Abstract
A field-collected Aphis gossypii clone [Kushima resistant (KR) clone] was resistant to neonicotinoid insecticides (23.8- to 394-fold). RNA-seq and next-generation sequence analyses were conducted to identify nine cytochrome P450 (CYP) genes that were significantly upregulated in the KR clone as compared with those in the insecticide-susceptible clone. A. gossypii P450s were transiently and efficiently expressed in S2 cell to show that CYP6CY22 (c21228) and CYP6CY13 (c21368), which were the most upregulated of the nine P450s in the KR clone, did not degrade sulfoxaflor, a new class of insecticides acting on insect nAChRs, but markedly metabolized all of the neonicotinoids tested. Hence, P450s are likely to underpin neonicotinoid resistance in other aphids as well in the future, and the P450 expression protocol established here will prompt studies on P450-medidated insecticide resistance and structural analyses of relevant metabolites.
Keywords: Aphis gossypii, neonicotinoid, insecticide resistance, metabolism, cytochrome P450
Introduction
The cotton aphid (Aphis gossypii) is an important sucking pest that causes severe crop losses in fields and greenhouses. Neonicotinoid insecticides (Fig. 1), which achieve excellent control of A. gossypii, target nicotinic acetylcholine (ACh) receptors (nAChRs) of insects.1–4) Resistance to neonicotinoid insecticides has developed relatively slowly; however, it is recognized as an important issue,5) particularly because of the resistance of numerous species such as whiteflies (Bemisia tabaci and Trialeurodes vaporariorum),5–7) brown planthoppers (Nilaparvata lugens),8) Colorado potato beetles (Leptinotarsa decemlineata),9) and western flower thrips (Frankliniella occidentalis).10) Resistance to neonicotinoid insecticides is attributed in some cases to mutations in nAChRs or to increased rates of insecticide detoxification. Target-site insensitivity occurs in field-caught peach potato aphids (Myzus persicae) and A. gossypii.11,12) These aphids harbor a point mutation (R81T) in the loop D region of the nAChR β1 subunit. Moreover, numerous studies indicate that R81 within this loop is a key determinant required for the binding of neonicotinoid insecticides to nAChRs.13–15)
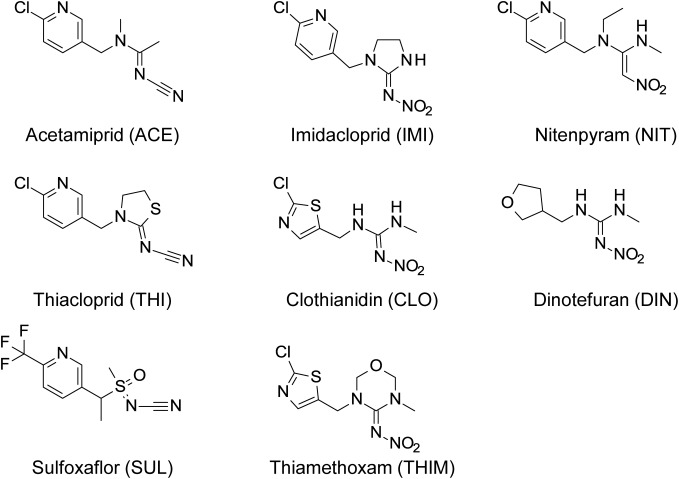
Fig. 1. Structure of acetamiprid, other neonicotinoids, and sulfoxaflor.
In most cases, cytochrome P450-mediated detoxification plays a primary role in the resistance to insecticides of diverse insects.9,16,17) Cytochrome P450 (CYP) occurs widely in nature and plays roles in many biological processes, such as hormone synthesis and the metabolism of xenobiotics. In insects, P450 is implicated in resistance to insecticides through the degradation of these foreign compounds to more soluble and less toxic forms.17) This is accomplished via an increase in P450 expression or structural changes that may alter substrate-specificity profiles or catalytic activities.18–23)
We previously characterized a field-isolated A. gossypii [Kushima resistant (KR) clone] that is resistant to nicotinoid insecticides (23.8- to 394-fold) conferred by the nAChR β1 subunit R81T mutation.12) Further, piperonyl butoxide (PBO) pretreatment reduces LC50 values of neonicotinoid insecticides through synergizing with acetamiprid and imidacloprid (synergistic factors=3.6 and 6.0, respectively). Therefore, we used next-generation sequencing (NGS) to identify P450 family members involved in the metabolism of neonicotinoid insecticides. Further, we molecularly cloned P450 cDNAs of A. gossypii KR clone and expressed. The functional recombinant P450s provide further evidence of the involvement of P450s in the metabolism of neonicotinoid insecticides.
Materials and Methods
1. Insects
KR and Miyazaki susceptible (MS) clones were collected from a green pepper and a cucumber, respectively, in Miyazaki Prefecture, Japan, in 2012.24) The Nippon soda susceptible clone (NS clone) has been maintained since 1993 at the Odawara Research Center, Nippon Soda Co., Ltd. (Odawara, Kanagawa, Japan). These clones were reared on cucumber seedlings at 25°C and 60% relative humidity under a 16:8-hr light:dark photoperiod in the absence of insecticides.
2. Chemicals
Acetamiprid and other neonicotinoids were synthesized at the Odawara Research Center. The purities of the test compounds (Fig. 1) were >99%.
