Abstract
HIV integrase mutation T97A emerges after suboptimal therapy with integrase strand transfer inhibitors (INSTIs), but the contribution of T97A to dolutegravir resistance remains uncertain. Here we report >10-fold increase in dolutegravir resistance after the single addition of T97A in 2 individuals with prior INSTI resistance receiving dolutegravir salvage therapy.
Keywords: antiretroviral therapy, dolutegravir, drug resistance, salvage therapy, T97A
Integrase strand transfer inhibitors (INSTIs) are recommended for most individuals with HIV infection initiating combination antiretroviral therapy (ART), and dolutegravir is now recommended as first-line therapy in resource-limited countries [1]. Dolutegravir (DTG) is also often used as part of salvage regimens. Antiretroviral resistance to INSTIs develops under suboptimal therapy, with several characteristic mutation pathways [2, 3], but the contribution of accessory mutations remains uncertain. T97A is an accessory mutation frequently co-selected with additional drug resistance mutations (DRMs) including Q148H and G140S in individuals failing raltegravir (RAL) or elvitegravir (EVG) therapy [4, 5] and contributes substantially to resistance to these first-generation INSTIs. However, the contribution of T97A to DTG resistance in the presence of other INSTI mutations has not been well described. We report 2 individuals with subtype B HIV and extensive multiple drug class resistance, including preexisting INSTI-associated mutations, who initiated DTG as salvage therapy. The T97A mutation emerged in both cases and was accompanied by a substantial decrease in DTG susceptibility.
CASES
Both participants enrolled in a research protocol at the National Institutes of Health (NIH) Clinical Center investigating the management of people living with HIV experiencing antiretroviral failure (NCT 01976715). The protocol includes inpatient directly observed therapy (DOT) to assess antiretroviral regimens. The study was approved by the Institutional Review Board, and written informed consent was obtained.
Participant #1 is a 52-year-old man with HIV diagnosed in 1989 who subsequently had an extensive history of antiretroviral drug (ARV) exposure and 4-class drug-resistant HIV-1 (Supplementary Tables 1 and 2). At enrollment in August 2014, his CD4 count was 259 cells/mm3 and plasma HIV-1 RNA was 136 476 copies/mL. He was receiving lopinavir/ritonavir + atazanavir + tenofovir disoproxil fumarate (DF)/emtricitabine. Interpretation of cumulative genotypic information (Standford/ANRS algorithms) indicated high-level resistance to all US Food and Drug Administration (FDA)–approved nucleoside reverse transcriptase inhibitors (NRTIs), non-nucleoside reverse transcriptase inhibitors (NNRTIs), protease inhibitors (PIs), and INSTIs, except for intermediate resistance to DTG with the G140S and Q148H mutations (Supplementary Table 3). T97A was not detected by Sanger sequencing or by next-generation sequencing (NGS; Illumina), which has a sensitivity of c. 1%. PhenoSense Integrase assay (Monogram Biosciences) revealed partial sensitivity to DTG (fold change [FC] 4.61), and tropism testing (Trofile ES Monogram Biosciences) revealed R5 tropism (Supplementary Tables 2 and 3).
