Abstract
The epithelial lateral membrane plays a central role in the integration of intercellular signals and, by doing so, is a principal determinant in the emerging properties of epithelial tissues. Mechanical force, when applied to the lateral cell–cell interface, can modulate the strength of adhesion and influence intercellular dynamics. Yet the relationship between mechanical force and epithelial cell behavior is complex and not completely understood. This commentary aims to provide an investigative look at the usage of cellular forces at the epithelial cell–cell adhesion interface.
Keywords: cell-cell, adhesion, force, actin, intercellular, lateral membrane, epithelial, contraction, protrusion, retraction
Introduction
Mechanical force, which is neither genetically encoded nor biochemically tractable, is being recognized as a significant determinant in the behaviors and functions of cells and tissues 1– 9. Whereas biochemical information is encoded in parameters such as post-translational modifications, biophysical information is encoded in parameters such as magnitude, frequency, and duration. Yet how these biophysical parameters of mechanical force contribute to the regulation of molecular and cellular events is not understood. Despite great efforts from biologists, physicists, and engineers to probe mechanical forces in living cells and tissues 10– 22, it remains tremendously difficult to measure the biophysical variables of mechanical forces inside cells. The purpose of this commentary is to explore, in the context of cell–cell interactions, the properties and usages of mechanical forces that can support cellular and molecular work 23– 26.
At the epithelial cell–cell adhesion interface, mechanical force is powered predominantly by actin dynamics and their associated molecular motors, including (a) protrusive force driven by actin polymerization and myosin activities 24, 27– 35; (b) retraction force contributed by cortical actin depolymerization, plasma membrane tension, retrograde actin flow, and myosin activities 36– 42; and (c) contraction force provided by actin dynamics and cross-linked actomyosin II activities 43– 48. This commentary will not discuss individual adhesion molecules and cytoskeletal regulators, which had been recently reviewed 49, 50.
The three-dimensional epithelial cell
The epithelial cell forms distinct membrane domains by using two sets of adhesion molecules: one set for adhesion to extracellular matrix and another set for adhesion between cells. By having two sets of adhesion systems, an epithelial cell establishes structural and spatial organization with (a) a lateral membrane organized by cell–cell adhesions, (b) a basal membrane organized by cell–matrix adhesions, and (c) a free surface at the apical membrane. Through interactions with its environment and neighbors, the epithelial cell acquires emergent properties integral to the functions of epithelial tissues, such as coordinated multi-cellular junctional constriction and collective movement 51– 59.
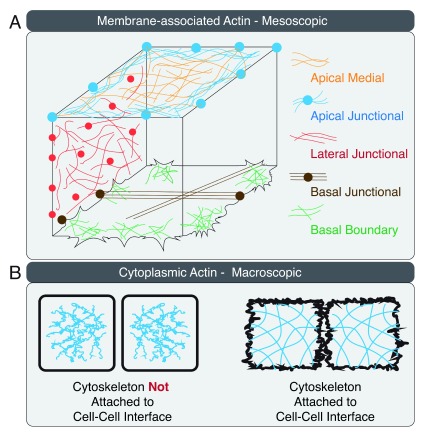
The three membrane domains of the epithelial cell contain actin nucleation factors, actin cross-linking proteins, and myosin II filaments that create distinct actomyosin II networks ( Figure 1A) 25, 60– 62. The lateral membrane is further organized into apical, lateral, and basal intercellular interactions with distinct protein compositions, actin dynamics, and contractile properties 63. Mechanical forces generated at the apical, lateral, and basal actomyosin II cytoskeleton are transmitted to cell–cell contacts via linkage to adhesion proteins or the plasma membrane ( Figure 1B).
Figure 1. Actin organization in a three-dimensional epithelial cell.
( A) Actin arrangement on the apical, lateral, and basal membranes of the epithelial cell is illustrated at the mesoscopic cellular level. ( B) Connectivity between cell–cell interface and the actin cytoskeleton is illustrated at the macroscopic multi-cellular level.
Direction of forces
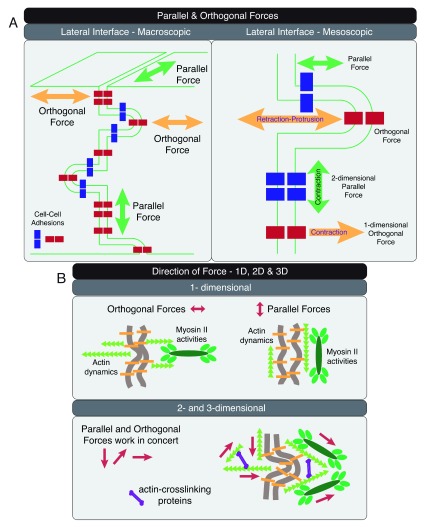
Unlike chemical signaling that is driven by random walks and does not have intrinsic directionality, mechanical force intrinsically has a direction that is important to guide biological processes 64– 68. The directionality of the mechanical force is heavily influenced by the spatial organization of the actomyosin II cytoskeleton. Mechanical forces on the lateral adhesion interface are frequently orthogonal or parallel to the cell boundary ( Figure 2A, left panel). One exception to this generalization is at folded membranes ( Figure 2A, right panel). When protrusions and retractions are formed on the lateral membrane, adhesion proteins are no longer in the same orientation as the cell–cell boundary. Consequently, the direction of force exerted on adhesion molecules would be dependent on their relative positions. Cell–cell adhesions formed at the tip of an intercellular membrane protrusion would experience orthogonal protrusion–retraction force, whereas cell–cell adhesions found at the trunk of an intercellular membrane protrusion would experience parallel drag force on the plane of the plasma membrane ( Figure 2A, right panel).
Figure 2. Direction of forces at the lateral cell–cell adhesion interface.
( A) Parallel and orthogonal forces are shown at the macroscopic multi-cellular (left panel) and mesoscopic cellular (right panel) scales. Cell–cell adhesions at the tip of a lateral membrane protrusion experience orthogonal force, whereas cell–cell adhesions at the trunk of the same membrane protrusion experience parallel force (right panel). ( B) Forces in one, two, and three dimensions can be generated through spatial organization of actin dynamics and actomyosin activities. Orthogonal and parallel one-dimensional forces can be generated with actin filaments arranged orthogonal and parallel to the plasma membrane, respectively (upper panel). Orthogonal and parallel two- and three-dimensional forces can be generated with a cross-linked actin network to support processes on the lateral cell–cell adhesion interface (lower panel).