3. RNA-seq and data analysis
Aphids were treated with acetamiprid (0.08 ppm for susceptible NS and MS clones and 5 ppm for the KR clone) or vehicle.12) Three biological replicates were performed using each condition. Total RNA of each biological replicate was extracted from the bodies of 10–15 wingless, viviparous females using TRizol reagent (Thermo Fisher Scientific K.K., Tokyo, Japan) and an RNeasy Mini Kit (QIAGEN K.K., Tokyo, Japan), in accordance with the manufacturer’s instructions.
cDNA libraries were prepared from the total RNAs and nucleotide sequencing using an Illumina HiSeq 2000 sequencer (paired-end, 101 bp) by Macrogen Japan Corp. (Kyoto, Japan). RNA-seq raw reads of each replicate were filtered using Trimmomatic.25) The filtered reads of the KR clone were merged and assembled de novo using Trinity.26) The contigs were used as reference transcript sequences for further analysis. Each contig was annotated with a description of the top protein hit in the NCBI nr database using Blastx (e-value threshold: 1e−03). Coding sequence (CDS) prediction was performed using TransDecoder bundled with Trinity, and the predicted CDSs were annotated with a description of the homologous domains of the Pfam protein family database acquired using an HMMER3 search.27)
The sequences of genes involved in insecticide resistance, such as those that encode detoxification enzymes (cytochrome P450s, glutathione S-transferases [GSTs], and carboxylesterases [COEs]), were identified according to the results of the Blastx and HMMER3 searches. For each sample, expression levels (tag count and TMM-normalized FPKM) of all genes (Trinity components) of the three replicates were calculated by aligning the filtered reads to the reference sequences using align_and_estimate_abundance.pl and abundance_estimates_to_matrix.pl bundled with Trinity. Differentially expressed gene (DEG) analysis of each susceptible and resistant clone was performed using iDEGES/edgeR, which is a statistical DEG analysis method that employs the tag counts of the two groups of RNA-seq data (three biological replicates for each group).28) Threshold values for identifying DEGs were a false discovery rate (FDR) of 0.05 and a fold change ≥2. The raw RNA-seq reads of the six samples were deposited in the DDBJ under accession number DRA005446.
4. Isolation of cDNA clones encoding A. gossypii P450s
Total RNA was isolated from adult aphids using ISOGEN and Spin Columns (Nippon Gene Co., Ltd., Tokyo, Japan), in accordance with the manufacturer’s instructions. First-strand cDNA was synthesized from total RNA using the Transcriptor First Strand cDNA Synthesis Kit (Roche Diagnostics K.K., Tokyo, Japan) with oligo-dT primers. PCR amplification was performed using KOD -Plus- Neo polymerase (TOYOBO Bio-Technology Co., Ltd., Osaka, Japan) and gene-specific primers as follows: c21228_g1-F (5′-ATA GGT ACC CAA AAT GAT ATC GTA TCT GAC CAA CTT GT-3′), c21228_g1-R (5′-ATA GGG CCC ATG TTC AAT GAT CGG TCT AAA TT-3′), c21368_g1-F (5′-ATA GAA TTC CAA AAT GAT TTC GTG GAC GAT CGA TTG-3′), and c21368_g1-R (5′-ATA GGG CCC AAC CGC AAC GAC TGG CTT TAG AC-3′). PCR amplification was performed using the following cycling conditions: 2 min at 94°C, followed by 35 cycles for 10 sec at 98°C and 50 sec at 68°C. Amplicons of the c21228_g1 gene were digested with KpnI and ApaI, and those of the c21368_g1 gene were digested using EcoRI and ApaI. Amplicons of the expected sizes were purified using the QIAquick Gel Extraction Kit (QIAGEN). Purified amplicons were ligated to the pAc5.1(+) vector (Thermo Fisher Scientific K.K., Tokyo, Japan). Each cDNA clone was sequenced using the BigDye Terminator V3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA). The sequences were confirmed using the following primers: pAc5.1-F (5′-ACA CAA AGC CGC TCC ATC AG-3′), c21228_g1-S1 (5′-TTC GTA CTT CAC CGA CCA CG-3′) and c21228_g1-S2 (5′-TGC AGG CGC GTA AAG AAT TG-3′) for c21228_g1, and c21368_g1-S1 (5′-AAG ACT TTG CGC ACT TCA CG-3′) and c21368_g1-S2 (5′-GGA ACG ATG TGG CAC AAA CA-3′) for c21368_g1.
5. Expression of A. gossypii P450 and metabolic studies
Drosophila melanogaster S2 cells were maintained at 28°C in a T-75 flask in Insectagro DS2 medium (Corning Inc., Corning, NY, USA) supplemented with 4 mM Ala-Glu (Sigma-Aldrich, Tokyo, Japan). The transient expression of A. gossypii P450 was performed using S2 cells seeded 24 hr before transfection in six-well plates (8×105 cells per well) incubated at 28°C. The transfection mixture in each well contained 3.6 µg of A. gossypii P450 DNA, 0.4 µg of D. melanogaster NADPH-cytochrome P450 reductase (CPR), and 4 µL of X-tremeGENE HP DNA Transfection Reagent (Roche Diagnostics K.K., Tokyo, Japan) in a final volume of 100 µL of Insectagro DS2 medium. D. melanogaster CPR (GenBank accession number Q27597) was synthesized by FASMAC (Kanagawa, Japan) and inserted into the pAc5.1 vector.
The medium was exchanged with serum-free CYP expression medium (supplemented with 0.1 mM ferric citrate, 0.1 mM 5-aminolevulinic acid, and 4 mM Ala-Glu in Insectagro DS2 medium) 6 hr after transfection. Next, the medium was exchanged with 2 mL of CYP expression medium 48 hr after transfection; test compounds (35 µL each) were added to a well and then sampled immediately (0 hr) and 72 hr later. All incubations were performed at 28°C. Test compounds were dissolved in dimethyl sulfoxide to prepare a 40,000 ppm stock, which was then diluted to 400 ppm with phosphate-buffered saline (final concentration=7 ppm).
After incubation, samples from the wells were transferred to 15-mL centrifuge tubes and diluted with an equal volume of acetonitrile (final volume 2 mL). The samples were vortexed for 10 sec, centrifuged at 14,000×g for 5 min at room temperature, and then filtered through a 0.45-µm filter. The extracts were analyzed using high-performance liquid chromatography (HPLC) (Shimadzu LC20A system; Shimadzu Corporation, Kyoto, Japan) with an Inertsil ODS-5, 150×4.6 mm, 5-µm particle column (GL Sciences Inc., Tokyo, Japan), with UV detection at 254 nm. Samples were separated using gradient elution with a mobile phase comprising 0.1% (v/v) formic acid, 5 mM ammonium acetate in HPLC-grade water, and methanol at 1.0 mL/min for 40 min. The gradient elution conditions were as follows: 0 min methanol : water 10 : 90, 10 min methanol : water 10 : 90, 30 min methanol : water 70 : 30, 32 min methanol : water 10 : 90, and 40 min methanol : water 10 : 90. The injection volume was 25 µL. Each well of a six-well plate was analyzed three times.
