Abstract
Bosentan is well-known to induce cholestatic liver toxicity in humans. The present study was set up to characterize the hepatotoxic effects of this drug at the transcriptomic, proteomic and metabolomic level. For this purpose, human hepatoma-derived HepaRG cells were exposed to a number of concentrations of bosentan during different periods of time. Bosentan was found to functionally and transcriptionally suppress the bile salt export pump as well as to alter bile acid levels. Pathway analysis of both transcriptomics and proteomics data identified cholestasis as a major toxicological event. Transcriptomics results further showed several gene changes related to the activation of the nuclear farnesoid X receptor. Induction of oxidative stress and inflammation were also observed. Metabolomics analysis indicated changes in the abundance of specific endogenous metabolites related to mitochondrial impairment. The outcome of this study may assist in the further optimization of adverse outcome pathway constructs that mechanistically describe the processes involved in cholestatic liver injury.
Keywords: bosentan, BSEP, HepaRG, cholestasis, transcriptomics, proteomics, metabolomics, adverse outcome pathway
1. Introduction
Bosentan is an endothelin receptor antagonist clinically used for the treatment of pulmonary arterial hypertension (Dingemanse and van Giersbergen, 2004; Muller and Milton, 2012). This drug is well-known to cause cholestatic liver toxicity as a major side effect, which is triggered, at least in part, by inhibition of the bile salt export pump (BSEP) (Dawson et al., 2011; Fattinger et al., 2001; Kis et al., 2012; Lepist et al., 2014). In fact, an adverse outcome pathway (AOP) was previously introduced to mechanistically describe the processes that underlie drug-induced cholestatic liver injury (Vinken et al., 2013). Functional BSEP inhibition is considered as the molecular initiating event (MIE) in this AOP. BSEP inhibition results in toxic concentrations of bile acids piled up in the cytosol of the hepatocytes or in the bile canaliculi, being a first key event (KE). These bile acids evoke the formation of the mitochondrial permeability transition pore, which leads to mitochondrial impairment and the production of reactive oxygen species. Simultaneously, 2 other KEs, namely oxidative stress and inflammation, are activated in hepatocytes, underlying the direct deteriorative response to BSEP inhibition. This ultimately burgeons into cell death, which is associated with the release of several cytosolic enzymes, such as alkaline phosphatase, in serum (Padda et al., 2011). In parallel, 3 nuclear receptors are activated, namely the farnesoid X receptor (FXR), the pregnane X receptor (PXR) and the constitutive androstane receptor (CAR). This KE activates an extensive set of transcriptional changes, targeted towards the increased removal of bile acids and their products from the organism (Wagner et al., 2009; Zollner and Trauner, 2006 and 2008) (Suppl. Fig. S1).
The objective of the present study is to characterize the hepatotoxic effects of bosentan at the transcriptomic, proteomic and metabolomic level, and to link this with the established AOP from BSEP inhibition to cholestatic liver injury (Vinken et al., 2013) by providing a number omics-based biomarkers that can substantiate existing or novel KEs. For this purpose, human hepatoma HepaRG cells, a well-established in vitro model to mechanistically study cholestatic responses at the cellular level (Bachour-El Azzi et al., 2015; Le Vee et al., 2006; Qiu et al., 2016), were used as experimental setting.
2. Materials and methods
2.1. Cell culture and chemical treatment
Three different batches of cryopreserved, differentiated human hepatoma-derived HepaRG cells were used (Biopredic International, France). The cells were cultivated in collagen-coated 24-well plates as described by the supplier’s instructions at a density of 0.24 million cells per cm2 unless otherwise specified. The cells were plated and cultured for 7 days with regular media change, after which the typical HepaRG cell morphology was obtained. All experiments were started with cells cultured for exactly the same time. Bosentan (Sequoia, UK) was dissolved in dimethyl sulfoxide (DMSO) to prepare a stock solution and was subsequently diluted (1:200) in HepaRG Induction Medium (Biopredic International, France) to obtain the working concentration. The amount of DMSO in all concentrations and their respective controls was 0.5%. For transcriptomics and metabolomics experiments, the cells were exposed for 1 hour (1 h), 24 hours (24 h) and 24 hours with a wash-out period of 72 hours (24 h+72 h) to 3 bosentan concentrations, namely inhibitory concentration of 10% (IC10), IC10/4 and IC10/10, based on assessment of cell viability as described in section 2.5. For proteomics experiments, only IC10 concentrations were tested at the 3 time points. For the determination of BSEP functionality, cells were seeded in 35 mm dishes (Ibidi, Belgium) at a density of 0.21 million cells per cm2 and further cultured following the supplier’s instructions. All exposure experiments were conducted using 3 different cell batches (n = 3) and tests were performed in triplicate (N = 3).