Mechanical force applied in a directional manner could elicit different molecular and cellular events on the cell–cell adhesion interface 65, 66, 69. At the microscopic scale ( Figure 2B, upper panel), actin dynamics can create one-dimensional (1D) protrusion and retraction forces orthogonal to the lateral membrane. Anti-parallel myosin II filaments on linear actin filaments can create 1D contraction force. Actin filaments arranged parallel to the membrane would support force generation parallel to the cell–cell interface. Organization of branched actin networks using actin cross-linking proteins would create parallel and orthogonal forces to drive 2D and 3D events ( Figure 2B, lower panel). Spatial and temporal control of forces in one, two, and three dimensions can support distinct force-dependent processes on the lateral membrane.
Pushing and pulling forces to drive kinetic processes
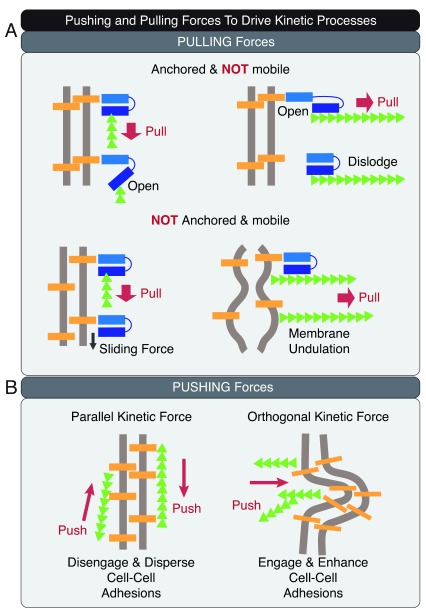
The kinetic energy produced by mechanical forces can be used to create movement, resulting in productive work such as pushing and pulling ( Figure 3) 41, 70– 76.
Figure 3. Pushing and pulling to drive kinetic processes.
( A) Pulling on adhesions and membrane can break intra-molecular and inter-molecular bonds, cause movement of adhesions on the membrane, and create undulations on the cell–cell adhesion interface. ( B) Pushing parallel to the lateral membrane can disengage and disperse adhesions, resulting in mixing of the membrane components. Pushing orthogonal to the lateral membrane can increase cell–cell contact area and time to enhance adhesion engagement.
Pulling forces can act on adhesions to unfold or disengage a protein by breaking intra-molecular or inter-molecular bonds, effectively changing the energy landscape of the cell–cell adhesion interface ( Figure 3A) 77– 82. Weaker interactions can be disrupted by weak forces but stronger interactions can resist weak forces and are selectively preserved. Thus, the input of different levels of kinetic energy would create different metastable states of the epithelial cell 83. For mobile and unanchored adhesions, pulling forces can result in sliding of the adhesion complex. Directional pushing can result in biased movement of the mobile fraction. Pulling orthogonally to the cell–cell interface can cause undulations of the lateral membrane, creating intercellular space and increasing the distance between apposing cell–cell adhesion molecules.
Pushing the lateral membrane can influence the distribution of protein complexes and membrane dynamics on the cell–cell adhesion interface ( Figure 3B) 70, 75, 84– 89. Pushing parallel to the membrane can disperse proteins and lipids, resulting in mixing of adhesion complexes and membrane components. By contrast, pushing orthogonal to the membrane can drive opposing cells together to increase contact time and area for adhesion engagement.
In summary, the spatial constraint created by organizing actin dynamics and actomyosin cytoskeleton can determine the magnitude, direction, and dimensionality of force. These biophysical properties of force are recognized by the 3D epithelial cells to drive different molecular and cellular events. Importantly, the orientation of the adhesion proteins on the plane of the plasma membrane determines the molecular outcome and ultimately the cellular effect of mechanical force on the cell–cell adhesion interface 90– 92.
Pushing and pulling forces to drive thermodynamic processes
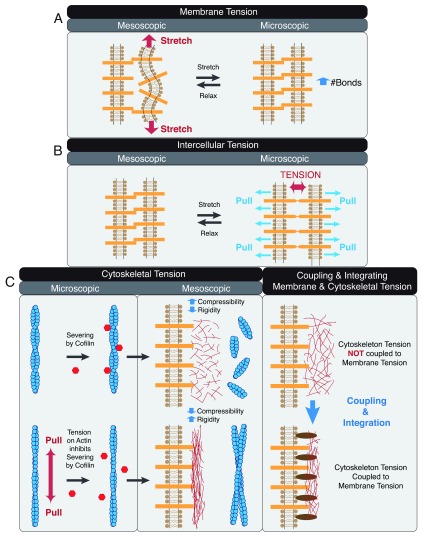
The energy supplied by mechanical forces can be stored as elastic energy to build up tension (a) on the plasma membrane ( Figure 4A) 93– 96, (b) on cell–cell adhesion molecules ( Figure 4B) 97, and (c) within the actin cytoskeleton ( Figure 4C) 98– 101. At the mesoscopic level, tension can be coupled and integrated to facilitate processes on the lateral interface 56, 102, 103.
Figure 4. Pushing and pulling to create tension.
( A) Stretching the membrane increases membrane tension at the mesoscopic lateral membrane level and can enhance intercellular interactions at the microscopic molecular level. ( B) Pulling orthogonal to the membrane increases intercellular tension on the mesoscopic lateral membrane level and can enhance tension at cell–cell adhesion molecules at the microscopic molecular level. ( C) Pulling actin filament alters actin dynamics and decreases cofilin severing at the microscopic molecular level (left panel). Changing the length distribution and organization of actin filaments alters cytoskeletal tension, ultimately affecting the compressibility and rigidity of cortical actin associated with the lateral membrane at the mesoscopic lateral membrane level (middle panel). Coupling membrane and cytoskeleton tension to adhesion proteins allows integration of intercellular, membrane, cytoskeletal forces on the lateral cell–cell adhesion interface (right panel).
Actin dynamics and actomyosin activities can increase membrane tension by stretching the membrane ( Figure 4A) 104– 106. Tension generated on the plane of the membrane will decrease undulation 107– 109, creating a taut region that supports lateral clustering of adhesion proteins 103, 110– 112. Orthogonal pulling force on the membrane can increase intercellular tension by stretching the extracellular domains of cell–cell adhesion proteins ( Figure 4B) 113.
Underneath the plasma membrane, tension can be generated in the cortical cytoskeleton ( Figure 4C) 114, 115. Pulling actin increases filament tension and decreases its susceptibility to severing by cofilin ( Figure 4C, left panel) 116 and results in changes in actin dynamics and filament stability 117, 118. Altering filament length and actin stability would affect connectivity of the actomyosin network and force transmission 119– 122. Pulling force on actin filaments could alter the spatial organization of the entire actomyosin network, resulting in mesoscopic differences in compressibility, stability, and rigidity ( Figure 4C, middle panel) 123– 127.
Plasma membrane and cytoskeletal tension can be coupled to create an integrated membrane–adhesion–cytoskeleton ensemble with properties that are more complex than the sum of the individual components ( Figure 4C, right panel) 56, 91, 128– 137. Thus, the energy input by pushing and pulling forces to generate tension on the lateral membrane, adhesion proteins, and cortical actin cytoskeleton ultimately results in the emergence of new properties and organization at the cell–cell adhesion interface 12, 138– 141.