Electrospray-ionization mass spectrometry (ESI-LC-MS spectra) was performed using a TSQ Vantage (Thermo Fisher Scientific K.K., Tokyo, Japan) operated in positive-ion mode. High-purity nitrogen (450°C) was used as the sheath gas, and argon served as the collision gas. After 72 hr incubation, potential metabolites of acetamiprid and imidacloprid were analyzed using LC-MS, as indicated by the detection of extracted ions of the parents (223+ for acetamiprid and 256+ for imidacloprid) and metabolites (209+ for acetamiprid and 272+ for imidacloprid). Separation was performed using an Inertsil ODS-5 column (150×2.1 mm, 5-µm-diameter particle) and a gradient (same conditions as for the HPLC analysis above, except that the mobile phase comprised 0.1% [v/v] formic acid and 5 mM ammonium acetate in HPLC water and acetonitrile) delivered at 1.0 mL/min. The injection volume was 25 µL.
An EzRIPA Lysis Kit (Atto Corporation, Tokyo, Japan) was used to prepare samples from the cells in each well for Western blotting analysis. The samples were added to equal volumes of 2×Laemmli sample buffer and heated at 100°C for 5 min. The samples were then separated using a 10% SDS-PAGE gel and transblotted onto a polyvinylidene fluoride membrane. The membrane was probed with an anti-V5 antibody, subsequently incubated with an anti-IgG2a-HRP antibody, and the immunocomplexes were visualized using EzWestBlue (Atto Corporation).
Results
1. RNA-seq and DEG analysis of A. gossypii genes encoding detoxification enzymes
The numbers of RNA-seq paired-end reads (raw reads, filtered reads, and reads mapped to reference transcriptome sequences) of each clone are shown in Table 1. Reference transcriptome sequences of 81,316 contigs (70,664,259 bp in total) were generated using de novo assembly of the RNA-seq paired-end reads of the KR clone (21,070,670,904 bp). The reference contigs represented 65,692 genes (Trinity components) encoding at least one isoform (contigs).
Table 1. The number of RNA-seq paired-end reads acquired for the three A. gossypii clones (NS, MS, and KR).
| Sample IDa) | Replicate ID | No. of RNA-seq paired-end readsb) | ||
|---|---|---|---|---|
| Raw | Filtered | Mapped to reference | ||
| NS (N) | rep1 | 36,807,970 | 29,952,366 | 23,732,526 |
| rep2 | 36,718,120 | 29,131,314 | 23,566,250 | |
| rep3 | 36,417,536 | 29,158,596 | 23,333,644 | |
| NS (T) | rep1 | 33,581,112 | 27,099,350 | 22,090,390 |
| rep2 | 34,283,634 | 27,316,198 | 22,119,944 | |
| rep3 | 37,757,450 | 30,506,432 | 23,567,262 | |
| MS (N) | rep1 | 38,883,482 | 30,185,406 | 26,480,438 |
| rep2 | 30,302,292 | 23,917,022 | 20,556,194 | |
| rep3 | 32,049,156 | 25,433,864 | 21,541,656 | |
| MS (T) | rep1 | 38,463,596 | 33,257,728 | 28,917,210 |
| rep2 | 36,832,066 | 31,746,908 | 27,596,414 | |
| rep3 | 35,566,136 | 30,776,962 | 26,170,864 | |
| KR (N) | rep1 | 35,392,952 | 30,615,400 | 25,700,126 |
| rep2 | 37,300,282 | 29,893,988 | 21,960,964 | |
| rep3 | 35,562,908 | 28,725,344 | 23,160,986 | |
| KR (T) | rep1 | 34,926,706 | 28,174,146 | 22,975,134 |
| rep2 | 32,963,336 | 28,548,450 | 23,747,834 | |
| rep3 | 32,474,320 | 28,474,746 | 22,322,760 | |
a) (N) means without acetamiprid treatment, and (T) means with acetamiprid treatment (0.08 ppm and 0.5 ppm for the susceptible and resistant clone, respectively). b) The number of raw reads, filtered reads, and reads mapped to the reference transcript sequences are shown for three biological replicates (rep1, rep2, and rep3) of each clone treated without or with acetamiprid.
Nine genes encoding A. gossypii detoxification enzymes were expressed at significantly higher levels by the KR clone as compared with those of the two insecticide-susceptible clones (Table 2). The nine genes encode cytochromes P450 according to annotation information acquired from the NCBI-nr and Pfam databases. In contrast, the transcriptional levels of genes encoding detoxification enzymes such as GST and COE were not significantly upregulated in the KR clone as compared with those of the susceptible clones. Transcriptional levels of the P450, GST, and COE genes were not significantly upregulated in the KR clone or the two insecticide-susceptible clones treated with acetamiprid vs. the control. Two of the nine P450 genes, c21228_g1 and c21368_g1 (Trinity IDs), were highly upregulated under the four conditions. The respective fold changes in the expression levels of the c21228_g1 and c21368_g1 genes were 46.21 and 7.46 for the KR/NS clones and 4.35 and 3.32 for the KR/MS clones in the absence of acetamiprid. Similarly, the respective values of the two genes were 38.05 and 8.22 for the KR/NS clones and 3.71 and 2.48 for the KR/MS clones in the presence of acetamiprid. The amino acid sequences of c21228_g1 (513 aa) and c21368_g1 (513 aa) are similar to those of CYP6CY22 (94.15% identical) and CYP6CY13 (99.61% identical) of Aphid gossypii, respectively.
