Abstract
Helicobacter pylori is a Gram-negative, microaerophilic bacterium colonising the gastric mucosa. Normally, this bacterium has a spiral shape, which is crucial for proper colonisation of the stomach and cork-screwing penetration of dense mucin covering this organ. However, H. pylori may also form curved/straight rods, filamentous forms and coccoid forms. This morphological variability affects nutrient transport and respiration processes, as well as motility, the ability to form aggregates/biofilms, and resistance to adverse environmental factors. For this reason, a more accurate understanding of the molecular determinants that control the morphology of H. pylori seems to be crucial in increasing the effectiveness of antibacterial therapies directed against this microorganism. This article focuses on the molecular factors responsible for peptidoglycan and cytoskeleton rearrangements affecting H. pylori morphology and survivability. In addition, the existence of proteins associated with modifications of H. pylori morphology as potential targets in therapies reducing the virulence of this bacterium has been suggested.
Keywords: coccoid form, filamentous form, murein, shape
Introduction
Helicobacter pylori is a Gram-negative, microaerophilic bacterium that inhabits the stomach mucosa [1–3]. Colonisation with this pathogen is often associated with the development of numerous digestive system disorders, i.e. gastritis, peptic ulcer disease (10–15%), gastric cancer (1–3%), and mucosa associated lymphoid tissue (MALT) lymphoma (< 0.1%) [4]. The type and severity of diseases depend on many factors, among them: the status of the host’s immune system, the pathogenicity of H. pylori strains, and the presence of environmental factors (diet, stress, hygiene level, or the presence of co-infections) [1]. Within these, the most attention is paid to the pathogenicity of H. pylori and the variety of virulence factors produced by these bacteria. The key determinants of virulence are cytotoxin-associated gene A (CagA) and vacuolating cytotoxin A (VacA), as well as the presence of numerous adhesins that facilitate adhesion to the gastric mucosa. Many researchers also point out that the spiral shape is crucial for proper colonisation of the stomach, and it determines the cork-screwing penetration of dense mucin [5, 6]. Nevertheless, the spiral shape is not the only morphological form created by H. pylori. The large heterogeneity of H. pylori morphological forms includes also the presence of curved/straight rods, filamentous forms, and coccoid forms [7–10]. Such a multitude of shapes suggests that morphological variability may be an important factor responsible for rapid response to changing external conditions and thus is a feature that allows survival in various environmental niches and facilitates modulation of pathogenicity. For this reason, understanding the molecular mechanisms that control these reactions, including the peptidoglycan and cytoskeleton rearrangements of H. pylori, seems to be justified.
Morphological forms of Helicobacter pylori
The shape of microbial cells is the product of billions of years of evolution [11]. Small dimensions are the resultant of a high surface-to-volume ratio, which translates into optimal transport of nutrients, effective respiration process, and structural integrity maintenance. The shape also affects many other physiological processes, including motility and dispersion, formation of microbial aggregates/biofilms, resistance to environmental stressors, and interactions with other organisms [12]. Over the last few years an increasing amount of evidence has highlighted the key role of morphogenesis in bacteria-bacteria and bacteria-host interactions.
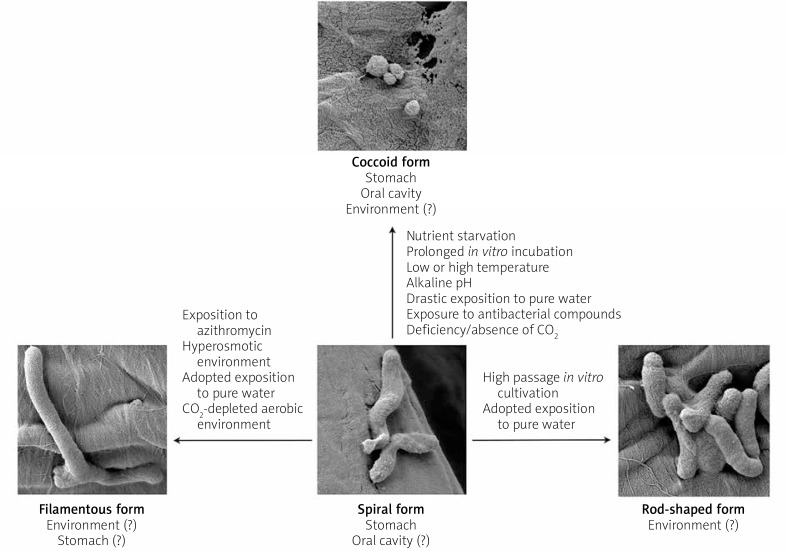
Morphological plasticity as a mechanism enabling the adaptation of microbes to changing environmental conditions may explain the high heterogeneity of the H. pylori population [13] (Figure 1). Classical classification of H. pylori morphological forms includes the division into: live, culturable spiral forms – related to the process of host colonisation, and live, non-culturable coccoid forms – associated with the viable but non-culturable (VBNC) process and the survival of this bacterium under unfavourable conditions [14]. In addition, the existence of rod-shaped and filamentous forms of H. pylori is also noted [7, 8, 15, 16].
Figure 1.
Factors stimulating morphological transformations of Helicobacter pylori. There are many factors responsible for the transition of H. pylori from spiral into spherical [19–38, 42, 43], rod-shaped [44, 45], and filamentous [15, 29, 45, 48–50] forms
Bacteria exposed to adverse conditions often become VBNC, and they are characterised by a lack of growth on standard media, despite maintaining metabolic activity (often lower than in physiologically active cells) [17, 18]. To date, scientists have demonstrated the ability to transit into VBNC forms in 85 species of bacteria, including Gram-negative: Campylobacter, Citrobacter, Escherichia, Helicobacter, Pseudomonas, Salmonella, Shigella, Vibrio and Yersinia, as well as Gram-positive: Bacillus, Clostridium, Enterococcus, Listeria, Mycobacterium and Staphylococcus. Cells in the VBNC state are not dead because intact cellular envelopes maintaining non-degraded genetic material are observed. These cells, despite being physiologically active, have a reduced level of nutrient absorption, slowed metabolism, and deceased amounts of proteins and cytoplasm. In VBNC cells a dwarfing process often occurs, which results in the formation of spherical forms with reduced dimensions. Such a strategy lowers the energy requirements of microorganisms. Moreover, compared to physiologically active cells, microbes in the VBNC state are characterised by higher resistance to physical and chemical factors, as well as tolerance against certain groups of antibiotics, which is conditioned by a decrease in the metabolic activity of microorganisms.
Coccoid forms
The transformation of H. pylori from culturable spiral forms into non-culturable coccoids occurs during exposition to adverse conditions, including: nutrient starvation [19, 20], prolonged in vitro incubation [21, 22], presence of low or high temperature [21, 23, 24], incubation at alkaline pH [25], drastic change of culture environment from a nutrient-rich medium to pure water [26, 27], incubation in the deficiency/absence of CO2 [28, 29], or exposure to antimicrobial compounds, e.g. antibiotics [30, 31], proton pump inhibitors (PPIs) [22, 32], and compounds secreted by bacteria of the genus Lactobacillus [33, 34] and Streptococcus [35] (Figure 1). Although coccoid forms are often observed during laboratory work with H. pylori, the presence of these forms has been reported in many in vivo cases [36–38]. Despite the reduced pathogenicity of spherical forms compared to spiral forms, these are still capable of producing urease, adhesion to epithelial cells, and inducing gastritis [26, 39]. In mice studies it was shown that coccoids may reverse in vivo into spiral forms and cause full-blown disease [40]. Most often, however, in the stomach environment spherical forms coexist with spirals [36, 37]. A report indicating the colonisation of stomach mucosa only by spherical forms also exists [38]. It was found that the number of coccoid H. pylori forms is higher in people with a gastric adenocarcinoma than in people with a peptic ulcer disease, suggesting the potential involvement of these forms in the initiation/progression of carcinogenesis [37]. This is in accordance with the observations of Loke et al., who showed in vitro a higher level of proteins associated with carcinogenesis promotion in spherical H. pylori forms [41]. In addition to the stomach, the oral cavity is a potential reservoir of this bacterium, in which H. pylori is predominantly in spherical form [42]. Reports on the possible coexistence of spherical forms with single spiral forms also exist [43]. It is suggested that the morphology of oral H. pylori corresponds to the conditions prevailing in this environment and the presence of physiological flora colonising this area [42].
