Abstract
Arsenic is a ubiquitous environmental toxic substance and a Class 1 human carcinogen. Arsenic methylation by the enzyme As(III) S-adenosylmethionine (SAM) methyltransferase (ArsM in microbes or AS3MT in animals) detoxifies As(III) in microbes but transforms it into more toxic and potentially more carcinogenic methylated species in humans. We previously proposed a reaction pathway for ArsM/AS3MT that involves initial 3-coordinate binding of As(III). To date, reported structures have had only 2-coordinately bound trivalent arsenicals. Here we report a crystal structure of CmArsM from Cyanidioschyzon sp.5508 in which As(III) is 3-coordinately bound to three conserved cysteine residues with a molecule of the product S-adenosyl-L-homocysteine bound in the SAM binding site. We propose that this structure represents the first step in the catalytic cycle. In a previously reported SAM-bound structure, a disulfide bond is formed between two conserved cysteine residues. Comparison of these two structures indicates that there is a conformational change in the N-terminal domain of CmArsM that moves a loop to allow formation of the 3-coordinate As(III) binding site. We propose that this conformational change is an initial step in the As(III) SAM methyltransferase catalytic cycle.
Graphical abstract
Arsenic, a Class 1 human carcinogen, is one of the most ubiquitous environmental toxins. It poses a serious threat to human health and, consequently, ranks first on the 2017 Environmental Protection Agency’s (EPA’s) and Agency for Toxic Substances and Disease Registry’s (ATSDR’s) Comprehensive Environmental Response, Compensation, and Liability Act (CERCLA) Substance Priority List of substances that pose the most significant potential threat to human health (http://www.atsdr.cdc.gov/spl/). Geochemical arsenic is biotransformed by microbes, creating an arsenic biogeocycle.1 Bacteria, plants, fungi, and animals methylate arsenite up to three times, producing primarly trivalent methylarsenite [MAs(III)] and dimethylarsenite [DMAs(III)], as well as smaller amounts of volatile trimethylarsine [TMAs(III)].1,2 As(III) SAM methyl- transferases (AsMTs) (EC 2.1.1.137) are termed ArsM in microbes and AS3MT in animals. In microbes, methylation detoxifies inorganic arsenic.3,4 The situation in animals, including humans, is less clear. Knockout mice exhibit hyperplasia when provided with arsenic in drinking water, consistent with methylation providing detoxification.5 In some human populations, methylation appears to be protective against exposure to environmental arsenic, perhaps because of more rapid clearance from the body.6 Paradoxically, production of MAs(III) and DMAs(III) has been proposed to increase reactivity, toxicity, and potentially carcinogenicity.7 This is an example of a Phase II enzyme-catalyzed biotransformation increasing toxicity and/or carcinogenicity.8
We have adopted eukaryotic CmArsM As(III) SAM methyltransferase (GenBank accession number ACN39191) from the acidothermoacidophilic red alga Cyanidioschyzon sp. 5508 as a thermostable model for human AS3MT.3 We previously determined the crystal structures of CmArsM with a final resolution of 1.6 Å with and without bound As(III) or SAM,9 as well as with MAs(III)10 and phenylarsenite [PhAs(III)].11 Nearly every ArsM/AS3MT ortholog has four conserved cysteine residues, which are Cys44, Cys72, Cys174, and Cys224 in CmArsM (Figure S1).12 In previously reported structures, As(III), MAs(III), and PhAs(III) are bound 2- coordinately to conserved cysteine residues Cys174 and Cys224. We proposed a model of the catalytic cycle of As(III) SAM methyltransferases that utilizes a disulfide bond cascade to reduce successive transient pentavalent arsenical inter- mediates.11,13 In this model, the substrates and products are all trivalent, but the transient pentavalent enzyme-bound intermediates are reduced by cysteine residues, forming a series of disulfide bonds that are reduced by thioredoxin, regenerating the cysteine residues in the enzyme. The initial step was proposed to be 3-coordinate binding of As(III) to three of the four conserved cysteine residues,12,13 but this has not been observed experimentally. Here we report the crystal structure of CmArsM with a final resolution of 2.84 Å in which As(III) is bound 3-coordinately to Cys44, Cys174, and Cys224, and SAH is bound in the SAM binding site [As(lIl)/SAH-bound CmArsM], consistent with our proposed reaction mechanism.
MATERIALS AND METHODS
Purification and Crystallization of CmArsM.
CmArsM lacking 31 residues from the N-terminus and 28 residues from the C-terminus with a C-terminal histidine tag (termed simply CmArsM) was expressed and purified as described previously.14 The purified enzyme was stored at −80 °C until it was used. Protein concentrations were estimated from the absorbance at 280 nm.15 Purified CmArsM at 18 mg/mL was mixed with 2 mM SAH and 1 mM As(III) on ice for 20 min, and crystals were obtained by mixing 2.0 μL of the protein solution with an equal volume of the reservoir solution consisting of 1.4 M K2HPO4, 0.4 M NaH2PO4, 0.1 M imidazole (pH 8.0), and 0.2 M NaCl. Crystals obtained at 293 K using Linbro 24-well plates (HR3–110) (Hampton Research, Aliso Viejo, CA) were transferred to a cryoprotectant solution [reservoir solution containing 10% 2-methyl-2,4-pentanediol (MPD)] and flash-cooled in liquid nitrogen for data collection.
X-ray Data Collection and Processing.
High-quality crystals with dimensions of 0.1 mm X 0.1 mm X 0.1 mm in a cryoprotectant solution were flash-cooled in liquid nitrogen for data collection. Data sets were collected at the Southeast Regional Collaborative Access Team (SER-CAT) facility at the Advanced Photon Source (APS), Argonne National Laboratory. Data were obtained from 240 image frames with a 1° rotation angle about φ using a MAR CCD 225 detector under standard cryogenic conditions (100 K) at synchrotron beamline 22-BM with a crystal-detector distance of 270 mm. The data sets were indexed, integrated, and scaled with the HKL2000 suite.16 The crystals diffracted at 2.84 Å Evaluation of crystal packing parameters indicated that the lattice can accommodate two molecules in the asymmetric unit with a solvent content of 56.83% and a Matthews coefficient of 2.83 Å Da−1.17
Structure Solution and Refinement.