Table 2. Selected genes identified by RNA-seq that were differentially transcribed at a significant level between the insecticide-resistant A. gossypii clone (KR) and the insecticide-susceptible clones (NS and MS).
| Gene IDa) | CDS length (bp) | ID of top-hit NCBI-nr protein | Description of top-hit NCBI-nr protein | Identity of top-hit (%) | Fold change of expression level | No. of tag countsb) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KR/NS (N) | KR/MS (N) | KR/NS (T) | KR/MS (T) | NS (N) | NS (T) | MS (N) | MS (T) | KR (N) | KR (T) | |||||||||||||||||
| rep1 | rep2 | rep3 | rep1 | rep2 | rep3 | rep1 | rep2 | rep3 | rep1 | rep2 | rep3 | rep1 | rep2 | rep3 | rep1 | rep2 | rep3 | |||||||||
| c21228_g1 | 1542 | XP_001948421 | PREDICTED: cytochrome P450 6k1-like [Acyrthosiphon pisum] | 78.35 | 46.21 | 4.35 | 38.05 | 3.71 | 53 | 10 | 23 | 50 | 23 | 25 | 411 | 374 | 262 | 612 | 542 | 243 | 1,166 | 1,654 | 943 | 1,516 | 1,086 | 1,521 |
| c21368_g1 | 1542 | XP_008184269 | PREDICTED: probable cytochrome P450 6a13 [Acyrthosiphon pisum] | 80.39 | 7.46 | 3.32 | 8.22 | 2.48 | 221 | 152 | 223 | 186 | 138 | 156 | 815 | 405 | 394 | 694 | 797 | 682 | 1,422 | 2,003 | 870 | 1,975 | 1,126 | 1,278 |
| c29523_g2 | 420 | XP_008184269 | PREDICTED: probable cytochrome P450 6a13 [Acyrthosiphon pisum] | 74.81 | 5.17 | 2.11 | 1.19 | 0.56 | 52 | 29 | 29 | 86 | 26 | 57 | 126 | 135 | 51 | 219 | 231 | 51 | 312 | 78 | 210 | 81 | 84 | 61 |
| c22273_g1 | 1533 | XP_001946384 | PREDICTED: probable cytochrome P450 6a13 [Acyrthosiphon pisum] | 91.59 | 3.68 | 9.06 | 1.21 | 1.99 | 161 | 153 | 168 | 83 | 50 | 79 | 80 | 72 | 77 | 61 | 62 | 52 | 598 | 429 | 752 | 65 | 119 | 99 |
| c26291_g2 | 1536 | XP_001952450 | PREDICTED: probable cytochrome P450 6a13 [Acyrthosiphon pisum] | 76.45 | 3.01 | 19.84 | 2.99 | 1.99 | 2,111 | 2,289 | 2,422 | 134 | 79 | 94 | 332 | 312 | 555 | 219 | 223 | 191 | 7,763 | 4,927 | 8,128 | 312 | 340 | 363 |
| c26291_g1 | 1518 | XP_001952450 | PREDICTED: probable cytochrome P450 6a13 [Acyrthosiphon pisum] | 81.98 | 2.66 | 45.25 | 1.93 | 1.88 | 465 | 384 | 546 | 52 | 27 | 29 | 44 | 28 | 26 | 55 | 78 | 21 | 1,727 | 341 | 1,842 | 67 | 71 | 92 |
| c4974_g1 | 1533 | XP_008188450 | PREDICTED: uncharacterized protein LOC100168454 [Acyrthosiphon pisum] | 88.34 | 2.43 | 3.94 | 5.39 | 1.84 | 478 | 407 | 484 | 153 | 112 | 95 | 502 | 218 | 296 | 629 | 476 | 352 | 1,188 | 1,158 | 943 | 768 | 729 | 663 |
| c27644_g1 | 1554 | XP_001947920 | PREDICTED: cytochrome P450 6a2-like [Acyrthosiphon pisum] | 74.81 | 1.59 | 4.26 | 3.43 | 0.83 | 785 | 699 | 974 | 113 | 56 | 72 | 490 | 413 | 189 | 622 | 665 | 112 | 1,495 | 1,015 | 1,447 | 310 | 255 | 347 |
| c23819_g1 | 288 | XP_001951034 | PREDICTED: cytochrome P450 4C1-like [Acyrthosiphon pisum] | 78.12 | 0.48 | 0.79 | 3.18 | 3.16 | 5 | 5 | 2 | 6 | 2 | 3 | 6 | 2 | 1 | 5 | 2 | 8 | 7 | 0 | 0 | 8 | 24 | 8 |
a) The amino acid sequence identities of each gene compared with Acyrthosiphon pisum P450 sequences acquired from the NCBI-nr protein sequence database are shown. b)(N), without acetamiprid treatment; (T), with acetamiprid treatment (0.08 ppm for the susceptible clones and 0.5 ppm for the resistant clone).Red cells indicate statistically significant differential upregulation (FDR <0.05 and fold change ≥2) between the two clones. The values of FDR and the fold changes of expression levels were calculated using the iDEGES/edgeR method with tag counts (mapped paired-end reads) of two groups of RNA-seq data. The numbers of tag counts (mapped paired-end reads) for each P450 gene are shown for three biological replicates (rep1, rep2, and rep3) of each clone treated with or without acetamiprid. Accession numbers and database sequence identifiers are as follows: c21228_g1 (LC223821), c21368_g1 (LC223822), c29523_g2 (LC223823), c22273_g1 (LC223824), c26291_g2 (LC223825), c26291_g1 (LC223826), c4974_g1 (LC223827), c27644_g1 (LC223828), and c23819_g1 (LC223829).
