Abstract
Background
Extended-spectrum β-lactamases (ESBLs)-producing Escherichia coli (E. coli) isolates in environment water become progressively a potential threat to public health, while the detailed information about the ESBL-producing E. coli isolates in the rivers and lakes in Northwest China is scarce. In the present study, it was aimed to characterize the ESBL-producing E. coli isolated from the surface waters in Northwest China.
Results
A total of 2686 E. coli isolates were obtained from eleven rivers and lakes in Northwest China to screen for ESBL producers. Seventy-six (2.8%) isolates were classified as ESBL producers, and phylogenic groups D and A accounted for 59.2% of the ESBL producers. CTX-Ms were the predominant ESBLs genotype, and they were represented by seven blaCTX-M subtypes. blaCTX-M-14 was the most prevalent specific CTX-M gene, followed by blaCTX-M-9, blaCTX-M-123, blaCTX-M-15, blaCTX-M-27, blaCTX-M-1 and blaCTX-M-65. Moreover, 54 of the 76 ESBL producers carried at least one plasmid-mediated quinolone resistance (PMQR) gene, and aac(6′)-Ib-cr was predominant. The overall occurrence of virulence factors ranged from 1.3% (eae) to 48.7% (traT). Thirty-seven sequence types (STs) were confirmed among the 76 ESBL producers, and the predominant was ST10, which was represented by 10 isolates; importantly, clone B2-ST131, associated with severe infections in humans and animals, was detected three times.
Conclusion
The prevalence of ESBL-producing E. coli from the rivers and lakes in Northwest China was low (2.8%), and the extraintestinal pathogenic E. coli (ExPEC) pathotype was the most commonly detected on the basis of the virulence factor profiles. 76.3% of ESBL producers harbored more than one β-lactamase gene, and blaCTX-M-14 was the predominant genotype. Notably, one ST131 isolate from Gaogan Canal simultaneously harbored blaCTX-M-9, blaCTX-M-15, blaCTX-M-123, blaKPC-2, blaNDM-1, blaOXA-2 as well as the PMQR genes qnrA, qnrS and aac(6′)-Ib-cr.
Keywords: Escherichia coli, Surface water, Antibiotic resistance, β-Lactamase, PMQR
Background
The use of a wide variety of antimicrobials in human medicine, veterinary clinics, livestock industries and aquaculture has resulted in the emergence and spread of antibiotic-resistant bacteria in different environments, particularly in many developing countries [1, 2]. It becomes evident that the resistance genes can be introduced into the natural bacterial community as the antibiotic-resistant bacteria in humans and animals entered the water bodies [3]. Hence, it is necessary to clarify the potential threat associated with the occurrence of antibiotic-resistant bacteria in water environments in order to further evaluate public health risk and prevent waterborne infections. As one of the most typical indicator bacterium of fecal contamination in the environments, Escherichia coli (E. coli) can easily acquire resistance to antibiotics consumption in humans and animals [4]. Generally, pathogenic E. coli isolates were categorized into several pathotypes based on the clinical symptoms of the patients and the distinct virulence traits of the bacteria. Therefore, E. coli isolates are characterized by their virulence properties and mechanisms of pathogenicity into the enteropathogenic E. coli (EPEC), enterotoxigenic E. coli (ETEC), shiga toxin-producing E. coli (STEC), enteroinvasive E. coli (EIEC), enteroaggregative E. coli (EAEC) as well as extraintestinal pathogenic E. coli (ExPEC) [5, 6]. STEC isolates are defined as E. coli isolates expressing either stx1 or stx2; EPEC isolates are defined as eae-harboring diarrheagenic E. coli isolates that do not possess the stx gene; ETEC isolates are characterized by estA and eltB; isolates carrying aggR and ipaH are referred to as EAEC and EIEC, respectively [7]. Lastly, ExPEC isolates are associated with fyuA, iutA, afa, papA, focG, sfaS, kpsMII, hlyD and traT. Thus, the pathotypes of the uncharacterized isolates can be inferred from their virulence properties.
Since the extended-spectrum β-lactamases (ESBLs) was firstly reported in 1979 [8], the prevalence of ESBL-producing bacteria have been frequently detected worldwide from clinical isolates due to the increasing use of β-lactam antibiotics and carbapenems; the latter are usually used as the last resort for most serious bacterial infections. Moreover, some ESBL-producing isolates have been recovered from surface waters, where contamination from unmetabolized antibiotics may exert a selective pressure on bacteria, resulting in the emergence and spread of antibiotic-resistant isolates, especially the multidrug-resistant (MDR) isolates during their migration in water resources [3]. Relatedly, plasmid-mediated quinolone resistance (PMQR) determinants also pose a serious threat to public health, and some PMQR genes are considered to be associated with the ESBLs encoding genes [9]. The spread of E. coli co-expressing quinolone resistance along with ESBLs into rivers and lakes is worrisome and contributes to the growing concerns about resistant E. coli and their potential hazards to the environment.
Until now, little data are available on the ESBL-producing E. coli isolates in the surface waters in Northwest China. Thus, the current study was designed to gain insight into the prevalence of ESBL-producing E. coli isolates obtained throughout March 2015 to November 2016 from the rivers and lakes in Shaanxi province, and to further analyze the molecular characteristics of the ESBL producers.
Methods
Collection of isolates
Between March 2015 and November 2016, a total of 2686 E. coli isolates were obtained from eleven water bodies located in Shaanxi province, Northwest China, including Hei River (n = 177), Ying Lake (n = 194), Xianyang Lake (n = 196), Qishui River (n = 264), East Lake of Fengxiang county (n = 154), Wei River (n = 343), Ba River (n = 256), Shichuan River (n = 294), Xiaowei River (n = 265), Qixing River (n = 276) and Gaogan Canal of Yangling (n = 267) (Fig. 1). Among these water bodies, Hei River functioned as a public water supply source, while the others were scenic spots or functioned as floodways of the cities and countryside. All sampling sites were sampled once or multiple times, and all samples were collected in sterile 500-ml polyethylene bottles without preservatives and transported at 4 °C to the Veterinary Pharmacology Laboratory in Northwest A&F University, where primary isolation of E. coli was performed. Briefly, multiple volumes of untreated water were membrane filtered directly through 0.45-μm pore size filters, and the filters were placed on MacConkey agar plates (Solarbio Science & Technology, Co., Ltd., Beijing, China) at 37 °C for the identification of E. coli isolates. All 2686 putative E. coli colonies on MacConkey agar were restreaked onto Eosin Methylene Blue agar (Solarbio Science & Technology, Co., Ltd., Beijing, China), and then the suspicious colonies of E. coli were further identified with standard biochemical tests. Finally, the confirmed isolates as E. coli were stored at − 80 °C in Tryptic Soy broth (Solarbio Science & Technology, Co., Ltd., Beijing, China) containing 30% glycerol until use.
Fig. 1.