Crystals of CmArsM with both bound As(III) and SAH [As(III)/SAH- bound CmArsM] belong to tetragonal space group P4222, with the following unit cell parameters: a = 101.80 Å, b = 101.80 Å, c = 175.01 Å, and α =β = γ = 90.0°. The structure was determined by the molecular replacement method using Phaser.18 The atomic coordinates of the ligand-free crystal structure of CmArsM [Protein Data Bank (PDB) entry 4FS8]9 were used as the starting model. Structural refinement of each data set was performed with REFMAC519 implemented with the CCP4 suite.20 Approximately 5% of the reflections were used for the test set. The refined model required manual adjustment to improve the fit to the experimental electron density using COOT.21 At this stage, restrained refinement was performed, and the final R factor and Rfree converged to 21.5 and 29.1%, respectively, for the CmArsM-bound As(III) and SAH structure. A Ramachandran plot calculated using PROCHECK22 indicated that 80.9% of the residues are in the most favored region, 16.2% of the residues are in the additionally allowed regions, 1.9% of the residues are in the generously allowed region, and 1.0% of the residues is in the disallowed region. Final data collection, processing, and refinement statistics and PDB entry are listed in Table 1. Figures were prepared using PyMOL.23 Ligplot+ was used to identify the hydrogen bonds and nonbonded interactions of CmArsM with As(III) and SAH,24 with the interactions shown using PyMOL.
Table 1.
Data Collection, Indexing, and Refinement Statistics for CmArsM with Bound Arsenic and SAHa
| Data Collection | |
| diffraction source | APS 22-BM |
| wavelength (Å) | 1.0 |
| Detector | MARCCD225 |
| space group | P4222 |
| a, b, c (Å) | 101.80, 101.80, 175.01 |
| α, β, γ (deg) | 90.0, 90.0, 90.0 |
| resolution range (Å) | 100−2.84 (2.94−2.84) |
| no. of unique reflections | 22216 (2058) |
| completeness (%) | 99.4 (95.7) |
| Redundancy | 18.7 (15.2) |
| (I)/σ(I) | 22.0 (2.8) |
| Rsymb (%) | 11.9 (67.4) |
| Rr.i.m. c (%) | 12.3 (69.5) |
| Rp.i.m. d (%) | 2.8 (16.5) |
| Wilson B factor (Å2) | 64.8 |
| Refinemente | |
| Rworkf (%) | 21.5 (29.7) |
| Rfreef (%) | 29.1 (38.5) |
| no. of atoms | 5275 |
| Macromolecules | 5226 |
| Ligands | 2 |
| protein residues | 663 |
| Waters | 47 |
| root-mean-square deviation for bonds (Å) | 0.011 |
| root-mean-square deviation for angles (deg) | 1.67 |
| Ramachandran favored (%) | 80.9 |
| Ramachandran allowed (%) | 16.2 |
| Ramachandran outliers (%) | 1.9 |
| average B factor (Å2) | 63.40 |
| Macromolecules | 63.50 |
| Ligands | 64.80 |
| Waters | 47.80 |
| PDB entry | 6CX6 |
Values in parentheses are for the highest-resolution bin.
Rmerge = ΣhklΣi|Ii,hkl − I̅hkl|/ΣhklΣi|Ii,hkl|, where Ii,hkl is the observed intensity and Ihkl is the average intensity over symmetry equivalent measurements.
Rr.i.m. = Σhkl[N/(N − 1)]1/2Σi|Ii,hkl − I̅hkl|/ΣhklΣi|Ii,hkl|.
Rp.i.m. = Σhkl[1/(N − 1)]1/2Σi|Ii,hkl − I̅hkl|/ΣhklΣi|Ii,hkl|.
Refinement using REFMAC.
Rwork = Σhkl||Fobs| − |Fcal||/Σhkl|Fobs|, where Rfree is calculated for a random chosen 5% of reflections that were not used for structure refinement and Rwork is calculated for the remaining reflections.
RESULTS
Structure of CmArsM with Bound As(III) and SAH.
Here we report the structure of CmArsM with bound As(III) and SAH [As(lIl)/SAH-bound CmArsM, PDB entry 6CX6]. Attempts to cystallize CmArsM with either As(III) and SAM or MAs(III) and SAH were not successful. The structural model of As(lIl)/SAH-bound CmArsM consists of a dimer containing 663 residues with both As(III) and SAH bound in both molecules. In chain A, residues Ser42-Ser371 are visible, and chain B shows residues Leu39-Ser371 (Figure S2). A structural model of chain A shows an overall structure that includes an N-terminal domain with the SAM binding site, a central domain with the As(III) binding site, and a C-terminal domain of unknown function (Figure 1). As(III) is bound to three conserved cysteine residues, Cys44, Cys174, and Cys224, and SAH is bound in the SAM binding site (Figure 1, close-up view).
Figure 1.
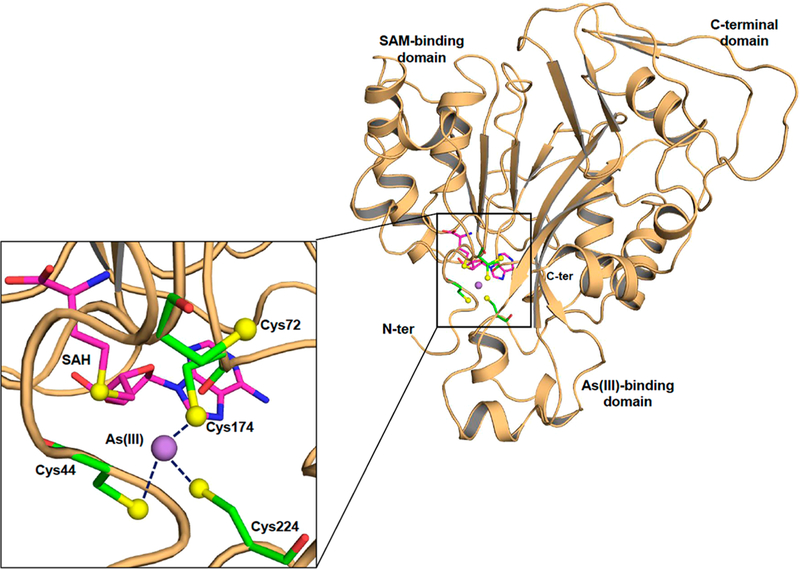
Structure of CmArsM with bound As(III) and SAH. Cartoon diagram (colored salmon) representation of As(III)/SAH-bound CmArsM (PDB entry 6CX6). The overall structure consists of an N-terminal domain, an As(III) binding domain, and a C-terminal domain. The inset shows a close-up of the active site showing the four conserved cysteine residues represented by balls and sticks and colored green (carbon), blue(nitrogen), red (oxygen), and yellow (sulfur). The purple sphere is the arsenic atom, and the SAH in the SAM binding site is represented by balls and sticks and colored magenta (carbon), blue (nitrogen), red (oxygen), and yellow (sulfur). As(III) is bound among conserved residues Cys44, Cys174, and Cys224.