2. A. gossypii P450 expression and CYP6-mediated metabolism
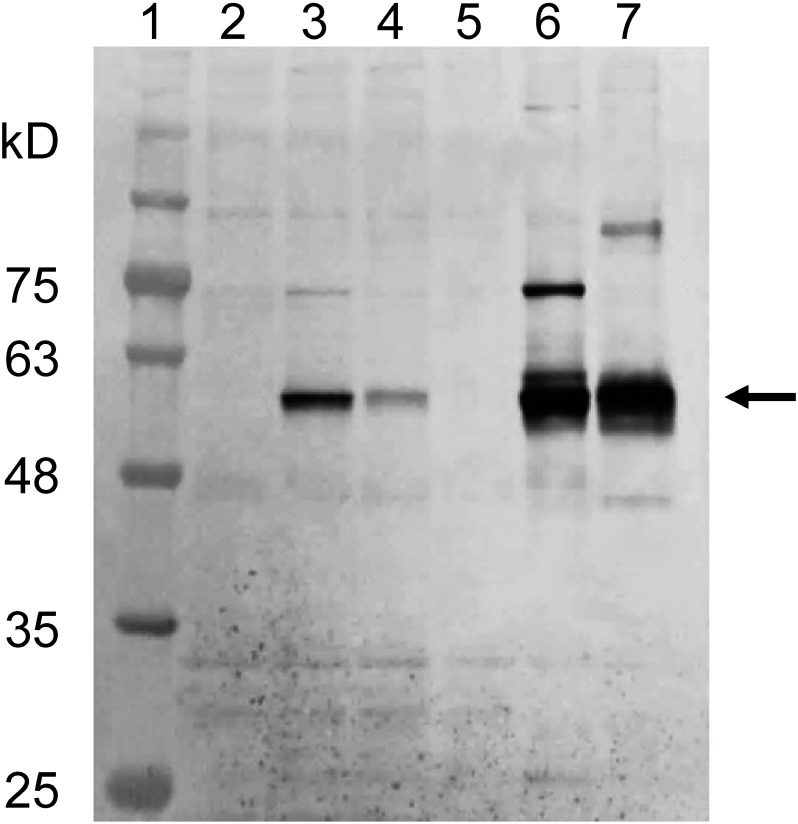
The expression levels of CYP6CY22 (c21228_g1) and CYP6CY13 (c21368_g1) were highly upregulated in the insecticide-resistant KR clone as compared with their levels in the insecticide-susceptible NS and MS clones (Table 2). We, therefore, attempted to express these two P450 genes using a Drosophila expression system to assess their metabolic activities. CYP6CY22 (c21228_g1) and CYP6A13 (c21368_g1) expression was detected in cells incubated in CYP expression medium 48 hr after transfection (Fig. 2); however, they were weakly expressed in normal medium (SDM containing 10% FBS) (Fig. 2).
Fig. 2. Expression of A. gossypii CYP6CY22 and CYP6CY13 genes in transfected D. melanogaster S2 cells. Lane 1=molecular weight standards. Lanes 2 and 5=negative control, cells transfected only with the pAc5.1 vector without inserted genes. Lanes 3 and 4=cells transfected with c21228_g1 and c21368_g1 genes and cultured in normal medium (Schneider’s Drosophila Medium with 10% FBS). Lanes 6 and 7=cells transfected with c21228_g1 and c21368_g1 genes (59 kDa) and cultured in CYP expression medium (Insectagro DS2 medium supplemented with 0.1 mM ferric citrate, 0.1 mM 5-aminolevulinic acid, and 4 mM Ala-Glu without FBS).
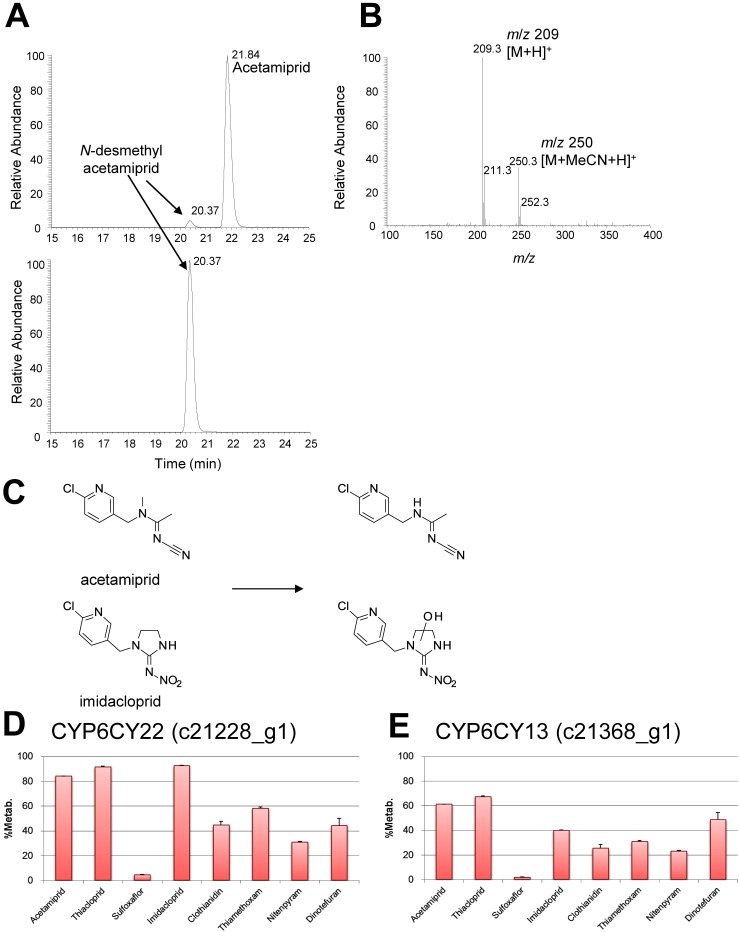
All neonicotinoid insecticides were almost completely recovered after they were incubated with untransfected D. melanogaster S2 cells (Table 2). However, when acetamiprid or imidacloprid was incubated with D. melanogaster S2 cells expressing CYP6CY22 (c21228_g1), little of the parent compound was recovered. In contrast, moderate amounts of the parent compounds were recovered from cultures of the CYP6CY13 (c21368_g1) transfectant. LC-MS analyses of D. melanogaster S2 cell extracts showed that acetamiprid was converted to N-desmethyl acetamiprid (Fig. 3A–C), and imidacloprid was converted to a hydroxylated metabolite (Fig. 3C).
