chao MC, et al. Noonan syndrome caused by germline KRAS mutation in Taiwan: report of two patients and a review of the literature. Eur J Pediatr. 2009;168:919–923. doi: 10.1007/s00431-008-0858-z. [DOI] [PubMed] [Google Scholar]
- 47.Niihori T, Aoki Y, Narumi Y, Neri G, Cavé H, Verloes A, et al. Germline KRAS and BRAF mutations in cardio-facio- cutaneous syndrome. Nat Genet. 2006;38:294–296. doi: 10.1038/ng1749. [DOI] [PubMed] [Google Scholar]
- 48.Rodriguez-Viciana P, Tetsu O, Tidyman WE, Estep AL, Conger BA, Cruz MS, et al. Germline mutations in genes within the MAPK pathway cause cardio-facio-cutaneous syndrome. Science. 2006;311:1287–1290. doi: 10.1126/science.1124642. [DOI] [PubMed] [Google Scholar]
- 49.Schulz AL, Albrecht B, Arici C, van der Burgt I, Buske A, Gillessen-Kaesbach G, et al. Mutation and phenotypic spectrum in patients with cardio-Facio-cutaneous and Costello syndrome. Clin Genet. 2008;73:62–70. doi: 10.1111/j.1399-0004.2007.00931.x. [DOI] [PubMed] [Google Scholar]
- 50.Sarkozy A, Carta C, Moretti S, Zampino G, Digilio MC, Pantaleoni F, et al. Germline BRAF mutations in Noonan, LEOPARD, and cardiofaciocutaneous syndromes: molecular diversity and associated phenotypic spectrum. Hum Mutat. 2009;30:695–702. doi: 10.1002/humu.20955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koudova M, Seemanova E, Zenker M. Novel BRAF mutation in a patient with LEOPARD syndrome and normal intelligence. Eur J Med Genet. 2009;52:337–340. doi: 10.1016/j.ejmg.2009.04.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated in the current study are available from the corresponding author on request.