Rod forms
An alternative phenotype for spiral and spherical is rod-shaped morphology [44, 45] (Figure 1). Little is known about the environmental conditions that stimulate the transition to rod forms, whereas it is observed that in freshly isolated strains of H. pylori about 10–15% of all cells become rod-like forms [44]. During high passage in vitro cultivation, the number of rods increases, with an inverse correlation observed in the amount of spiral forms. A drastic change in the culture environment from a nutrient-rich medium to pure water/saline solution contributes to the transformation to spherical H. pylori forms [26, 27]. A contrary observation was made by Fernandes et al., who, by the process of extended adaptation of H. pylori to incubation in pure water, increased the period of the isolation of culturable H. pylori [45]. During microscopic examination it was noted that the adapted forms were dominated by rod-shaped cells and the presence of single cells with a very elongated shape (filamentous cells). These results are in line with the hypothesis of the potential role of water as the source of H. pylori transmission [46, 47].
Filamentous forms
Another morphological form presented by H. pylori is the filamentous, elongated phenotype [15, 16, 48–50] (Figure 1). Normally, filamentation contributes to the increase of adhesion to the mucosal surfaces, promoting slow, ligand-dependent uptake of filamentous microbes into the interior of eukaryotic cells (invasion), and by multiplying the cell length this process is also responsible for phagocytosis avoidance [13]. It is suggested that filamentation may play a protective role against the action of genotoxic antibiotics. Multinucleated filamentous forms increase the chance of repairing damaged genetic material, and in the process of mutation accumulation they promote the intensity of recombination and selection of favourable traits. In the case of H. pylori, the filamentation process is triggered, among others, by exposure to azithromycin [15] or incubation in a hyperosmotic environment [48, 49]. The presence of individual filament-shaped cells was also noticed when incubating in a CO2-free aerobic environment [29] and in adapted H. pylori strains incubated in pure water [45]. There was also a case of isolation of filamentous H. pylori from gastric biopsies [50]. The function of elongated H. pylori cells has not been determined, whereas it seems that, unlike in other bacteria, filamentation does not participate in host colonisation. It was demonstrated that the ability to migrate and colonise C57/BL6J mice was reduced in filamentous H. pylori [16]. However, they are assigned a role in antibiotic resistance because the presence of such forms leads to a drastic increase in the MBC value of amoxicillin, i.e. 0.06 vs. 32 μg/ml. Thus, the filamentation may be considered as a protective mechanism against the antibacterial action of this antibiotic against H. pylori. Based on these few reports (the induction of filamentation by the aerobic environment [29] and hyperosmotic conditions [48, 49]), it seems that the filamentous forms of H. pylori may be more important in the survival/ transmission of these microbes in the environment outside the human body.
Cytoskeletal proteins
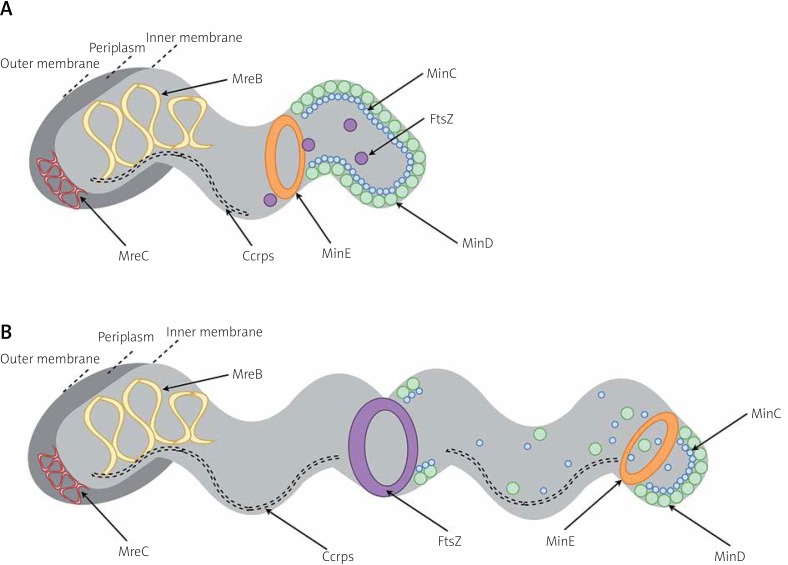
For a long time, it was thought that bacterial cells, unlike eukaryotic cells, did not have adequate compartmentation because of their small size and lack of organelles [51]. Studies have shown, however, that microorganisms also have well-organised cellular interiors [52]. The bacterial cytoskeleton participates in many important physiological functions, i.e. cell division, morphogenesis, and DNA segregation. It seems that in bacteria the cytoskeleton is most probably used as a system conditioning the proper spatial organisation of peptidoglycan synthesis-associated proteins, rather than as a mechanical scaffold maintaining the shape of cells. In eukaryotic cells three main cytoskeletal systems are distinguished: microfilaments, microtubules, and intermediate filaments (IF) [53]. These systems have their counterparts in bacterial cells, and these are: filamenting temperature sensitive mutant Z (FtsZ, responsible for septal peptidoglycan synthesis), mecillinam resistance-like proteins (Mre-like proteins, responsible for sidewall peptidoglycan synthesis), and intermediate filaments-like proteins (IF-like proteins, associated with an auxiliary function), respectively [52, 54] (Figure 2).
Figure 2.
Cytoskeleton of Helicobacter pylori. This picture shows non-dividing (A) and dividing (B) H. pylori. FtsZ has an ability to form Z-ring, which is associated with cell division and peptidoglycan remodelling. Localisation of FtsZ ring is conditioned by Min proteins. MinCD are most abundant at cell poles. Because these proteins have an inhibitory effect on FtsZ, such localisation prevents the formation of FtsZ ring at polar cell regions and keeps FtsZ in the form of free-diffusing monomers. MinE subunits form a ring-like structure and localize near the middle of the cell. The MinE ring, via an inhibitory effect on the MinC and MinD proteins, facilitates the construction of FtsZ ring at the mid-cell site. During the later stages of cell division, MinE gradually displaces MinCD from the cell membrane, leading to a transfer of MinC and MinD subunits to the opposite cell pole. This action is accompanied by a new polar MinCD localisation, inhibition of the FtsZ-dependent division, and separation of a daughter cell [57, 59]. The complex of Mre-like proteins is called elongasome, and in H. pylori it consists of two components: MreB and MreC. They are involved in the chromosome segregation and cellular length modulation [44, 55]. Coiled coil rich proteins (Ccrps) form extended filament structures. In H. pylori Ccrp58, Ccrp59, Ccrp1142, and Ccrp1143 are present, all of which are involved in maintaining the spiral shape of this bacterium [44, 60]. Localisation of Mre-like proteins and Ccrps is symmetrical in the cell
Mre-like proteins
The rod shape of bacteria is determined by the presence of mreBCD genes, of which mreB plays the most important role, including cellular polarity maintenance, segregation of chromosomes, and participation in motility [53, 54]. The complex of Mre-like proteins is called elongasome [12]. In most bacteria mreBCD mutations contribute to the loss of rod shape, transformation into spherical cells, and/or death [54]. Only two genes, mreB and mreC, are present in H. pylori [44, 55] (Figure 2). In H. pylori MreB does not affect the shape of cells, but it is involved in the chromosomes segregation, cellular length modulation, and pathogenicity of these bacteria [44]. In ΔmreB mutants the presence of significantly longer cells (even three times longer than wild-type strains), non-segregated chromosomes and decreased urease activity was demonstrated, whereas the deletion of this gene was not lethal. In another study using the two-hybrid bacterial system, interaction between MreB and various H. pylori virulence factors was demonstrated, i.e. VacA (vacuolating cytotoxin; eukaryotic cells degradation), UreB (urease subunit; alkalisation of the acidic environment), HydB (hydrogenase subunit; maintenance of redox homeostasis), HylB (haemolysin secretion protein precursor; eukaryotic cells degradation), and AddA (recB-like nuclease-helicase; DNA repair and recombination) [56]. What is more, the interaction of MreB with proteins involved in the H. pylori morphogenesis process was not observed. In the mreC deletion mutants the transformation of these bacteria from spiral into coccoid forms was noticed, whereas in mreC-overexpression mutants the appearance of filamentous phenotype was observed [55]. Therefore, the participation of MreC in the course of the proper elongation process is indicated.
FtsZ protein
FtsZ is a tubulin homologue [12, 51, 54, 57]. This protein localising at the site of septation forms a Z-ring and polymerises in linear protofilaments in a GTP-dependent manner recruiting other proteins (e.g. FtsA, ZipA, and ZapA) associated with cell division and peptidoglycan remodelling, referred to as the divisome. In H. pylori most of these proteins are absent, while Min proteins are responsible for the coordination of FtsZ functioning [57] (Figure 2). In most bacteria the Min protein system consists of MinC, MinD, and MinE [58]. MinCD proteins protect against FtsZ polymerisation and cell division. For this reason, in ΔminCD mutants, hyperactivity of divisome proteins, intensification of septation, and formation of very short cells (mini cells) are observed. In mutants with overexpression of minCD, however, disruption of Z-ring formation and induction of filamentation are noticed. The opposite phenotype is presented by cells with altered minE expression, a minCD inhibitor, i.e. the formation of filamentous cells in ΔminE mutants and creation of mini cells in minE over-expressing microbes. In H. pylori a different mechanism is observed because the deletion of any of the genes (minC, minD, and/ or minE) leads to filamentous cell formation [57]. It is concluded that MinCD in H. pylori are responsible for the process of proper cell division, affecting FtsZ assembly and/ or Z-ring conformation [57, 59]. MinE, in turn, may play a role in morphological transformation into a coccoidal form, because ΔminE mutants presented an extremely low number of spherical forms [57].