Fig. 3. ESI-LC-MS spectra of acetamiprid and imidacloprid, structures of the metabolites, and percentages of metabolites. (A) LC elution profile of acetamiprid in extracts of D. melanogaster S2 cells expressing the c21228_g1 gene (12 hr after test compound was added). (B) LC-MS spectrum of acetamiprid in an extract of D. melanogaster S2 cells expressing the c21368_g1 gene. (C) Structures of the metabolites of acetamiprid and imidacloprid. (D) Metabolism of test compounds in cultures of cells expressing the c21228_g1 gene. (E) Metabolism of test compounds in cultures of cells expressing the c21368_g1 gene. Data are the mean±SD (n=3).
LC-MS analyses identified acetamiprid and its main metabolite N-desmethyl acetamiprid (Fig. 3A, B). The retention times and m/z values of the components detected were consistent with those of the reference substance. Recoveries of thiacloprid, clothianidin, thiamethoxam, nitenpyram, and dinotefuran from cultures of cells expressing CYP6CY22 (c21228_g1) were 8.5%, 55.2%, 42.8%, 69.3%, and 55.8%, respectively (Table 3). Recoveries of thiacloprid, clothianidin, thiamethoxam, nitenpyram, and dinotefuran from cultures of cells expressing CYP6CY13 (c21368_g1) were 32.8%, 74.5%, 69.3%, 77.1%, and 51.2%, respectively (Table 3). In contrast, sulfoxaflor was almost completely recovered from cultures of cells expressing CYP6CY22 (c21228_g1) or CYP6CY13 (c21368_g1). Lower and higher concentrations of the insecticides and their metabolites, respectively, were detected in cultures of D. melanogaster S2 cells that expressed CYP6CY22 (c21228_g1) vs. CYP6CY13 (c21368_g1) (Table 3).
Table 3. Metabolism of test compounds by D. melanogaster S2 cells expressing CYP6CY22(c21228_g1) and CYP6CY13 (c21368_g1) genes.
| c21228_g1 (CYP6CY22) | c21368_g1 (CYP6CY13) | RF1b) | RF2c) | |||||
|---|---|---|---|---|---|---|---|---|
| %Recovery (avg.±S.D.)a) | %Metab. | %Recovery (avg.±S.D.)a) | %Metab. | |||||
| Control | +transfection | Control | +transfection | |||||
| Acetamiprid | 102.2±0.2 | 26.0±0.1 | 84.0 | 99.5±1.0 | 38.8±1.0 | 61.2 | 65.4 | 104 |
| Thiacloprid | 99.8±0.7 | 8.5±0.7 | 91.5 | 95.6±0.3 | 32.8±0.8 | 67.2 | 23.8 | 17 |
| Sulfoxaflor | 99.6±0.6 | 95.6±0.6 | 4.4 | 101.2±1.9 | 98.3±2.8 | 1.7 | 22.6 | |
| Imidacloprid | 101.5±0.4 | 7.5±0.4 | 92.5 | 98.5±1.2 | 60.0±1.0 | 40.0 | 216 | 253 |
| Clothianidin | 98.2±1.5 | 55.2±3.0 | 44.8 | 96.5±0.4 | 74.5± 1.1 | 25.5 | 394 | 687 |
| Thiamethoxam | 95.6±1.1 | 42.8±1.3 | 57.2 | 98.2±0.2 | 69.3±0.3 | 30.7 | 295 | 246 |
| Nitenpyrum | 97.5±1.2 | 69.3±1.0 | 30.7 | 96.5±1.3 | 77.1±2.3 | 22.9 | 253 | 43 |
| Dinotefuran | 103.4±1.2 | 55.8±5.9 | 44.2 | 101.3±2.1 | 51.2±7.5 | 48.8 | 237 | 199 |
Discussion
In the present study, we performed RNA-seq analysis and NGS to identify P450 genes involved in the metabolism of neonicotinoid insecticides by A. gossypii. We identified nine upregulated cytochrome P450 genes that were expressed at significantly higher levels by the insecticide-resistant KR clone as compared with those of insecticide-susceptible clones (Table 2). Genes encoding detoxification enzymes such as GST and COE were not similarly upregulated. Moreover, we established an A. gossypii P450 expression method in the S2 insect cell expression system to generate functionally active P450 by improving transient expression methods. This approach will enable the generation of ample quantities of insect P450 for insecticide resistance studies and structural analysis of metabolites of insecticides.
The P450 genes c21228_g1 and c21368_g1 were highly upregulated in the KR clone in the presence or absence of acetamiprid, as compared with the levels of upregulation in the insecticide-susceptible clones NS and MS (Table 2). The sequences of the c21228_g1 and c21368_g1 genes are similar to those of the A. gossypii genes encoding CYP6CY22 and CYP6CY13, respectively. Numerous P450s catalyze a highly restricted set of reactions, although some metabolize diverse compounds. In insects, members of the CYP4, -6, -9, and -12 families are implicated in biological detoxifying functions in the environment, and members of the CYP4 and CYP6 subfamilies are most commonly involved in the metabolism of and resistance to xenobiotics.29,30)
The CYP6CY22 (c21228_g1) gene was overexpressed 46-fold in the KR clone, as compared with the NS clone, and 4-fold, as compared with the MS clone when aphids were treated without acetamiprid. Similarly, the CYP6CY13 (c21368_g1) gene was expressed at a 7-fold higher level by the KR clone vs. the NS clone and a 3-fold higher level by the KR clone vs. the MS clone under the same conditions. The findings were similar for acetamiprid treatment. The levels of the two P450 genes expressed by the KR clone were significantly increased as compared with those of the NS clone. P450 genes were not upregulated in the KR clone in the presence or absence of acetamiprid, indicating constitutive upregulation, as compared with the insecticide-susceptible clones.