The electron densities of bound As(III), the four conserved cysteine residues, and SAH in chains A and B are well-defined in the 2F0 — Fc electron density map contoured at 1.0σ (Figure S3). The N-terminal domain containing conserved cysteine residue Cys72 has two small helices (residues 52–59 and 63–67) followed by a loop (residues 69–85). In the pyramidal site, the distances between the central arsenic atom and the thiolates of Cys44, Cys174, and Cys224 are 2.4, 2.3, and 2.0 Å, respectively, in chain A (Figure 2A) and2.6, 2.2, and 2.0 Å, respectively, in chain B (Figure 2B). The thiolates of Cys44, Cys174, and Cys224 are at an average distance of 3.6 Å from each other in chain A and 3.3 Å in chain B. The distance between the sulfur atom of SAH and bound As(III) is approximately 4.9 Å (shown for chain A in Figure 3A). Chains A and B can be superimposed with a root-mean-square deviation (RMSD) of 0.24 Å over 322 residues. The structure of As(lIl)/SAH-bound CmArsM was superimposed with the SAM-bound structure (PDB entry 4FR0)9 with an RMSD of 0.72 Å. The SAM and SAH molecules are superimposable in the two structures. We can infer that the distance from the S- methyl group of SAM to the As atom is 2.9 Å when both are bound, indicating that the methyl group is poised for transfer from SAM to As (Figure 3B), which is the precursor to a postulated four-coordinate positively charged As(V) enzyme- bound intermediate.13
Figure 2.
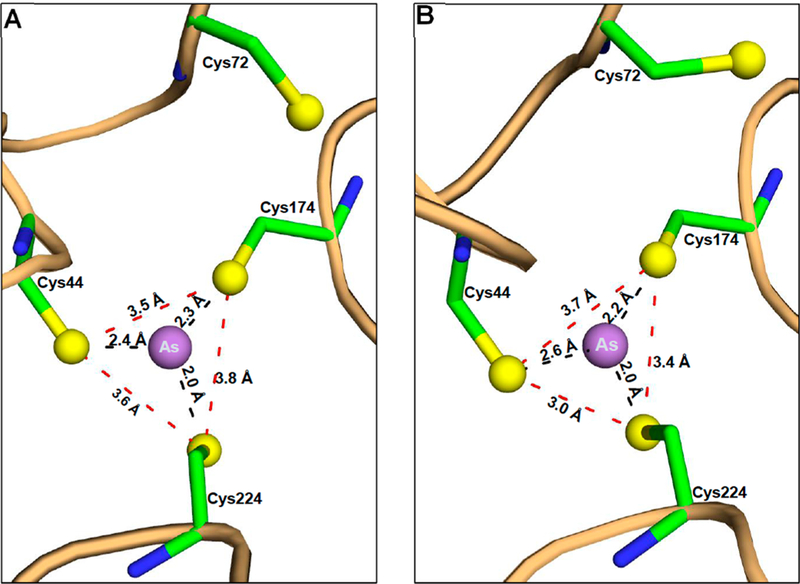
Detailed analysis of the As binding site. The As atom is located among conserved residues Cys44, Cys174, and Cys224 in both (A) chain A and (B) chain B of the As(III)/SAH-bound CmArsM structure. The atoms are represented by balls and sticks and colored as in Figure 1, and the distances between atoms are indicated.
Figure 3.
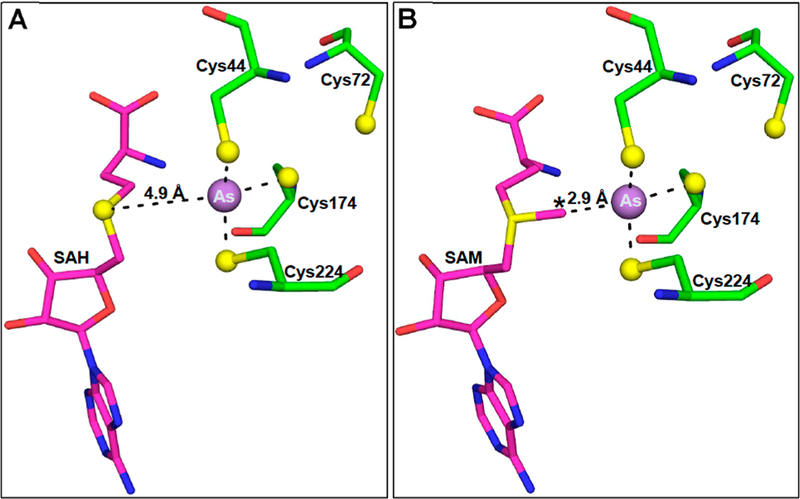
Distances between the As atom and the sulfur atoms of SAH or SAM in the SAM binding site. (A) The As atom in the binding site consisting of Cys44, Cys174, and Cys224 is positioned near SAH in the SAM binding site of chain A. The distance between the sulfur atom of SAH and the As atom is 4.9 Å. (B) SAM was placed in the binding site by superimposition of the SAM-bound structure with chain A of the As(III)/ SAH-bound structure and substitution of SAM for SAH. At a distance of 2.9 Å, the S-methyl group is poised for electron transfer from SAM to As(III).
Although CmArsM is a monomer in solution, the crystal structure has two molecules in the asymmetric unit. The crystallographic contacts between chains A and B appear to facilitate formation of a 3-coordinate complex with As(III). Residues in the C-terminal domain from each chain are involved in hydrogen bonds and nonbonded interactions with residues in the central arsenical binding domain from the opposite molecule (Figure S4). The side chain OHTyr290 from chain A and the side chain NH1Arg209 of chain B form a 2.9 Å hydrogen bond. The reciprocal distance between chain A NH1Arg209 and OHTyr290 is slightly longer at 3.1Å The side chain hydroxyl atoms of Glu284 from chain A and nitrogen atoms of Arg209 from chain B are involved in hydrogen bond formation (OE1Glu284-NH1Arg209, 3.2 Å; OE2Glu284-NH2Arg209, 2.9 Å). The reciprocal interactions in the other interface are at distances of 3.2 Å (OE1Glu284-NH1Arg209) and 2.7Å (OE2Glu284-NH2Arg209). Lys350 from each molecule and Glu367 from the other are at a distance of 3.1 Å. There are hydrogen bond interactions that are present at only the interface of the C-terminal domain of chain A and the central As(III) binding domain of chain B. The hydroxyl group of OE1Glu344 in chain A is hydrogen-bonded with the hydroxyl group (OGSer211) from chain B at a distance of 2.50 Å. The side chain of Tyr230 from chain B interacts with the side chains of chain A Leu348 (3.1 Å) and Glu288 (2.3 and 3.1 Å). These last two interactions are missing in the other interface.