Magnitude of force
The reason to consider the magnitude of force is twofold. First, the magnitude of force dictates which process it can perform ( Table 1, upper panel) 27, 40, 78, 142– 148. Whether the mechanical force is meant to create tension, unfold protein, cause movement, or mix components on the membrane would depend on the energy required for the specific process. The magnitude of force can have a differential effect on proteins; weak force will break weaker bonds and preserve stronger bonds, whereas strong force will break both weak and strong bonds. For example, unfolding alpha-catenin would require less force than detaching an E-cadherin homophilic bond. Thus, the magnitude of force, along with the intrinsic transition states of the cell–cell adhesion system, dictates the kinetic and thermodynamic pathways for epithelial processes such as adhesion remodeling and strengthening.
Table 1. Magnitude of forces.
( Upper) Different processes require different levels of force. ( Lower) Different molecular mechanisms produce different levels of force.
| Process | Force |
|---|---|
| Filopodium extension | < 3 pN |
| Unfolding vinculin binding site on α-catenin | 5-10-15 pN |
| Unfolding intramolecular auto-inhibitory sites on ZO-1 | 5-20 pN |
| Detaching N-WASP from Arp2/3-F-actin complex | 6-7 pN |
| Lamellipodia pushing force | 10-20 pN |
| Breaking single E-cadherin-E-cadherin bond (without actin
binding) |
15-25 pN |
| Breaking single E-cadherin-E-cadherin bond | 35-50 pN |
| Filopodium Retraction force (with actin and plasma membrane) | 50-2000 pN |
| Machine | Force |
| Actin Polymerization | 1 pN |
| Formin-driven Barbed-end Actin Polymerization | 1-2 pN |
| Myosin I motor on actin | 1-2 pN |
| Myosin VI motor on actin | 2 pN |
| Myosin II single motor head on actin | 1-6 pN |
| Myosin II minifilament on actin | 20-50 pN |
| Myosin II minifilament on α-actinin-crosslinked actin bundle | 100 pN |
| Myosin II minifilament on α-actinin-crosslinked actin network | 1 micro N |
Second, the magnitude of force dictates which mechanism can supply the force ( Table 1, lower panel) 130, 149– 152. Polymerization of a single actin filament supplies less force than a bundle of parallel actin filaments. To generate protrusion, a bundle of actin must polymerize in concert and work together 153. However, if force is needed only to push an adhesion protein closer to its binding partner on the membrane, a single actin filament may supply enough energy to promote biased diffusion. A myosin pulling on an actin filament produces less force than a bundle of myosin pulling on a bundle of parallel actin filaments. Anti-parallel myosin filaments pulling on parallel bundles of actin filaments produce less force than pulling on a cross-linked network of actin filaments. Thus, epithelial cells control the amount of force available by regulating the spatial distribution of actin dynamics and actomyosin II activities to dictate which molecular process is being executed at the lateral cell–cell adhesion interface 154.
Conclusions
Mechanical force plays an important role at the cell–cell adhesion interface. How mechanical force regulates cellular and molecular processes depends on its encoded properties, including directionality and magnitude. Additional biophysical parameters such as frequency, duration, and waveform that are instructive signals for other biological regulatory systems such as calcium signaling and neuronal firing are also encoded in cellular mechanical forces, but their contribution to epithelial cell–cell interaction is currently unknown because of a lack of experimental data and approaches. However, if epithelial cells could encode and tune frequency, duration, and waveform in mechanical forces, it would greatly increase the parameter space, allowing epithelial cells to build complexity. Future development of techniques to measure cell-generated mechanical force will provide formal assessment of these biophysical variables in regulating epithelial cell–cell interactions and tissue behaviors.
Acknowledgments
The author apologizes for not being able to cite all pertinent literature.
Editorial Note on the Review Process
F1000 Faculty Reviews are commissioned from members of the prestigious F1000 Faculty and are edited as a service to readers. In order to make these reviews as comprehensive and accessible as possible, the referees provide input before publication and only the final, revised version is published. The referees who approved the final version are listed with their names and affiliations but without their reports on earlier versions (any comments will already have been addressed in the published version).
The referees who approved this article are:
Alpha Yap, Division of Cell Biology and Molecular Medicine, Institute for Molecular Bioscience, The University of Queensland, Brisbane, QLD, Australia
Alex Mogilner, Courant Institute and Department of Biology, New York University, New York, NY, USA
Funding Statement
The author is supported by the National Institute of Diabetes and Digestive and Kidney Diseases, the National Institutes of Health (R01 DK098398).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; referees: 2 approved]
References
- 1. Kim Y, Hazar M, Vijayraghavan DS, et al. : Mechanochemical actuators of embryonic epithelial contractility. Proc Natl Acad Sci U S A. 2014;111(40):14366–71. 10.1073/pnas.1405209111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jackson TR, Kim HY, Balakrishnan UL, et al. : Spatiotemporally Controlled Mechanical Cues Drive Progenitor Mesenchymal-to-Epithelial Transition Enabling Proper Heart Formation and Function. Curr Biol. 2017;27(9):1326–35. 10.1016/j.cub.2017.03.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Acharya BR, Yap AS: Pli Selon Pli: Mechanochemical Feedback and the Morphogenetic Role of Contractility at Cadherin Cell-Cell Junctions. Curr Top Dev Biol. 2016;117:631–46. 10.1016/bs.ctdb.2015.10.021 [DOI] [PubMed] [Google Scholar]
- 4. Dasbiswas K, Hannezo E, Gov NS: Theory of Epithelial Cell Shape Transitions Induced by Mechanoactive Chemical Gradients. Biophys J. 2018;114(4):968–77. 10.1016/j.bpj.2017.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gross P, Kumar KV, Grill SW: How Active Mechanics and Regulatory Biochemistry Combine to Form Patterns in Development. Annu Rev Biophys. 2017;46:337–56. 10.1146/annurev-biophys-070816-033602 [DOI] [PubMed] [Google Scholar]
- 6. Petridou NI, Spiró Z, Heisenberg CP: Multiscale force sensing in development. Nat Cell Biol. 2017;19(6):581–8. 10.1038/ncb3524 [DOI] [PubMed] [Google Scholar]