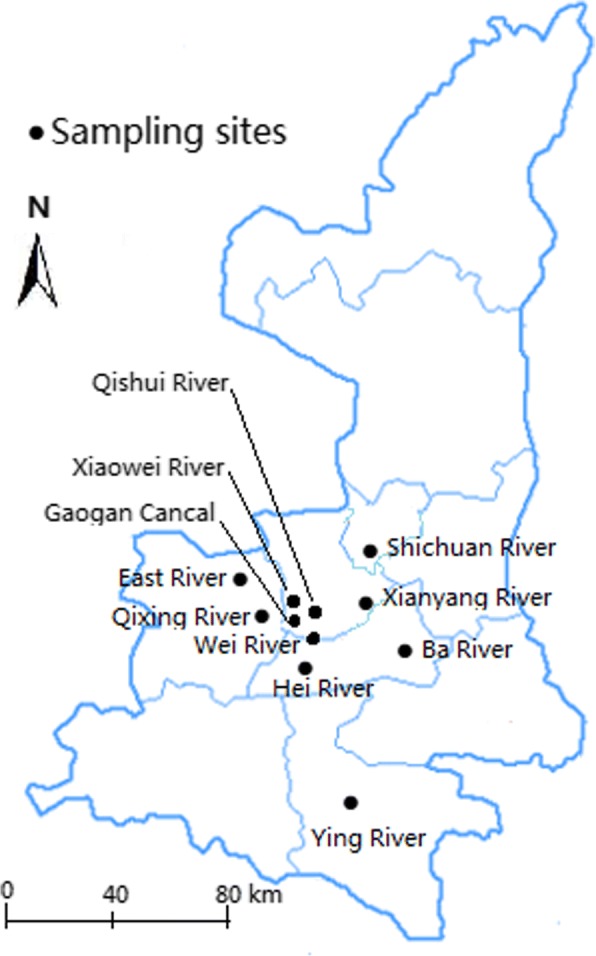
The map of sample locations
Antimicrobial susceptibility testing
The broth microdilution procedure recommended by Clinical Laboratories Standards Institute (CLSI) [10] was performed to determine the antimicrobial susceptibility of all E. coli isolates against 16 antimicrobials representing six antimicrobial classes: β-lactams, including penicillins (ampicillin, amoxicillin-clavulanic acid and ticarcillin-clavulanic acid), the first-generation cephalosporins (cephalothin), the third-generation cephalosporins (cefotaxime, ceftazidime and ceftriaxone), cephamycins (cefoxitin), and carbapenems (meropenem); tetracyclines (tetracycline); amphenicols (thiamphenicol); quinolones (nalidixic acid and ciprofloxacin); aminoglycosides (gentamicin and amikacin); sulfonamides (sulfamethoxazole-trimethoprim). The control strain for susceptibility testing was E. coli ATCC 25922.
Moreover, ESBL production among the E. coli isolates resistant to the third-generation cephalosporins was detected phenotypically by the double disk synergy test with disks supplemented with cefotaxime and ceftazidime alone or coupled with clavulanic acid [10]. Initial screening analyses indicated that 2.8% (n = 76) E. coli isolates were phenotypic ESBL-positive isolates, and these isolates were used for further analysis.
Phylogenetic typing and determination of virulence factors
Total DNA was isolated from the ESBL producers by using the boiling method. Phylogenetic grouping was determined for the ESBL-producing isolates according to the novel quadruplex PCR method [11]. Meanwhile, seven virulence factor genes known to be characteristic of intestinal pathogenic E. coli (IPEC), including aggR for EAEC, stx1 and stx2 for STEC; eae for EPEC, estA and eltB for ETEC, EIEC-specific gene ipaH; as well as seven markers of virulence associated with uropathogenic E. coli (UPEC), including traT, fyuA, papC, chuA, afa/dra, iutA and PAI [12], were performed by PCR.
Characterization of β-lactamase and PMQR genes
PCR detection and gene identification were performed for β-lactamase genes (TEM, SHV, CTX-Ms), plasmid-mediated AmpC β-lactamase (CMY-2) and carbapenemase genes (class A, KPC-2; class B, NDM-1; class D, OXA) in ESBL-producing E. coli. blaCTX-M group-specific primers for CTX-M-1, CTX-M-2, CTX-M-8 and CTX-M-9 were used to detect of blaCTX-M genes. The PCR products were purified and sequenced by Sangon Biotech (Shanghai, China), and then the β-lactamase genes were identified using the β-lactamase database (http://www.lahey.org/studies/webt.asp) after all the sequences were analyzed online using BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Moreover, all the 76 ESBL-producing E. coli isolates were screened by PCR for PMQR genes (qnrA, qnrB, qnrD, qnrS, aac(6′)-Ib-cr, oqxAB and qepA) as described previously [13, 14].
Conjugation experiments
Potential horizontal transferability of β-lactamase and PMQR genes from 15 randomly selected ESBL-producing E. coli isolates (at least one isolate per sampling site) was assessed by conjugation studies (broth mating method) using E. coli J53 AZr as the recipient [15]. The Mueller-Hinton agar supplemented with 150 μg/ml sodium azide and 2 μg/ml cefotaxime were used to select the transconjugants, which were subsequently analyzed by PCR to determine the transferability of β-lactamase and PMQR genes. In addition, the resistance patterns of the recipient and all transconjugants were analyzed.
Multilocus sequence typing (MLST) determination
Internal fragments of seven conserved housekeeping genes (adk, fumC, gyrB, icd, mdh, purA and recA) of each ESBL-producing E. coli isolate were amplified by PCR. A detailed scheme of the MLST procedure, including the primers, PCR conditions, allelic type and sequence type assignment methods, is available at MLST database website (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli).
Statistical analysis
Pearson′s Chi-squared test was used for statistical analysis, and the statistical significance level was established at P < 0.05.
Results
Antimicrobial susceptibility
Among the 2686 E. coli isolates collected, 76 (2.8%) isolates were identified as the ESBL-producing isolates, which were unevenly distributed in 11 sampling sites at levels ranging from 1.1 to 6.4%. Moreover, 64 of the 76 (84.2%) isolates expressed the MDR phenotype. The 76 ESBL-producing isolates showed high resistance to tetracycline (97.3%), followed by ticarcillin-clavulanic acid (90.8%), cephalothin (89.5%), nalidixic acid (81.6%), cefotaxime (77.6%), ciprofloxacin (69.7%), sulfamethoxazole-trimethoprim (69.7%), thiamphenicol (63.2%), and cefoxitin (57.6%), whereas they exhibited high susceptibility to meropenem (96.1%).
Phylogenetic groups and the virulence genes distribution
Phylogenetic analysis showed that the 76 ESBL-producing isolates were composed of phylogenetic groups D (n = 24), A (n = 21), B2 (n = 15), B1 (n = 10), C (n = 4), and E (n = 2). Overall, 78.9% (60/76) of ESBL-producing isolates harbored as least one virulence factor, and the prevalence of individual virulence genes ranged from 1.3% (eae) to 52.6% (traT). estA and aggR were detected in ten and two isolates, respectively, while stx1, stx2 and ipaH were not detected. The virulence genes associated with UPEC isolates were detected throughout the sources, whereas the virulence genes associated with STEC and EIEC isolates were not detected.
Distribution of β-lactamase and PMQR genes
As shown in Table 1, blaSHV, blaTEM and blaCTX-M were detected in 36.8% (n = 28), 43.4% (n = 33) and 76.3% (n = 58) of ESBL producers, respectively, and 58 of the 76 isolates possessed more than one β-lactamase gene. It is interesting that the number of the β-lactamase genes in an E. coli isolate was positively correlated the prevalence of the ESBL producer in each sampling site. For the blaCTX-M positive isolates, blaCTX-M-14 (n = 35) was the predominant genotype, followed by blaCTX-M-9 (n = 17), blaCTX-M-123 (n = 15), blaCTX-M-15 (n = 7), blaCTX-M-27 (n = 4), blaCTX-M-1 (n = 3) and blaCTX-M-65 (n = 3). On the other hand, blaOXA-2, blaKPC-2, blaCMY-2 and blaNDM-1 were detected in five, four, two and one isolate, respectively. It is noteworthy that 80% (4/5) of blaOXA-2 positive isolates were isolated from Gaogan Canal. Among the 33 TEM-positive isolates, two were blaTEM-3 and the rest were non-ESBL gene blaTEM-1. The blaSHV genes were represented by blaSHV-2 (n = 7) and blaSHV-12 (n = 21), and it is interesting to note that ESBL gene blaSHV-12 and non-ESBL gene blaTEM-1 simultaneously appeared in 20 isolates. Furthermore, 54 of 76 (71.1%) ESBL-producing isolates harbored at least one PMQR gene, which was co-located in the ESBL producers with β-lactamase genes. Aac(6′)-Ib-cr (n = 46) was the most dominant PMQR gene, followed by the qnr genes (n = 34). Moreover, one isolate harbored the qepA gene, while the oqxAB gene was not detected in any isolate.
Table 1.