SAH Interactions in the SAM Binding Site.
The product SAH is bound in the β1-α2 glycine rich long loop of the N- terminal domain. Ligplot+ was used to identify and display hydrogen bond and hydrophobic interactions between the protein and SAH in the SAM binding site (Figure S5). The adenine ring is sandwiched between the Ile151 on one side and Met116 on the other. The adenine ring forms a hydrogen bond with Glu152 at a distance of 3.0 Å in chain A (Figure S5A) and 2.94 Å in chain B (Figure S5B), similar to those in bound SAM. The hydroxyl groups of ribose O2 and O3 are hydrogen- bonded to the side chains of Asp115 (OD1-O2, 2.50 Å; OD2-O3, 2.80 Å) and Gln120 (OE1-O3, 2.80 Å) in chain A. These hydrogen bond interactions (OD1-O2, 2.40 Å; OD2- O3, 2.40 Å) and that of Gln120 (OE1-O3, 2.70 Å) are stronger in chain B. Other hydrogen bonds and nonbonded interactions are shown in Figure S5.
Conformational Movements of Conserved Cysteine Residues during Ligand Binding.
Conserved cysteine residues frequently play essential roles in protein function by forming disulfide bonds that stabilize specific conformations.25,26 In the unliganded, As(lll)-bound (PDB entry 4FSD)9 and MAs(lIl)-bound CmArsM structures (PDB entry 5JWN),10 only three of the four conserved cysteines are visible in the refined models. In the CmArsM structure with bound PhAs(lIl) (PDB entry 4KW7),11 which is the aromatic equivalent of MAs(III) in its product-bound conformation, all four conserved cysteines are visible, with Cys44 and Cys72 forming a disulfide bond in an N-terminal loop (residues 50—77) that connects the SAM binding domain to the As(III) binding domain.
Three regions of the As(III)/SAH-bound structure were superimposed with the corresponding regions of unliganded CmArsM, PhAs(III)-bound, and SAM-bound structures to examine movement of the large loop containing conserved cysteines Cys44 and Cys72 (residues 50—77) and the two smaller loops containing Cys174 (residues 169—177) and Cys224 (residues 219—230). Those three structures superimposed on the As(III)/SAH structure with RMSDs of 1.32, 1.42, and 0.44 Å, respectively. The first loop containing Cys72 in the As(III)/SAH-bound structure is 5.3 Å (Cα—Cα) closer to the As(III) binding site relative to unliganded CmArsM (Figure 4A). This relative movement brings the thiols of Cys44, Cys174, and Cys224 close enough together to become the three ligands to the arsenic atom. The Cα—Cα distances of Cys174 and Cys224 of As(III)/SAH-bound CmArsM and ligand-free CmArsM are 1.0 and 1.2 Å, respectively. The second loop containing Cys174 is immobile, and the third loop containing Cys224 is between 310-helices and has limited flexibility. These constraints ensure that the As(III) binding site remains intact. The distances between the sulfur atoms of Cys174 and Cys224 of As(III)/SAH-bound CmArsM are 1.7 and 1.2 Å, respectively, from the sulfur atoms of Cys174 and Cys224 of ligand-free CmArsM. This reflects free rotation of the sulfur atoms of Cys72, Cys174, and Cys224 in unbound CmArsM around the As binding site of As(III)/SAH-bound CmArsM (Figure 4A). A similar analysis comparing As(III)/ SAH-bound CmArsM with PhAs(III)-bound CmArsM shows that the loop containing Cys72 is displaced 5.4 Å from As(III)/SAH-bound CmArsM (Figure 4B), which clearly shows that the loop moves when either SAM or SAH fills the SAM binding site relative to when the site is unoccupied. The take-home message from these results is that the ligand- free, SAM-bound, and SAH-bound enzymes have three different conformations that we predict reflect the conformation of ArsM during three steps of the catalytic cycle. In contrast, there are no significant changes in the position of the loop when the ligand-free structure is compared with any structure that has a bound arsenical, indicating that arsenical binding per se does not alter the conformation of the enzyme. The C—C distances of Cys44, Cys174, and Cys224 are 1.3, 0.8, and 1.3 Å, respectively, comparing the As(lIl)/SAH-bound and PhAs(lIl)-bound structures. The sulfur atoms of Cys174 and Cys224 in PhAs(III)-bound CmArsM are 1.0 and 1.4 Å from the sulfur atoms of Cys174 and Cys224, respectively, in As(III)/SAH-bound CmArsM (Figure 4B). In PhAs(III)- bound CmArsM, the sulfur atom of Cys44 is 4.4 Å from its position, permitting it to form a disulfide bond with Cys72, consistent with our proposed disulfide cascade mechanism.
Figure 4.