- 7. Wickström SA, Niessen CM: Cell adhesion and mechanics as drivers of tissue organization and differentiation: local cues for large scale organization. Curr Opin Cell Biol. 2018;54:89–97. 10.1016/j.ceb.2018.05.003 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 8. Martino F, Perestrelo AR, Vinarský V, et al. : Cellular Mechanotransduction: From Tension to Function. Front Physiol. 2018;9: 824. 10.3389/fphys.2018.00824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yusko EC, Asbury CL: Force is a signal that cells cannot ignore. Mol Biol Cell. 2014;25(23):3717–25. 10.1091/mbc.E13-12-0707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Siedlik MJ, Varner VD, Nelson CM: Pushing, pulling, and squeezing our way to understanding mechanotransduction. Methods. 2016;94:4–12. 10.1016/j.ymeth.2015.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Haase K, Pelling AE: Investigating cell mechanics with atomic force microscopy. J R Soc Interface. 2015;12(104): 20140970. 10.1098/rsif.2014.0970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Harris AR, Daeden A, Charras GT: Formation of adherens junctions leads to the emergence of a tissue-level tension in epithelial monolayers. J Cell Sci. 2014;127(Pt 11):2507–17. 10.1242/jcs.142349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Collins C, Denisin AK, Pruitt BL, et al. : Changes in E-cadherin rigidity sensing regulate cell adhesion. Proc Natl Acad Sci U S A. 2017;114(29):E5835–E5844. 10.1073/pnas.1618676114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Przybyla L, Lakins JN, Sunyer R, et al. : Monitoring developmental force distributions in reconstituted embryonic epithelia. Methods. 2016;94:101–13. 10.1016/j.ymeth.2015.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stapleton SC, Chopra A, Chen CS: Force measurement tools to explore cadherin mechanotransduction. Cell Commun Adhes. 2014;21(3):193–205. 10.3109/15419061.2014.905929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Campàs O, Mammoto T, Hasso S, et al. : Quantifying cell-generated mechanical forces within living embryonic tissues. Nat Methods. 2014;11(2):183–9. 10.1038/nmeth.2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Campàs O: A toolbox to explore the mechanics of living embryonic tissues. Semin Cell Dev Biol. 2016;55:119–30. 10.1016/j.semcdb.2016.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Serwane F, Mongera A, Rowghanian P, et al. : In vivo quantification of spatially varying mechanical properties in developing tissues. Nat Methods. 2017;14(2):181–6. 10.1038/nmeth.4101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rübsam M, Mertz AF, Kubo A, et al. : E-cadherin integrates mechanotransduction and EGFR signaling to control junctional tissue polarization and tight junction positioning. Nat Commun. 2017;8(1): 1250. 10.1038/s41467-017-01170-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cartagena-Rivera AX, Van Itallie CM, Anderson JM, et al. : Apical surface supracellular mechanical properties in polarized epithelium using noninvasive acoustic force spectroscopy. Nat Commun. 2017;8(1): 1030. 10.1038/s41467-017-01145-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Freikamp A, Mehlich A, Klingner C, et al. : Investigating piconewton forces in cells by FRET-based molecular force microscopy. J Struct Biol. 2017;197(1):37–42. 10.1016/j.jsb.2016.03.011 [DOI] [PubMed] [Google Scholar]
- 22. Tang X, Tofangchi A, Anand SV, et al. : A novel cell traction force microscopy to study multi-cellular system. PLoS Comput Biol. 2014;10(6):e1003631. 10.1371/journal.pcbi.1003631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bosch-Fortea M, Martín-Belmonte F: Mechanosensitive adhesion complexes in epithelial architecture and cancer onset. Curr Opin Cell Biol. 2018;50:42–9. 10.1016/j.ceb.2018.01.013 [DOI] [PubMed] [Google Scholar]
- 24. Malinova TS, Huveneers S: Sensing of Cytoskeletal Forces by Asymmetric Adherens Junctions. Trends Cell Biol. 2018;28(4):328–41. 10.1016/j.tcb.2017.11.002 [DOI] [PubMed] [Google Scholar]
- 25. Steinbacher T, Ebnet K: The regulation of junctional actin dynamics by cell adhesion receptors. Histochem Cell Biol. 2018;150(4):341–350. 10.1007/s00418-018-1691-8 [DOI] [PubMed] [Google Scholar]
- 26. Leckband DE, de Rooij J: Cadherin adhesion and mechanotransduction. Annu Rev Cell Dev Biol. 2014;30:291–315. 10.1146/annurev-cellbio-100913-013212 [DOI] [PubMed] [Google Scholar]
- 27. Sayyad WA, Amin L, Fabris P, et al. : The role of myosin-II in force generation of DRG filopodia and lamellipodia. Sci Rep. 2015;5: 7842. 10.1038/srep07842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Prass M, Jacobson K, Mogilner A, et al. : Direct measurement of the lamellipodial protrusive force in a migrating cell. J Cell Biol. 2006;174(6):767–72. 10.1083/jcb.200601159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Footer MJ, Kerssemakers JW, Theriot JA, et al. : Direct measurement of force generation by actin filament polymerization using an optical trap. Proc Natl Acad Sci U S A. 2007;104(7):2181–6. 10.1073/pnas.0607052104 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 30. Krendel MF, Bonder EM: Analysis of actin filament bundle dynamics during contact formation in live epithelial cells. Cell Motil Cytoskeleton. 1999;43(4):296–309. [DOI] [PubMed] [Google Scholar]
- 31. Homem CCF, Peifer M: Diaphanous regulates myosin and adherens junctions to control cell contractility and protrusive behavior during morphogenesis. Development. 2008;135(6 ):1005–18. 10.1242/dev.016337 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 32. Campàs O, Mahadevan L, Joanny JF: Actin network growth under load. Biophys J. 2012;102(5):1049–58. 10.1016/j.bpj.2012.01.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhu J, Mogilner A: Mesoscopic model of actin-based propulsion. PLoS Comput Biol. 2012;8(11):e1002764. 10.1371/journal.pcbi.1002764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schaus TE, Borisy GG: Performance of a population of independent filaments in lamellipodial protrusion. Biophys J. 2008;95(3):1393–411. 10.1529/biophysj.107.125005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dimchev G, Steffen A, Kage F, et al. : Efficiency of lamellipodia protrusion is determined by the extent of cytosolic actin assembly. Mol Biol Cell. 2017;28(10):1311–25. 10.1091/mbc.E16-05-0334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Reymann AC, Suarez C, Guérin C, et al. : Turnover of branched actin filament networks by stochastic fragmentation with ADF/cofilin. Mol Biol Cell. 2011;22(14):2541–50. 