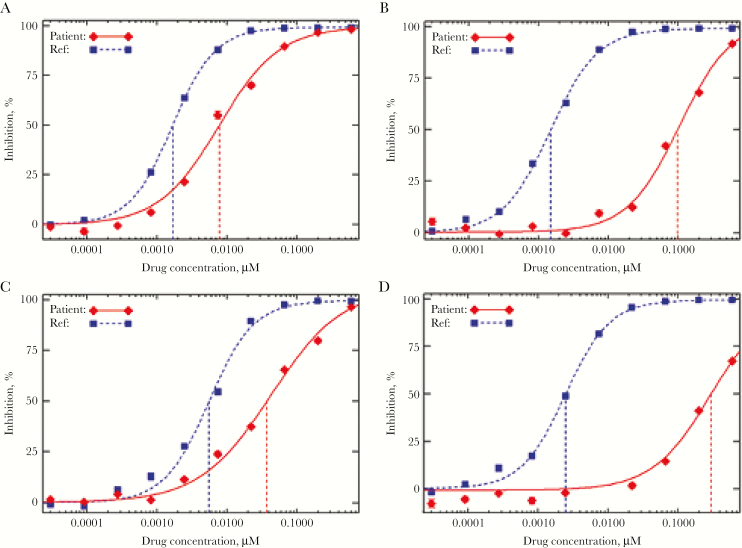
In October 2014, his ART was changed to DTG (twice daily) + darunavir/ritonavir (twice daily) + tenofovir DF/emtricitabine + maraviroc (genotypic susceptibility score [GSS] 1.25). His plasma HIV RNA levels declined according to well-described first- and second-phase kinetics and were <40 copies/mL at 12 weeks. At 24 weeks (Supplementary Figure 1), however, his HIV-1 RNA rebounded to 1638 copies/mL and remained elevated thereafter. His darunavir and DTG concentrations, performed at 6 study visits, had consistently been within therapeutic range, including on the day of viral rebound. A repeat INSTI genotype noted T97A (Supplementary Tables 2 and 3). Several polymorphisms were also detected in NGS that were not detected at baseline and were not previously reported or associated with INSTI resistance (Supplementary Table 4). Phenotyping noted an increase in DTG FC from 4.61 to 66 (Figure 1; Supplementary Table 3). Repeat tropism again revealed R5-tropic HIV.
Figure 1. .
Impact of T97A mutation on phenotypic sensitivity to dolutegravir in HIV-1 isolates with baseline partial sensitivity to dolutegravir. (A) Participant #1 pre-T97A (Mutations – G140S, Q148H; DTG FC 4.61); (B) participant #1 post-T97A (Mutations – G140S, Q148H, T97A; DTG FC 66); (C) participant #2 pre-T97A (Mutations – G140S, Q148H, E138T; DTG FC 6.71); (D) participant #2 post-T97A (Mutations – G140S, Q148H, E138T, T97A; DTG FC 119). HIV-1 phenotypic inhibition curves were obtained from PhenoSense™ Integrase and PhenoSense GT™ plus Integrase (Courtesy of Monogram Biosciences, South San Francisco, CA).
Participant #2 is a 52-year-old man diagnosed with HIV in 1993 who also had an extensive history of ARV exposure and resistance to all FDA-approved antiretrovirals (Supplementary Tables 1 and 2). In November 2014, a PhenoSense Integrase GT assay detected DRMs to NRTI, NNRTI, PI, and the INSTI-associated mutations E138T, G140S, and Q148H. It demonstrated partial sensitivity to DTG (FC 6.71) (Supplementary Table 3) and sensitivity to tenofovir DF and abacavir. Retrospective NGS analysis of a plasma sample from November 2015 (while the individual was on RAL) did not reveal the T97A mutation. In January 2016, a new regimen of DTG/abacavir/lamivudine + tenofovir DF was initiated, all taken once daily (GSS 0) to reduce pill burden. Despite the presence of preexisting INSTI DRMs, the DTG dose was not increased to twice daily as recommended. He took the new regimen intermittently for 11 months. In November 2016, his HIV-1 RNA level was 54443 copies/mL, and NGS sequencing analysis detected no additional INSTI-associated DRMs. The DTG/abacavir/lamivudine + tenofovir DF regimen was continued; he reported adherence from November 2016 to the time of NIH study enrollment in January 2017.
At enrollment at the NIH, his HIV-1 RNA level and CD4 count were 44 186 copies/mL and 51 cells/mm3, respectively (Supplementary Figure 2). NGS detected T97A in addition to the previously detected integrase mutations (Supplementary Table 2). PhenoSense Integrase assay showed DTG resistance, with an increase in FC from 6.71 to 119 (Figure 1; Supplementary Table 3). Thus T97A emerged within 8 weeks of consistent DTG therapy. Several polymorphisms were detected by NGS (Supplementary Table 4) that have not been previously associated with DTG resistance.