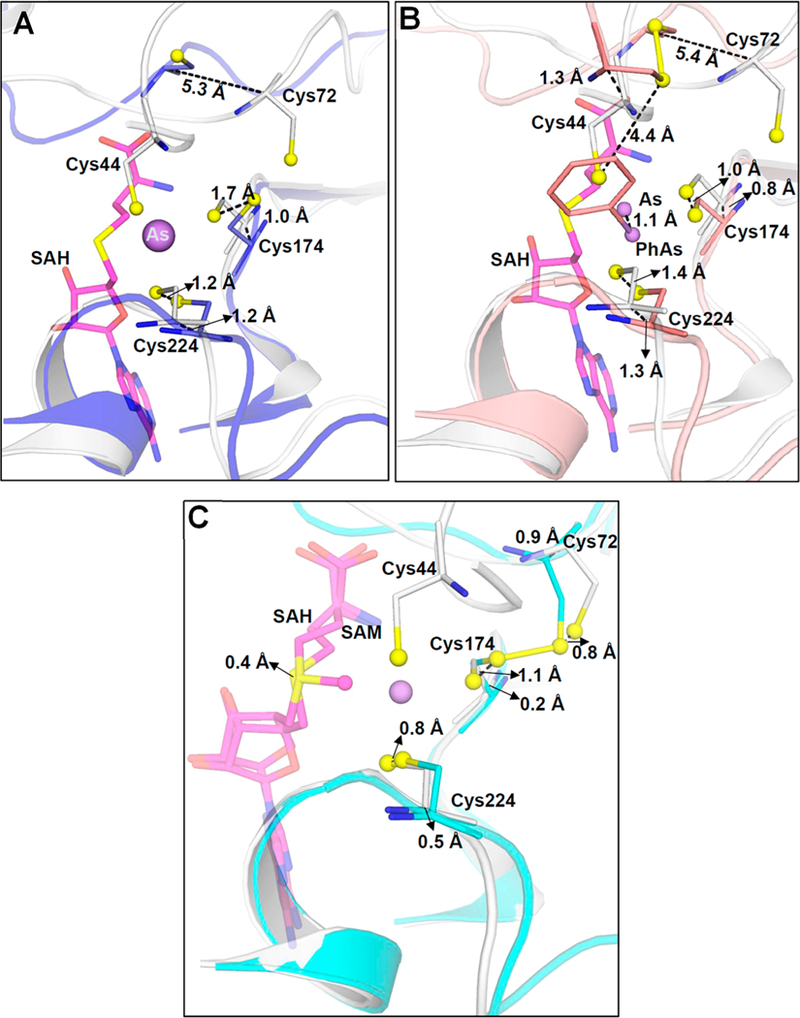
Movement of conserved cysteine thiols during substrate and/or product binding. (A) Superimposition of As(III)/SAH-bound CmArsM (gray) (PDB entry 6CX6) with unliganded CmArsM (blue) (PDB entry 4FS8) shows that the loop containing Cys72 moves 5.3 Å toward the As binding site when As(III) and SAH are bound. (B) Superimposition of As(III)/SAH-bound CmArsM (gray) with PhAs(III)-bound CmArsM (salmon) (PDB entry 4KW7) shows that the loop moves 5.4 Å in the direction of the As binding site relative to the site with bound PhAs(III). (C) Superimposition of As(III)/SAH-bound CmArsM (gray) with SAM-bound CmArsM (cyan) (PDB entry 4FR0) shows that the loop is 0.9 Å closer to the As binding site when SAH and As(III) are bound compared to when SAM is bound. Conserved cysteine residues, SAM, and SAH are represented as balls and sticks. Cα−Cα distances are indicated.
In the 2-coordinately As(III)- or PhAs(III)-bound structures, the large N-terminal loop with a short 310-helix is located near a strand in the β-sheet (Figure 5A,B). In the SAM-bound and As(III)/SAH-bound structures, the 310-helix becomes a long turn, and the strand moves toward the As(III) binding site (Figure 5C,D). In addition, the hydroxyphenyl ring of Tyr70 is oriented toward the SAM site, but the arsenic binding site is open (Figure 5A,B). In the SAM-bound and As(III)/ SAH-bound structures, the Tyr70 side chain is oriented away from the SAM binding site (Figure 5C). In this conformation, the SAM site is open but the arsenic binding site is occluded because of the formation of a disulfide bond between Cys72 and Cys174. In the As(III)/SAH-bound structure, the Tyr70 side chain is flipped away from the SAM binding site, similar to the SAM-bound form (Figure 5D). These results suggest that the orientation of the side chain of Tyr70 gates entry and/or release of substrates and/or products.
Figure 5.
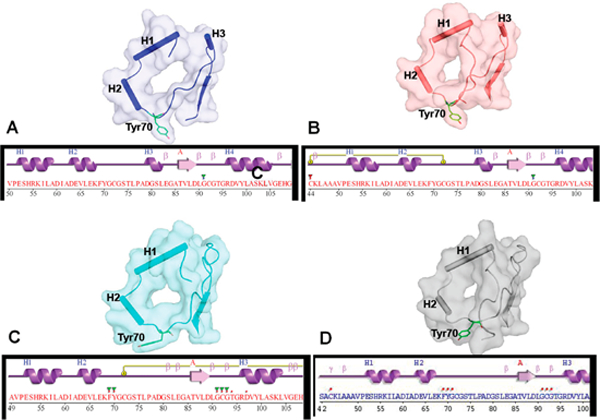
Conformational changes in the ternary structure of CmArsM of the N-terminal domain upon SAM or SAH binding. (A) Unliganded CmArsM (PDB entry 4FS8) and (B) PhAs(III)-bound CmArsM (PDB entry 4KW7) structures have helix H3 between the loop containing Tyr70. (C) SAM-bound CmArsM (PDB entry 4RF0) and (D) As(III)/SAH-bound CmArsM (PDB entry 6CX6) structures lack helix H3. In its place, the residues are part of a large loop that is moved in the direction of the arsenic binding site relative to structures without SAM or SAH. In addition, the position of Tyr70 is oriented away from the SAM binding site, allowing SAM or SAH to access a solvent channel.
Structural Comparison of ArsMs.
Superposition of the entire As(III)/SAH-bound structure with ligand-free (PDB entry 4FS8, 311 Cα residues) and PhAs(III)-bound (PDB entry 4KW7, 311 amino acid residues) structures yields RMSDs of 1.46 and 1.48 A, respectively. Superposition of As(III)/SAH-bound and SAM-bound (PDB entry 4FR0, 288 residues) structures yields an RMSD of 0.72 Å. The overall conformations of the structures are similar. The Rossman fold of the central parallel seven-stranded β-sheets superimposes well in all CmArsM structures. The C-terminal domain residues from position 281 to 372 of unbound, As(III)- bound, PhAs(III)-bound, and SAM-bound CmArsM structures superimpose well with those of the As(III)/SAH-bound CmArsM structure, with RMSDs of 0.39, 0.38, 0.39, and 0.38, respectively. These comparisons clearly show that the C- terminal domain of unknown function is superimposable in all the CmArsM structures. The conformational changes that likely occur during catalysis are in either the N-terminal or As(III) binding domains. Cys72 in the N-terminal loop of the As(III)/SAH-bound structure appears to move toward the As(III) binding site during catalysis by 6.3 Å compared with its position in ligand-free or PhAs(III)-bound CmArsM, and helices α1 and α2 appear to move approximately 3.0 Å toward the As(III) binding site compared with their positions in the other two structures (Figure 6A). Superposition of the N- terminal domain of the As(III)/SAH-bound and SAM-bound structures shows the loop shifts by 0.1 Å between helices α1 and α2 and 0.5 Å closer to the loop containing Cys72 (Figure 6B). The ligand-free and PhAs(III)-bound structures are superimposable, and the N-terminal domains of the As(III)/ SAH-bound and SAM-bound structures also superimpose well (Figure 6C).
