Abstract
Background
Acinetobacter baumannii strains with multiple antimicrobial resistance are primarily known as opportunistic nosocomial bacteria but they may also be regarded as emerging bacterial contaminants of food samples of animal origin. Here we aimed to study the molecular characteristics of the A. baumanni strains isolated from raw meat samples.
Methods
A total of 22 A. baumanni strains were isolated from 126 animal meat samples and were genotyped by ERIC-PCR method and by PCR detection of their virulence and antimicrobial resistance determinants. A. baumannii strains with 80% and more similarities were considered as one cluster.
Results
Sixteen different genetic clusters were found amongst the 22 A. baumanni strains. Of the 22 strains, 12 (54.54%) had similar genetic cluster. A. baumannii strains exhibited the highest percentage of resistance against tetracycline (90.90%), trimethoprim (59.09%), cotrimoxazole (54.54%) and gentamicin (50.00%). TetA (81.81%), tetB (72.72%), dfrA1 (63.63%), aac(3)-IV (63.63%), sul1 (63.63%) and aadA1 (45.45%) were the most commonly detected antibiotic resistance genes. FimH (81.81%), afa/draBC (63.63%), csgA (63.63%), cnf1 (59.09%), cnf2 (54.54%) and iutA (50.00%) were the most commonly detected virulence factors. A. baumannii strains isolated from the chicken meat samples had the highest similarities in the genetic cluster.
Conclusions
A. baumannii strains with similar genetic cluster (ERIC-Type) had the same prevalence of antibiotic resistance, antibiotic resistance genes and virulence factors. Genetic cluster of the A. baumannii strains is the main factor affected the similarities in the genotypic and phenotypic properties of the A. baumannii strains.
Keywords: Acinetobacter baumannii, ERIC-PCR, Virulence genes, Antibiotic resistance pattern, Raw meat
Background
Consumption of raw or undercooked meat has been associated with several outbreaks of bacterial foodborne disease all-around the world [1–5]. Bacterial strains isolated from meat are both pathogenic and commensal such as Staphylococcus aureus, Streptococcus species, Listeria monocytogenes, Bacillus spp., Salmonella spp., Campylobacter spp., Escherichia coli, Yersinia enterocolitica, Acinetobacter spp., Aeromonas spp., Pseudomonas spp. and etc. [2, 3, 6–9]. Acinetobacter species are saprophytic, ubiquitous and have emerged as an important nosocomial pathogen due to its ability for survival in the hospital environment on a wide range of dry and moist surfaces [10, 11]. Human infections caused by Acinetobacter species include pneumonia, which is most often related to endotracheal tubes or tracheostomies, endocarditis, meningitis, skin and wound infections, peritonitis in patients receiving peritoneal dialysis, UTI and bacteremia.[10, 11] Acinetobacter spp. and especially Acinetobacter baumannii are one of the newly emerged bacteria all-around the world [10, 11]. A. baumannii colonizes the respiratory tract, skin, urinary and gastrointestinal systems, and frequently leads to pneumonia, surgical site infections, catheter-related blood circulatory infections, and urinary tract infections and rarely community acquired pneumonia, meningitis, mediastinitis, osteomyelitis and cholangitis [10, 11].
A. baumannii strains isolated from clinical infections harbored the high prevalence of certain virulence factors including pathogenicity islands (PAI), P fimbriae (pap genes), S fimbriae adhesion and F1C (sfa), a fimbriae adhesion and Dr. antigen family (afa/dra), type 1 fimbriae (fimH), fibronectin receptor (fbn), serum resistance (traT), curli fibers (csg), aerobactin (iutA), yersiniabactin (fyuA), polysaccharide coatings as group II and III capsules (kpsMT), cytotoxic necrotizing factors (cnf), invasins factors (IbeA) and colicin V production (cvaC). Moreover, these factors are responsible for adhesion, colonization and invasion of the A. baumannii strains into the different target organs [12, 13].
A. baumannii strains are also known to harbor multiple resistance against different types of antibiotics. Documented data revealed that the A. baumannii strains isolated from human and animal clinical infections and food stuffs harbored the high prevalence of resistance against different types of antibiotics [14, 15]. The genes that encode resistance against antibiotics and especially aadA1 (streptomycin), aac(3)-IV (gentamicin), sul1 (sulfonamide), blaSHV and blaCTX-M (cephalosporins), cat1 and cmlA (chloramphenicol), tetA and tetB (tetracycline), dfrA1 (trimethoprim), qnr (fluoroquinolones),imp, sim and vim (carbenicillin), and Oxa-51-like(oxacillinases) are mainly responsible for occurrence of severe antibiotic resistance [14–16]. In keeping with this, genotyping is one of the best techniques to find the molecular relationship between bacterial strains isolated from different sources. Enterobacterial Repetitive Intergenic Consensus Polymerase Chain Reaction (ERIC-PCR) is one of the best typing methods of DNA fingerprints of bacterial strains. This method has sporadically been used to characterize A. baumannii strains isolated from clinical samples [17].
Studies regarding the association of Acinetobacter with foodborne illnesses are somewhat limited [10, 11].Therefore, the present investigation was done to study the phenotypic and genotypic characterization of antibiotic resistance and molecular typing of the A. baumannii strains isolated from raw meat samples.
Methods
A. baumannii strains
From January 2016 to May 2017, a total of 22 A. baumannii strains were isolated from 126 samples of raw chicken, turkey, bovine, ovine and camel meat samples. Meat samples were randomly collected (simple random sampling procedure) from the butchers and shopping center of different parts of the Chaharmahal Va Bakhtiari province, Iran. All isolates were transferred to the Microbiology Research Center of the Islamic Azad University of Shahrekord in cooler with ice-packs.
A. baumanni strains were isolated based on the method described by the Cappuccino and Welsh (2016) [18]. Briefly, the specimens were inoculated initially on blood agar (Merck) and MacConkey agar (Merck) medium and incubated for 24 h at 37 °C. Conventional biochemical methods such as oxidase, citrate, urea urease, malonate consumption, oxidation and fermentation of sugars, motility and indole production were used to identify A. baumannii. Additionally, genus Acinetobacter was identified by Gram staining, cell and colony morphology, positive catalase test, negative oxidase test and absence of motility. Speciation of Acinetobacter was performed on the basis of glucose oxidation, gelatin liquefaction, beta hemolysis, growth at 37 °C and 42 °C, arginine hydrolysis and susceptibility to chloramphenicol [18]. The isolates were stored in BHI medium containing 15% glycerol at − 20 °C.