Bacterial IF-like proteins
Bacterial IF-like proteins can also play a role in modulating the morphology of microbial cells, potentially by affecting peptidoglycan biosynthesis [12]. This group includes bactofilins, cytoskeletal-like scaffolding proteins, and coiled coil rich proteins (Ccrps). The most extensively characterised group of IF-like proteins in H. pylori is the last group [44, 60, 61] (Figure 2). Ccrps are proteins that form extended filament structures [44, 60]. In H. pylori, four genes encoding Ccrps were detected: ccrp58, ccrp59, ccrp1142, and ccrp1143, all of which are involved in maintaining the spiral shape of this bacterium. The deletion of these genes was associated with the formation of straight rod-shaped cells, with the lowest intensity in the case of Δccrp1142 mutants (~40%) and the highest in Δccrp59 strains (85–100%). In addition, deletion mutants also had noticeable defects in motility. It is speculated that Ccrps, due to their high heterogeneity of sequences, have different degrees of curvature, which then may result in the large diversity of H. pylori morphological forms [60]. In the mutants Δccrp58/59 (but not Δccrp1142/1143), the reduced effectiveness of the cag-T4SS system, i.e. the system related to the promotion of inflammatory reactions and destruction of eukaryotic cells, has been demonstrated [61]. This is another example, next to Mre-like proteins, of a direct/indirect link between the functioning of cytoskeletal proteins and H. pylori virulence.
Because many cytoskeletal proteins are not encoded by H. pylori or do not participate in morphogenesis processes, it seems that there are other, more important proteins involved in the morphological processes of this bacterium.
Peptidoglycan-modifying enzymes of Helicobacter pylori
Most bacteria have a cell wall that, due to its covalently closed, net-like structure, maintains a specific shape and thus also imposes it on cells [12, 53]. Peptidoglycan (sometimes referred to as murein, from Latin murus [wall]) consists of alternating subunits of N-acetylglucosamine and N-acetylmuramic acid connected by β1,4-glycosidic linkages [53, 62]. The presence of rigid sugar chains, cross-linked additionally with short-chain peptide bridges, affects the strength and stiffness of this polymer while maintaining flexibility [53, 63]. Peptidoglycan was once considered a static structure, but it is now known that this highly dynamic macromolecule is involved in many important physiological functions of bacteria [64].
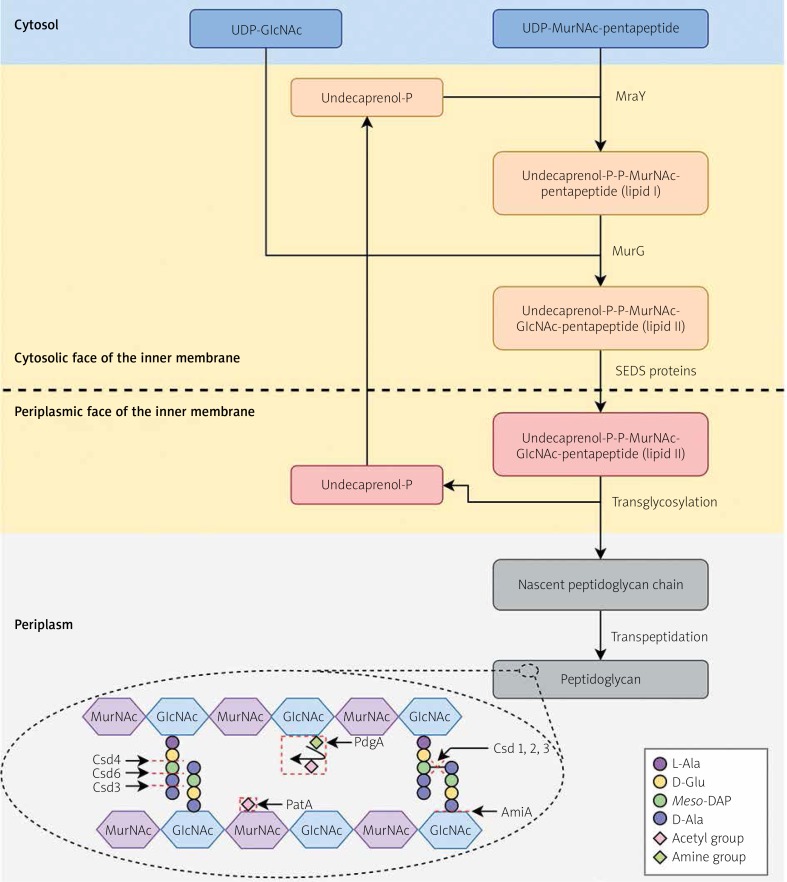
The peptidoglycan biosynthesis process involves several steps [12, 63, 65] (Figure 3). Within the cytoplasm, peptidoglycan precursors are synthesised, i.e. UDP-N-acetylglucosamine (UDP-GlcNAc) and UDP-N-acetylmuramylpentapeptide (UDP-N-MurNAc-pentapeptide). In the action of MraY, membrane-anchored lipid I (MurNAc-(pentapeptide)-pyrophosphoryl-undecaprenol) is synthesised by linking UDP-N-MurNAc-pentapeptide to the lipid carrier, undecaprenyl phosphate. Then, UDP-GlcNAc is attached, which together with lipid I forms lipid II (GlcNAc-β-[1,4]-MurNAc-[pentapeptide]-pyrophosphoryl-undecaprenol). This process is carried out by MurG. The formation of a complex with undecaprenyl phosphate, a molecule with a hydrophobic character, allows the translocation of hydrophilic precursors from the aqueous environment of the cytoplasm by a hydrophobic inner membrane. The translocation of the complex occurs through the action of shape, elongation, division, and sporulation (SEDS) proteins, among which RodA and FtsW are distinguished. Such a molecule in the periplasmic environment is then biochemically processed by glycosyltransferases (linear polymerisation) and transpeptidases (peptide crosslinking). Undecaprenyl pyrophosphate undergoes dephosphorylation, which affects its availability and the possibility of carrying out subsequent translocations of peptidoglycan synthesis precursors.
Figure 3.
Peptidoglycan biosynthesis and modifications in Helicobacter pylori. Within the cytoplasm, peptidoglycan precursors are synthesised, i.e. UDP-GlcNAc and UDP-N-MurNAc-pentapeptide. In the action of MraY, membrane-anchored lipid I is synthesised by linking UDP-N-MurNAc-pentapeptide to the lipid carrier, undecaprenyl phosphate. Then, UDP-GlcNAc is attached, which together with lipid I forms lipid II. This process is carried out by MurG. The formation of a complex with undecaprenyl phosphate allows the translocation of hydrophilic precursors by a hydrophobic inner membrane. The translocation of the complex occurs through the action of SEDS (shape, elongation, division, and sporulation) proteins. Such a molecule in the periplasmic environment is then biochemically processed by glycosyltransferases (linear polymerisation) and transpeptidases (peptide crosslinking). Undecaprenyl pyrophosphate undergoes dephosphorylation, which affects its availability and the possibility of carrying out subsequent translocations of peptidoglycan synthesis precursors. The Csd1, Csd2, and Csd3 proteins have D,D-endopeptidase activity, cleaving a 4-3 peptide bond (or DD-cross link) linking a muropeptide dimer. Furthermore, Csd3 also functions as a D,D-carboxypeptidase whose targets are pentapeptide monomers. The resulting GM-tetrapeptides are processed by Csd6, forming GM-tripeptides, subsequently trimmed to GM-dipeptides by the Csd4 activity. The AmiA, by an activity of N-acetylmuramoyl-L-alanyl amidase, cleavages the link between N-acetylmuramoyl residue and L-alanine. The PgdA and PatA lead to the N-deacetylation of N-acetylglucosamine and O-acetylation of N-acetylmuramic acid, respectively. Based on [63, 83] with minor modifications
L-Ala – L-alanine, D-Ala – D-alanine, meso-DAP – meso-diaminopimelic acid, D-Glu – D-glutamic acid, GlcNAc – N-acetylglucosamine, MurNAc – N-acetylmuramic acid.