Figure 6.
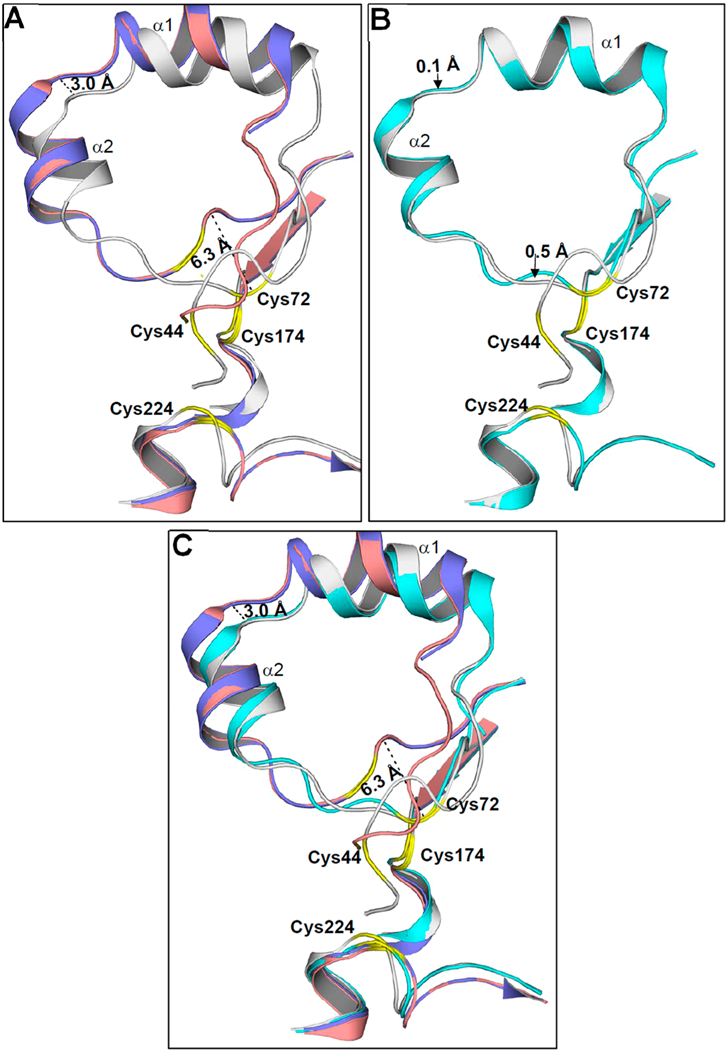
Superimposition of the N-terminal domains of CmArsM structures. (A) The N-terminal domain of As(III)/SAH-bound CmArsM (gray) and unliganded CmArsM (blue) shows a 6.3 Å movement toward the arsenical binding and a 3.0 Å shift in the loop between helices α1 and α2. The N-terminal domain of PhAs(III)-bound CmArsM (salmon) showed a similar movement relative to the N-terminal domain of the unliganded structure. (B) Superimposition of the N-terminal domain of As(III)/SAH-bound CmArsM (gray) and SAM-bound CmArsM (cyan) shows a 0.5 Å shift in the loop between helices α1 and α2. (C) The N-terminal domains of unliganded (blue) and PhAs(III)-bound (salmon) CmArsM structures are superimposable. Similarly, the N-terminal domains of As(III)/SAH-bound CmArsM (gray) and SAM-bound CmArsM (cyan) structures superimpose well with each other. The conserved cysteines are labeled and colored yellow in the loop of all the structures.
Movement of the Arsenic Atom.
In contrast to the 2- coordinate ligation of the arsenic atom in the As(III)-, MAs(III)-, and PhAs(III)-bound structures, As(III) is bound among three conserved cysteine residues in the current structure. The position of the As atom differs by 1.1 Å via comparison of the As(III)/SAH-bound structure with the structure with only As(III) bound (Figure 7A). Comparison with the MAs(III)- and PhAs(III)-bound structures indicates that the As atom differs by 1.6 and 1.1 Å, respectively (Figure 7B,C). PhAs(III) appears to restrict movement of the N- terminal loop, perhaps because of the disulfide bond between Cys44 and Cys72. Thus, in the As/SAH-bound structure, the arsenic atom is displaced 1.1–1.6 Å toward the SAM site, as would be predicted for initiation of S-methyl group transfer.
Figure 7.
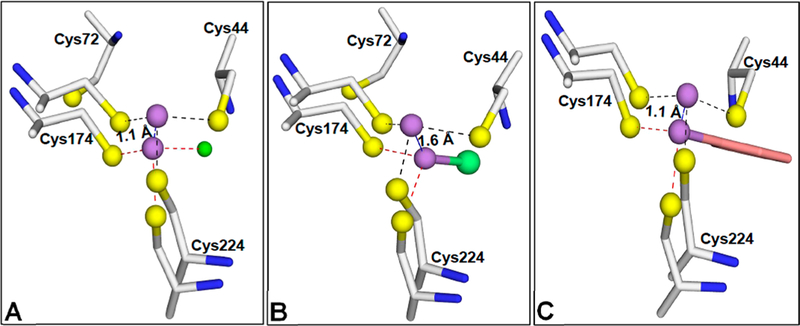
Movement of the As binding site during binding of arsenicals. (A) The position of the arsenic atom was analyzed by superposition of the structure of As(III)/SAH-bound CmArsM (PDB entry 6CX6) with that of As(III)-bound CmArsM (PDB entry 4FSD). The arsenic atom in As(III)/SAH bound CmArsM is liganded to Cys44, Cys174, and Cys224 (dashed lines). In the As(III)-bound structure, the arsenic atom is liganded to only Cys174 and Cys224, with a chlorine atom as the third ligand (green sphere). The arsenic atom is 1.1 Å closer to the SAM binding site in the As(III)/SAH-bound structure than in the As(III)-bound structure. (B) Superimposition of the As(III)/SAH-bound CmArsM structure with the MAs(III)-bound structure shows that the arsenic atom in MAs(III) is 1.6 Å distant from the SAM binding site and the methyl group of MAs(III) is facing the solvent channel of the SAM binding site. (C) Similarly, the arsenic atom in the PhAs(III)-bound structure is displaced 1.1 Å relative to its position in the As(III)/SAH-bound structure.