PCR confirmation of the A. baumannii isolates
Confirmation of A. baumannii isolates were done using the PCR method. Presence of the16S–23S ribosomal RNA (16S–23SrRNA) gene was analyzed using the PCR method described previously [19]. A. baumannii isolates were sub-cultured on TSB media (Merck, Germany) and incubated for 48 h at 37 °C. Genomic DNA was extracted from bacterial colonies using the DNA extraction kit (Cinnagen, Iran) according to manufacturer’s instruction. A pair of primers of P-Ab-ITSF (5’-CATTATCACGGTAATTAGTG-3′) and PAb-ITSB (5’-AGAGCACTGTGCACTTAAG-3′) (208 bp) was used for PCR amplification of the 16S–23SrDNA gene of the A. baumanni according to the method described by Chiang et al., (2011) [19]. Confirmed strains were maintained in slant nutrient agar (Merck, Germany) and stored at − 70 °C on 20% sterile buffered glycerol.
Antibiotic susceptibility testing
Patterns of antimicrobial resistance of the A. baumanni strains were studied using the simple disk diffusion technique. The Mueller–Hinton agar (Merck, Germany) medium was used for this purpose. Susceptibility of A. baumanniisolates were tested against several types of antibiotics with appropriate disks containing azithromycin (15 μg), erythromycin (15 μg), rifampin (5 μg), nitrofurantoin (300 μg), chloramphenicol (30 μg), imipenem (30 μg), meropenem (10 μg), levofloxacin (5 μg), ciprofloxacin (5 μg), trimethoprim (5 μg), tetracycline (30 μg), ceftazidime (30 μg), cephalotin (30 μg), trimethoprim/sulphamethoxazole(25 μg), tobramycin (10 μg), amikacin (30 μg), gentamicin (10 μg) and streptomycin (10 μg) (produced by PadTan-Teb, Iran), according to the instruction of Clinical and Laboratory Standards Institute [20]. A. baumanni ATCC 19606 was used for quality control organism in antimicrobial susceptibility determination.
Detection of antibiotic resistance genes and virulence factors
Thermo-cycler (Eppendorf Mastercycler 5330, Eppendorf-Nethel-Hinz GmbH, Hamburg, Germany) was used in all PCR reactions. Table 1 represents the list of primers and PCR conditions used for amplification of virulence factors and antibiotic resistance genes in the A. baumannii strains.
Table 1.
Oligonucleotide primers and PCR conditions used for amplification of the antibiotic resistance genes and virulence factors in the A. baumannii strains of animal origin [19, 34–46]
| Target gene | Primer Sequence (5′-3′) | Size of product (bp) | PCR conditions | Volume (50 μl) |
|---|---|---|---|---|
| draBC | GCTGGGCAGCAAACTGATAACTCTC CATCAAGCTGTTTGTTCGTCCGCCG |
750 | 1 cycle: 95 0C ------------ 4 min. 30 cycle: 95 0C ------------ 50 s 58 0C ------------ 60 s 72 0C ------------ 45 s 1 cycle: 72 0C ------------ 8 min |
5 μL PCR buffer 10X 1.5 mM Mgcl2 200 μM dNTP (Fermentas) 0.5 μM of each primers F & R 1.25 U Taq DNA polymerase (Fermentas) 2.5 μL DNA template |
| cnf1 | AAGATGGAGTTTCCTATGCAGGAG CATTCAGAGTCCTGCCCTCATTATT |
498 | ||
| csgA | ACTCTGACTTGACTATTACC AGATGCAGTCTGGTCAAC |
200 | ||
| cvaC | CACACACAAACGGGAGCTGTT CTTCCCGCAGCATAGTTCCAT |
680 | ||
| iutA | GGCTGGACATCATGGGAACTGG CGTCGGGAACGGGTAGAATCG |
300 | ||
| fyuA | TGATTAACCCCGCGACGGGAA CGCAGTAGGCACGATGTTGTA |
880 | ||
| cnf2 | AATCTAATTAAAGAGAAC CATGCTTTGTATATCTA |
543 | 1 cycle: 94 0C ------------ 6 min. 34 cycle: 95 0C ------------ 50 s 58 0C ------------ 70 s 72 0C ------------ 55 s 1 cycle: 72 0C ------------ 10 min |
5 μL PCR buffer 10X 2 mM Mgcl2 150 μM dNTP (Fermentas) 0.75 μM of each primers F & R 1.5 U Taq DNA polymerase (Fermentas) 3 μL DNA template |
| kpsMT II | GCGCATTTGCTGATACTGTTG CATCCAGACGATAAGCATGAGCA |
272 | ||
| PAI | GGACATCCTGTTACAGCGCGCA TCGCCACCAATCACAGCCGAAC |
930 | ||
| papC | GACGGCTGTACTGCAGGGTGTGGCG ATATCCTTTCTGCAGGGATGCAATA |
328 | ||
| fimH | TGCAGAACGGATAAGCCGTGG GCAGTCACCTGCCCTCCGGTA |
508 | 1 cycle: 95 0C ------------ 4 min. 34 cycle: 94 0C ------------ 60 s 56 0C ------------ 45 s 72 0C ------------ 60 s 1 cycle: 72 0C ------------ 10 min |
5 μL PCR buffer 10X 2 mM Mgcl2 200 μM dNTP (Fermentas) 0.5 μM of each primers F & R 1.5 U Taq DNA polymerase (Fermentas) 5 μL DNA template |
| ibeA | AGGCAGGTGTGCGCCGCGTAC TGGTGCTCCGGCAAACCATGC |
170 | ||
| PapG II-III | CTGTAATTACGGAAGTGATTTCTG ACTATCCGGCTCCGGATAAACCAT |
1070 | ||
| sfa/focDE | CTCCGGAGAACTGGGTGCATCTTAC CGGAGGAGTAATTACAAACCTGGCA |
410 | ||
| traT | GGTGTGGTGCGATGAGCACAG CACGGTTCAGCCATCCCTGAG |
290 | ||
| aadA1 | TATCCAGCTAAGCGCGAACT ATTTGCCGACTACCTTGGTC |
447 | 1 cycle: 94 0C ------------ 6 min. 33 cycle: 95 0C ------------ 70 s 55 0C ------------ 65 s 72 0C ------------ 90 s 1 cycle: 72 0C ------------ 8 min |
5 μL PCR buffer 10X 2 mM Mgcl2 150 μM dNTP (Fermentas) 0. 5 μM of each primers F & R 1.5 U Taq DNA polymerase (Fermentas) 2 μL DNA template |
| aac(3)-IV | CTTCAGGATGGCAAGTTGGT TCATCTCGTTCTCCGCTCAT |
286 | ||
| sul1 | TTCGGCATTCTGAATCTCAC ATGATCTAACCCTCGGTCTC |
822 | ||