10.1091/mbc.E11-01-0052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Leijnse N, Oddershede LB, Bendix PM: Helical buckling of actin inside filopodia generates traction. Proc Natl Acad Sci U S A. 2015;112(1):136–41. 10.1073/pnas.1411761112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ryan GL, Holz D, Yamashiro S, et al. : Cell protrusion and retraction driven by fluctuations in actin polymerization: A two-dimensional model. Cytoskeleton (Hoboken). 2017;74(12):490–503. 10.1002/cm.21389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Peukes J, Betz T: Direct measurement of the cortical tension during the growth of membrane blebs. Biophys J. 2014;107(8):1810–20. 10.1016/j.bpj.2014.07.076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bornschlögl T, Romero S, Vestergaard CL, et al. : Filopodial retraction force is generated by cortical actin dynamics and controlled by reversible tethering at the tip. Proc Natl Acad Sci U S A. 2013;110(47):18928–33. 10.1073/pnas.1316572110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mège RM, Gavard J, Lambert M: Regulation of cell-cell junctions by the cytoskeleton. Curr Opin Cell Biol. 2006;18(5):541–8. 10.1016/j.ceb.2006.08.004 [DOI] [PubMed] [Google Scholar]
- 42. Ashdown GW, Burn GL, Williamson DJ, et al. : Live-Cell Super-resolution Reveals F-Actin and Plasma Membrane Dynamics at the T Cell Synapse. Biophys J. 2017;112(8):1703–13. 10.1016/j.bpj.2017.01.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chew TG, Huang J, Palani S, et al. : Actin turnover maintains actin filament homeostasis during cytokinetic ring contraction. J Cell Biol. 2017;216(9):2657–67. 10.1083/jcb.201701104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bidone TC, Jung W, Maruri D, et al. : Morphological Transformation and Force Generation of Active Cytoskeletal Networks. PLoS Comput Biol. 2017;13(1):e1005277. 10.1371/journal.pcbi.1005277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Oelz D, Mogilner A: Actomyosin contraction, aggregation and traveling waves in a treadmilling actin array. Physica D. 2016;318–319:70–83. 10.1016/j.physd.2015.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Oelz DB, Rubinstein BY, Mogilner A: A Combination of Actin Treadmilling and Cross-Linking Drives Contraction of Random Actomyosin Arrays. Biophys J. 2015;109(9):1818–29. 10.1016/j.bpj.2015.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zimmermann D, Homa KE, Hocky GM, et al. : Mechanoregulated inhibition of formin facilitates contractile actomyosin ring assembly. Nat Commun. 2017;8(1): 703. 10.1038/s41467-017-00445-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Luxenburg C, Heller E, Pasolli HA, et al. : Wdr1-mediated cell shape dynamics and cortical tension are essential for epidermal planar cell polarity. Nat Cell Biol. 2015;17(5):592–604. 10.1038/ncb3146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Charras G, Yap AS: Tensile Forces and Mechanotransduction at Cell-Cell Junctions. Curr Biol. 2018;28(8):R445–R457. 10.1016/j.cub.2018.02.003 [DOI] [PubMed] [Google Scholar]
- 50. Sluysmans S, Vasileva E, Spadaro D, et al. : The role of apical cell-cell junctions and associated cytoskeleton in mechanotransduction. Biol Cell. 2017;109(4):139–61. 10.1111/boc.201600075 [DOI] [PubMed] [Google Scholar]
- 51. Wyatt T, Baum B, Charras G: A question of time: tissue adaptation to mechanical forces. Curr Opin Cell Biol. 2016;38:68–73. 10.1016/j.ceb.2016.02.012 [DOI] [PubMed] [Google Scholar]
- 52. Khalilgharibi N, Fouchard J, Recho P, et al. : The dynamic mechanical properties of cellularised aggregates. Curr Opin Cell Biol. 2016;42:113–20. 10.1016/j.ceb.2016.06.003 [DOI] [PubMed] [Google Scholar]
- 53. Tambe DT, Hardin CC, Angelini TE, et al. : Collective cell guidance by cooperative intercellular forces. Nat Mater. 2011;10(6):469–75. 10.1038/nmat3025 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 54. Friedl P, Mayor R: Tuning Collective Cell Migration by Cell-Cell Junction Regulation. Cold Spring Harb Perspect Biol. 2017;9(4): pii: a029199. 10.1101/cshperspect.a029199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Danjo Y, Gipson IK: Actin 'purse string' filaments are anchored by E-cadherin-mediated adherens junctions at the leading edge of the epithelial wound, providing coordinated cell movement. J Cell Sci. 1998;111(Pt 22):3323–32. [DOI] [PubMed] [Google Scholar]
- 56. Duque J, Gorfinkiel N: Integration of actomyosin contractility with cell-cell adhesion during dorsal closure. Development. 2016;143(24):4676–86. 10.1242/dev.136127 [DOI] [PubMed] [Google Scholar]
- 57. Zulueta-Coarasa T, Fernandez-Gonzalez R: Tension (re)builds: Biophysical mechanisms of embryonic wound repair. Mech Dev. 2017;144(Pt A):43–52. 10.1016/j.mod.2016.11.004 [DOI] [PubMed] [Google Scholar]
- 58. Begnaud S, Chen T, Delacour D, et al. : Mechanics of epithelial tissues during gap closure. Curr Opin Cell Biol. 2016;42:52–62. 10.1016/j.ceb.2016.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Fadul J, Rosenblatt J: The forces and fates of extruding cells. Curr Opin Cell Biol. 2018;54:66–71. 10.1016/j.ceb.2018.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yano T, Kanoh H, Tamura A, et al. : Apical cytoskeletons and junctional complexes as a combined system in epithelial cell sheets. Ann N Y Acad Sci. 2017;1405(1):32–43. 10.1111/nyas.13432 [DOI] [PubMed] [Google Scholar]
- 61. Arnold TR, Stephenson RE, Miller AL: Rho GTPases and actomyosin: Partners in regulating epithelial cell-cell junction structure and function. Exp Cell Res. 2017;358(1):20–30. 10.1016/j.yexcr.2017.03.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Grikscheit K, Grosse R: Formins at the Junction. Trends Biochem Sci. 2016;41(2):148–59. 10.1016/j.tibs.2015.12.002 [DOI] [PubMed] [Google Scholar]
- 63. Tang V: Cell-cell adhesion interface: rise of the lateral membrane [version 1; referees: 2 approved]. F1000Res. 2017;6(F1000 Faculty Rev):276. 10.12688/f1000research.10680.1 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 64. Vahey MD, Fletcher DA: The biology of boundary conditions: cellular reconstitution in one, two, and three dimensions. Curr Opin Cell Biol. 2014;26:60–8. 10.1016/j.ceb.2013.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hadjivasiliou Z, Hunter GL, Baum B: A new mechanism for spatial pattern formation via lateral and protrusion-mediated lateral signalling. J R Soc Interface. 2016;13(124): pii: 20160484. 10.1098/rsif.2016.0484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Vasilopoulos G, Painter KJ: Pattern formation in discrete cell tissues under long range filopodia-based direct cell to cell contact. Math Biosci. 2016;273:1–15. 10.1016/j.mbs.2015.12.008 [DOI] [PubMed] [Google Scholar]