The NS clone has been maintained at the Odawara Research Center without insecticide selection since 1993. In contrast, the levels of the two P450 genes expressed by the KR clone were moderately increased as compared with those of the insecticide-susceptible MS clone, and the levels of the latter were higher as compared with those of the NS clone. The MS clone was collected in a field in Miyazaki Prefecture, Japan, and is likely to have been exposed to more insecticides, as compared with the NS clone. This may explain the difference in expression levels between the two insecticide-susceptible clones, as insecticide exposure likely induced the expression of P450 genes.31) Furthermore, differences in expression levels between these two insecticide-susceptible clones are explained by the exchange of host plants.32,33) However, although the expression levels of the two P450 genes of the two susceptible strains were significantly different, the resistance factors (LC50 resistant clone/LC50 susceptible clone) for neonicotinoid insecticides of the KR/NS clones are similar to those of the KR/MS clones.12,24)
The contribution of P450-mediated detoxification to metabolism is indicated by the use of PBO, an inhibitor of metabolic enzymes, including P450s. Further, PBO pretreatment reduces the LC50 values of neonicotinoid insecticides, and the synergistic factors of acetamiprid and imidacloprid=3.6 and 6.0, respectively.12) Furthermore, the two A. gossypii P450 genes metabolize neonicotinoid insecticides (Fig. 3, Table 3). These findings suggest that these P450s affect neonicotinoid resistance.
The neonicotinoids examined here were metabolized to varying degrees by D. melanogaster S2 cells expressing A. gossypii CYP6CY22 (c21228_g1) and CYP6CY13 (c21368_g1) (Table 3, Fig. 3D, E). Under the conditions applied here, acetamiprid, thiacloprid, and imidacloprid were more extensively metabolized than the other neonicotinoids (Table 3, Fig. 3), which is consistent with the findings of a previous study (D. melanogaster CYP6G1).34) However, these results differ from those observed for the monooxygenase CYP6CM1vQ, which is associated with the imidacloprid resistance of B. tabaci.35)
Imidacloprid, thiacloprid, and clothianidin are metabolized by CYP6CM1vQ, but not acetamiprid and thiamethoxam.35) In the present study, all compounds were metabolized, although sulfoxaflor was a poor substrate for A. gossypii CYP6CY22 and CYP6CY13 (Table 3, Fig. 3D, E), which is consistent with results obtained for D. melanogaster CYP6G1.34) Furthermore, CYP6G1 metabolizes structurally diverse insecticides such as DDT, malathion, and pyrethroids.36) Thus, A. gossypii CYP6CY22 and CYP6CY13 may have broader substrate specificities. The incubation of D. melanogaster S2 cells expressing A. gossypii CYP6CY22 or CYP6CY13 with acetamiprid and imidacloprid produced a metabolite with a hydroxyl group on the imidazolidine ring and N-desmethyl acetamiprid, respectively (Fig. 3A–C).37–39) Hydroxylation and N-dealkylation are likely the key reactions catalyzed by A. gossypii CYP6CY22 and CYP6CY13, respectively.
In conclusion, we used RNA-seq to identify nine differentially upregulated P450 genes involved in the metabolism of neonicotinoids by the A. gossypii KR clone. Among them, genes encoding CYP6CY22 and CYP6CY13 were the most highly upregulated. Furthermore, we constructed A. gossypii CYP6CY22 and CYP6CY13 expression vectors and used them to transfect an insect cell line to study P450-mediated insecticide metabolism, although it was difficult to determine the catalytic constants (e.g., Km and Kcat) of these recombinant P450s. Moreover, we did not determine the P450 activities of microsomal membrane fractions. Nevertheless, our findings contribute compelling evidence that these P450s may contribute to aphids’ resistance to neonicotinoids in the future.
Acknowledgments
This work was supported by a grant from the Ministry of Agriculture, Forestry, and Fisheries of Japan (Genomics-based Technology for Agricultural Improvement, PRM-03). We are grateful for comments and advice from Dr. David Nelson.
References
- 1).K. Matsuda, S. D. Buckingham, D. Kleier, J. J. Rauh, M. Grauso and D. B. Sattelle: Trends Pharmacol. Sci. 22, 573–580 (2001). [DOI] [PubMed] [Google Scholar]
- 2).K. Matsuda, M. Shimomura, M. Ihara, M. Akamatsu and D. B. Sattelle: Biosci. Biotechnol. Biochem. 69, 1442–1452 (2005). [DOI] [PubMed] [Google Scholar]
- 3).M. Tomizawa and J. E. Casida: Annu. Rev. Pharmacol. 45, 247–268 (2005). [DOI] [PubMed] [Google Scholar]
- 4).S. H. Thany, G. Lenaers, V. Raymond-Delpech, D. B. Sattelle and B. Lapied: Trends Pharmacol. Sci. 28, 14–22 (2007). [DOI] [PubMed] [Google Scholar]
- 5).R. Nauen and I. Denholm: Arch. Insect Biochem. Physiol. 58, 200–215 (2005). [DOI] [PubMed] [Google Scholar]
- 6).A. Elbert and R. Nauen: Pest Manag. Sci. 56, 60–64 (2000). [Google Scholar]