DISCUSSION
In humans, exposure to inorganic arsenic is a major contributor to arsenic-related diseases,27,28 including bladder,22 lung,29 and skin cancers.30 A major question is whether the increased risk results from exposure to inorganic arsenic or to the products of methylation. The answer should depend in large part on the enzymatic mechanism of this class of enzymes. We have proposed a disulfide bond cascade pathway of methylation by As(III) SAM methyltransferases.12,13 Figure 8 recapitulates the first four steps, from binding of substrates to formation of the first product, enzyme-bound MAs(III). The first step was proposed to be binding of inorganic As(III) to three conserved cysteine residues. However, in the previous structure with only bound As(III), only two of the conserved cysteine residues, Cys174 and Cys224, are arsenic ligands. In the new As(III)/ SAH-bound structure, we identify the third As(III) ligand as Cys44. In the second step, the methyl group of SAM is transferred to trivalent As(III), oxidizing it to a proposed positively charged pentavalent MAs(V) species that would remain bound to Cys44, Cys174, and Cys224. This intermediate would be predicted to be unstable and, as yet, has not been observed. In the third step, the pentavalent arsenic would be rapidly reduced to enzyme-bound MAs(III), with electrons donated by formation of a disulfide bond between Cys44 and Cys72. In the fourth step, the disulfide bond would subsequently be reduced by thioredoxin, allowing the second methylation step to proceed.
Figure 8.
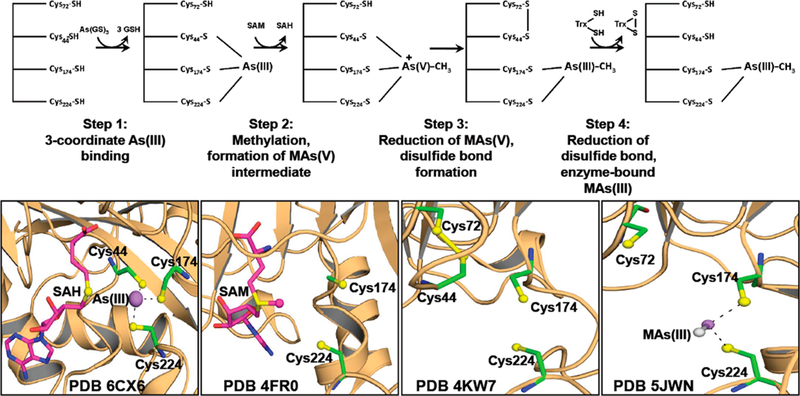
Initial steps in the methylation pathway. The first four steps of the pathway leading to methylation of As(III) to MAs(III). In step 1, As(III) is bound to Cys44, Cys174, and Cys224. In step 2, As(III) is oxidatively methylated to form a hypothetical pentavelent intermediate. In step 3, the transient pentavelent intermediate is reduced to enzyme-bound MAs(III) bound to Cys174 and Cys224 with electrons from Cys72, which forms a disulfide bond with Cys44. In step 4, the disulfide bond is reduced by thioredoxin. The bottom panels show structures related to the first four steps: (1) As(III)/SAH-bound CmArsM (PDB entry 6CX6), showing As(III) bound to Cys44, Cys174, and Cys224; (2) SAM-bound CmArsM (PDB entry 4FR0), showing the orientation of the S-methyl group toward the arsenic binding site; (3) formation of the Cys44−Cys72 disulfide bond (PDB entry 4KW7); and (4) the reduced disulfide bond, regenerating Cys44 and Cys72, in MAs(III)-bound CmArsM (PDB entry 5JWN).
In summary, the demonstration of 3-coordinately bound As(III) fills in a major gap in our understanding of the enzymatic mechanism, showing the conformational change that brings Cys44 into proximity of Cys174 and Cys224. This allows Cys44 to become the third As(III) ligand and brings it close enough to bound SAM for S-methyl group transfer. In addition, we propose that the orientation of the aromatic side chain of Tyr70 controls access to the active site. When the SAM binding site is empty, Tyr70 gates access to the arsenic binding site, but when the SAM binding site is filled, As(III) can enter its binding site to form a 3-coordinate complex, which facilitates S-methyl transfer.
Supplementary Material
ACKNOWLEDGMENTS
This project utilized the Southeast Regional Collaborative Access Team (SERCAT) 22-ID beamline of the Advanced Photon Source, Argonne National Laboratory. Use of the Advanced Photon Source was supported by the U.S. Department of Energy (DOE), Office of Science, Office of Basic Energy Sciences, under Contract W-31–109-Eng-38. The authors thank Dr. Banumathi Sankaran (Berkeley Center for Structural Biology, Lawrence Berkeley National Laboratory) for help in the initial screening of crystals at the Advanced Light Source with support by the Director, Office of Science, Office of Basic Energy Sciences, of the DOE under Contract DE-AC02–05CH11231.
Funding
This work was supported by National Institutes of Health Grant R01 GM55425 and pilot project (Grant 800008538) funding from the Herbert Wertheim College of Medicine to C.P.
ABBREVIATIONS
- SAM
S-adenosylmethionine
- SAH
S-adenosylhomocysteine
- MAs(III)
methylarsenite
- MAs(V)
methylarsenate
- DMAs- (V)
dimethylarsenate
- DMAs(III)
dimethylarsenite
Footnotes
Notes
The authors declare no competing financial interest.
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.bio- chem.8b00457.