| blaSHV | TCGCCTGTGTATTATCTCCC CGCAGATAAATCACCACAATG |
768 | ||
| CITM | TGGCCAGAACTGACAGGCAAA TTTCTCCTGAACGTGGCTGGC |
462 | ||
| cat1 | AGTTGCTCAATGTACCTATAACC TTGTAATTCATTAAGCATTCTGCC |
547 | ||
| cmlA | CCGCCACGGTGTTGTTGTTATC CACCTTGCCTGCCCATCATTAG |
698 | ||
| tet(A) | GGTTCACTCGAACGACGTCA CTGTCCGACAAGTTGCATGA |
577 | ||
| tet(B) | CCTCAGCTTCTCAACGCGTG GCACCTTGCTGATGACTCTT |
634 | ||
| dfrA1 | GGAGTGCCAAAGGTGAACAGC GAGGCGAAGTCTTGGGTAAAAAC |
367 | ||
| qnr | GGGTATGGATATTATTGATAAAG CTAATCCGGCAGCACTATTTA |
670 | ||
| imp | GAATAGAATGGTTAACTCTC CCAAACCACTAGGTTATC |
188 | 1 cycle: 95 0C ------------ 4 min. 30 cycle: 95 0C ------------ 45 s 58 0C ------------ 60s 72 0C ------------ 40 s 1 cycle: 72 0C ------------ 5 min |
5 μL PCR buffer 10X 1.5 mM Mgcl2 100 μM dNTP (Fermentas) 1 μM of each primers F & R 1 U Taq DNA polymerase (Fermentas) 2.5 μL DNA template |
| vim | GTTTGGTCGCATATCGCAAC AATGCGCAGCACCAGGATAG |
382 | ||
| sim | GTACAAGGGATTCGGCATCG GTACAAGGGATTCGGCATCG |
569 | ||
| Oxa-23-like | GATCGGATTGGAGAACCAGA ATTTCTGACCGCATTTCCAT |
501 | 1 cycle: 94 0C ------------ 5 min. 32 cycle: 95 0C ------------ 50 s 60 0C ------------ 60 s 72 0C ------------ 70 s 1 cycle: 72 0C ------------ 10 min |
5 μL PCR buffer 10X 2.5 mM Mgcl2 200 μM dNTP (Fermentas) 0.5 μM of each primers F & R 1.5 U Taq DNA polymerase (Fermentas) 2 μL DNA template |
| Oxa-24-like | GGTTAGTTGGCCCCCTTAAA AGTTGAGCGAAAAGGGGATT |
246 | ||
| Oxa-51-like | TAATGCTTTGATCGGCCTTG TGGATTGCACTTCATCTTGG |
353 | ||
| Oxa-58-like | AAGTATTGGGGCTTGTGCTG CCCCTCTGCGCTCTACATAC |
599 |
Molecular typing of A. baumannii isolates using the ERIC-PCR
The primer pair ERIC1: 5′-ATGTAAGCTCCTGGGGATTCAC-3′ and ERIC2: 5′- AAGTAAGTGACTGGGGTGAGCG-3′ were used to amplify intervening fragments of ERIC in the genomic DNA extracted from the bacterial colonies [17]. Amplification reactions were performed in a final volume of 25 μL including 150 μM dNTP mixture, 1.5 U Taq enzyme, 0.5 μM each primer and 1 μL DNA template. The PCR conditions were as follows: initial denaturation (5 min at 94 °C), followed by four cycles of denaturation (1 min at 94 °C), annealing (1 min at 26 °C) and extension (1 min at 72 °C) and 40 cycles of denaturation (30 s at 94 °C), annealing (30 s at 40 °C) and extension (1 min at 72 °C), followed by the final extension at 72 °C for 10 min [17]. Amplified products of each sample were subjected to electrophoresis in 2% agarose gel containing DNA Safe Stain (CinnaGen, Iran).
Data analysis
Electrophoretic patterns were analyzed either visually or by using the Bionumerics software (Applied Maths, Sint-Martems-Latem, Belgium). The BioNumerics analysis was performed using the Dice coefficient and the unweighted pair group method of averages (UPGMA) with a 1% tolerance limit and 1% optimization. Isolates that clustered with ≥80% similarity were considered to belong to the same ERIC type, respectively.
Statistical analysis
Data were transferred to a Microsoft Excel spreadsheet (version 15; Microsoft Corp., Redmond, WA, USA) for analysis. Using statistical software (version 16; SPSS Inc., Chicago, USA), Chi-square test and Fisher’s exact two-tailed test analysis was performed and differences were considered significant at values of p < 0.05.
Results
Of 22 tested A. baumannii strains, 10 strains were originated from the chicken meat, 4 strains from bovine meat, 3 strains from camel meat, 3 strains from turkey meat and 2 strains from ovine meat.
Figure 1 represent the electrophoretic patterns of the ERIC types of A. baumannii strains No 1-17 isolated from raw meat samples. Figure 2 represents the electrophoretic patterns of the ERIC types of A. baumannii strains No 18-22 isolated from raw meat samples. Figure 3 shows the results of the molecular typing of the A. baumannii strains isolated from different animal origins. A. baumannii strains with 80% and more than 80% similarity in the molecular typing were considered as one cluster. In keeping with this, a total of 16 ERIC types were identified. We found that strains no 19 and 22 (originated from chicken meat), 12 and 15 (originated from chicken meat), 6 (originated from bovine meat) and 14 (originated from chicken meat), 1 and 3 (originated from chicken meat), 4 (originated from bovine meat) and 5 (originated from turkey meat) and finally 2 (originated from chicken meat) and 10 (originated from ovine meat) had similar genetic cluster and were related to the 1, 4, 9, 13, 15 and 16 genetic cluster of the A. baumannii strains, respectively.
Fig. 1.
Electrophoretic patterns of the ERIC types of A. baumannii strains No 1–17 isolated from raw meat samples, NC=Negative control, M = DNA Ladder 100 bp, 1Kb
Fig. 2.

Electrophoretic patterns of the ERIC types of A. baumannii strains No 18–22 isolated from raw meat samples, NC=Negative control, M = DNA Ladder 100 bp, 1Kb
Fig. 3.