DISCUSSION
Although selected by DTG in RAL- or EVG-experienced individuals [4, 6, 7], there are limited data on the impact of the sole emergence of T97A to DTG in the presence of other INSTI-associated resistence mutations in B or non-B subtypes. However, a retrospective analysis reported the probability of having a detectable HIV-1 RNA at the last follow up study time point was higher in INSTI experienced patients with non-B viral subtypes and detectable HIV-1 RNA at the time of DTG initiation [8]. T97A is infrequently present (1%–5%) in INSTI-naïve individuals, though it is reported at a higher rate in HIV-1 subtypes A, J, and P [2, 3]. In site-directed mutagenesis studies, T97A alone has minimal impact on INSTI susceptibility or HIV replication capacity [9–11]. Although amino acid 97 is within the catalytic core domain of the enzyme, it is not positioned near the active site but may interact with portions of INSTI [10, 12]. In an analysis of clinical isolates from INSTI-treated individuals, it has been associated with 5–10-fold reduced susceptibility to RAL and EVG when present with other accessory resistance mutations or minority variant primary resistance mutations [13]. T97A has also been reported in pooled clinical trial data of RAL and EVG from ARV-naïve and INSTI-naïve ARV-experienced individuals [2]. Of 3881 individuals enrolled across 16 clinical trials, the T97A mutation emerged alone in 8 individuals (EVG, n = 6; RAL, n = 2) who were treatment experienced, without primary INSTI-associated DRMs and after at least a year on therapy.
Here we describe emergence of T97A in 2 heavily treatment-experienced individuals within 8–24 weeks of receiving DTG-containing salvage therapy, which was accompanied by a >10-fold increase in IC50 to DTG. In both individuals, NGS did not detect T97A before DTG therapy, but the mutation was readily detectable (NGS, in ≥95% of sample) within weeks of starting or restarting DTG. In participant #1, the mutation emerged at viral rebound while on an optimized regimen containing DTG 50 mg twice daily, darunavir 600 mg/ritonavir 100 mg twice daily, maraviroc 150 mg twice daily, and emtricitabine/tenofovir DF 200/300 mg daily. It should be noted that the participant’s therapeutic DTG and darunavir levels do not preclude intermittent adherence between clinic visits. Participant #2 developed the T97A mutation while on a suboptimal regimen including once-daily DTG/ABC/3TC and tenofovir DF. It is likely the once-daily DTG, rather than recommended twice-daily, may have contributed to the emergence of the T97A mutation, which highlights the importance of ensuring appropriate dosing for highly treatment-experienced patients receiving DTG as part of salvage therapy. We noted several additional polymorphisms emerged during DTG therapy (Supplementary Table 4) which have not been previously reported as INSTI-associated DRMs; however, none were common to both individuals. Additional study will be necessary to determine their significance.
Emergence of the T97A mutation leading to reduced susceptibility to DTG in INSTI-experienced individuals was first reported in the VIKING trials [4, 6]. Individuals developed high-level resistance within 4–24 weeks of DTG, and T97A was usually accompanied by additional mutations. Data from our 2 participants suggest that the single emergence of T97A in HIV with preexisting major integrase DRMs may be rapid and lead to more profound reduction in DTG antiviral activity. In the VIKING phase IIb trial, 2 subjects received twice-daily DTG after DTG functional monotherapy and subsequently developed the T97A mutation. One experienced protocol defined virologic failure (PDVF) at week 8 (DTG FC 42.32), and the second at week 16 (DTG FC 93) [6]. In these cases, genotypic analysis revealed that T97A emerged in addition to other mutations, including E92E/Q, E138E/K, and N155H. It is possible that these VIKING participants developed T97A quickly but it was not detected until after they had acquired additional mutations. In VIKING-4, 2 subjects with baseline integrase substitutions at positions 140 and 148 developed T97A as the only new INSTI-associated DRM. T97A emerged at week 32 (DTG FC 55) and day 28 (DTG FC 32) [4]. The observed increases in DTG FC were higher in our 2 participants (66 and 119, respectively) after the sole emergence of T97A compared with reported increases in DTG FC in the VIKING trials.