ESBL-producing E. coli isolates from rivers and lakes in the Northwest China
| Sampling sites | Isolates No. | PG | Antimicrobial resistance profiles | β-lactamase genes | PMQR genes | Virulence genes | MLST |
|---|---|---|---|---|---|---|---|
| Hei River | HH1609014 | B1 | AMP AMC TIM CEP CTX CEX FOX TEC GEM AMK SXT | CTX-M-14 | fyuA, traT | ST155 | |
| HH1510025 | B2 | AMP CEP TPH GEM SXT | TEM-1, SHV-12 | qnrB | ST1587 | ||
| Ying Lake | YH1507022 | A | AMP AMC TIM CEP CTX CAZ CEX FOX TEC TPH NAC CIP SXT | TEM-1, CTX-M-14 | aac(6′)-Ib-cr | traT, papC, chuA | ST617 |
| YH1606032 | A | AMP AMC TIM CEP CTX CAZ CEX FOX TEC NAC CIP GEM AMK SXT | CTX-M-14 | aac(6′)-Ib-cr | afa/dra, PAI | ST44 | |
| YH1607018 | D | AMP AMC TIM CEP CAZ TEC TPH GEM AMK | TEM-1, SHV-12 | traT, chuA | ST2148 | ||
| Xianyang Lake | XY1608045 | B2 | AMP AMC TIM CEP CTX CAZ TEC TPH NAC CIP GEM AMK SXT | CTX-M-14 | aac(6′)-Ib-cr | traT, iutA | ST602 |
| XY1605044 | D | AMP AMC TIM CEP CAZ CEX FOX TEC | CTX-M-14 | ST393 | |||
| XY1605033 | D | AMP AMC TIM CEP CEX TEC NAC CIP SXT | TEM-1, SHV-12 | qnrB, aac(6′)-Ib-cr | traT, chuA | ST393 | |
| XY1507042 | E | AMP AMC CTX CAZ TPH NAC CIP | TEM-1, SHV-12 | fyuA, traT | ST1301 | ||
| Qishui River | QS1608021 | A | AMP AMC TIM CEP CTX CEX FOX TEC NAC CIP SXT | TEM-1, CTX-M-1 | aac(6′)-Ib-cr | estA | ST10 |
| QS1607026 | A | AMP AMC TIM CEP CTX CAZ TEC NAC CIP GEM SXT | CTX-M-9 | traT, chuA | ST4429 | ||
| QS1608034 | B2 | AMP AMC TIM CEP CEX FOX TEC TPH NAC CIP SXT | CTX-M-9 | qnrB, qnrS | traT, chuA | ST331 | |
| QS1610030 | C | AMP AMC TIM CEP CEX TEC TPH NAC CIP | TEM-1, SHV-12 | qnrB, aac(6′)-Ib-cr | estA | ST23 | |
| East Lake | EH1507029 | A | AMP AMC CTX CAZ TEC NAC CIP SXT | CTX-M-1 | qnrB, aac(6′)-Ib-cr | ST10 | |
| EH1607033 | A | AMP AMC TIM CTX CAZ CEX FOX TEC SXT | CTX-M-14 | aac(6′)-Ib-cr | traT, chuA, papC, | ST10 | |
| EH1607014 | A | AMP AMC TIM CEP CTX CAZ CEX FOX TEC TPH NAC CIP GEM SXT | TEM-1, CTX-M-14 | aac(6′)-Ib-cr | traT, PAI | ST167 | |
| EH1608016 | A | AMP AMC TIM CEP CAZ TEC NAC CIP | SHV-12 | aac(6′)-Ib-cr | ST167 | ||
| Wei River | WH1606023 | A | AMP AMC CEP CEX FOX TEC TPH | TEM-1, SHV-12 | aac(6′)-Ib-cr | fyuA | ST10 |
| WH1508055 | B1 | AMP AMC TIM CTX CAZ TEC NAC CIP GEM AMK SXT | CTX-M-27 | aac(6′)-Ib-cr | traT, afa/dra | ST58 | |
| WH1606078 | D | AMP AMC TIM CEP CEX FOX TEC TPH NAC CIP GEM AMK SXT | CTX-M-14 | qnrS | traT, papC, afa/dra | ST609 | |
| WH1510002 | D | AMP AMC TIM CEP CTX CAZ CEX | CTX-M-9 | qnrB, aac(6′)-Ib-cr | fyuA, traT, papC | ST38 | |
| WH1607120 | E | AMP AMC TIM CEP CTX CAZ TEC NAC CIP GEM AMK SXT | CTX-M-14 | aac(6′)-Ib-cr | traT | ST1301 | |
| Ba River | BA1605012 | A | AMP AMC TIM CEP TEC TPH NAC CIP SXT | TEM-1, SHV-12 | aac(6′)-Ib-cr | ST44 | |
| BA1605022 | B1 | AMP AMC TIM CEP CTX CEX TEC NAC CIP SXT | CTX-M-9, CTX-M-14 | qnrS, aac(6′)-Ib-cr | traT, afa/dra | ST155 | |
| BA1508024 | D | AMP AMC TIM CEP CAZ TEC NAC | TEM-1, SHV-12 | ST4068 | |||
| BA1510031 | D | AMP AMC TIM CEP CTX CAZ CEX FOX TEC TPH SXT | CTX-M-9, CTX-M-14 | traT, papC, sfaS | ST2003 | ||
| BA1509025 | D | AMP AMC TIM CTX CAZ CEX TEC TPH NAC CIP GEM AMK | CTX-M-14, CTX-M-15 | qnrB, aac(6′)-Ib-cr | fyuA, traT, papC | ST69 | |
| BA1509015 | D | AMP AMC TIM CTX CEX FOX TEC TPH | CTX-M-14, CTX-M-15 | qnrS, aac(6′)-Ib-cr | fyuA, iutA | ST405 | |
| Shichuan River | SC1608022 | A | AMP AMC TIM CEP CTX CAZ TEC SXT | SHV-12, CTX-M-123 | traT, chuA | ST93 | |
| SC1506012 | A | AMP AMC TIM CEP CTX CAZ CEX FOX TEC TPH NAC SXT | CTX-M-14 | ST746 | |||
| SC1507014 | A | AMP AMC TIM CEP CTX CAZ CEX FOX TEC NAC CIP SXT | TEM-3, CTX-M-123 | aac(6′)-Ib-cr | traT, papC, hlyD | ST2376 | |
| SC1604029 | B1 | AMP AMC TIM CEX FOX TEC TPH NAC CIP GEM AMK | TEM-1, SHV-12 | qnrS, aac(6′)-Ib-cr | ST155 | ||
| SC1607063 | B2 | AMP AMC TIM CEP CTX CEX FOX MEM TEC TPH NAC CIP GEM AMK SXT | CTX-M-15, CTX-M-123 | qnrS, aac(6′)-Ib-cr | fyuA, traT, papC | ST131 | |
| SC1608102 | B2 | AMP AMC TIM CEP CTX CAZ CEX TIC TPH NAC | TEM-1, SHV-2 | fyuA | ST95 | ||
| SC1610005 | D | AMP AMC TIM CEP CTX CEX FOX TEC NAC CIP GEM AMK SXT | TEM-1, SHV-12, CTX-M-15 | qnrS, aac(6′)-Ib-cr | estA | ST38 | |
| SC1609081 | D | AMP AMC TIM CEP CTX CEX TEC NAC CIP GEM SXT | TEM-1, CTX-M-14 | qnrB, aac(6′)-Ib-cr | estA | ST405 | |
| Xiaowei River | XW1608112 | A | AMP AMC TIM CEP CTX CEX FOX TEC TPH NAC CIP GEM AMK SXT | TEM-1, SHV-12 | qnrB, aac(6′)-Ib-cr | iutA, afa/dra | ST10 |
| XW1608047 | A | AMP AMC TIM CEP CTX CAZ CEX FOX TEC NAC SXT | TEM-1, SHV-12, CTX-M-14 | ST44 | |||
| XW1609034 | B1 | AMP AMC TIM CEP CTX CAZ CEX TEC TPH NAC CIP SXT | CTX-M-14, CTX-M-65 | qnrS | fyuA, PAI | ST75 | |
| XW1608023 | B2 | AMP AMC TIM CEP CTX CEX TEC TPH NAC | TEM-1, SHV-2 | traT, chuA | ST95 | ||
| XW1607012 | B2 | AMP AMC TIM CEP CTX CAZ CEX FOX TEC TPH NAC CIP SXT | CTX-M-9, CTX-M-14, CTX-M-123 | aac(6′)-Ib-cr | fyuA, traT, iutA | ST12 | |
| XW1607055 | B2 | AMP AMC TIM CEP CTX CAZ FOX TEC TPH NAC CIP SXT | CTX-M-14 | ST2855 | |||
| XW1609057 | D | AMP AMC TIM CEP CTX CAZ TEC TPH SXT | TEM-1, SHV-12 | ST5164 | |||
| XW1608026 | D | AMP AMC TIM CEP CTX CAZ CEX FOX TEC NAC CIP GEM AMK SXT | CTX-M-1 | aac(6′)-Ib-cr | traT, hlyD | ST3880 | |
| XW1607034 | D | AMP AMC TIM CEP CTX CAZ CEX FOX TEC NAC CIP SXT | CTX-M-14, CTX-M-123 | aac(6′)-Ib-cr | fyuA, traT | ST38 | |