- 67. Ladoux B, Nelson WJ, Yan J, et al. : The mechanotransduction machinery at work at adherens junctions. Integr Biol (Camb). 2015;7(10):1109–19. 10.1039/c5ib00070j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Mao Y, Baum B: Tug of war--the influence of opposing physical forces on epithelial cell morphology. Dev Biol. 2015;401(1):92–102. 10.1016/j.ydbio.2014.12.030 [DOI] [PubMed] [Google Scholar]
- 69. Lappalainen P: Actin-binding proteins: the long road to understanding the dynamic landscape of cellular actin networks. Mol Biol Cell. 2016;27(16):2519–22. 10.1091/mbc.E15-10-0728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Biswas KH, Hartman KL, Yu CH, et al. : E-cadherin junction formation involves an active kinetic nucleation process. Proc Natl Acad Sci U S A. 2015;112(35):10932–7. 10.1073/pnas.1513775112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yang C, Hoelzle M, Disanza A, et al. : Coordination of membrane and actin cytoskeleton dynamics during filopodia protrusion. PLoS One. 2009;4(5):e5678. 10.1371/journal.pone.0005678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Pyrpassopoulos S, Feeser EA, Mazerik JN, et al. : Membrane-bound myo1c powers asymmetric motility of actin filaments. Curr Biol. 2012;22(18):1688–92. 10.1016/j.cub.2012.06.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ricca BL, Venugopalan G, Fletcher DA: To pull or be pulled: parsing the multiple modes of mechanotransduction. Curr Opin Cell Biol. 2013;25(5):558–64. 10.1016/j.ceb.2013.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Vogel SK, Greiss F, Khmelinskaia A, et al. : Control of lipid domain organization by a biomimetic contractile actomyosin cortex. eLife. 2017;6: pii: e24350. 10.7554/eLife.24350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Efimova N, Svitkina TM: Branched actin networks push against each other at adherens junctions to maintain cell-cell adhesion. J Cell Biol. 2018;217(5):1827–45. 10.1083/jcb.201708103 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 76. Carlsson AE: Membrane bending by actin polymerization. Curr Opin Cell Biol. 2018;50:1–7. 10.1016/j.ceb.2017.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Mykuliak VV, Haining AWM, von Essen M, et al. : Mechanical unfolding reveals stable 3-helix intermediates in talin and α-catenin. PLoS Comput Biol. 2018;14(4):e1006126. 10.1371/journal.pcbi.1006126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Yao M, Qiu W, Liu R, et al. : Force-dependent conformational switch of α-catenin controls vinculin binding. Nat Commun. 2014;5: 4525. 10.1038/ncomms5525 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 79. Manibog K, Yen CF, Sivasankar S: Measuring Force-Induced Dissociation Kinetics of Protein Complexes Using Single-Molecule Atomic Force Microscopy. Meth Enzymol. 2017;582:297–320. 10.1016/bs.mie.2016.08.009 [DOI] [PubMed] [Google Scholar]
- 80. Rakshit S, Zhang Y, Manibog K, et al. : Ideal, catch, and slip bonds in cadherin adhesion. Proc Natl Acad Sci U S A. 2012;109(46):18815–20. 10.1073/pnas.1208349109 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 81. Nunes-Alves A, Arantes GM: Mechanical Unfolding of Macromolecules Coupled to Bond Dissociation. J Chem Theory Comput. 2018;14(1):282–90. 10.1021/acs.jctc.7b00805 [DOI] [PubMed] [Google Scholar]
- 82. Seifert C, Gräter F: Protein mechanics: How force regulates molecular function. Biochim Biophys Acta. 2013;1830(10):4762–8. 10.1016/j.bbagen.2013.06.005 [DOI] [PubMed] [Google Scholar]
- 83. Agosti A, Ambrosi D, Turzi S: Strain energy storage and dissipation rate in active cell mechanics. Phys Rev E. 2018;97(5–1):052410. 10.1103/PhysRevE.97.052410 [DOI] [PubMed] [Google Scholar]
- 84. Breslin JW, Zhang XE, Worthylake RA, et al. : Involvement of local lamellipodia in endothelial barrier function. PLoS One. 2015;10(2):e0117970. 10.1371/journal.pone.0117970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Petersen EN, Chung HW, Nayebosadri A, et al. : Kinetic disruption of lipid rafts is a mechanosensor for phospholipase D. Nat Commun. 2016;7: 13873. 10.1038/ncomms13873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Gauvin TJ, Young LE, Higgs HN: The formin FMNL3 assembles plasma membrane protrusions that participate in cell-cell adhesion. Mol Biol Cell. 2015;26(3):467–77. 10.1091/mbc.E14-07-1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Köster DV, Husain K, Iljazi E, et al. : Actomyosin dynamics drive local membrane component organization in an in vitro active composite layer. Proc Natl Acad Sci U S A. 2016;113(12):E1645–54. 10.1073/pnas.1514030113 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 88. Lemière J, Valentino F, Campillo C, et al. : How cellular membrane properties are affected by the actin cytoskeleton. Biochimie. 2016;130:33–40. 10.1016/j.biochi.2016.09.019 [DOI] [PubMed] [Google Scholar]
- 89. Mesarec L, Góźdź W, Kralj S, et al. : On the role of external force of actin filaments in the formation of tubular protrusions of closed membrane shapes with anisotropic membrane components. Eur Biophys J. 2017;46(8):705–18. 10.1007/s00249-017-1212-z [DOI] [PubMed] [Google Scholar]
- 90. Letort G, Politi AZ, Ennomani H, et al. : Geometrical and mechanical properties control actin filament organization. PLoS Comput Biol. 2015;11(5):e1004245. 10.1371/journal.pcbi.1004245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Dmitrieff S, Nédélec F: Amplification of actin polymerization forces. J Cell Biol. 2016;212(7):763–6. 10.1083/jcb.201512019 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 92. Boujemaa-Paterski R, Galland R, Suarez C, et al. : Directed actin assembly and motility. Methods Enzymol. 2014;540:283–300. 10.1016/B978-0-12-397924-7.00016-9 [DOI] [PubMed] [Google Scholar]
- 93. Pontes B, Monzo P, Gauthier NC: Membrane tension: A challenging but universal physical parameter in cell biology. Semin Cell Dev Biol. 2017;71:30–41. 10.1016/j.semcdb.2017.08.030 [DOI] [PubMed] [Google Scholar]