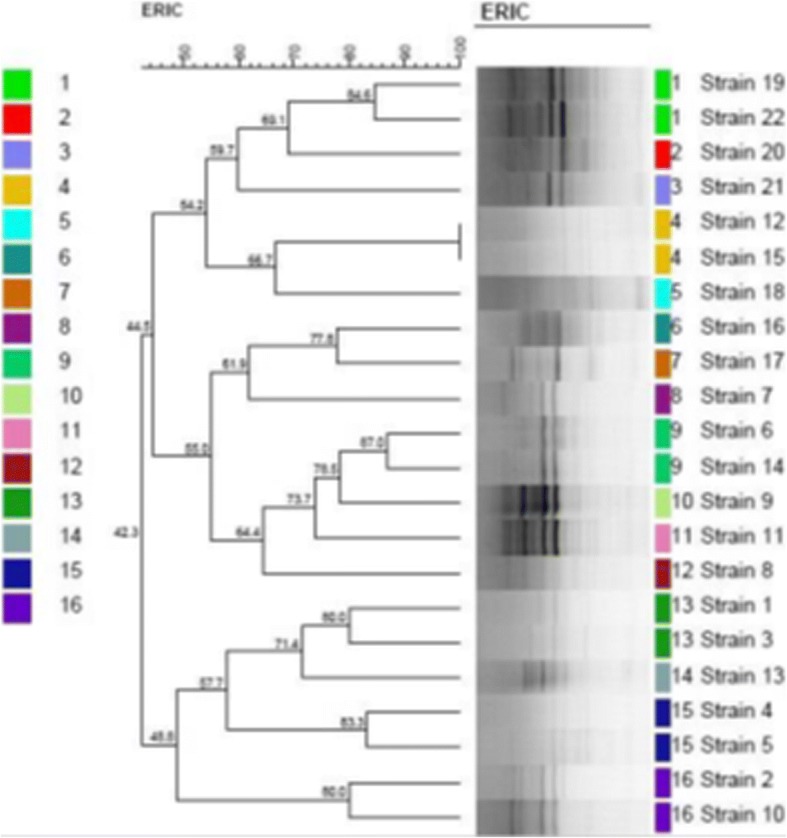
Dendrogram of genetic relatedness of the A. baumannii strains isolated from meat of animal origins using the ERIC-PCR
Table 2 represents the antibiotic resistance pattern of the A. baumannii strains isolated from different animal origins. A. baumannii strains harbored the highest prevalence of resistance against tetracycline (90.90%), trimethoprim (59.09%), co-trimoxazole (54.54%) and gentamicin (50.00%) antibiotics. A. baumannii strains of similar molecular cluster (ERIC-type) showed the same pattern of antibiotic resistance (Table 2). TetA (81.81%), tetB (72.72%), dfrA1 (63.63%), aac(3)-IV (63.63%), sul1 (63.63%) and aadA1 (45.45%) were the most commonly detected antibiotic resistance genes. A. baumannii strains of the same molecular cluster had the same antibiotic resistance genes (Table 2).
Table 2.
Profiles obtained by ERIC-PCR, virulence factors and antimicrobial resistance pattern in A. baumannii strains isolated from animal origins
| ERIC typesa | Numbers and source of strains | Antibiotic resistance phenotypes | Antibiotic resistance genes | ® |
|---|---|---|---|---|
| I | 19,22 (Chicken meat) | SXT25, TE30, F300 | aadA1, sul1, tetB, sim, oxa-58-like | cnf1, csgA, fimH |
| II | 20 (Chicken meat) | GM10, TE30, TMP5, CIP5, C30, F300 | sul1, tetA, tetB, dfrA1, qnr, cat1, vim | ibeA, sfa/focDE, afa/draBC |
| III | 21 (Chicken meat) | CL30, TE30, TMP5, E15 |
tetB, dfrA1, qnr, cmlA,
oxa-23-like |
kpsMTII, PAI, ibeA, traT, afa/draBC, cnf2, csgA, fyuA, fimH |
| IV | 12,15 (Chicken meat) | S10, GM10, SXT25, TE30, TMP5 | aadA1, aac(3)-IV, sul1, blaSHV, tetA, dfrA1 | kpsMTII, sfa/focDE, cnf1, iutA |
| V | 18 (Turkey meat) | TOB10, CL30, CAZ30, TE30, AZM15 | aac(3)-IV, blaSHV, ClTM, tetA, tetB | cvaC, fimH |
| VI | 16 (Ovine meat) | GM10, AK30, SXT25, CL30, CAZ30, TE30, TMP5, CIP5, LEV5, MEM10, C30, F300, E15 | aadA1, aac(3)-IV, sul1, blaSHV, ClTM, tetA, tetB, dfrA1, cat1, cmlA, sim, oxa-51-like, oxa-23-like | kpsMTII, ibeA, papC, papGII, papGIII, traT, afa/draBC, cnf1, cnf2, csgA, iutA, fimH |
| VII | 17 (Bovine meat) | S10, CAZ30, TE30, TMP5, IPM10, RA5 | aadA1, aac(3)-IV, ClTM, dfrA1, imp | PAI, papC, papGIII, traT, afa/draBC, cnf1, cnf2, csgA, cvaA, iutA, fyuA, fimH |
| VIII | 7 (Bovine meat) | F300, RA5 |
aadA1, ClTM, tetA, tetB, dfrA1, cat1, imp,
oxa-51-like, oxa-24-like |
kpsMTII, PAI, papC, papGIII, traT, afa/draBC, cnf1, cnf2, csgA, iutA, fyuA, fimH |
| IX | 14 (Chicken meat) 6 (Bovine meat) |
GM10, AK30, TOB10, CL30, TE30, CIP5, C30 | aac(3)-IV, sul1, blaSHV, tetA, tetB, dfrA1 | papGII, papGIII, afa/draBC, cnf1, cnf2, cvaC, fyuA, fimH |
| X | 9 (Camel meat) | S10, AZM15 | aadA1, tetA | PAI, papC, papGII, sfa/focDE, afa/draBC, csgA, cvaC |
| XI | 11 (Camel meat) | GM10, TOB10, SXT25, TE30, C30, RA5 | aadA1, aac(3)-IV, sul1, tetA, tetB, cmlA | ibeA, papGII, sfa/focDE, traT, afa/draBC, cnf2, csgA, cvaC, fimH |
| XII | 8 (Chicken meat) | S10, AK30, CAZ30, TE30, LEV5, IPM10, C30 |
sul1, ClTM, tetA, tetB, qnr, cat1, cmlA, vim, oxa-51-like, oxa-23-like, oxa-24-like |
PAI, ibeA, papGII, sfa/focDE, cnf2, iutA, fimH |
| XIII | 1,3 (Chicken meat) | SXT25, TE30, TMP5, CIP5, MEM10, E15 | aac(3)-IV, sul1, ClTM, tetA, tetB, dfrA1 | cnf2, csgA, iutA, fimH |
| XIV | 13 (Turkey meat) | TOB10, TE30, TMP5, CIP5, LEV5, E15 |
aadA1, tetA, tetB, dfrA1, oxa-51-like,
oxa-24-like |
ibeA, papC, sfa/focDE, fimH |
| XV | 5 (Turkey meat) 4 (Bovine meat) |
GM10, TOB10, SXT25, TE30, TEP5, E15 | aac(3)-IV, sul1, tetA, tetB | afa/draBC, cnf1, cnf2, csgA, cvaC, fimH |
| XVI | 2 (Camel meat) 10 (Ovine meat) |
S10, GM10, TE30, TMP5 |
aac(3)-IV, tetA, dfrA1,
oxa-58-like |
sfa/focDE, afa/draBC, cnf1, csgA, cvaC, iutA, fyuA, fimH |
aIn this table: S10 = Streptomycin, GM10 = Gentamicin, AK30 = Amikacin, TOB10 = Tobramycin, SXT25 = Trimethoprim/sulphamethoxazole, CL30 = Cephalothin, CAZ30 = Ceftazidime, TE30 = Tetracycline, TMP5 = Trimethoprim, CIP5 = Ciprofloxacin, LEV5 = Levofloxacin, IPM10 = Imipenem, MEM10 = Meropenem, C30 = Chloramphenicol, F300 = Nitrofurantoin, AZM15 = Azithromycin, RA5 = Rifampin, E15 = Erythromycin
Table 2 also represents the distribution of virulence factors among the A. baumannii strains isolated from meat samples of animals. FimH (81.81%), afa/draBC (63.63%), csgA (63.63%), cnf1 (59.09%), cnf2 (54.54%) and iutA (50.00%) were the most commonly detected virulence factors amongst the A. baumannii strains isolated from animal origins. A. baumannii strains of same molecular cluster (ERIC-type) had the same virulence factors.