Many researchers focus their attention on understanding the mechanisms of cell wall modifications resulting in the morphological variability of microorganisms [13]. Variations in cellular shape can be obtained by changes in the thickness of peptidoglycan as well as by modifications in the chemical composition or cross-linking of this polymer. In H. pylori there are several proteins determining cell shape; these are Csd1-6 (cell shape determinant 1-6) [7–9, 66–70], CcmA (curved cell morphology A) [7, 67], and AmiA (amidase A) [15, 16] (Table I, Figure 3). The Csd1, Csd2, and Csd3 proteins have D,D-endopeptidase activity, cleaving a 4-3 peptide bond (or DD-cross link) linking a muropeptide dimer. Furthermore, Csd3 also functions as a D,D-carboxypeptidase whose targets are pentapeptide monomers. The resulting GM-tetrapeptides (GlcNAc-MurNAc-tetrapeptide) are processed by Csd6, forming GM-tripeptides, subsequently trimmed to GM-dipeptides by the Csd4 activity. The Csd5 and CcmA proteins are potentially regulatory factors affecting the function of other proteins involved in the peptidoglycan turnover. The AmiA, by an activity of N-acetylmuramoyl-L-alanyl amidase and the cleavage of the link between a N-acetylmuramoyl residue and L-alanine, also plays an important role in the maintenance of cell morphology by supervising the correct separation of daughter cells from mother cells.
Table I.
Proteins involved in modifications of Helicobacter pylori peptidoglycan
| Protein | Enzymatic activity | Action | Function | Phenotype of cells with a defective protein(s) | Literature |
|---|---|---|---|---|---|
| Csd1 | D,D-endopeptidase | Cleaves muropeptide dimers to monomers | Cell helicity maintenance | Loss of helicity and formation of curved cells | [7, 67] |
| Csd2 | |||||
| Csd3 | D,D-endopeptidase | Formation of phenotypically diverse cells, i.e. C-shaped forms or forms with a straight or slightly curved morphology | [7, 9] | ||
| D,D-carboxypeptidase | Trims GM-pentapeptides into GM-tetrapeptides | ||||
| Csd6 | L,D-carboxypeptidase | Trims GM-tetrapeptides into GM-tripeptides | Cell curvature maintenance | Loss of curvature and formation of cells with rod-shaped morphology | [68, 70] |
| Csd4 | D,L-carboxypeptidase | Trims GM-tripeptides into GM-dipeptides | [8, 66] | ||
| Csd5 | Unknown | Regulation of the activity of peptidoglycan metabolism proteins | Influence on the asymmetric location and functioning of proteins involved in the peptidoglycan turnover | Loss of cellular helicity (similar as in Δcsd1, Δcsd2, and Δcsd1csd2 mutants) | [8] |
| CcmA | Loss of cellular curvature (similar as in Δcsd4, Δcsd6, and Δcsd4csd6 mutants) | [7, 67] | |||
| AmiA | N-acetylmuramoyl-L-alanyl amidase | Trims the link between N-acetylmuramoyl residue and L-alanine | Release of daughter cells after cell division (septum-cleaving enzyme) | Formation of filamentous cells | [15, 16] |
| PgdA | Peptidoglycan N-deacetylase | N-deacetylation of N-acetylglucosamine | Protection against lysozyme and oxidative stress, and mitigation of immune system activity | Increased sensitivity of cells to the host’s anti-bacterial proteins (lysozyme and lactoferrin) activity | [80, 81] |
| PatA | Peptidoglycan O-acetyltransferase | O-acetylation of N-acetylmuramic acid |
The Csd1-3 endopeptidases
The Csd1-3 proteins, through D,D-endopeptidase activity, contribute to the cleavage of tetra-pentapeptide crosslinks, local relaxation of the coherence of rigid sugar strands, and the relaxing-dependent process of cell helicity obtaining [7, 12, 63] (Table I). These proteins by cleaving muropeptide dimers to monomers, provide substrates for the Csd6 activity [68, 70].
It has been shown that Csd1 and Csd2 form heterodimers (1 : 1) [67]. This is consistent with the in vitro observations showing that a defect in the production of one of these proteins, or both simultaneously, contributes to similar consequences [7]. The lack of these proteins affects the loss of helicity and the formation of curved cells with slightly increased width. The most noticeable change is the increase in the amount of tetra-pentapeptide dimers, while reducing the amount of tetrapeptide monomers. In the case of csd3 deletion, the loss of helicity was also observed, resulting in the formation of C-shaped cells and numerous forms with a straight or slightly curved morphology. These mutants have been shown to have reduced amounts of tetra-tetrapeptides and tetra-tripeptide dimers.
In semi-solid agar tests, after four-day H. pylori incubation, it was demonstrated that the deletion mutants of these genes had reduced motility (the Δcsd1 and Δcsd3 mutants had a halo zone lower by 11% and 25%, respectively) [8]. Defects in motility probably have a negative impact on the host colonisation because these mutants have been found to possess significantly reduced effectiveness of this process [7, 69]. In a comparative experiment between Δcsd3 mutants and H. pylori wild strains, a significant reduction in mice colonisation capacity was found in mutants [69]. One week post-infection the number of deletion mutants was four log lower than in wild strains. Additionally, in the csd3 mutants (but not csd1 and csd2), changes in morphological transformation ability were observed, i.e. in the Δcsd3 strains the transition into coccoid forms was slightly delayed, whereas in the csd3-overexpressing strains the transformation into spherical forms was intensified [7, 69]. This observation is consistent with the results of Hung et al., who during coccoid-inducing stressogenic culture of Vibrio parahaemolyticus showed an increase of expression in 15 of 17 selected genes associated with cell wall synthesis/modifications [71]. The highest increase (~18-fold) was observed for dacB, a gene coding for the production of D,D-carboxypeptidase, a homolog of csd3 in H. pylori. In H. pylori in the logarithmic growth phase (with a spiral shape) high levels of GM-pentapeptides and low levels of GM-tripeptides and GM-dipeptides were observed [16]. During the H. pylori morphological transition from spiral to spherical form, GM-dipeptide motifs accumulate with a simultaneous reduction of the GM-tetrapeptides and GM-tripeptides amount [16, 72]. Based on these results, it can be concluded that D,D-carboxypeptidase activity of Csd3 and Csd3-dependent generation of GM-tetrapeptide monomers is an important factor in the H. pylori morphological transition into coccoid forms. The substrates provided in this way can be further processed to GM-tripeptides and GM-dipeptides via Csd6 and Csd4 proteins, respectively. The accumulation of GM-dipeptides in the cell wall determines the peptidoglycan relaxation and the formation of spherical forms by H. pylori.
The Csd6 carboxypeptidase
The Csd6 protein possesses L,D-carboxypeptidase activity and trims GM-tetrapeptides into GM-tripeptides [68, 70] (Table I). It has been shown that both the lack of expression and overexpression contribute to the disturbance of H. pylori helical shape and the formation of straight rod morphology [68]. The Δcsd6 mutants had practically undetectable amounts of GM-tripeptides and GM-dipeptides, and elevated levels of tetrapeptide monomers and tetra-tetrapeptide dimers. Therefore, it is suggested that the latter are a substrate for this enzyme. A Campylobacter spp. homolog of the Csd6 is Pgp2, which, as in H. pylori, contributes to spiral cell shape maintenance [73, 74].
The Csd4 carboxypeptidase
Another protein in the H. pylori peptidoglycan modification cascade is Csd4 [8, 66] (Table I). It is an enzyme with the activity of D,L-carboxypeptidase, the target and product of which are GM-tripeptides and GM-dipeptides, respectively. Defects in the production of this protein contribute, similarly as in Δcsd6 mutants, to the disturbance of the helical shape of H. pylori and the formation of cells with straight rod-like morphology. The deletion mutants showed a significantly higher level of GM-tripeptides and virtually no GM-dipeptides. Changes in the amount of muropeptide dimers have also been noticed, i.e. an increase in tetra-tripeptide dimers and a reduction in tetra-tetrapeptides and tetra-pentapeptides motifs. In semi-solid agar tests, after 4-day H. pylori incubation, the Δcsd4 strains were shown to have 17% reduced motility, suggesting the importance of a spiral shape in the efficient movement in an environment of increased density. The straight rod morphology also negatively influenced the efficacy of mice colonisation, whereas it was not important in sensitivity to polymyxin, acidic pH, or increased osmotic pressure. A Campylobacter spp. homolog of the Csd4 is Pgp1. As in H. pylori, this protein contributes to maintaining spiral cell shape. The deletion of pgp1 affects numerous virulence traits, including motility, biofilm formation, and poultry colonisation [73, 75].