- 94. Pontes B, Monzo P, Gole L, et al. : Membrane tension controls adhesion positioning at the leading edge of cells. J Cell Biol. 2017;216(9):2959–77. 10.1083/jcb.201611117 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 95. Diz-Muñoz A, Fletcher DA, Weiner OD: Use the force: membrane tension as an organizer of cell shape and motility. Trends Cell Biol. 2013;23(2):47–53. 10.1016/j.tcb.2012.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Anishkin A, Loukin SH, Teng J, et al. : Feeling the hidden mechanical forces in lipid bilayer is an original sense. Proc Natl Acad Sci U S A. 2014;111(22):7898–905. 10.1073/pnas.1313364111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Priya R, Yap AS: Active tension: the role of cadherin adhesion and signaling in generating junctional contractility. Curr Top Dev Biol. 2015;112:65–102. 10.1016/bs.ctdb.2014.11.016 [DOI] [PubMed] [Google Scholar]
- 98. Ma R, Berro J: Structural organization and energy storage in crosslinked actin assemblies. PLoS Comput Biol. 2018;14(5):e1006150. 10.1371/journal.pcbi.1006150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Mak M, Zaman MH, Kamm RD, et al. : Interplay of active processes modulates tension and drives phase transition in self-renewing, motor-driven cytoskeletal networks. Nat Commun. 2016;7: 10323. 10.1038/ncomms10323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Romet-Lemonne G, Jégou A: Mechanotransduction down to individual actin filaments. Eur J Cell Biol. 2013;92(10–11):333–8. 10.1016/j.ejcb.2013.10.011 [DOI] [PubMed] [Google Scholar]
- 101. Jégou A, Romet-Lemonne G: Single Filaments to Reveal the Multiple Flavors of Actin. Biophys J. 2016;110(10):2138–46. 10.1016/j.bpj.2016.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Sadhu RK, Chatterjee S: Actin filaments growing against an elastic membrane: Effect of membrane tension. Phys Rev E. 2018;97(3–1):032408. 10.1103/PhysRevE.97.032408 [DOI] [PubMed] [Google Scholar]
- 103. Johannes L, Pezeshkian W, Ipsen JH, et al. : Clustering on Membranes: Fluctuations and More. Trends Cell Biol. 2018;28(5):405–15. 10.1016/j.tcb.2018.01.009 [DOI] [PubMed] [Google Scholar]
- 104. Lieber AD, Yehudai-Resheff S, Barnhart EL, et al. : Membrane tension in rapidly moving cells is determined by cytoskeletal forces. Curr Biol. 2013;23(15):1409–17. 10.1016/j.cub.2013.05.063 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 105. Chugh P, Clark AG, Smith MB, et al. : Actin cortex architecture regulates cell surface tension. Nat Cell Biol. 2017;19(6):689–97. 10.1038/ncb3525 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 106. Venit T, Kalendová A, Petr M, et al. : Nuclear myosin I regulates cell membrane tension. Sci Rep. 2016;6: 30864. 10.1038/srep30864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Smith AS, Nowak RB, Zhou S, et al. : Myosin IIA interacts with the spectrin-actin membrane skeleton to control red blood cell membrane curvature and deformability. Proc Natl Acad Sci U S A. 2018;115(19):E4377–E4385. 10.1073/pnas.1718285115 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 108. Simon C, Caorsi V, Campillo C, et al. : Interplay between membrane tension and the actin cytoskeleton determines shape changes. Phys Biol. 2018;15(6):065004. 10.1088/1478-3975/aad1ab [DOI] [PubMed] [Google Scholar]
- 109. Biswas A, Alex A, Sinha B: Mapping Cell Membrane Fluctuations Reveals Their Active Regulation and Transient Heterogeneities. Biophys J. 2017;113(8):1768–81. 10.1016/j.bpj.2017.08.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Delanoë-Ayari H, Al Kurdi R, Vallade M, et al. : Membrane and acto-myosin tension promote clustering of adhesion proteins. Proc Natl Acad Sci U S A. 2004;101(8):2229–34. 10.1073/pnas.0304297101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Pontani LL, Jorjadze I, Brujic J: Cis and Trans Cooperativity of E-Cadherin Mediates Adhesion in Biomimetic Lipid Droplets. Biophys J. 2016;110(2):391–9. 10.1016/j.bpj.2015.11.3514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Strale PO, Duchesne L, Peyret G, et al. : The formation of ordered nanoclusters controls cadherin anchoring to actin and cell-cell contact fluidity. J Cell Biol. 2015;210(2):333–46. 10.1083/jcb.201410111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Kannan N, Tang VW: Myosin-1c promotes E-cadherin tension and force-dependent recruitment of α-actinin to the epithelial cell junction. J Cell Sci. 2018;131(12): pii: jcs211334. 10.1242/jcs.211334 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 114. Weirich KL, Banerjee S, Dasbiswas K, et al. : Liquid behavior of cross-linked actin bundles. Proc Natl Acad Sci U S A. 2017;114(9):2131–6. 10.1073/pnas.1616133114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Carvalho K, Lemière J, Faqir F, et al. : Actin polymerization or myosin contraction: two ways to build up cortical tension for symmetry breaking. Philos Trans R Soc Lond B Biol Sci. 2013;368(1629): 20130005. 10.1098/rstb.2013.0005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Hayakawa K, Tatsumi H, Sokabe M: Actin filaments function as a tension sensor by tension-dependent binding of cofilin to the filament. J Cell Biol. 2011;195(5):721–7. 10.1083/jcb.201102039 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 117. Yamashiro S, Tanaka S, McMillen LM, et al. : Myosin-dependent actin stabilization as revealed by single-molecule imaging of actin turnover. Mol Biol Cell. 2018;29(16):1941–7. 10.1091/mbc.E18-01-0061 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 118. De La Cruz EM, Gardel ML: Actin Mechanics and Fragmentation. J Biol Chem. 2015;290(28):17137–44. 10.1074/jbc.R115.636472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Reymann AC, Boujemaa-Paterski R, Martiel JL, et al. : Actin network architecture can determine myosin motor activity. Science. 2012;336(6086):1310–4. 10.1126/science.1221708 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 120. Ennomani H, Letort G, Guérin C, et al. : Architecture and Connectivity Govern Actin Network Contractility. Curr Biol. 2016;26(5):616–26. 10.1016/j.cub.2015.12.069 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 121. Vasquez CG, Martin AC: Force transmission in epithelial tissues. Dev Dyn. 2016;245(3):361–71. 10.1002/dvdy.24384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. McFadden WM, McCall PM, Gardel ML, et al. : Filament turnover tunes both force generation and dissipation to control long-range flows in a model actomyosin cortex. PLoS Comput Biol. 2017;13(12):e1005811. 10.1371/journal.pcbi.1005811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Rottner K, Kage F: Actin Networks: Adapting to Load through Geometry. Curr Biol. 2017;27(23):R1274–R1277. 10.1016/j.cub.2017.10.042 [DOI] [PubMed] [Google Scholar]
- 124. Kobb AB, Zulueta-Coarasa T, Fernandez-Gonzalez R: Tension regulates myosin dynamics during Drosophila embryonic wound repair. J Cell Sci. 2017;130(4):689–96. 10.1242/jcs.196139 [DOI] [PubMed] [Google Scholar]