Discussion
Acinetobacter is a nosocomial pathogen. Its ability to infect healthy hosts and its propensity to develop antimicrobial drug resistance is a cause for concern among infectious disease specialty. Although the ubiquitous existence of A. baumannii in hospital environment has been considered a routine delusion by several investigations [10, 12–14, 16, 17, 19, 21], different recent reports have undeniably highlighted the presence of the bacterium in different types of animal origins [11, 22, 23]. In recent years, nosocomial infections of A. baumannii, as an opportunistic pathogen, are increasing. Treatment of this bacteria especially multi-drug resistant and broad-spectrum beta-lactamases strains is of major concern [11, 22, 23]. These recent works have mainly been done only on the prevalence rate of A. baumannii and in some cases, on antibiotic susceptibility patterns. Reversely, to the best of our knowledge, the study presented here is the first report of the molecular typing, on the distribution of virulence factors and genotypic evaluation of antibiotic resistance of the A. baumannii strains isolated from different animal origins. A total of 22 strains of A. baumannii were assessed and 16 different genetic cluster were detected. Meat-derived A. baumannii strains may originate in the slaughterhouses, butchers and shopping centers especially due to the manipulation of the meat samples.
A. baumannii has previously been recognized as an animal colonizer with diverse distributions in different countries including in Scotland, 1.20% [22] and Senegal, 5.10% [24]. Rafei et al. (2015) [25] reported the high prevalence of A. baumannii strains in food samples with animal origins including raw meat, raw milk, and dairy products. The A. baumannii strains of the present research were frequently resistant to clinically relevant antibiotics. However, a few isolates displayed low levels of resistance against imipenem, azithromycin, meropenem, rifampin, levofloxacin, ceftazidime and tobramycin. This latter is still among the drugs of choice for the treatment of A. baumannii infections in humans and animals in Iran.
A. baumannii strains of the same molecular cluster (ERIC-type) had the same profile of the antibiotic resistance pattern. We also found that all of A. baumannii strains originated from chicken meat samples were susceptible to azithromycin and rifampin and resistant against tetracycline. These findings have been supported by several studies conducted on Lebanon [25], Switzerland [26], and France [27].
We found that molecular similarities of 19 and 22, 12 and 15, 6 and 14, 1 and 3, 4 and 5 and finally 2 and 10 A. baumannii strains were 84.60%, 100%, 87%, 80%, 83.30% and 80% with each other, respectively. All the other A. baumannii strains were classified as differing ERIC types. A. baumannii strains recovered from the bovine, camel, turkey and ovine meat samples didn’t show any similarity in their molecular typing and there were no similar antibiotic resistance patterns between them. Our findings showed that they also had similar phenotypic pattern of antibiotic resistance. One possible explanation for the similar molecular types of the A. baumannii strains recovered from different animal origins is their common source of infection. It may also be due to the close contact between bovine and chicken, bovine and turkey and between chicken and ovine species, facilitating transmission of the A. baumannii strains with same molecular type. Unfortunately, Iranian ranchers frequently maintain and breed different species of animals and especially bovine, ovine and caprine species in contact to each other. This may cause easy transmission of the A. baumannii strains between different species. The high genetic diversity of strains isolated from meat samples has also previously been reported by Lupo et al. (2014) [26] and Carvalheira et al. (2017) [28]. Recent research [21] revealed the similar antibiotic resistance pattern of the A. baumannii strains isolated from different types of raw meat samples. They showed that the prevalence of resistance against trimethoprim-sulfamethoxazole, tetracycline, amikacin, tobramycin, ampicillin-sulbactam, meropenem and imipenem were 23.20%, 23.20%, 14.30%, 12.50%, 12.50%, 8.30% and1.20%, respectively which was different to our findings. The prevalence of antibiotic resistant bacteria in meat samples has been attributed, at least partially, to the extensive use of antimicrobials for treatment, prevention and control of diseases and finally for growth stimulate in food-producing animals, since this enhances the antimicrobial selective pressure for strains present. Otherwise, using antibiotics for growth stimulation is allowed in Iran. Low prevalence rate of resistance against carbapenems is due to the fact that these antibiotics are not allowed to treat food-producing animals.
Phenotypic characterization of antibiotic resistance was also confirmed by genotyping analysis of antibiotic resistance. We identified the genes for resistances to aminoglycosides (aadA1 and aac(3)-IV), beta-lactams (blaSHV and blaCTM), chloramphenicol (cat1 and cmlA), tetracyclines (tetA and tetB), sulfonamides (sul1 and dfrA1), carbenicilins (imp, sim and vim) and to fluoroquinolones (qnr). Furthermore, A. baumannii strains harbored their own specific antibiotic resistance genes (Table 2). This part of our study was in agreement with previous researches [21, 29]. Additionally, we found that A. baumannii strains of the same molecular cluster (ERIC-type) had the same profile of the antibiotic resistance genes., as follows: strains no 1 and 3 of the same ERIC-type (type XIII) were positive for CITM,dfrA1, tetA, tetB, aac(3)-IV and sul1 which showed their same genetic pattern of antibiotic resistance. Strains no 12 and 15 of the same ERIC-type (type IV) were positive for tetA, aadA1, aac(3)-IV, blaSHV, dfrA1 and sul1. Strains no 19 and 22 of the same ERIC-type (type I) were positive for tetB, aadA1, sim, oxa-58-like and sul1. Despite of the different animal origin of the A. baumannii strains no 6 and 14 of the same ERIC-type (type IX), were positive for tetA, tetB, aac(3)-IV, sul1, dfrA1 and blaSHV antibiotic resistance genes. Additionally, A. baumannii strains no 4 and 5 of the same ERIC-type (type XV) had different origins but they harbored tetA, tetB, sul1 and aac(3)-IV antibiotic resistance genes. Finally, A. baumannii strains no 2 and 10 of the same ERIC-type (type XVI) were positive for tetA, aac(3)-IV, dfrA1 and oxa-58-like antibiotic resistance. A. baumannii strains of other ERIC types had unique pattern of antibiotic resistance.