We discontinued DTG in our 2 participants to avoid selection of additional integrase mutations, in anticipation of the future development of newer INSTIs that attempt to optimize strand transfer inhibition in the setting of major resistance mutations [14]. Bictegravir and cabotegravir are not likely to be effective treatment options for these individuals. In an analysis of individual derived isolates with E138K, G140S, and Q148H, the FC to bictegravir is reported to be as high as 19-fold (2–19), whereas baseline DTG FC was 63 (3–63) [13]. Several INSTIs under development retain potent in vitro antiviral activity against mutations at positions N155, G140, Q148, and T97A, and may show promise as candidates for drug development [15]. Although the utility of recently approved and investigational ARVs in these and similar cases remains to be determined, particularly in the setting of prior medication nonadherence, it is important for providers to consider last-line and investigational treatment options. Participant #1 declined enfuviritide, and participant #2 had previous treatment failure on enfuviritide. The novel CD4 postattachment inhibitor ibalizumab may be considered for heavily treatment-experienced individuals with an optimized background regimen that includes at least 1 fully active agent. The gp120 attachment inhibitor fostemsavir is also currently under investigation in treatment-experienced individuals with ≤2 active agents and may offer an additional option in the future [16].
These cases highlight the complex challenges in the management of heavily treatment-experienced individuals with HIV, many of whom have problems with medication adherence and are on deep salvage therapy with limited effective treatment options. DTG represents a strong antiretroviral choice, but our experience with these 2 individuals suggests that the emergence of T97A in HIV with preexisting major integrase DRMs may be relatively rapid and lead to profound reduction in antiviral activity of DTG. Close clinical evaluation with more frequent viral RNA monitoring is prudent in these patients.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
We would like to thank the individuals who participated in our study and the clinical staff at the NIAID OP-8 Clinic. The study team would also like to thank Monogram Biosciences, South San Francisco, California, for providing the HIV-1 phenotypic curves from the PhenoSense Integrase and PhenoSense GT plus Integrase assays.
Disclaimer. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does the mention of trade names, commercial products, or organizations imply endorsements by the US Government. The views expressed are those of the authors and do not necessarily reflect the official views or policies of the Uniformed Services University of the Health Sciences, Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc., the US Department of Defense or the Departments of the Army, Navy, or Air Force. Mention of trade names, commercial products, or organizations does not imply endorsement by the US Government.
Financial support. The work was supported in part by the Division of Intramural Research (A1000585) of the National Institute of Allergy and Infectious Disease, National Institutes of Health. The project has also been funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract number HHSN261200800001E. Support for this work (IDCRP-000-33) was provided by the Infectious Disease Clinical Research Program (IDCRP), a Department of Defense (DoD) program executed through Uniformed Services University of the Health Sciences. This project was funded with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, under Inter-Agency Agreement Y1-AI-5072.
HIV Sequences. Resistance sequences from study participants are deposited in GenBank with accession numbers MH785272 and MH785273.
Institutional review board approval. The research drawn on in this case report was determined to be exempt by the Uniformed Services University of the Health Sciences Institutional Review Board.
Copyright statement. Some of the authors are employees of the US Government. This work was prepared as part of their official duties. Title 17 U.S.C. 105 provides that “copyright protection under this title is not available for any work of the United States Government.” Title 17 U.S.C. 101 defines a United States Government work as a work prepared by a military service member or employee of the United States Government as part of that person’s official duties.