- 125. Yap AS, Duszyc K, Viasnoff V: Mechanosensing and Mechanotransduction at Cell-Cell Junctions. Cold Spring Harb Perspect Biol. 2018;10(8): pii: a028761. 10.1101/cshperspect.a028761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Wu SK, Budnar S, Yap AS, et al. : Pulsatile contractility of actomyosin networks organizes the cellular cortex at lateral cadherin junctions. Eur J Cell Biol. 2014;93(10–12):396–404. 10.1016/j.ejcb.2014.09.001 [DOI] [PubMed] [Google Scholar]
- 127. Dasanayake NL, Carlsson AE: Stress generation by myosin minifilaments in actin bundles. Phys Biol. 2013;10(3):36006. 10.1088/1478-3975/10/3/036006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Haase K, Pelling AE: Resiliency of the plasma membrane and actin cortex to large-scale deformation. Cytoskeleton (Hoboken). 2013;70(9):494–514. 10.1002/cm.21129 [DOI] [PubMed] [Google Scholar]
- 129. Tsai FC, Koenderink GH: Shape control of lipid bilayer membranes by confined actin bundles. Soft Matter. 2015;11(45):8834–47. 10.1039/c5sm01583a [DOI] [PubMed] [Google Scholar]
- 130. Pyrpassopoulos S, Arpağ G, Feeser EA, et al. : Force Generation by Membrane-Associated Myosin-I. Sci Rep. 2016;6: 25524. 10.1038/srep25524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Pontes B, Ayala Y, Fonseca AC, et al. : Membrane elastic properties and cell function. PLoS One. 2013;8(7):e67708. 10.1371/journal.pone.0067708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Pontes B, Viana NB, Salgado LT, et al. : Cell cytoskeleton and tether extraction. Biophys J. 2011;101(1):43–52. 10.1016/j.bpj.2011.05.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Trichet L, Campàs O, Sykes C, et al. : VASP governs actin dynamics by modulating filament anchoring. Biophys J. 2007;92(3):1081–9. 10.1529/biophysj.106.091884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Simunovic M, Voth GA: Membrane tension controls the assembly of curvature-generating proteins. Nat Commun. 2015;6: 7219. 10.1038/ncomms8219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. McConnell RE, Tyska MJ: Leveraging the membrane - cytoskeleton interface with myosin-1. Trends Cell Biol. 2010;20(7):418–26. 10.1016/j.tcb.2010.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Kaya M, Tani Y, Washio T, et al. : Coordinated force generation of skeletal myosins in myofilaments through motor coupling. Nat Commun. 2017;8: 16036. 10.1038/ncomms16036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Bendix PM, Koenderink GH, Cuvelier D, et al. : A quantitative analysis of contractility in active cytoskeletal protein networks. Biophys J. 2008;94(8):3126–36. 10.1529/biophysj.107.117960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Dumont S, Prakash M: Emergent mechanics of biological structures. Mol Biol Cell. 2014;25(22):3461–5. 10.1091/mbc.E14-03-0784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Lecuit T, Yap AS: E-cadherin junctions as active mechanical integrators in tissue dynamics. Nat Cell Biol. 2015;17(5):533–9. 10.1038/ncb3136 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 140. Liu Z, Tan JL, Cohen DM, et al. : Mechanical tugging force regulates the size of cell-cell junctions. Proc Natl Acad Sci U S A. 2010;107(22):9944–9. 10.1073/pnas.0914547107 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 141. Leerberg JM, Gomez GA, Verma S, et al. : Tension-sensitive actin assembly supports contractility at the epithelial zonula adherens. Curr Biol. 2014;24(15):1689–99. 10.1016/j.cub.2014.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Cojoc D, Difato F, Ferrari E, et al. : Properties of the force exerted by filopodia and lamellipodia and the involvement of cytoskeletal components. PLoS One. 2007;2(10):e1072. 10.1371/journal.pone.0001072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Shahapure R, Difato F, Laio A, et al. : Force generation in lamellipodia is a probabilistic process with fast growth and retraction events. Biophys J. 2010;98(6):979–88. 10.1016/j.bpj.2009.11.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Spadaro D, Le S, Laroche T, et al. : Tension-Dependent Stretching Activates ZO-1 to Control the Junctional Localization of Its Interactors. Curr Biol. 2017;27(24):3783–3795.e8. 10.1016/j.cub.2017.11.014 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 145. Fujiwara I, Suetsugu S, Uemura S, et al. : Visualization and force measurement of branching by Arp2/3 complex and N-WASP in actin filament. Biochem Biophys Res Commun. 2002;293(5):1550–5. 10.1016/S0006-291X(02)00421-7 [DOI] [PubMed] [Google Scholar]
- 146. Bajpai S, Feng Y, Wirtz D, et al. : β-Catenin serves as a clutch between low and high intercellular E-cadherin bond strengths. Biophys J. 2013;105(10):2289–300. 10.1016/j.bpj.2013.09.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Bajpai S, Feng Y, Krishnamurthy R, et al. : Loss of alpha-catenin decreases the strength of single E-cadherin bonds between human cancer cells. J Biol Chem. 2009;284(27):18252–9. 10.1074/jbc.M109.000661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Yao M, Goult BT, Chen H, et al. : Mechanical activation of vinculin binding to talin locks talin in an unfolded conformation. Sci Rep. 2014;4: 4610. 10.1038/srep04610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Kovar DR, Pollard TD: Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc Natl Acad Sci U S A. 2004;101(41):14725–30. 10.1073/pnas.0405902101 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 150. Stadler RV, White LA, Hu K, et al. : Direct measurement of cortical force generation and polarization in a living parasite. Mol Biol Cell. 2017;28(14):1912–23. 10.1091/mbc.E16-07-0518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Chuan P, Spudich JA, Dunn AR: Robust mechanosensing and tension generation by myosin VI. J Mol Biol. 2011;405(1):105–12. 10.1016/j.jmb.2010.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Koenderink GH, Dogic Z, Nakamura F, et al. : An active biopolymer network controlled by molecular motors. Proc Natl Acad Sci U S A. 2009;106(36):15192–7. 10.1073/pnas.0903974106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Démoulin D, Carlier MF, Bibette J, et al. : Power transduction of actin filaments ratcheting in vitro against a load. Proc Natl Acad Sci U S A. 2014;111(50):17845–50. 10.1073/pnas.1414184111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Fritzsche M, Li D, Colin-York H, et al. : Self-organizing actin patterns shape membrane architecture but not cell mechanics. Nat Commun. 2017;8: 14347. 10.1038/ncomms14347 [DOI] [PMC free article] [PubMed] [Google Scholar]