Virulence of A. baumannii is dependent on several other attributes (i.e. “slime”, LPS production and etc.) beside the selected virulence genes listed here [30, 31].
Another notable finding is the high prevalence of certain virulence factors in the A. baumannii strains isolated from meat of different animals. The most commonly detected virulence genes amongst these A. baumannii strains were fimH, afa/draBC, csgA, cnf1, cnf2 and iutA. Prevalence of non-adhesive virulence factors including traT (serum resistance), cvaC (colicin V), ibeA (invasion), fyuA (yersiniabactin) and PAI (indicator gene) were low. This finding is in agreement with those of previous researches [30, 31]. Non-adhesive virulence factors play an important role in bacterial survival in special conditions such as in human blood and exposure to serum and poor iron environments contributing to the pathogenesis of extra instestinal diseases. However, presence of these non-adhesive virulence factors may not be essential for pathogenicity of food-borne enteric diseases. The gene iutA (aerobactin) has a high prevalence (50.00%) in our strains. Darvishi (2016) [32] reported that the prevalence of cnf1, csgA, cvaC and iutA virulence factors amongst the A. baumannii strains isolated from hospitalized patients were 50%, 70%, 10% and 25%, respectively. Daryanavard and Safaei (2015) [33] reported that the total prevalence of csga, cnf1, cvaC and iutA virulence genes among the samples of UTIs were 55%, 40%, 10% and 30%, respectively which was similar to our findings. Momtaz et al. (2015) [13] reported that the prevalence of csga, cnf1, cvaC and iutA virulence genes among the A. baumannii strains of clinical infections in Iran were 12.39%, 35.53%, 21.48% and 19%, respectively which was lower than our results.
We also found that the A. baumannii strains of the same molecular cluster (ERIC-type) had the same virulence factors. Despite of the different origin of the A. baumannii strains no 6 and 14 of the same ERIC-type (type IX), were both positive for fimH, fyuA, cvaC, cnf1, cnf2, papGII, papGIII, and afa/draBC virulence factors. Furthermore, A. baumannii strains no 4 and 5 of the same ERIC-type (molecular type XV) had different origins but they both harbored fimH, cvaC, csgA, cnf1, afa/draBC and cnf2 virulence factors. Finally, A. baumannii strains no 2 and 10 of the same ERIC-type (type XVI) were positive for fimH, fyuA, csgA, afa/draBC, cvaC, iutA, cnf1 and sfa/focDE virulence factors. These high similarities in the pattern of virulence factors of the A. baumannii strains of same ERIC types recovered from different origins showed that genetic cluster of bacterial strains is closely related to their virulence determinants.
A. baumannii strains isolated from the chicken meat samples had similar molecular type. A. baumannii strains isolated from other meat samples did not fall into common molecular types. Two A. baumannii strains of bovine origins had similar molecular type with turkey and chicken. A camel and ovine A. baumannii strains had also similar molecular type.
Conclusions
In conclusion, we studied the phenotypic and genotypic patterns of antibiotic resistance of the A. baumannii strains isolated from meat samples of different animals. High prevalence of resistance against tetracycline, trimethoprim, co-trimoxazole and gentamicin antibiotics and high distribution of tetA, tetB, dfrA1, aac(3)-IV, sul1 and aadA1 antibiotic resistance genes was established. Besides, fimH, afa/draBC, csgA, cnf1, cnf2 and iutA were the most commonly detected virulence factors amongst the A. baumannii strains of our study. Twenty-two A. baumannii strains were classified into the 16 different genetic clusters according to the results of the ERIC-PCR. We also found that genetic cluster of the strains was closely related to their antibiotic resistance genes and virulence factors. Similarities in the genetic cluster of some strains was also recorded which may represent their similar sources of contamination.
Acknowledgements
The authors would like to thank Dr. Abbas Farahani for his assistance in sample collection and PCR genetic alignments. This work was financially supported by the Islamic Azad University, Shahrekord Branch, Shahrekord, Iran.
Funding
Hassan Momtaz received Research grants for Research at Islamic Azad University, Shahrekord Branch with grant number 96/1046.
Availability of data and materials
All data analysed during this study are included in this published article.
Abbreviations
- CLSI
Clinical and Laboratory Standards Institute
- ERIC-PCR
Enterobacterial Repetitive Intergenic Consensus-Polymerase Chain Reaction
Authors’ contributions
MT, HM and LS carried out the molecular genetic studies, participated in the primers sequence alignment and drafted the manuscript. PM and ET carried out the sampling and culture method. HM and MT participated in the design of the study, performed the statistical analysis and writing the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study was done on meat samples collected from raw animal meats, so there have no ethical issue in this work.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Marziyeh Tavakol, Email: marziyeh.tavakol@yahoo.com.
Hassan Momtaz, Phone: +983833361064, Email: hamomtaz@yahoo.com, Email: hamomtaz@iaushk.ac.ir.
Parviz Mohajeri, Email: p_mohajeri@yahoo.com.
Leili Shokoohizadeh, Email: Shokoohizadeh@yahoo.com.
Elahe Tajbakhsh, Email: ee_tajbakhsh@yahoo.com.