| XW1609038 | D | AMP AMC TIM CEP CTX CEX FOX MEM TEC NAC CIP GEM AMK SXT | CTX-M-15 | qnrB, aac(6′)-Ib-cr | traT, iutA, papC | ST69 | |
| XW1608041 | D | AMP AMC CEP CTX CEX TEC TPH NAC CIP SXT | TEM-1, SHV-2, CTX-M-14 | traT, chuA | ST609 | ||
| Qixing River | QX1608021 | A | AMP AMC TIM CEP CTX CAZ TEC TPH NAC SXT | TEM-1, SHV-12 | ST10 | ||
| QX1608013 | A | AMP AMC TIM CEP CEX TEC TPH SXT | TEM-1, SHV-12 | aac(6′)-Ib-cr | traT, papC, PAI | ST10 | |
| QX1509072 | A | AMP AMC TIM CEP CTX CAZ FOX TEC NAC GEM AMK SXT | TEM-1, SHV-2 | qnrS, aac(6′)-Ib-cr | estA | ST10 | |
| QX1608015 | A | AMP AMC TIM CEP CTX CEX FOX TEC TPH NAC CIP GEM AMK SXT | CTX-M-9, CTX-M-27 | qnrS | traT, papC | ST3902 | |
| QX1605083 | B1 | AMP AMC TIM CEP CTX CAZ CEX FOX TEC TPH NAC CIP GEM AMK SXT | TEM-1, CTX-M-9, KPC-2 | fyuA, papC, traT | ST3160 | ||
| QX1608005 | B1 | AMP AMC TIM CEP CTX CAZ CEX TEC NAC CIP SXT GEM SXT | CTX-M-14 | qnrB | estA | ST75 | |
| QX1507055 | B2 | AMP AMC TIM CEP CTX CAZ CEX FOX TEC TPH NAC CIP GEM SXT | CTX-M-27 | qnrS | aggR | ST1304 | |
| QX1508112 | B2 | AMP AMC TIM CEP CTX CAZ FOX TEC TPH NAC CIP GEM AMK SXT | TEM-1, SHV-12, CTX-M-9, OXA-2 | qnrB, qnrS, aac(6′)-Ib-cr | traT, iutA, PAI | ST12 | |
| QX1608059 | B2 | AMP AMC CEP CAZ CEX FOX TEC TPH NAC CIP GEM AMK | CTX-M-14, CTX-M-123 | estA | ST2077 | ||
| QX1510043 | C | AMP AMC TIM CEP CTX CAZ CEX FOX TEC NAC CIP GEM SXT | CTX-M-9, CTX-M-14 | aac(6′)-Ib-cr | traT, papC | ST23 | |
| QX1608046 | D | AMP AMC TIM CEP CTX CAZ CEX FOX TEC TPH NAC CIP SXT | CTX-M-14, CTX-M-123, CTX-M-65 | afa/dra, hlyD | ST3880 | ||
| QX1604103 | D | AMP AMC TIM CEP CEX FOX TEC TPH NAC CIP GEM AMK SXT | TEM-1, SHV-2, CTX-M-14, CTX-M-123 | qnrB, aac(6′)-Ib-cr | fyuA, iutA, PAI | ST609 | |
| QX1609108 | D | AMP AMC TIM CEP CTX CAZ CEX FOX MEM TEC TPH NAC CIP SXT | TEM-3, CTX-M-14 | fyuA, afa/dra | ST2148 | ||
| Gaogan Canal | GG1505017 | A | AMP AMC TIM CEP CTX CEX TEC NAC CIP GEM AMK SXT | TEM-1, SHV-2, CTX-M-14 | aac(6′)-Ib-cr | ST10 | |
| GG1509025 | A | AMP AMC TIM CEP CEX TEC TPH GEM AMK SXT | TEM-1, SHV-12, CTX-M-65 | aac(6′)-Ib-cr | estA | ST10 | |
| GG1508074 | B1 | AMP AMC TIM CEP CTX CAZ CEX MEM TEC TPH NAC CIP SXT | TEM-1, SHV-12, CTX-M-9 | aac(6′)-Ib-cr | eae | ST58 | |
| GG1609024 | B1 | AMP AMC TIM CEP CTX CAZ CEX MEM TEC TPH NAC CIP SXT | CTX-M-9, CTX-M-123 | qnrB | traT, papC, afa/dra | ST155 | |
| GG1609158 | B1 | AMP AMC CEP CEX FOX TEC TPH N GEM SXT | TEM-1, SHV-12 | ST1049 | |||
| GG1609019 | B2 | AMP AMC TIM CEP CTX CAZ FOX TEC TPH SXT | CTX-M-9, CTX-M-14 | iutA, afa/dra | ST3252 | ||
| GG1609022 | B2 | AMP AMC TIM CEP CTX CAZ CEX FOX TEC NAC CIP | CTX-M-9, CTX-M-14 | qnrA, aac(6′)-Ib-cr | fyuA, traT, iutA, PAI | ST12 | |
| GG1609068 | B2 | AMP AMC CEP CTX CEX FOX TEC NAC CIP SXT | CTX-M-14, KPC-2, OXA-2 | qnrB, qnrS, aac(6′)-Ib-cr | fyuA, papC, traT, iutA | ST131 | |
| GG1610109 | B2 | AMP AMC TIM CEP CTX CAZ CEX FOX MEM TEC TPH NAC CIP GEM AMK SXT | CTX-M-9, CTX-M-15, CTX-M-123, KPC-2, NDM-1, OXA-2 |
qepA, qnrS, aac(6′)-Ib-cr | fyuA, papC, traT, chuA, iutA | ST131 | |
| GG1609086 | C | AMP AMC TIM CEP CTX CAZ FOX TEC TPH NAC CIP SXT | CTX-M-9, CTX-M-14, CTX-M-123 | qnrS, aac(6′)-Ib-cr | traT, afa/dra, papC, PAI | ST410 | |
| GG1607066 | C | AMP AMC TIM CEP CTX CAZ CEX TEC TPH NAC CIP GEM SXT | TEM-1, SHV-12, CTX-M-123 | qnrB, aac(6′)-Ib-cr | traT, chuA | ST88 | |
| GG1609121 | D | AMP AMC TIM CEP CTX CAZ TEC TPH NAC CIP GEM AMK SXT | CTX-M-15, CTX-M-123 | aac(6′)-Ib-cr | estA | ST38 | |
| GG1609016 | D | AMP AMC TIM CEP CTX CAZ CEX FOX TEC TPH NAC SXT | CTX-M-14, CMY-2 | aac(6′)-Ib-cr | fyuA, traT | ST69 | |
| GG1604028 | D | AMP AMC TIM CTX CEX FOX TEC NAC CIP GEM AMK | CTX-M-14, CTX-M-123 | aac(6′)-Ib-cr | aggR | ST69 | |
| GG1506027 | D | AMP AMC TIM CEP CTX CAZ CEX FOX TEC TPH NAC CIP GEM AMK SXT | CTX-M-9, CTX-M-123, KPC-2, OXA-2 | qnrB, aac(6′)-Ib-cr | fyuA, traT, chuA, iutA | ST405 | |
| GG1608063 | D | AMP AMC TIM CEP CTX CAZ CEX FOX MEM TEC TPH NAC CIP SXT | CTX-M-14, CTX-M-27, CMY-2, OXA-2 | qnrS, aac(6′)-Ib-cr | fyuA, traT, chuA, iutA, PAI | ST405 |
AMP ampicillin, AMC amoxicillin-clavulanic acid, TIM ticarcillin-clavulanic acid, CEP cephalothin, CTX cefotaxime, CAZ ceftazidime, CEX ceftriaxone, FOX cefoxitin, MEM meropenem, TEC tetracycline, TPH thiamphenicol, NAC nalidixic acid, CIP ciprofloxacin, GEN gentamicin, AMK amikacin, SXT sulfamethoxazole-trimethoprim
Conjugation experiments
Ten out of fifteen ESBL producers were horizontally transferred to recipient strain E. coli J53 AZr. PCR demonstrated the presence of β-lactamase and PMQR genes in transconjugants (Table 2). Antimicrobial susceptibility patterns revealed that all transconjugants kept the similar antibiotic resistance profiles to ampicillin, amoxicillin-clavulanic acid, ticarcillin-clavulanic acid, cefotaxime, ceftazidime, ceftriaxone and cefoxitin compared with the donors, and all transconjugants exhibited at least 8-fold increase in MICs compared with the recipient. The ciprofloxacin MICs for eight transconjugants harboring PMQRs ranged from 0.125 to 1 μg/ml, representing an increase of 2-fold to 16-fold compared with the recipient (Table 2). However, the transconjugants were still susceptible to meropenem, tetracycline, ciprofloxacin, gentamicin, thiamphenicol and sulfamethoxazole-trimethoprim.