REFERENCES
- (1).Zhu YG, Yoshinaga M, Zhao FJ, and Rosen BP (2014) Earth abides arsenic biotransformations. Annu. Rev. Earth Planet. Sci. 42, 443–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2).Thomas DJ, and Rosen BP (2013) Arsenic methyl- transferases In Encyclopedia of Metalloproteins (Kretsinger RH., Uversky VN., and Permyakov EA, Eds.) pp 138–143, Springer, New York. [Google Scholar]
- 3).Qin J, Lehr CR, Yuan C, Le XC, McDermott TR, and Rosen BP (2009) Biotransformation of arsenic by a Yellowstone thermoacidophilic eukaryotic alga. Proc. Natl. Acad. Sci. U. S. A. 106 (13), 5213–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4).Qin J, Rosen BP, Zhang Y, Wang G, Franke S, and Rensing C (2006) Arsenic detoxification and evolution of trimethylarsine gas by a microbial arsenite S-adenosylmethionine methyltransferase. Proc. Natl. Acad. Sci. U. S. A. 103 (7), 2075–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5).Yokohira M, Arnold LL, Pennington KL, Suzuki S, Kakiuchi-Kiyota S, Herbin-Davis K, Thomas DJ, and Cohen SM (2011) Effect of sodium arsenite dose administered in the drinking water on the urinary bladder epithelium of female arsenic (+3 oxidation state) methyltransferase knockout mice. Toxicol. Sci. 121 (2), 257–66. [DOI] [PubMed] [Google Scholar]
- 6).Schlebusch CM, Gattepaille LM, Engstrom K, Vahter M, Jakobsson M, and Broberg K (2015) Human adaptation to arsenic- rich environments. Mol. Biol. Evol 32 (6), 1544–55. [DOI] [PubMed] [Google Scholar]
- 7).Thomas DJ, Li J, Waters SB, Xing W, Adair BM, Drobna Z, Devesa V, and Styblo M (2007) Arsenic (+3 oxidation state) methyltransferase and the methylation of arsenicals. Exp. Biol. Med. (London, U. K.) 232 (1), 3–13. [PMC free article] [PubMed] [Google Scholar]
- 8).Guengerich FP (2000) Metabolism of chemical carcinogens. Carcinogenesis 21 (3), 345–51. [DOI] [PubMed] [Google Scholar]
- 9).Ajees AA, Marapakala K, Packianathan C, Sankaran B, and Rosen BP (2012) Structure of an As(III) S-adenosylmethionine methyltransferase: insights into the mechanism of arsenic biotransformation. Biochemistry 51 (27), 5476–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10).Packianathan C, Li J, Kandavelu P, Sankaran B, and Rosen BP (2018) Reorientation of the methyl group in MAs(III) is the rate-limiting step in the ArsM As(III) S-adenosylmethionine methyltransferase reaction. ACS omega 3 (3), 3104–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11).Marapakala K, Packianathan C, Ajees AA, Dheeman DS, Sankaran B, Kandavelu P, and Rosen BP (2015) A disulfide-bond cascade mechanism for As(III) S-adenosylmethionine methyltransfer- ase. Acta Crystallogr., Sect. D: Biol. Crystallogr. 71 (3), 505–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12).Marapakala K, Qin J, and Rosen BP (2012) Identification of catalytic residues in the As(III) S-adenosylmethionine methyl- transferase. Biochemistry 51 (5), 944–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13).Dheeman DS, Packianathan C, Pillai JK, and Rosen BP (2014) Pathway of human AS3MT arsenic methylation. Chem. Res. Toxicol. 27 (11), 1979–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14).Marapakala K, Ajees AA, Qin J, Sankaran B, and Rosen BP (2010) Crystallization and preliminary X-ray crystallographic analysis of the ArsM arsenic(III) S-adenosylmethionine methyltrans- ferase. Acta Crystallogr, Sect. F: Struct. Biol. Cryst. Commun. 66 (9), 1050–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15).Gill SC, and von Hippel PH (1989) Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182, 319–326. [DOI] [PubMed] [Google Scholar]
- 16).Otwinowski Z, and Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276, 307–26. [DOI] [PubMed] [Google Scholar]
- 17).Matthews BW (1968) Solvent content of protein crystals. J. Mol. Biol. 33 (2), 491–497. [DOI] [PubMed] [Google Scholar]
- 18).McCoy AJ (2007) Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr., Sect. D: Biol. Crystallogr. 63 (1), 32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19).Vagin AA, Steiner RA, Lebedev AA, Potterton L, McNicholas S, Long F, and Murshudov GN (2004) REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr., Sect. D: Biol. Crystallogr. 60 (12), 2184–2195. [DOI] [PubMed] [Google Scholar]
- 20).Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, and Wilson KS (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. Sect. D: Biol. Crystallogr. 67 (4), 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21).Emsley P, and Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr., Sect. D: Biol. Crystallogr. 60 (12), 2126–2132. [DOI] [PubMed] [Google Scholar]
- 22).Chen YC, Su HJ, Guo YL, Hsueh YM, Smith TJ, Ryan LM, Lee MS, and Christiani DC (2003) Arsenic methylation and bladder cancer risk in Taiwan. Cancer, Causes Control Pap. Symp. 14 (4), 303–10. [DOI] [PubMed] [Google Scholar]
- 23).Peters B, Moad C, Youn E, Buffington K, Heiland R, and Mooney S (2006) Identification of similar regions of protein structures using integrated sequence and structure analysis tools. BMC Struct. Biol. 6, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24).Hughes MF (2006) Biomarkers of exposure: a case study with inorganic arsenic. Environ. Health Perspect. 114 (11), 1790–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25).Fomenko DE, Xing W, Adair BM, Thomas DJ, and Gladyshev VN (2007) High-throughput identification of catalytic redox-active cysteine residues. Science 315 (5810), 387–389. [DOI] [PubMed] [Google Scholar]
- 26).Beeby M, O’Connor BD, Ryttersgaard C, Boutz DR, Perry LJ, and Yeates TO (2005) The genomics of disulfide bonding and protein stabilization in thermophiles. PLoS Biol. 3 (9), e309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27).Abernathy CO, Thomas DJ, and Calderon RL (2003).Health effects and risk assessment of arsenic. J. Nutr. 133 (5), 1536S–1538S. [DOI] [PubMed] [Google Scholar]
- 28).Tchounwou PB, Patlolla AK, and Centeno JA (2003) Carcinogenic and systemic health effects associated with arsenic exposure-a critical review. Toxicol. Pathol. 31 (6), 575–588. [DOI] [PubMed] [Google Scholar]
- 29).Putila JJ, and Guo NL (2011) Association of arsenic exposure with lung cancer incidence rates in the United States. PLoS One 6 (10), e25886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30).Rossman TG, Uddin AN, Burns FJ, and Bosland MC (2001) Arsenite is a cocarcinogen with solar ultraviolet radiation for mouse skin: an animal model for arsenic carcinogenesis. Toxicol. Appl. Pharmacol.176(1),64–71. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