References
- 1.Dehkordi FS, Parsaei P, Saberian S, Moshkelani S, Hajshafiei P, Hoseini SR, et al. Prevalence study of theileria annulata by comparison of four diagnostict techniques in Southwest Iran. Bulgarian J Vet Med. 2012;15(2):123–130. [Google Scholar]
- 2.Momtaz H, Davood Rahimian M, Safarpoor Dehkordi F. Identification and characterization of Yersinia enterocolitica isolated from raw chicken meat based on molecular and biological techniques. J Appl Poult Res. 2013;22(1):137–145. doi: 10.3382/japr.2012-00549. [DOI] [Google Scholar]
- 3.Momtaz H, Dehkordi FS, Rahimi E, Asgarifar A, Momeni M. Virulence genes and antimicrobial resistance profiles of Staphylococcus aureus isolated from chicken meat in Isfahan province, Iran. J Appl Poult Res. 2013;22(4):913–921. doi: 10.3382/japr.2012-00673. [DOI] [Google Scholar]
- 4.Ghorbani F, Gheisari E, Dehkordi FS. Genotyping of vacA alleles of helicobacter pylori strains recovered from some Iranian food items. Trop J Pharm Res. 2016;15(8):1631–1636. doi: 10.4314/tjpr.v15i8.5. [DOI] [Google Scholar]
- 5.Dehkordi FS, Gandomi H, Basti AA, Misaghi A, Rahimi E. Phenotypic and genotypic characterization of antibiotic resistance of methicillin-resistant Staphylococcus aureus isolated from hospital food. Antimicrob Resist Infect Control. 2017;6(1):104. doi: 10.1186/s13756-017-0257-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Momtaz H, Safarpoor Dehkordi F, Taktaz T, Rezvani A, Yarali S. Shiga toxin-producing Escherichia coli isolated from bovine mastitic milk: serogroups, virulence factors, and antibiotic resistance properties. Sci World J. 2012;2012. [DOI] [PMC free article] [PubMed]
- 7.Rahimi E, Sepehri S, Dehkordi FS, Shaygan S, Momtaz H. Prevalence of Yersinia species in traditional and commercial dairy products in Isfahan Province, Iran. Jundishapur J Microbiol. 2014;7(4). [DOI] [PMC free article] [PubMed]
- 8.Madahi H, Rostami F, Rahimi E, Dehkordi FS. Prevalence of enterotoxigenic Staphylococcus aureus isolated from chicken nugget in Iran. Jundishapur J Microbiol 2014;7(8). [DOI] [PMC free article] [PubMed]
- 9.Ranjbar R, Masoudimanesh M, Dehkordi FS, Jonaidi-Jafari N, Rahimi E. Shiga (Vero)-toxin producing Escherichia coli isolated from the hospital foods; virulence factors, o-serogroups and antimicrobial resistance properties. Antimicrob Resist Infect Control. 2017;6(1):4. doi: 10.1186/s13756-016-0163-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engür D, Çetinkaya Çakmak B, Kaynak Türkmen M, Telli M, Eyigör M, Güzünler M. A milk pump as a source for spreading Acinetobacter baumannii in a neonatal intensive care unit. Breastfeeding Med. 2014;9(10):551–554. doi: 10.1089/bfm.2014.0054. [DOI] [PubMed] [Google Scholar]
- 11.Gurung M, Nam HM, Tamang MD, Chae MH, Jang GC, Jung SC, et al. Prevalence and antimicrobial susceptibility of Acinetobacter from raw bulk tank milk in Korea. J Dairy Sci. 2013;96(4):1997–2002. doi: 10.3168/jds.2012-5965. [DOI] [PubMed] [Google Scholar]
- 12.Eijkelkamp BA, Stroeher UH, Hassan KA, Paulsen IT, Brown MH. Comparative analysis of surface-exposed virulence factors of Acinetobacter baumannii. BMC Gen. 2014;15(1):1020. doi: 10.1186/1471-2164-15-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Momtaz H, Seifati SM, Tavakol M. Determining the prevalence and detection of the most prevalent virulence genes in Acinetobacter baumannii isolated from hospital infections. Int J Med Lab. 2015;2(2):87–97. [Google Scholar]
- 14.Asadollahi P, Akbari M, Soroush S, Taherikalani M, Asadollahi K, Sayehmiri K, et al. Antimicrobial resistance patterns and their encoding genes among Acinetobacter baumannii strains isolated from burned patients. Burns. 2012;38(8):1198–1203. doi: 10.1016/j.burns.2012.04.008. [DOI] [PubMed] [Google Scholar]
- 15.Pourhajibagher M, Hashemi FB, Pourakbari B, Aziemzadeh M, Bahador A. Antimicrobial resistance of Acinetobacter baumannii to imipenem in Iran: a systematic review and meta-analysis. Open Microbiol J. 2016;10:32. doi: 10.2174/1874285801610010032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang H, Wang J, Yu P, Ge P, Jiang Y, Xu R, et al. Identification of antibiotic resistance genes in the multidrug-resistant Acinetobacter baumannii strain, MDR-SHH02, using whole-genome sequencing. Int J Mol Med. 2017;39(2):364–372. doi: 10.3892/ijmm.2016.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ying C, Li Y, Wang Y, Zheng B, Yang C. Investigation of the molecular epidemiology of Acinetobacter baumannii isolated from patients and environmental contamination. J Antibiot. 2015;68(9):562. doi: 10.1038/ja.2015.30. [DOI] [PubMed] [Google Scholar]
- 18.Cappuccino JG, Welsh CT. Microbiology: a laboratory manual. 11. London: Pearson Education; 2016. [Google Scholar]
- 19.Chiang MC, Kuo SC, Chen YC, Lee YT, Chen TL, Fung CP. Polymerase chain reaction assay for the detection of Acinetobacter baumannii in endotracheal aspirates from patients in the intensive care unit. J Microbiol, Immunol Infect. 2011;44(2):106–110. doi: 10.1016/j.jmii.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 20.CLSI. Performance standards for antimicrobial susceptibility testing; twenty-fifth informational supplement. CLSI document M100-S25. Wayne: Clinical and Laboratory Standards Institute; 2017.