Regulatory proteins CcmA and Csd5
The CcmA and Csd5 proteins have the potential to function as regulatory factors [67] (Table I). The observations show that defects in the production of these proteins are associated with the formation of cells with the phenotype corresponding to the deletion of csd1 and csd2, or csd6 and csd4, respectively [7, 8]. In the ΔccmA strains loss of helicity and formation of curved cells is noticed, whereas in Δcsd5 mutants an opposite morphology is recorded (loss of curvature and generation of straight cells). On this basis it was concluded that CcmA and Csd5 may influence the asymmetric location and functioning of other proteins involved in the peptidoglycan turnover. Similarly in Proteus mirabilis, CcmA plays a key role in interactions with proteins associated with murein metabolism and the maintenance of their stability/location [76].
The AmiA amidase
The AmiA is a protein with the activity of N-ace-tylmuramoyl-L-alanyl amidase [15, 16] (Table I). In ΔamiA mutants the presence of a filamentous phenotype was demonstrated, suggesting the involvement of this protein in maintaining of the proper shape of H. pylori. Such cells had completely formed division sites, while the daughter cells remained unseparated. Mutant defective in the production of AmiA had a reduced amount of muropeptides with 1,6-anhydro-N-acetylmuramate linkages. In a wild-type strain, an increase in the amount of anhydromuropeptides was observed along with the time of cultivation (1.7% and 23.3% after 8 h and 48 h, respectively), while in the case of the ΔamiA strain an opposite trend was detected (12.2% and 6.7% after 8 h and 48 h, respectively) [16]. In addition, wild-type strain accumulates 2-3-times faster GM-dipeptides than the ΔamiA mutant [15]. The slowed accumulation of GM-dipeptides correlates with disturbances in morphological transformation into coccoid forms, i.e. after a week of incubation the amount of these forms accounted for 55.8% and 6.3% in wild-type and ΔamiA strains, respectively. What is more, despite the presence of intact flagella, the majority of ΔamiA mutants were defective in the ability to effectively migrate in and colonise the stomach of C57/BL6J mice [16]. On the basis of the presented results, it can be concluded that AmiA plays an important role in maintaining cell morphology, virulence, and survival in the host organism [15, 16].
The influence of H. pylori cell shape on motility
Numerous studies on the Csd proteins of H. pylori have shown that defects in the production of these factors affect the morphology of cells, and often the motility and ability to colonise the host [7, 8, 68, 69]. It was therefore concluded that the helical shape of H. pylori may be crucial for the physiological functioning of this bacterium. In the single-cell analysis of the H. pylori phenotype, the existence of a large heterogeneity of shapes, sizes, and number of flagella was observed [10]. Such variability directly affected the bacterial swimming speed in broth and viscous gastric mucin media. Both factors (shape and number of flagella) influenced the motility of H. pylori, because the cells with straight-rod morphology and fewer flagella had a lower speed of movement. These results are consistent with previous observations suggesting the importance of the spiral shape in the efficient movement of H. pylori [5, 6]. An alternative hypothesis was proposed by Constantino et al., who showed that the movement speed of flagellated bacteria is only slightly modified by the cell shape, i.e. the participation in the total propulsive thrust and swimming speed is estimated in this case for less than 15% [77]. Also, Celli et al. demonstrated that the spiral shape of H. pylori is not sufficient to penetrate a thick layer of gastric mucosa and is not responsible for the corkscrew movement [78]. In this case a more important factor was the production of urease and the alkalisation-dependent transition of mucus from gel to viscous solution. Such reduction of the viscosity of this polymer facilitates free movement of these bacteria, regardless of cell morphology. Therefore, it is difficult to determine the significance of the H. pylori spiral shape, and the reason for the existence of the entire spectrum of proteins involved in maintaining such morphology. There is still a lot of research needed to finally resolve this issue.
Peptidoglycan deacetylase PgdA and acetyltransferase PatA
The intense accumulation of GM-dipeptide motifs correlates in H. pylori with a morphological transition from spiral to coccoid forms [15]. Spherical forms induce a significantly lower level of proinflammatory factors, i.e. IL-8 cytokines and NF-κB transcription factors. This may suggest that morphological transformation is one way to avoid an attack from the host immune system.
There are also other mechanisms of immune escape, independent of the peptide bridge shortening. One of them is N-deacetylation of murein. The enzyme responsible for this process is peptidoglycan deacetylase A (PgdA) (Table I). The pgdA expression is induced during oxidative stress exposure, as demonstrated during H. pylori culture at various oxygen concentrations (three-fold higher at 12% oxygen than at 2%) and co-incubation with macrophages (3.5-fold increase) [79]. In ΔpgdA mutants, increased sensitivity to lysozyme and decreased mice colonisation capacity (nine-fold lower titre of deletion mutants than wild strains at 9 weeks post-infection) were demonstrated. In mice infected with the ΔpgdA strains an increased level of MIP-2 (macrophage inflammatory protein-2; IL-8 analog), IL-10, and TNF-α was also noticed, suggesting the participation of PgdA in the reduction of the host’s immune activity [80]. The post-translational regulator of PgdA functioning is AcnB aconitase, which is expressed in response to oxidative stress and changes in the iron level. The second way to mitigate the immune response to H. pylori, obtained by modifying murein, is the ability to O-acetylate this polymer. The enzyme responsible for this activity is peptidoglycan acetyltransferase A (PatA) (Table I). The synergistic activity of PatA and PgdA, contributing to increased resistance to lysozyme and the effectiveness of host colonisation, was noted [81]. After six-hour incubation of H. pylori (109 CFU/ml) in the presence of 30 mg/ml lysozyme, a much greater decrease in the ΔpgdA or ΔpatA (104 CFU/ ml) and ΔpgdApatA (< 10 CFU/ml) mutants was observed compared to wild-type strains (106 CFU/ml). A similar susceptibility gradation was observed when treating these strains with a mixture of lysozyme (0.3 mg/ml) and lactoferrin (3 mg/ml). In addition, the H. pylori double mutants had strongly reduced mice colonisation capacity; the number of deletion mutants was 10-fold lower than in wild-type strains (60 ±52 CFU/mg stomach vs. 691 ±103 CFU/mg stomach). On this basis, it was concluded that the action of both enzymes is needed for a full tolerance to lysozyme and effective host colonisation.
Peptidoglycan-modifying proteins of Helicobacter pylori as potential targets for antimicrobial therapies
The shape of bacterial cells is a factor that affects the ability of microbes to colonise specific niches (including the host organism), escape from immune clearance, or cause disease development [12]. For this reason, the shape seems to be an interesting target in creating new pathogen-focused eradication therapies. Antibiotics with antimicrobial activity, whose target is the cell wall, have been widely used for over 70 years [82]. The compounds that interfere with the process of peptidoglycan synthesis include: β-lactams, antibiotics targeting D,D-transpeptidases activity, and glycopeptides, antibiotics targeting D-Ala-D-Ala residues of lipid II precursors [82, 83]. Helicobacter pylori exhibits natural resistance to glycopeptides, while it is sensitive to β-lactams, of which amoxicillin is used in routine antibiotic therapies [84]. Currently, resistance to amoxicillin is satisfactorily low (< 2%) [85]. However, there are reports indicating the possibility of amoxicillin resistance spreading, which is achieved by the activity of β-lactamases [86] or pbp1A mutation [86–88]. For this reason, the search for alternative methods interfering with peptidoglycan biosynthesis is an interesting direction of research aimed at increasing the degree of H. pylori eradication.
One of the ways to reduce the pathogenicity of H. pylori is the possibility of disrupting helical cell formation. An example of a substance with such properties is a phosphonic acid-based pseudopeptide, which has the ability to bind to the active site of Csd4 (or Pgp1 in Campylobacter jejuni) and mediate protein activity disturbances [89]. The millimolar concentrations of this compound contributed to the straightening of H. pylori and C. jejuni cells.
The other option is to inhibit the transformation of H. pylori from antibiotic-susceptible spiral forms into more resistant coccoid forms. Because the overproduction of Csd3 contributed to the intensification of spherical morphotype formation, and the dysfunction of Csd3 leads to delay of the transformation process, this protein is another interesting target for anti-H. pylori therapy [9]. Another potential target is AmiA. Bacteria defective in the production of this protein have reduced ability to form spherical forms and impaired motility, colonisation of the host’s stomach, and evasion of immune responses [15, 16].
An alternative approach to reduce the protective properties of these microbes could be to create substances that would interfere with the functioning of PatA and/or PgdA. This would increase the sensitivity of these bacteria to the effect of physiological concentrations of the host’s anti-bacterial proteins (e.g. lysozyme and lactoferrin) and promote the intensification of the immune system’s activity to the ongoing infection [80, 81]. Similarly, in Gram-positive Staphylococcus, Streptococcus, and Enterococcus, a disruption of peptidoglycan acetyltransferase activity as an anti-virulent mechanism promoting increased immunity protection is suggested [90].