Table 2.
Antimicrobial susceptibility profiles of ESBL-producing E. coli isolates used in the conjugation experiments
| Isolates | MIC (μg/ml) of antimicrobials | Presence of | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | AMC | TIM | CTX | CAZ | CEX | FOX | MEM | TEC | TPH | CIP | GEN | SXT | β-lactamase genes | PMQR genes | |
| Donors | |||||||||||||||
| HH1609014 | 256 | 32 | 32 | 32 | 4 | 32 | 8 | 0.03 | 32 | 2 | 0.5 | 32 | 128 | CTX-M-14 | |
| XY1608045 | 512 | 32 | 16 | 32 | 64 | 4 | 2 | 0.125 | 64 | 32 | 64 | 32 | 32 | CTX-M-14 | aac(6′)-Ib-cr |
| EH1607014 | 256 | 32 | 32 | 64 | 64 | 128 | 16 | 0.063 | 32 | 64 | 128 | 16 | 64 | TEM-1, CTX-M-14 | aac(6′)-Ib-cr |
| WH1510002 | 256 | 32 | 32 | 32 | 128 | 64 | 1 | 0.063 | 0.25 | 1 | 2 | 4 | 16 | CTX-M-9 | qnrB, aac(6′)-Ib-cr |
| BA1605022 | 512 | 64 | 16 | 64 | 8 | 64 | 2 | 0.063 | 64 | 0.25 | 32 | 2 | 64 | CTX-M-9, CTX-M-14 | qnrS, aac(6′)-Ib-cr |
| QX1604103 | 256 | 64 | 32 | 4 | 256 | 128 | 32 | 0.03 | 128 | 128 | 128 | 64 | 256 | TEM-1, SHV-2, CTX-M-14, CTX-M-123 | qnrB, aac(6′)-Ib-cr |
| SC1610005 | 512 | 64 | 32 | 32 | 8 | 64 | 32 | 0.03 | 32 | 2 | 16 | 32 | 128 | TEM-1, SHV-12, CTX-M-15 | qnrS, aac(6′)-Ib-cr |
| XW1609038 | 256 | 32 | 32 | 32 | 4 | 64 | 16 | 4 | 0.25 | 0.5 | 64 | 128 | 64 | CTX-M-15 | qnrB, aac(6′)-Ib-cr |
| GG1509025 | 256 | 64 | 16 | 4 | 2 | 32 | 2 | 0.125 | 128 | 64 | 2 | 64 | 64 | TEM-1, SHV-12, CTX-M-65 | aac(6′)-Ib-cr |
| GG1610109 | 512 | 64 | 32 | 128 | 64 | 128 | 32 | 16 | 128 | 128 | 128 | 128 | 128 | CTX-M-9, CTX-M-15, CTX-M-123, KPC-2, NDM-1, OXA-2 | qepA, qnrS, aac(6′)-Ib-cr |
| Recipient J53AZr | 4 | 1 | 1 | 0.125 | 0.063 | 0.063 | 0.125 | 0.03 | 0.25 | 0.125 | 0.063 | 0.25 | 0.25 | ||
| Transformants | |||||||||||||||
| Trans-HH1609014 | 128 | 16 | 16 | 16 | 1 | 8 | 8 | 0.03 | 0.5 | 0.25 | 0.125 | 0.25 | 0.5 | CTX-M-14 | |
| Trans-XY1608045 | 256 | 32 | 16 | 16 | 32 | 0.5 | 0.5 | 0.063 | 0.25 | 0.125 | 0.125 | 0.125 | 0.25 | CTX-M-14 | aac(6′)-Ib-cr |
| Trans-EH1607014 | 256 | 16 | 16 | 32 | 32 | 64 | 8 | 0.03 | 0.125 | 0.125 | 0.5 | 0.125 | 1 | CTX-M-14 | aac(6′)-Ib-cr |
| Trans-WH1510002 | 128 | 32 | 16 | 32 | 32 | 64 | 0.5 | 0.03 | 0.063 | 0.063 | 0.063 | 0.063 | 0.25 | CTX-M-9 | aac(6′)-Ib-cr |
| Trans-BA1605022 | 128 | 32 | 32 | 64 | 1 | 16 | 0.5 | 0.03 | 0.5 | 0.063 | 0.125 | 0.03 | 0.5 | CTX-M-9, CTX-M-14 | qnrS, aac(6′)-Ib-cr |
| Trans-QX1604103 | 128 | 32 | 32 | 1 | 64 | 64 | 16 | 0.125 | 0.125 | 0.125 | 0.5 | 0.125 | 2 | TEM-1, CTX-M-14 | aac(6′)-Ib-cr |
| Trans-SC1610005 | 128 | 16 | 16 | 16 | 1 | 16 | 4 | 0.03 | 0.5 | 0.25 | 0.5 | 0.5 | 1 | TEM-1, SHV-12, CTX-M-15 | qnrS, aac(6′)-Ib-cr |
| Trans-XW1609038 | 128 | 16 | 16 | 16 | 1 | 32 | 16 | 0.063 | 0.063 | 0.063 | 0.125 | 0.25 | 0.5 | CTX-M-15 | qnrB, aac(6′)-Ib-cr |
| Trans-GG1509025 | 256 | 32 | 16 | 1 | 0.5 | 32 | 1 | 0.063 | 0.125 | 0.125 | 0.125 | 0.125 | 0.25 | SHV-12, CTX-M-65 | aac(6′)-Ib-cr |
| Trans-GG1610109 | 256 | 32 | 32 | 32 | 32 | 64 | 16 | 0.03 | 0.25 | 0.063 | 1 | 0.25 | 0.5 | CTX-M-15, CTX-M-123, KPC-2, NDM-1 | qepA, qnrS, aac(6′)-Ib-cr |
MLST determination
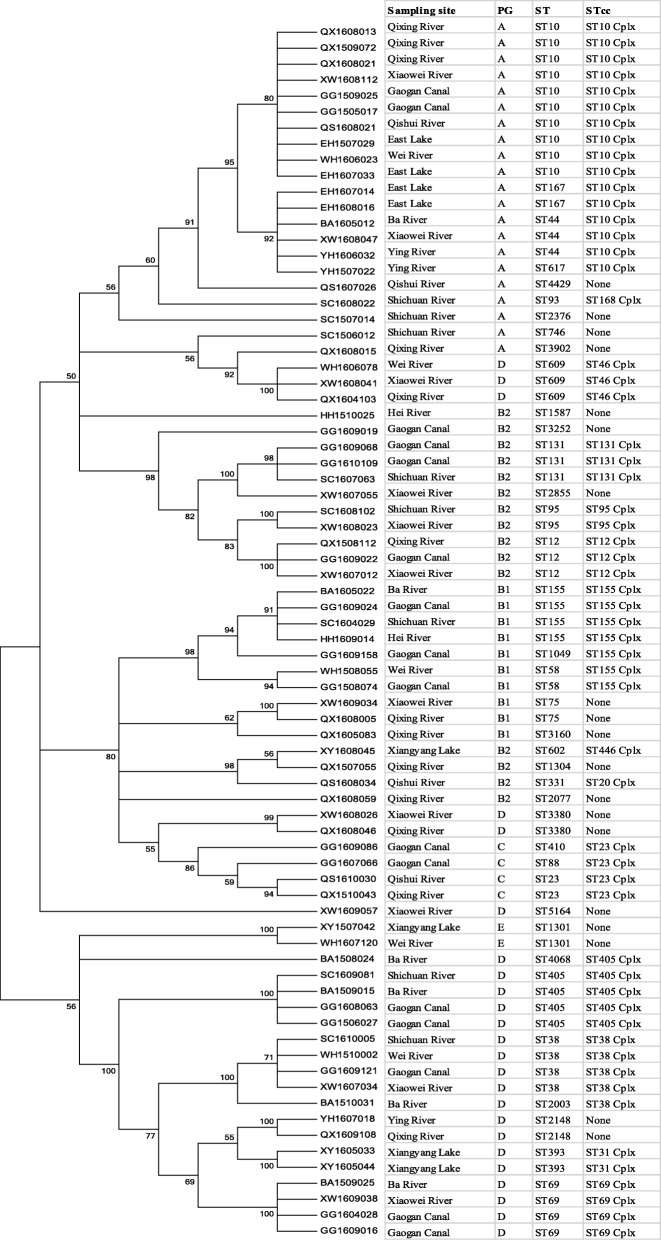
The diversity and phylogenetic relationships of the ESBL-producing E. coli isolates were evaluated by MLST. MEGA 6.0 software was used to construct the phylogenetic tree for 76 ESBL-producing E. coli isolates using the maximum likelihood approach with on the basis of the Tamura-Nei model and seven concatenated housekeeping gene sequences (Fig. 2). The 76 ESBL producers belonged to 37 STs (Fig.1 and Table 1). Among of them, 19 STs were represented by more than two isolates, and the other 18 STs represented a single isolate each. ST10 (n = 10) was more prevalent compared with other STs (P < 0.001). It is difficult to infer a significant correlation between the water bodies and the STs because of the limited number of ESBL producers. Nevertheless, we found that some ESBL producers from different water bodies shared the same STs, and some STs, e.g., ST10, ST38, ST69, ST405, identified in this study were also found among the E. coli isolates from dogs in Shannxi province. Three ST131 isolates were from Shichuan River and Gaogan Canal, which flowed through several cities and villages. Furthermore, the ST131 isolate from Shichuan River simultaneously harbored blaCTX-M-15 and blaCTX-M-123; one ST131 isolate from Gaogan Canal harbored blaCTX-M-9, blaCTX-M-15, blaCTX-M-123, blaKPC-2, blaNDM-1, blaOXA-2 as well as PMQR genes qnrA, qnrS and aac(6′)-Ib-cr, while another ST131 isolate from Gaogan Canal harbored blaCTX-M-14, blaKPC-2, blaOXA-2 as well as qnrB, qnrS and aac(6′)-Ib-cr.