- 21.Kiani S, Momtaz H, Serajian AA, Tajbakhsh E. Detection of integrons in Acinetobacter baumannii strains isolated from the nosocomial infections of Ahvaz city and their relation with the resistance pattern. Int J Med Lab. 2016;3(1):50–63. [Google Scholar]
- 22.Hamouda A, Findlay J, Al Hassan L, Amyes SG. Epidemiology of Acinetobacter baumannii of animal origin. Int J Antimicrob Agents. 2011;38(4):314–318. doi: 10.1016/j.ijantimicag.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 23.Zordan S, Prenger-Berninghoff E, Weiss R, van der Reijden T, van den Broek P, Baljer G, et al. Multidrug-resistant Acinetobacter baumannii in veterinary clinics, Germany. Emerg Infect Dis. 2011;17(9):1751. doi: 10.3201/eid1709.101931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kempf M, Rolain JM, Diatta G, Azza S, Samb B, Mediannikov O, et al. Carbapenem resistance and Acinetobacter baumannii in Senegal: the paradigm of a common phenomenon in natural reservoirs. PLoS One. 2012;7(6):e39495. doi: 10.1371/journal.pone.0039495. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Rafei R, Hamze M, Pailhoriès H, Eveillard M, Marsollier L, Joly-Guillou ML, et al. Extra-human epidemiology of Acinetobacter baumannii in Lebanon. Appl Environ Microbiol 2015:AEM-03824. [DOI] [PMC free article] [PubMed]
- 26.Lupo A, Vogt D, Seiffert SN, Endimiani A, Perreten V. Antibiotic resistance and phylogenetic characterization of Acinetobacter baumannii strains isolated from commercial raw meat in Switzerland. J Food Protect. 2014;77(11):1976–1981. doi: 10.4315/0362-028X.JFP-14-073. [DOI] [PubMed] [Google Scholar]
- 27.Hauck Y, Soler C, Jault P, Mérens A, Gérome P, Mac Nab C, et al. Diversity of Acinetobacter baumannii in four French military hospitals, as assessed by multiple locus variable number of tandem repeats analysis. PLoS One. 2012;7(9):e44597. doi: 10.1371/journal.pone.0044597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carvalheira A, Casquete R, Silva J, Teixeira P. Prevalence and antimicrobial susceptibility of Acinetobacter spp. isolated from meat. Int J Food Microbiol. 2017;243:58–63. doi: 10.1016/j.ijfoodmicro.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 29.Farahani Kheltabadi R, Moniri R, Shajari GR, Shirazi N, Hossein M, Musavi SG, et al. Antimicrobial susceptibility patterns and the distribution of resistance genes among Acinetobacter species isolated from patients in shahid Beheshti hospital, Kashan. KAUMS J (FEYZ) 2009;12(4):61–67. [Google Scholar]
- 30.Farahami A, Khodarahmi R. Frequency of adhesive virulence factors in Carbapenemase-producing Acinetobacter baumannii isolated from clinical samples. Asian J Biol Sci. 2014;7(4):158–164. doi: 10.3923/ajbs.2014.158.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mohajeri P, Sharbati S, Farahani A, Rezaei Z. Evaluate the frequency distribution of nonadhesive virulence factors in carbapenemase-producing Acinetobacter baumannii isolated from clinical samples in Kermanshah. J Nat Sci Biol Med. 2016;7(1):58. doi: 10.4103/0976-9668.175071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Darvishi M. Virulence factors profile and antimicrobial resistance of Acinetobacter baumannii strains isolated from various infections recovered from immunosuppressive patients. Biomed Pharmacol J. 2016;9(3):1057–1062. doi: 10.13005/bpj/1048. [DOI] [Google Scholar]
- 33.Daryanavard R, Safaei HR. Virulence genes and antimicrobial resistance properties of Acinetobacter baumannii isolated from pediatrics suffered from UTIs. Int J Adv Res Biol Sci. 2015;2(11):272–279. [Google Scholar]
- 34.Randall LP, Cooles SW, Osborn MK, Piddock LJ, Woodward MJ. Antibiotic resistance genes, integrons and multiple antibiotic resistance in thirty-five serotypes of Salmonella enterica isolated from humans and animals in the UK. J Antimicrob Chemother. 2004;53(2):208–216. doi: 10.1093/jac/dkh070. [DOI] [PubMed] [Google Scholar]
- 35.Toro CS, Farfán M, Contreras I, Flores O, Navarro N, Mora GC, et al. Genetic analysis of antibiotic-resistance determinants in multidrug-resistant Shigella strains isolated from Chilean children. Epidemiol Infect. 2005;133(1):81–86. doi: 10.1017/S0950268804003048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van TT, Chin J, Chapman T, Tran LT, Coloe PJ. Safety of raw meat and shellfish in Vietnam: an analysis of Escherichia coli isolations for antibiotic resistance and virulence genes. Int J Food Microbiol. 2008;124(3):217–223. doi: 10.1016/j.ijfoodmicro.2008.03.029. [DOI] [PubMed] [Google Scholar]
- 37.Woodford N, Ellington MJ, Coelho JM, Turton JF, Ward ME, Brown S, et al. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents. 2006;27(4):351–353. doi: 10.1016/j.ijantimicag.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 38.Mammeri H, Van De Loo M, Poirel L, Martinez-Martinez L, Nordmann P. Emergence of plasmid-mediated quinolone resistance in Escherichia coli in Europe. Antimicrob Agents Chemother. 2005;49(1):71–76. doi: 10.1128/AAC.49.1.71-76.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mendes RE, Kiyota KA, Monteiro J, Castanheira M, Andrade SS, Gales AC, et al. Rapid detection and identification of metallo-β-lactamase-encoding genes by multiplex real-time PCR assay and melt curve analysis. J Clin Microbiol. 2007;45(2):544–547. doi: 10.1128/JCM.01728-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marklund BL, Tennent JM, Garcia E, Hamers A, Baga M, Lindberg F, et al. Horizontal gene transfer of the Escherichia coli pap and prs pili operons as a mechanism for the development of tissue-specific adhesive properties. Mol Microbiol. 1992;6(16):2225–2242. doi: 10.1111/j.1365-2958.1992.tb01399.x. [DOI] [PubMed] [Google Scholar]
- 41.Le Bouguenec C, Archambaud M, Labigne A. Rapid and specific detection of the pap, afa, and sfa adhesin-encoding operons in uropathogenic Escherichia coli strains by polymerase chain reaction. J Clin Microbiol 1992;30(5):1189–1193. [DOI] [PMC free article] [PubMed]
- 42.Johnson JR, Brown JJ, Carlino UB, Russo TA. Colonization with and acquisition of uropathogenic Escherichia coli as revealed by polymerase chain reaction-based detection. J Infect Dis. 1998;177(4):1120–1124. doi: 10.1086/517409. [DOI] [PubMed] [Google Scholar]
- 43.Johnson JR, Stell AL. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis. 2000;181(1):261–272. doi: 10.1086/315217. [DOI] [PubMed] [Google Scholar]
- 44.Huang SH, Wass C, Fu QI, Prasadarao NV, Stins M, Kim KS. Escherichia coli invasion of brain microvascular endothelial cells in vitro and in vivo: molecular cloning and characterization of invasion gene ibe10. Infect Immun. 1995;63(11):4470–4475. doi: 10.1128/iai.63.11.4470-4475.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamamoto S, Terai A, Yuri K, Kurazono H, Takeda Y, Yoshida O. Detection of urovirulence factors in Escherichia coli by multiplex polymerase chain reaction. FEMS Immunol Med Microbiol. 1995;12(2):85–90. doi: 10.1111/j.1574-695X.1995.tb00179.x. [DOI] [PubMed] [Google Scholar]
- 46.Schubert S, Rakin A, Karch H, Carniel E, Heesemann J. Prevalence of the “high-Pathogenicity Island” of Yersinia species among Escherichia coliStrains that are pathogenic to humans. Infect Immun. 1998;66(2):480–485. doi: 10.1128/iai.66.2.480-485.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data analysed during this study are included in this published article.