Summary
Helicobacter pylori is one of the model bacteria in research on the mechanisms responsible for the morphological variability of microbes. In many available scientific data, attention is focused on environmental factors affecting the morphogenesis of this microorganism [19–23, 25, 28, 29, 48, 49]. Nonetheless, relatively little is known about the molecular determinants that govern these processes.
The bacterial cytoskeleton participates in many important physiological functions, i.e. cell division, morphogenesis, and DNA segregation. Among the proteins of H. pylori cytoskeleton, Mre-like proteins are associated with the chromosomes segregation, cellular length modulation, and pathogenicity of this bacteria, Min proteins are responsible for the process of proper cell division, while Ccrps are involved in maintaining the spiral shape of this bacterium. Because many cytoskeletal proteins are not encoded by H. pylori or do not participate in morphogenesis processes, it seems that there are other, more important proteins involved in the morphological processes of this bacterium.
There are at least two networks of peptidoglycan-modifying enzymes that affect the shape of this bacterium; these are: helicity-generating Csd1, Csd2, and CcmA, and curvature-determining Csd4, Csd5, and Csd6. The Csd3 protein is considered a link between both cascades of murein modifications. Additionally, AmiA amidase also plays an important role in the maintenance of cell morphology by supervising the correct separation of daughter cells from mother cells.
To date, the function of the spiral shape in motility and the ability to colonise the stomach by H. pylori has not been unambiguously determined. There are studies confirming the key role of this morphological form in host colonisation, while others deny such dependence. Perhaps increased swimming speed of this bacteria, although insignificant in research models, is crucial for efficient stomach mucosa colonisation and protection against the lethal effect of acidic pH. Alternatively, motility in a high-density environment may occur equally well in H. pylori spiral and rod-shaped forms, while the energy expenditure of movement is more beneficial for spiral-shaped forms. Nevertheless, the spiral shape is not the only morphological form produced by H. pylori. The large heterogeneity of H. pylori morphological forms includes also the presence of curved/straight rods, filamentous forms, and coccoid forms. Such a multitude of shapes suggests that morphological variability may be an important factor responsible for rapid response to changing external conditions and thus is a feature that allows survival in various environmental niches and facilitates modulation of pathogenicity.
Based on the presented literature data, the number of studies focusing on molecular factors modifying the composition of H. pylori peptidoglycan/cytoskeleton should be increased. It seems that understanding the function and structure of these proteins would be of great importance in the development of new therapies aimed at disrupting the motility, colonisation, and ability of persistent stomach colonisation by H. pylori.
Acknowledgments
The study was supported by Wroclaw Medical University grant No.: STA130.16.031. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We would like to thank Dr. Jakub Grzesiak (Electron Microscopy Laboratory, Wroclaw Research Centre EIT+, Poland) for performing electron microscopic observations.
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Robinson K, Letley DP, Kaneko K. The human stomach in health and disease: infection strategies by Helicobacter pylori. Curr Top Microbiol Immunol. 2017;400:1–26. doi: 10.1007/978-3-319-50520-6_1. [DOI] [PubMed] [Google Scholar]
- 2.Malfertheiner P, Link A, Selgrad M. Helicobacter pylori: perspectives and time trends. Nat Rev Gastroenterol Hepatol. 2014;11:628–38. doi: 10.1038/nrgastro.2014.99. [DOI] [PubMed] [Google Scholar]
- 3.Venerito M, Vasapolli R, Malfertheiner P. Helicobacter pylori and gastric cancer: timing and impact of preventive measures. Adv Exp Med Biol. 2016;908:409–18. doi: 10.1007/978-3-319-41388-4_20. [DOI] [PubMed] [Google Scholar]
- 4.Dunne C, Dolan B, Clyne M. Factors that mediate colonization of the human stomach by Helicobacter pylori. World J Gastroenterol. 2014;20:5610–24. doi: 10.3748/wjg.v20.i19.5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hazell SL, Lee A, Brady L, et al. Campylobacter pyloridis and gastritis: association with intercellular spaces and adaptation to an environment of mucus as important factors in colonization of the gastric epithelium. J Infect Dis. 1986;153:658–63. doi: 10.1093/infdis/153.4.658. [DOI] [PubMed] [Google Scholar]
- 6.Keshavarz T, Walker MM, Karim QN, et al. The relationship between Helicobacter pylori motility, morphology and phase of growth: implications for gastric colonization and pathology. Microbiology. 1999;145:2803–11. doi: 10.1099/00221287-145-10-2803. [DOI] [PubMed] [Google Scholar]
- 7.Sycuro LK, Pincus Z, Gutierrez KD, et al. Peptidoglycan crosslinking relaxation promotes Helicobacter pylori’s helical shape and stomach colonization. Cell. 2010;141:822–33. doi: 10.1016/j.cell.2010.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sycuro LK, Wyckoff TJ, Biboy J, et al. Multiple peptidoglycan modification networks modulate Helicobacter pylori’s cell shape, motility, and colonization potential. PLoS Pathog. 2012;8:e1002603. doi: 10.1371/journal.ppat.1002603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.An DR, Kim HS, Kim J, et al. Structure of Csd3 from Helicobacter pylori, a cell shape-determining metallopeptidase. Acta Crystallogr Sect D Biol Crystallogr. 2015;71:675–86. doi: 10.1107/S1399004715000152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martínez LE, Hardcastle JM, Wang J, et al. Helicobacter pylori strains vary cell shape and flagellum number to maintain robust motility in viscous environments. Mol Microbiol. 2016;99:88–110. doi: 10.1111/mmi.13218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.French S, Côté JP, Stokes JM, et al. Bacteria getting into shape: genetic determinants of E. coli morphology. MBio. 2017;8:e01977–16. doi: 10.1128/mBio.01977-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Teeseling MCF, de Pedro MA, Cava F. Determinants of bacterial morphology: from fundamentals to possibilities for antimicrobial targeting. Front Microbiol. 2017;8:1264. doi: 10.3389/fmicb.2017.01264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang DC, Blair KM, Salama NR. Staying in shape: the impact of cell shape on bacterial survival in diverse environments. Microbiol Mol Biol Rev. 2016;80:187–203. doi: 10.1128/MMBR.00031-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krzyżek P, Gościniak G. A proposed role for diffusible signal factors in the biofilm formation and morphological transformation of Helicobacter pylori. Turk J Gastroenterol. 2018;29:7–13. doi: 10.5152/tjg.2017.17349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chaput C, Ecobichon C, Cayet N, et al. Role of AmiA in the morphological transition of Helicobacter pylori and in immune escape. PLoS Pathog. 2006;2:e97. doi: 10.1371/journal.ppat.0020097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaput C, Ecobichon C, Pouradier N, et al. Role of the N-acetylmuramoyl-l-alanyl amidase, AmiA, of Helicobacter pylori in peptidoglycan metabolism, daughter cell separation, and virulence. Microb Drug Resist. 2016;22:477–86. doi: 10.1089/mdr.2016.0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao X, Zhong J, Wei C, et al. Current perspectives on viable but non-culturable state in foodborne pathogens. Front Microbiol. 2017;8:580. doi: 10.3389/fmicb.2017.00580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li L, Mendis N, Trigui H, et al. The importance of the viable but non-culturable state in human bacterial pathogens. Front Microbiol. 2014;5:258. doi: 10.3389/fmicb.2014.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Enroth H, Engstrand L. Egg passage of rodshaped and coccoid forms of Helicobacter pylori: preliminary studies. Helicobacter. 1996;1:183–6. doi: 10.1111/j.1523-5378.1996.tb00036.x. [DOI] [PubMed] [Google Scholar]