Fig. 2.
Phylogenetic tree showing the relationship of 76 ESBL-producing E. coli isolates. The dendrogram was constructed by using the nucleotide sequences of the seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA and recA) of 76 ESBL-producing E. coli isolates from rivers and lakes in Northwest China with the maximum likelihood method. Sampling sites, phylogenetic group (PG), sequence type (ST) and ST clonal complex (STcc) were displayed the right of the dendrogram
Discussion
The spread of ESBL-producing E. coli isolates in the environment, especially in water is worrisome both in developing and developed countries as they pose potential risks to public health [16–18]. Rivers and lakes are usually considered to be of special importance as a reservoir of resistance genes because they can collect the surface waters containing contaminants from different origins, e.g., municipal wastewater, agricultural activities, or the sewage from the hospitals and livestock, which include abundant antibiotic-resistant bacteria. In this study, 2686 E. coli isolates were collected from 11 water bodies between March 2015 and November 2016, with 90.9% (10/11) of sampling sites located in Guanzhong region, an economically developed and densely populated area in Shaanxi province. Generally, the prevalence rate of ESBL producers was 2.8%, which was much lower than the prevalence of ESBL producers among the E. coli isolated from dogs (24.2%), retail meat (22.3%) and pigs (9.6%, unpublished data from our group) in Shaanxi province [19, 20]. Meanwhile, the frequency of ESBL-producing E. coli varied significantly at different sampling sites, and it was more frequently isolated in the Gaogan Canal (6.4%), Qixing River (4.3%) and Xiaowei River (4.2%) compared with Hei River (1.1%) (P < 0.01). It is noteworthy that a blaNDM-1-producing ST131 clone, and four of the five blaOXA-2-producing isolates were isolated from Gaogan Canal. There is a high probability that the Gaogan Canal, Qixing River and Xiaowei River were contaminated by the wastewater from the hospitals, pharmaceutical manufactures or livestock farms, which are located in or adjacent to cities or rural villages. However, ESBL-producing E. coli isolates were seldom detected in the Hei River, Ying Lake and Qishui River, which belong to public water supply source or scenic spots. The results indicate that there is a positive linear relationship between the occurrence of ESBL producers and discharge of wasterwater, such as the sewage of the hospitals and the livestock farms.
It is of particular concern that the majority (84.2%) of 76 ESBL-producing isolates included in this study expressed the MDR phenotype and showed high resistance rates to amoxicillin-clavulanic acid (98.7%), tetracycline (97.3%) and ticarcillin-clavulanic acid (90.8%). Moreover, it is worrisome that most ESBL producers were commonly located on conjugative plasmids that also harbor genes conferring cross-resistance to non-β-lactam antibiotics [21]. Traditionally, phylogroups A and B1 contain commensal isolates, while groups B2 and D are considered to be opportunistic ExPEC isolates. The 76 ESBL-producing E. coli isolates surveyed belonged mainly to phylogroups D and A (59.2%), followed by group B2 (19.7%). Normally, virulence factors are ideal targets for determining the pathogenic potential of a given E. coli isolate. Most of our ESBL-producing isolates (65.8%) possessed UPEC-related virulence factors, followed by estA, which is associated with the ETEC. Our results generally agree with a previous study that found ExPEC as the main pathotype in E. coli isolates from other water sources [6]. However, our findings tend to strongly disagree with the previous finding of significantly higher prevalence of ETEC isolates in surface waters of developing countries [22, 23], which may be due to the large differences in the sampling environments. It has been shown that ExPEC isolates can exist as commensals in the guts of healthy animals and humans, where they may gain or lose virulence genes through genetic exchange [6]. Moreover, UPEC isolates, the primary ExPEC associated with urinary tract infections, are also an important source of ESBLs entering the water system [24].