Potential conflicts of interest. All authors: no reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. World Health Organization; Update on Antiretroviral Regimens for Treating and Preventing HIV Infection and Update on Early Infant Diagnosis of HIV: Interim Guidance. Geneva: World Health Organization; 2018. [Google Scholar]
- 2. Abram ME, Ram RR, Margot NA, et al. . Lack of impact of pre-existing T97A HIV-1 integrase mutation on integrase strand transfer inhibitor resistance and treatment outcome. PLoS One 2017; 12:e0172206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stanford University. HIVdb (Integrated Genotypic Resistance Interpretation Systems). https://hivdb.stanford.edu/. Accessed 4 May 2017. [Google Scholar]
- 4. Naeger LK, Harrington P, Komatsu T, Deming D. Effect of dolutegravir functional monotherapy on HIV-1 virological response in integrase strand transfer inhibitor resistant patients. Antivir Ther 2016; 21:481–8. [DOI] [PubMed] [Google Scholar]
- 5. Varghese V, Liu TF, Rhee SY, et al. . HIV-1 integrase sequence variability in antiretroviral naïve patients and in triple-class experienced patients subsequently treated with raltegravir. AIDS Res Hum Retroviruses 2010; 26:1323–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Eron JJ, Clotet B, Durant J, et al. ; VIKING Study Group Safety and efficacy of dolutegravir in treatment-experienced subjects with raltegravir-resistant HIV type 1 infection: 24-week results of the VIKING study. J Infect Dis 2013; 207:740–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hardy I, Brenner B, Quashie P, et al. . Evolution of a novel pathway leading to dolutegravir resistance in a patient harbouring N155H and multiclass drug resistance. J Antimicrob Chemother 2015; 70:405–11. [DOI] [PubMed] [Google Scholar]
- 8. Rusconi S, Adorni F, Tau P, et al. ; ARCA (Antiviral Response Cohort Analysis) Dolutegravir (DTG)-containing regimens after receiving raltegravir (RAL) or elvitegravir (EVG): durability and virological response in a large Italian HIV drug resistance network (ARCA). J Clin Virol 2018; 105:112–7. [DOI] [PubMed] [Google Scholar]
- 9. Kulkarni R, Hodder SL, Cao H, et al. . Week 48 resistance analysis of elvitegravir/cobicistat/emtricitabine/tenofovir DF versus atazanavir + ritonavir + emtricitabine/tenofovir DF in HIV-1 infected women (WAVES study GS-US-236-0128). HIV Clin Trials 2017; 18:164–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Abram ME, Hluhanich RM, Goodman DD, et al. . Impact of primary elvitegravir resistance-associated mutations in HIV-1 integrase on drug susceptibility and viral replication fitness. Antimicrob Agents Chemother 2013; 57:2654–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ceccherini-Silberstein F, Van Baelen K, Armenia D, et al. . Secondary integrase resistance mutations found in HIV-1 minority quasispecies in integrase therapy-naive patients have little or no effect on susceptibility to integrase inhibitors. Antimicrob Agents Chemother 2010; 54:3938–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Reigadas S, Masquelier B, Calmels C, et al. . Structure-analysis of the HIV-1 integrase Y143C/R raltegravir resistance mutation in association with the secondary mutation T97A. Antimicrob Agents Chemother 2011; 55:3187–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tsiang M, Jones GS, Goldsmith J, et al. . Antiviral activity of Bictegravir (GS-9883), a novel potent HIV-1 integrase strand transfer inhibitor with an improved resistance profile. Antimicrob Agents Chemother 2016; 60:7086–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhao XZ, Smith SJ, Maskell DP, et al. . Structure-guided optimization of HIV integrase strand transfer inhibitors. J Med Chem 2017; 60:7315–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Smith SJ, Zhao XZ, Burke TR, Hughes SH. HIV-1 integrase inhibitors that are broadly effective against drug resistant mutants. Antimicrob Agents Chemother. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Department of Health and Human Services; Drugs. https://www.aidsinfo.nih.gov/drugs. Accessed 3 May 2018. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.