- 20.Shao C, Sun Y, Wang N, et al. Changes of proteome components of Helicobacter pylori biofilms induced by serum starvation. Mol Med Rep. 2013;8:1761–6. doi: 10.3892/mmr.2013.1712. [DOI] [PubMed] [Google Scholar]
- 21.Nilsson HO, Blom J, Abu-Al-Soud W, et al. Effect of cold starvation, acid stress, and nutrients on metabolic activity of Helicobacter pylori. Appl Environ Microbiol. 2002;68:11–9. doi: 10.1128/AEM.68.1.11-19.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cellini L, Allocati N, Di Campli E, et al. Helicobacter pylori: a fickle germ. Microbiol Immunol. 1994;38:25–30. doi: 10.1111/j.1348-0421.1994.tb01740.x. [DOI] [PubMed] [Google Scholar]
- 23.Tominaga K, Hamasaki N, Watanabe T, et al. Effect of culture conditions on morphological changes of Helicobacter pylori. J Gastroenterol. 1999;34(Suppl):28–31. [PubMed] [Google Scholar]
- 24.Jiang X, Doyle MP. Effect of environmental and substrate factors on survival and growth of Helicobacter pylori. J Food Prot. 1998;61:929–33. doi: 10.4315/0362-028x-61.8.929. [DOI] [PubMed] [Google Scholar]
- 25.Catrenich CE, Makin KM. Characterization of the morphologic conversion of Helicobacter pylori from bacillary to coccoid forms. Scand J Gastroenterol. 1991;26:58–64. [PubMed] [Google Scholar]
- 26.She FF, Lin JY, Liu JY, et al. Virulence of water-induced coccoid Helicobacter pylori and its experimental infection in mice. World J Gastroenterol. 2003;9:516–20. doi: 10.3748/wjg.v9.i3.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.West AP, Millar MR, Tompkins DS. Survival of Helicobacter pylori in water and saline. J Clin Pathol. 1990;43:609. doi: 10.1136/jcp.43.7.609-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bury-Mone S, Kaakoush NO, Asencio C, et al. Is Helicobacter pylori a true microaerophile? Helicobacter. 2006;11:296–303. doi: 10.1111/j.1523-5378.2006.00413.x. [DOI] [PubMed] [Google Scholar]
- 29.Park SA, Ko A, Lee NG. Stimulation of growth of the human gastric pathogen Helicobacter pylori by atmospheric level of oxygen under high carbon dioxide tension. BMC Microbiol. 2011;11:96. doi: 10.1186/1471-2180-11-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Faghri J, Poursina F, Moghim S, et al. Morphological and bactericidal effects of different antibiotics on Helicobacter pylori. Jundishapur J Microbiol. 2014;7:e8704. doi: 10.5812/jjm.8704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.She FF, Su DH, Lin JY, et al. Virulence and potential pathogenicity of coccoid Helicobacter pylori induced by antibiotics. World J Gastroenterol. 2001;7:254–8. doi: 10.3748/wjg.v7.i2.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saniee P, Shahreza S, Siavoshi F. Negative effect of proton-pump inhibitors (PPIs) on Helicobacter pylori growth, morphology, and urease test and recovery after PPI removal – an in vitro study. Helicobacter. 2016;21:143–52. doi: 10.1111/hel.12246. [DOI] [PubMed] [Google Scholar]
- 33.Kim TS, Hur JW, Yu MA, et al. Antagonism of Helicobacter pylori by bacteriocins of lactic acid bacteria. J Food Prot. 2003;66:3–12. doi: 10.4315/0362-028x-66.1.3. [DOI] [PubMed] [Google Scholar]
- 34.Le Moal VL, Fayol-Messaoudi D, Servin AL. Compound(s) secreted by Lactobacillus casei strain Shirota YIT9029 irreversibly and reversibly impair the swimming motility of Helicobacter pylori and Salmonella enterica serovar Typhimurium, respectively. Microbiology. 2013;159:1956–71. doi: 10.1099/mic.0.067678-0. [DOI] [PubMed] [Google Scholar]
- 35.Khosravi Y, Dieye Y, Loke MF, et al. Streptococcus mitis induces conversion of Helicobacter pylori to coccoid cells during co-culture in vitro. PLoS One. 2014;9:e112214. doi: 10.1371/journal.pone.0112214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Janas B, Czkwianianc E, Bak-Romaniszyn L, et al. Electron microscopic study of association between coccoid forms of Helicobacter pylori and gastric epithelial cells. Am J Gastroenterol. 1995;90:1829–33. [PubMed] [Google Scholar]
- 37.Chan WY, Hui PK, Leung KM, et al. Coccoid forms of Helicobacter pylori in the human stomach. Am J Clin Pathol. 1994;102:503–7. doi: 10.1093/ajcp/102.4.503. [DOI] [PubMed] [Google Scholar]
- 38.Balakrishna JP, Filatov A. Coccoid forms of Helicobacter pylori causing active gastritis. Am J Clin Pathol. 2013;140:A101. [Google Scholar]
- 39.Wang X, Sturegard E, Rupar R, et al. Infection of BALB/c A mice by spiral and coccoid forms of Helicobacter pylori. J Med Microbiol. 1997;46:657–63. doi: 10.1099/00222615-46-8-657. [DOI] [PubMed] [Google Scholar]
- 40.Cellini L, Allocati N, Angelucci D, et al. Coccoid Helicobacter pylori not culturable in vitro reverts in mice. Microbiol Immunol. 1994;38:843–50. doi: 10.1111/j.1348-0421.1994.tb02136.x. [DOI] [PubMed] [Google Scholar]
- 41.Loke MF, Ng CG, Vilashni Y, et al. Understanding the dimorphic lifestyles of human gastric pathogen Helicobacter pylori using the SWATH-based proteomics approach. Sci Rep. 2016;6:26784. doi: 10.1038/srep26784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krzyżek P, Gościniak G. Oral Helicobacter pylori: interactions with host and microbial flora of the oral cavity. Dent Med Probl. 2018;55:75–82. doi: 10.17219/dmp/81259. [DOI] [PubMed] [Google Scholar]
- 43.Hirsch C, Tegtmeyer N, Rohde M, et al. Live Helicobacter pylori in the root canal of endodontic-infected deciduous teeth. J Gastroenterol. 2012;47:936–40. doi: 10.1007/s00535-012-0618-8. [DOI] [PubMed] [Google Scholar]
- 44.Waidner B, Specht M, Dempwolff F, et al. A novel system of cytoskeletal elements in the human pathogen Helicobacter pylori. PLoS Pathog. 2009;5:e1000669. doi: 10.1371/journal.ppat.1000669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fernandes RM, Silva H, Oliveira R, et al. Morphological transition of Helicobacter pylori adapted to water. Future Microbiol. 2017;12:1167–79. doi: 10.2217/fmb-2016-0174. [DOI] [PubMed] [Google Scholar]
- 46.Aziz RK, Khalifa MM, Sharaf RR. Contaminated water as a source of Helicobacter pylori infection: a review. J Adv Res. 2015;6:539–47. doi: 10.1016/j.jare.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ranjbar R, Khamesipour F, Jonaidi-Jafari N, et al. Helicobacter pylori in bottled mineral water: genotyping and antimicrobial resistance properties. BMC Microbiol. 2016;16:40. doi: 10.1186/s12866-016-0647-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gancz H, Jones KR, Merrell DS. Sodium chloride affects Helicobacter pylori growth and gene expression. J Bacteriol. 2008;190:4100–5. doi: 10.1128/JB.01728-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takeuchi H, Nakazawa T, Okamoto T, et al. Cell elongation and cell death of Helicobacter pylori is modulated by the disruption of cdrA (cell division-related gene A) Microbiol Immunol. 2006;50:487–97. doi: 10.1111/j.1348-0421.2006.tb03819.x. [DOI] [PubMed] [Google Scholar]
- 50.Singh M, Prasad K, Yachha S. Elongated Helicobacter pylori in gastric mucosa of children associated with gastric disease. J Pediatr Infect Dis. 2015;3:35–9. [Google Scholar]
- 51.Ingerson-Mahar M, Gitai Z. A growing family: the expanding universe of the bacterial cytoskeleton. FEMS Microbiol Rev. 2012;36:256–66. doi: 10.1111/j.1574-6976.2011.00316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cho H. The role of cytoskeletal elements in shaping bacterial cells. J Microbiol Biotechnol. 2015;25:307–16. doi: 10.4014/jmb.1409.09047. [DOI] [PubMed] [Google Scholar]
- 53.Cabeen MT, Jacobs-Wagner C. Bacterial cell shape. Nat Rev Microbiol. 2005;3:601–10. doi: 10.1038/nrmicro1205. [DOI] [PubMed] [Google Scholar]
- 54.Celler K, Koning RI, Koster AJ, et al. Multidimensional view of the bacterial cytoskeleton. J Bacteriol. 2013;195:1627–36. doi: 10.1128/JB.02194-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.El Ghachi M, Matteï PJ, Ecobichon C, et al. Characterization of the elongasome core PBP2:MreC complex of Helicobacter pylori. Mol Microbiol. 2011;82:68–86. doi: 10.1111/j.1365-2958.2011.07791.x. [DOI] [PubMed] [Google Scholar]
- 56.Zepeda G, Reyna C, Fu Y, et al. Novel protein interactions with an actin homolog (MreB) of Helicobacter pylori determined by bacterial two-hybrid system. Microbiol Res. 2017;201:39–45. doi: 10.1016/j.micres.2017.04.008. [DOI] [PubMed] [Google Scholar]
- 57.Nishida Y, Takeuchi H, Morimoto N, et al. Intrinsic characteristics of Min proteins on the cell division of Helicobacter pylori. FEMS Microbiol Lett. 2016;363:fnw025. doi: 10.1093/femsle/fnw025. [DOI] [PubMed] [Google Scholar]