In recent years, CTX-M subtypes of the CTX-M-1 and CTX-M-9 groups have become the most prevalent ESBL-encoding genes among the E. coli from clinical and aquatic environments [4]. In the present study, CTX-Ms were represented by seven blaCTX-M subtypes that mostly expressed blaCTX-M-14. Two recent studies in our laboratory revealed that the predominant blaCTX-M subtypes in the ESBL-producing E. coli isolated from dogs and pigs, respectively, in the Guanzhong region of Shaanxi province [20, 25]. blaCTX-M-15 and blaCTX-M-14 were also prevalence in humans in Asia [26]. We identified three isolates that harbored blaCTX-M-65, which has not been reported before in Northwest China, although it has been frequently reported in other places in China [27–29]. All 76 ESBL-producing isolates were assigned to 37 STs, with ST10 as the most predominant. In contrast to the genetic characteristics of the ESBL-producing E. coli isolates from other sources, all the ESBL producers were much more diverse compared to the isolates from pigs and dogs in Shaanxi province. The emergence of clone ST131 represents a major challenge to public health worldwide since it was first discovered in human clinical samples. Subsequently, it has disseminated to various animal species and environments [4]. Our study indicated that three (3.9%, 3/76) ST131 isolates were detected in Shichuan River and Gaogan Canal, of which two ST131 isolates harbored blaCTX-M-15 and one harbored blaCTX-M-14, blaKPC-2 and blaOXA-2. The previous study suggested that the worldwide pandemic B2-ST131 E. coli isolates harboring blaCTX-M-27-producing have been closely associated with underlying severe infections in human and animal medicine [30]. We also detected four blaCTX-M-27-producing E. coli isolates, although these were not of the ST131 clone. Hence, further studies will need to be performed to explore these isolates, while at the same time, appropriate measures urgently need to be enforced to alleviate the stress posed by antibiotic resistance in the environments.
We found that almost all blaSHV-12 genes mainly co-existed with non-ESBL gene blaTEM-1 but not the other β-lactamase genes (Table 1). With respect to PMQR genes, their prevalence among E. coli isolates from humans and animals has been described frequently. However, there are few reports on the presence of PMQR genes in the ESBL-producing E. coli in water bodies. Our surface water E. coli isolates yielded one or more PMQR genes in 71.1% of the ESBL-producing isolates tested, with aac(6′)-Ib-cr as the most prevalent (63.2%), which was similar with a previous study in our laboratory that showed aac(6′)-Ib-cr as the most prevalent PMQR gene in extended-spectrum cephalosporin-resistant E. coli isolates from dogs in Shaanxi [20]. However, a previous study in Heilongjiang province showed that the oqxAB gene was the most dominant in the ESBL-producing E. coli from piglets [31]. All the PMQR genes co-localized with blaCTX-M in our E. coli isolates. The emergence of PMQRs indicates that quinolone resistance can also be acquired through horizontal gene transfer, and PMQR genes qnr and aac-(6′)-Ib-cr were co-transferred with β-lactamase genes, which were confirmed by the conjugation experiments in the present study. Notably in this study, one ST131 isolate from Gaogan Canal simultaneously harbored blaCTX-M-9, blaCTX-M-15, blaCTX-M-123, blaKPC-2, blaNDM-1, blaOXA-2 as well as the PMQR genes qnrA, qnrS and aac(6′)-Ib-cr. To our knowledge, this is the first description of the coexistence of so many resistance genes in one E. coli isolate from water. Hence, more studies should be carried out in the future in order to judge if these genes are located on the same plasmid.
Conclusion
In conclusion, the prevalence of ESBL-producing E. coli from the rivers and lakes in Northwest China was 2.8%, and the ExPEC pathotype was the most frequently detected depending on the virulence factor profiles. 76.3% of ESBL producers harbored more than one β-lactamase gene, and blaCTX-M-14 was the predominant genotype; the most dominant PMQR gene was aac(6′)-Ib-cr. The ESBL producers showed a high degree of overlaps in terms of resistance phenotypes, β-lactamases, PMQR genes and other genetic characteristics. The most prevalent sequence type was ST10, and three ST131 clones were detected.
Acknowledgements
The authors are thankful to Haohao Feng, Jinglong Ye, Runan Zuo and Yuyang Miao for their assistance in sample collection.
Funding
This study was supported by the Key Research and Development Project of Shaanxi Province (No. 2018NY-109, No. 2018NY-005), and the Agricultural Science and Technology Promotion Project of Yangling Demonstration Zone (No. TS-2016-12). The funding bodies are play role in provide research funding of the study. They have no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
All the data supporting our findings is contained within the manuscript.
Authors′ contributions
XL and HL conceived and guided the experiments. HL also drafted the manuscript. HZ, QZ and QP participated in the identification of the isolates, and performed the antimicrobial susceptibility assays. QZ, JW and QL participated in the conjugation experiments, and contributed to the manuscrip revision. HL, XL, QZ and QL performed the molecular studies, and analyzed the experimental data. All authors have read and approved the final manuscript.
Abbreviations
- EAEC
Enteroaggregative E. coli
- EIEC
Enteroinvasive E. coli
- EPEC
Enteropathogenic E. coli
- ESBL
Extended-spectrum β-lactamase
- ETEC
Enterotoxigenic E. coli
- ExPEC
Extraintestinal pathogenic E. coli
- PMQR
Plasmid-mediated quinolone resistance
- STEC
Shiga toxin-producing E. coli
- UPEC
Uropathogenic E. coli
Ethics approval and consent to participate
In this study, informed consent was not necessary because the isolates included in the study were obtained from surface waters. Ethics approval and consent to participate.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Haixia Liu, Email: liuhaixia209@163.com.
Hongchao Zhou, Email: lxqcpl@163.com.
Qinfan Li, Email: 438098745@qq.com.
Qian Peng, Email: 2008115855@nwafu.edu.cn.
Qian Zhao, Email: 46633885@qq.com.
Jin Wang, Email: 809070563@qq.com.
Xiaoqiang Liu, Email: liuxiaoqiang142@163.com.