- 7).K. Gorman, G. Devine, J. Bennison, P. Coussons, N. Punchard and I. Denholm: Pest Manag. Sci. 63, 555–558 (2007). [DOI] [PubMed] [Google Scholar]
- 8).L. Zewen, H. Zhaojun, W. Yinchang, Z. Lingchun, Z. Hongwei and L. Chengjun: Pest Manag. Sci. 59, 1355–1359 (2003). [DOI] [PubMed] [Google Scholar]
- 9).J.-Z. Zhao, B. A. Bishop and E. J. Grafius: J. Econ. Entomol. 93, 1508–1514 (2000). [DOI] [PubMed] [Google Scholar]
- 10).G. Zhao, W. Liu, J. M. Brown and C. O. Knowles: J. Econ. Entomol. 88, 1164–1170 (1995). [Google Scholar]
- 11).C. Bass, A. M. Puinean, M. Andrews, P. Cutler, M. Daniels, J. Elias, V. L. Paul, A. J. Crossthwaite, I. Denholm, L. M. Field, S. P. Foster, R. Lind, M. S. Williamson and R. Slater: BMC Neurosci. 12, 51 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12).K. Hirata, R. Kiyota, A. Matsuura, S. Toda, A. Yamamoto and T. Iwasa: J. Pestic. Sci. 40, 25–31 (2015). [Google Scholar]
- 13).M. Shimomura, M. Yokota, M. Ihara, M. Akamatsu, D. B. Sattelle and K. Matsuda: Mol. Pharmacol. 70, 1255–1263 (2006). [DOI] [PubMed] [Google Scholar]
- 14).T. T. Talley, M. Harel, R. E. Hibbs, Z. Radic, M. Tomizawa, J. E. Casida and P. Taylor: Proc. Natl. Acad. Sci. U.S.A. 105, 7606–7611 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15).M. Ihara, T. Okajima, A. Yamashita, T. Oda, K. Hirata, H. Nishiwaki, T. Morimoto, M. Akamatsu, Y. Ashikawa, S. Kuroda, R. Mega, S. Kuramitsu, D. B. Sattelle and K. Matsuda: Invert. Neurosci. 8, 71–81 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16).N. Rauch and R. Nauen: Arch. Insect Biochem. Physiol. 54, 165–176 (2003). [DOI] [PubMed] [Google Scholar]
- 17).J. G. Scott: Insect Biochem. Mol. Biol. 29, 757–777 (1999). [DOI] [PubMed] [Google Scholar]
- 18).D. Nikou, H. Ranson and J. Hemingway: Gene 318, 91–102 (2003). [DOI] [PubMed] [Google Scholar]
- 19).R. Feyereisen, J. F. Koener, D. E. Farnsworth and D. W. Nebert: Proc. Natl. Acad. Sci. U.S.A. 86, 1465–1469 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20).M. R. Bogwitz, H. Chung, L. Magoc, S. Rigby, W. Wong, M. O’Keefe, J. A. McKenzie, P. Batterham and P. J. Daborn: Proc. Natl. Acad. Sci. U.S.A. 102, 12807–12812 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21).J. W. Pridgeon and N. Liu: Insect Biochem. Mol. Biol. 33, 1043–1048 (2003). [DOI] [PubMed] [Google Scholar]
- 22).M. Amichot, S. Tarès, A. Brun-Barale, L. Arthaud, J.-M. Bride and J.-B. Bergé: Eur. J. Biochem. 271, 1250–1257 (2004). [DOI] [PubMed] [Google Scholar]
- 23).J. Bergé, R. Feyereisen and M. Amichot: Philos. Trans. R. Soc. Lond. B Biol. Sci. 353, 1701–1705 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24).A. Matsuura and M. Nakamura: Appl. Entomol. Zool. (Jpn.) 49, 535–540 (2014). [Google Scholar]
- 25).A. M. Bolger, M. Lohse and B. Usadel: Bioinformat. 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26).M. G. Grabherr, B. J. Haas, M. Yassour, J. Z. Levin, D. A. Thompson, I. Amit, X. Adiconis, L. Fan, R. Raychowdhury, Q. Zeng, Z. Chen, E. Mauceli, N. Hacohen, A. Gnirke, N. Rhind, F. di Palma, B. W. Birren, C. Nusbaum, K. Lindblad-Toh, N. Friedman and A. Regev: Nat. Biotechnol. 29, 644–652 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27).S. R. Eddy: PLOS Comput. Biol. 7, e1002195 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28).J. Sun, T. Nishiyama, K. Shimizu and K. Kadota: BMC Bioinformatics 14, 219 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29).M. R. Berenbaum: J. Chem. Ecol. 28, 873–896 (2002). [DOI] [PubMed] [Google Scholar]
- 30).X. Li, M. A. Schuler and M. R. Berenbaum: Annu. Rev. Entomol. 52, 231–253 (2007). [DOI] [PubMed] [Google Scholar]
- 31).M. D. K. Markussen and M. Kristensen: Pestic. Biochem. Physiol. 98, 50–58 (2010). [Google Scholar]
- 32).X. Li, M. R. Berenbaum and M. A. Schuler: Insect Mol. Biol. 11, 343–351 (2002). [DOI] [PubMed] [Google Scholar]
- 33).J. G. Scott, N. Liu and Z. Wen: Comp. Biochem. Physiol. C Pharmacol. Toxicol. Endocrinol. 121, 147–155 (1998). [DOI] [PubMed] [Google Scholar]
- 34).T. C. Sparks, G. J. DeBoer, N. X. Wang, J. M. Hasler, M. R. Loso and G. B. Watson: Pestic. Biochem. Physiol. 3, 159–165 (2012). [Google Scholar]
- 35).E. Roditakis, E. Morou, A. Tsagkarakou, M. Riga, R. Nauen, M. Paine, S. Morin and J. Vontas: Insect Sci. 8, 23–29 (2011). [Google Scholar]
- 36).R. T. Jones, S. E. Bakker, D. Stone, S. N. Shuttleworth, S. Boundy, C. McCart, P. J. Daborn, R. H. ffrench-Constant and J. M. H. van den Elsen: Pest Manag. Sci. 66, 1106–1115 (2010). [DOI] [PubMed] [Google Scholar]
- 37).R. Nauen, U. Reckmann, S. Armborst, H.-P. Stupp and A. Elbert: Pestic. Sci. 55, 265–271 (1999). [Google Scholar]
- 38).H. Nishiwaki, K. Sato, Y. Nakagawa, M. Miyashita and H. Miyagawa: J. Pestic. Sci. 29, 110–116 (2004). [Google Scholar]
- 39).J.-L. Brunet, A. Badiou and L. P. Belzunces: Pest Manag. Sci. 61, 742–748 (2005). [DOI] [PubMed] [Google Scholar]