- 58.Yu XC, Margolin W. Deletion of the min operon results in increased thermosensitivity of an ftsZ84 mutant and abnormal FtsZ ring assembly, placement, and disassembly. J Bacteriol. 2000;182:6203–13. doi: 10.1128/jb.182.21.6203-6213.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chiou PY, Luo CH, Chang KC, et al. Maintenance of the cell morphology by MinC in Helicobacter pylori. PLoS One. 2013;8:e71208. doi: 10.1371/journal.pone.0071208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Specht M, Schatzle S, Graumann PL, et al. Helicobacter pylori possesses four coiled-coil-rich proteins that form extended filamentous structures and control cell shape and motility. J Bacteriol. 2011;193:4523–30. doi: 10.1128/JB.00231-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schätzle S, Specht M, Waidner B. Coiled coil rich proteins (Ccrp) influence molecular pathogenicity of Helicobacter pylori. PLoS One. 2015;10:e0121463. doi: 10.1371/journal.pone.0121463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vollmer W. Structural variation in the glycan strands of bacterial peptidoglycan. FEMS Microbiol Rev. 2008;32:287–306. doi: 10.1111/j.1574-6976.2007.00088.x. [DOI] [PubMed] [Google Scholar]
- 63.Typas A, Banzhaf M, Gross CA, et al. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol. 2011;10:123–36. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cava F, de Pedro MA. Peptidoglycan plasticity in bacteria: Emerging variability of the murein sacculus and their associated biological functions. Curr Opin Microbiol. 2014;18:46–53. doi: 10.1016/j.mib.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 65.Scheffers DJ, Pinho MG. Bacterial cell wall synthesis: new insights from localization studies. Microbiol Mol Biol Rev. 2005;69:585–607. doi: 10.1128/MMBR.69.4.585-607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim HS, Kim J, Im HN, et al. Structural basis for the recognition of muramyltripeptide by Helicobacter pylori Csd4, a D,L-carboxypeptidase controlling the helical cell shape. Acta Crystallogr Sect D Biol Crystallogr. 2014;70:2800–12. doi: 10.1107/S1399004714018732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.An DR, Im HN, Jang JY, et al. Structural basis of the heterodimer formation between cell shape-determining proteins Csd1 and Csd2 from Helicobacter pylori. PLoS One. 2016;11:e0164243. doi: 10.1371/journal.pone.0164243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sycuro LK, Rule CS, Petersen TW, et al. Flow cytometry-based enrichment for cell shape mutants identifies multiple genes that influence Helicobacter pylori morphology. Mol Microbiol. 2013;90:869–83. doi: 10.1111/mmi.12405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bonis M, Ecobichon C, Guadagnini S, et al. A M23B family metallopeptidase of Helicobacter pylori required for cell shape, pole formation and virulence. Mol Microbiol. 2010;78:809–19. doi: 10.1111/j.1365-2958.2010.07383.x. [DOI] [PubMed] [Google Scholar]
- 70.Kim HS, Im HN, An DR, et al. The cell shape-determining Csd6 protein from Helicobacter pylori constitutes a new family of L,D-carboxypeptidase. 2015;290:25103–17. doi: 10.1074/jbc.M115.658781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hung WC, Jane WN, Wong HC. Association of a D-alanyl-D-alanine carboxypeptidase gene with the formation of aberrantly shaped cells during the induction of viable but nonculturable Vibrio parahaemolyticus. Appl Environ Microbiol. 2013;79:7305–12. doi: 10.1128/AEM.01723-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Costa K, Bacher G, Allmaier G, et al. The morphological transition of Helicobacter pylori cells from spiral to coccoid is preceded by a substantial modification of the cell wall. J Bacteriol. 1999;181:3710–5. doi: 10.1128/jb.181.12.3710-3715.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Esson D, Mather AE, Scanlan E, et al. Genomic variations leading to alterations in cell morphology of Campylobacter spp. Sci Rep. 2016;6:38303. doi: 10.1038/srep38303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Frirdich E, Vermeulen J, Biboy J, et al. Peptidoglycan LD-carboxypeptidase Pgp2 influences Campylobacter jejuni helical cell shape and pathogenic properties and provides the substrate for the DL-carboxypeptidase Pgp1. J Biol Chem. 2014;289:8007–18. doi: 10.1074/jbc.M113.491829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Frirdich E, Biboy J, Adams C, et al. Peptidoglycan-modifying enzyme Pgp1 Is required for helical cell shape and pathogenicity traits in Campylobacter jejuni. PLoS Pathog. 2012;8:e1002602. doi: 10.1371/journal.ppat.1002602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hay NA, Tipper DJ, Gygi D, et al. A novel membrane protein influencing cell shape and multicellular swarming of Proteus mirabilis. J Bacteriol. 1999;181:2008–16. doi: 10.1128/jb.181.7.2008-2016.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Constantino MA, Jabbarzadeh M, Fu HC, et al. Helical and rod-shaped bacteria swim in helical trajectories with little additional propulsion from helical shape. Sci Adv. 2016;2:e1601661. doi: 10.1126/sciadv.1601661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Celli JP, Turner BS, Afdhal NH, et al. Helicobacter pylori moves through mucus by reducing mucin viscoelasticity. Proc Natl Acad Sci. 2009;106:14321–6. doi: 10.1073/pnas.0903438106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Austin CM, Maier RJ. Aconitase-mediated posttranscriptional regulation of Helicobacter pylori peptidoglycan deacetylase. J Bacteriol. 2013;195:5316–22. doi: 10.1128/JB.00720-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang G, Maier SE, Lo LF, et al. Peptidoglycan deacetylation in Helicobacter pylori contributes to bacterial survival by mitigating host immune responses. Infect Immun. 2010;78:4660–6. doi: 10.1128/IAI.00307-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang G, Lo LF, Forsberg LS, et al. Helicobacter pylori peptidoglycan modifications confer lysozyme resistance and contribute to survival in the host. MBio. 2012;3:e00409–12. doi: 10.1128/mBio.00409-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bush K. Antimicrobial agents targeting bacterial cell walls and cell membranes. Rev Sci Tech. 2012;31:43–56. doi: 10.20506/rst.31.1.2096. [DOI] [PubMed] [Google Scholar]
- 83.Jovetic S, Zhu Y, Marcone GL, et al. Beta-Lactam and glycopeptide antibiotics: first and last line of defense? Trends Biotechnol. 2010;28:596–604. doi: 10.1016/j.tibtech.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 84.Mégraud F. Resistance of Helicobacter pylori to antibiotics. Aliment Pharmacol Ther. 1997;11:43–53. doi: 10.1046/j.1365-2036.11.s1.11.x. [DOI] [PubMed] [Google Scholar]
- 85.Nishizawa T, Suzuki H. Mechanisms of Helicobacter pylori antibiotic resistance and molecular testing. Front Mol Biosci. 2014;1:19. doi: 10.3389/fmolb.2014.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tseng YS, Wu DC, Chang CY, et al. Amoxicillin resistance with beta-lactamase production in Helicobacter pylori. Eur J Clin Invest. 2009;39:807–12. doi: 10.1111/j.1365-2362.2009.02166.x. [DOI] [PubMed] [Google Scholar]
- 87.Matteo MJ, Granados G, Olmos M, et al. Helicobacter pylori amoxicillin heteroresistance due to point mutations in PBP-1A in isogenic isolates. J Antimicrob Chemother. 2008;61:474–7. doi: 10.1093/jac/dkm504. [DOI] [PubMed] [Google Scholar]
- 88.Gerrits MM, Godoy APO, Kuipers EJ, et al. Multiple mutations in or adjacent to the conserved penicillin-binding protein motifs of the penicillin-binding protein 1A confer amoxicillin resistance to Helicobacter pylori. Helicobacter. 2006;11:181–7. doi: 10.1111/j.1523-5378.2006.00398.x. [DOI] [PubMed] [Google Scholar]
- 89.Liu Y, Frirdich E, Taylor JA, et al. A bacterial cell shape-determining inhibitor. ACS Chem Biol. 2016;11:981–91. doi: 10.1021/acschembio.5b01039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sychantha D, Jones CS, Little DJ, et al. In vitro characterization of the antivirulence target of Gram-positive pathogens, peptidoglycan O-acetyltransferase A (OatA) PLoS Pathog. 2017;13:e1006667. doi: 10.1371/journal.ppat.1006667. [DOI] [PMC free article] [PubMed] [Google Scholar]