References
- 1.Gundogdu A, Jennison AV, Smith HV, Stratton H, Katouli M. Extended-spectrum beta-lactamase producing Escherichia coli in hospital wastewaters and sewage treatment plants in Queensland, Australia. Can J Microbiol. 2013;59(11):737–745. doi: 10.1139/cjm-2013-0515. [DOI] [PubMed] [Google Scholar]
- 2.Tausova D, Dolejska M, Cizek A, Hanusova L, Hrusakova J, Svoboda O, Camlik G, Literak I. Escherichia coli with extended-spectrum beta-lactamase and plasmid-mediated quinolone resistance genes in great cormorants and mallards in Central Europe. J Antimicrob Chemoth. 2012;67(5):1103–1107. doi: 10.1093/jac/dks017. [DOI] [PubMed] [Google Scholar]
- 3.Amaya E, Reyes D, Paniagua M, Calderon S, Rashid MU, Colque P, Kuhn I, Mollby R, Weintraub A, Nord CE. Antibiotic resistance patterns of Escherichia coli isolates from different aquatic environmental sources in Leon, Nicaragua. Clin Microbiol Infec. 2012;18(9):E347–E354. doi: 10.1111/j.1469-0691.2012.03930.x. [DOI] [PubMed] [Google Scholar]
- 4.Hu YY, Cai JC, Zhou HW, Chi D, Zhang XF, Chen WL, Zhang R, Chen GX. Molecular typing of CTX-M-producing Escherichia coli isolates from environmental water, swine feces, specimens from healthy humans, and human patients. Appl Environ Microbiol. 2013;79(19):5988–5996. doi: 10.1128/AEM.01740-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chandran A, Mazumder A. Pathogenic potential, genetic diversity, and population structure of Escherichia coli strains isolated from a forest-dominated watershed (Comox Lake) in British Columbia, Canada (vol 81, pg 1788, 2015) Appl Environ Microb. 2016;82(2):767. doi: 10.1128/AEM.03528-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamelin K, Bruant G, El-Shaarawi A, Hill S, Edge TA, Bekal S, Fairbrother JM, Harel J, Maynard C, Masson L, et al. A virulence and antimicrobial resistance DNA microarray detects a high frequency of virulence genes in Escherichia coli isolates from Great Lakes recreational waters. Appl Environ Microb. 2006;72(6):4200–4206. doi: 10.1128/AEM.00137-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nataro JP, Kaper JB. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11(1):142. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanders CC, Sanders WE., Jr Emergence of resistance to cefamandole: possible role of cefoxitin-inducible beta-lactamases. Antimicrob Agents Chemother. 1979;15(6):792–797. doi: 10.1128/AAC.15.6.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, He T, Han J, Wang J, Foley SL, Yang GY, Wan SX, Shen JZ, Wu CM. Prevalence of ESBLs and PMQR genes in fecal Escherichia coli isolated from the non-human primates in six zoos in China. Vet Microbiol. 2012;159(1–2):53–59. doi: 10.1016/j.vetmic.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 10.CLSI . Performance standards for antimicrobial susceptibility testing; twenty-first informational supplement, CLSI document M100-S21. Wayne: Clinical and Laboratory Standards Institute; 2011. [Google Scholar]
- 11.Clermont O, Christenson JK, Denamur E, Gordon DM. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Env Microbiol Rep. 2013;5(1):58–65. doi: 10.1111/1758-2229.12019. [DOI] [PubMed] [Google Scholar]
- 12.Muller A, Stephan R, Nuesch-Inderbinen M. Distribution of virulence factors in ESBL-producing Escherichia coli isolated from the environment, livestock, food and humans. Sci Total Environ. 2016;541:667–672. doi: 10.1016/j.scitotenv.2015.09.135. [DOI] [PubMed] [Google Scholar]
- 13.Liu X, Boothe DM, Thungrat K, Aly S. Mechanisms accounting for fluoroquinolone multidrug resistance Escherichia coli isolated from companion animals. Vet Microbiol. 2012;161(1–2):159–168. doi: 10.1016/j.vetmic.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 14.Kim HB, Wang M, Park CH, Kim EC, Jacoby GA, Hooper DC. oqxAB encoding a multidrug efflux pump in human clinical isolates of Enterobacteriaceae. Antimicrob Agents Chemother. 2009;53(8):3582–3584. doi: 10.1128/AAC.01574-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shaheen BW, Nayak R, Foley SL, Kweon O, Deck J, Park M, Rafii F, Boothe DM. Molecular characterization of resistance to extended-spectrum cephalosporins in clinical Escherichia coli isolates from companion animals in the United States. Antimicrob Agents Chemother. 2011;55(12):5666–5675. doi: 10.1128/AAC.00656-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blaak H, Lynch G, Italiaander R, Hamidjaja RA, Schets FM, AMD H. Multidrug-resistant and extended spectrum beta-lactamase-producing Escherichia coli in Dutch Surface Water and Wastewater. PLoS One. 2015;10(6):e0127752. doi: 10.1371/journal.pone.0127752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haque A, Yoshizumi A, Saga T, Ishii Y, Tateda K. ESBL-producing Enterobacteriaceae in environmental water in Dhaka, Bangladesh. J Infect Chemother. 2014;20(11):735–737. doi: 10.1016/j.jiac.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 18.Li S, Zhu ZC, Wang L, Zhou YF, Tang YJ, Miao ZM. Prevalence and characterization of extended-spectrum beta-lactamase-producing Enterobacteriaceae in spring waters. Lett Appl Microbiol. 2015;61(6):544–548. doi: 10.1111/lam.12489. [DOI] [PubMed] [Google Scholar]
- 19.Xi M, Wu Q, Wang X, Yang B, Xia X, Li D. Characterization of extended-spectrum beta-lactamase-producing Escherichia coli strains isolated from retail foods in Shaanxi Province, China. J Food Prot. 2015;78(5):1018–1023. doi: 10.4315/0362-028X.JFP-14-490. [DOI] [PubMed] [Google Scholar]
- 20.Liu XQ, Liu HX, Li YQ, Hao CJ. High prevalence of beta-lactamase and plasmid-mediated quinolone resistance genes in extended-spectrum cephalosporin-resistant Escherichia coli from dogs in Shaanxi, China. Front Microbiol. 2016;7:1843. doi: 10.3389/fmicb.2016.01843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zurfluh K, Hachler H, Nuesch-Inderbinen M, Stephan R. Characteristics of extended-spectrum beta-lactamase- and carbapenemase-producing Enterobacteriaceae isolates from rivers and lakes in Switzerland. Appl Environ Microb. 2013;79(9):3021–3026. doi: 10.1128/AEM.00054-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brennan FP, Abram F, Chinalia FA, Richards KG, O’Flaherty V. Characterization of environmentally persistent Escherichia coli isolates leached from an Irish soil. Appl Environ Microbiol. 2010;76(7):2175–2180. doi: 10.1128/AEM.01944-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Titilawo Y, Obi L, Okoh A. Occurrence of virulence gene signatures associated with diarrhoeagenic and non-diarrhoeagenic pathovars of Escherichia coli isolates from some selected rivers in South-Western Nigeria. BMC Microbiol. 2015;15:204. doi: 10.1186/s12866-015-0540-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zarfel G, Galler H, Feierl G, Haas D, Kittinger C, Leitner E, Grisold AJ, Mascher F, Posch J, Pertschy B, et al. Comparison of extended-spectrum-beta-lactamase (ESBL) carrying Escherichia coli from sewage sludge and human urinary tract infection. Environ Pollut. 2013;173:192–199. doi: 10.1016/j.envpol.2012.09.019. [DOI] [PubMed] [Google Scholar]
- 25.Liu XQ, Liu HX, Wang L, Peng Q, Li YQ, Zhou HC, Li QF. Molecular characterization of extended-spectrum beta-lactamase-producing multidrug resistant Escherichia coli from swine in Northwest China. Front Microbiol. 2018;9:1756. doi: 10.3389/fmicb.2018.01756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ewers C, Bethe A, Semmler T, Guenther S, Wieler LH. Extended-spectrum ss-lactamase-producing and AmpC-producing Escherichia coli from livestock and companion animals, and their putative impact on public health: a global perspective. Clin Microbiol Infec. 2012;18(7):646–655. doi: 10.1111/j.1469-0691.2012.03850.x. [DOI] [PubMed] [Google Scholar]
- 27.Yin J, Cheng J, Sun Z, Ye Y, Gao YF, Li JB, Zhang XJ. Characterization of two plasmid-encoded cefotaximases found in clinical Escherichia coli isolates: CTX-M-65 and a novel enzyme, CTX-M-87. J Med Microbiol. 2009;58(6):811–815. doi: 10.1099/jmm.0.006007-0. [DOI] [PubMed] [Google Scholar]
- 28.Yang X, Liu W, Liu Y, Wang J, Lv L, Chen X, He D, Yang T, Hou J, Tan Y, et al. F33: A-: B-, IncHI2/ST3, and IncI1/ST71 plasmids drive the dissemination of fosA3 and blaCTX-M-55/−14/−65 in Escherichia coli from chickens in China. Front Microbiol. 2014;5:688. doi: 10.3389/fmicb.2014.00688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rao L, Lv L, Zeng Z, Chen S, He D, Chen X, Wu C, Wang Y, Yang T, Wu P, et al. Increasing prevalence of extended-spectrum cephalosporin-resistant Escherichia coli in food animals and the diversity of CTX-M genotypes during 2003-2012. Vet Microbiol. 2014;172(3–4):534–541. doi: 10.1016/j.vetmic.2014.06.013. [DOI] [PubMed] [Google Scholar]
- 30.Rogers BA, Sidjabat HE, Paterson DL. Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J Antimicrob Chemoth. 2011;66(1):1–14. doi: 10.1093/jac/dkq415. [DOI] [PubMed] [Google Scholar]
- 31.Xu G, An W, Wang H, Zhang X. Prevalence and characteristics of extended-spectrum beta-lactamase genes in Escherichia coli isolated from piglets with post-weaning diarrhea in Heilongjiang province, China. Front Microbiol. 2015;6:1103. doi: 10.3389/fmicb.2015.01103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the data supporting our findings is contained within the manuscript.