2.2. BSEP immunolocalization
HepaRG cells were fixated with 4% paraformaldehyde (Sigma, Belgium) for 10 min and treated with 100 mM glycine (Merck, Belgium) for 15 min. After permeabilization with 0.1% Triton X (Sigma, Belgium) and blocking with 10% donkey serum (Jackson Immuno Research, Belgium), cells were incubated overnight at 4°C with anti-BSEP primary antibody (74500c) (Santa Cruz, Belgium) (1:200 dilution) followed by incubation with secondary antibody anti-mouse DyLight 488 (Abcam, Belgium) (1:500 dilution) for 1.5 h at room temperature. Finally, the cells were mounted with Vectashield containing 4’,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, Belgium) for nuclear staining and bleaching protection. Fluorescent images were obtained by fluorescence microscopy at 40x magnification (Nikon Eclipse Ti-S, Belgium).
2.3. Determination of BSEP functional activity
For the determination of the functional activity of BSEP, HepaRG cells were exposed for 24 h to IC10 concentration of bosentan prior to exposure for 30 min to the specific BSEP probe tauronor-THCA-24-DBD (excitation/emission wavelength 450/570 nm). The dishes were placed under a Zeiss LSM780 confocal microscope with a temperature control unit set at 37°C equipped with an EC Plan-Neofluar 40x/1.30 NA Oil DIC M27. Fluorescence images were made at 20x magnification (Zeiss, Belgium). Image analyses for the quantification of fluorescence intensity and area was conducted using Zeiss Zen Imaging Software.
2.4. Determination of cholic acid and glycocholic acid
For the quantification of bile acids, HepaRG cells (i.e. 0.24 million cells per cm2), whether or not exposed to bosentan, were rinsed with cold Hanks' balanced salt solution (HBSS) (Thermo Fisher, Belgium) and subsequently collected using 250 µL cold methanol per well. The cells were kept at -20°C until further analyses. Five samples per condition were pooled and centrifuged at 15000 rpm for 15 min at 4°C using a Hettich 220R centrifuge (Geldersmalsen, the Netherlands). Then, samples were evaporated to dryness using a Savant Speedvac concentrator (Thermo Scientific, USA) and reconstituted in 100 µL 50/50 MeOH/ammonium buffer (5 mM adjusted to pH 3.6 with acidic acid). The samples were transferred into autosampler vials for subsequent HPLC and MS/MS analysis. The specification of these analyses can be found in supplementary materials and methods 1 (Suppl. MM1).
2.5. Viability assay
Subcytotoxic concentrations of bosentan were determined using a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide viability assay (MTT), (Sigma, Belgium) (Mosmann, 1983). MTT was dissolved in phosphate-buffered saline (PBS) (Sigma, Belgium) at a concentration of 5 mg/ml (pH = 7.65) and subsequently diluted 10 times with cell culture medium. HepaRG cells were exposed for 24 h to 8 concentrations of bosentan ranging from 20 µM to 1000 µM. Thereafter, the cells were rinsed with PBS and incubated with the MTT solution for 1.5 h at 37°C in a 5% CO2 atmosphere. The resulting dark blue formazan crystals were dissolved in DMSO while shaken on an orbital shaker (VWR, Belgium) for 10 min at room temperature in the absence of light. The solution was measured with a spectrophotometer (PerkinElmer, Belgium) at 560 ± 10 nm. Cytotoxicity was conversely correlated with the measured absorbance. The IC10 concentration value at 24 h was determined using a 5 parameter logistic nonlinear regression analysis of the obtained dose-response curves. This analysis was performed with Masterplex Readerfit 2010 software (Hitashi Solutions, USA). The resulting IC10 or IC10/4 and IC10/10 concentrations were used in all further experiments.
2.6. Transcriptomics analysis
At the time of sampling, HepaRG cells were collected in a RNA protecting solution composed of RNA stabilization reagent (Qiagen, Belgium) and cell culture medium (5:1). Total RNA was extracted from all samples using a GenElute Mammalian Total RNA Purification Miniprep Kit (Sigma, Belgium) according to the manufacturer’s instructions. Purity and quantification of the isolated RNA were determined by spectrophotometric analysis using a Nanodrop spectrophotometer (Thermo Scientific, Belgium). Microarray technologies from Affymetrix (Germany), including reagents and instrumentation, were used for whole genome expression analysis. As such, 100 ng total RNA per sample was amplified using a Genechip 3’ IVT Express Kit following the manufacturer's instructions (Affymetrix, Germany). Amplified RNA was purified with magnetic beads and 15 mg biotin-amplified RNA was treated with the fragmentation reagent. Subsequently, 12.5 µg fragmented amplified RNA was hybridized to Affymetrix Human Genome U133 plus 2.0 arrays and placed in a Genechip Hybridization Oven-645 (Affymetrix, Germany) rotating at 60 rpm at 45°C for 16 h. After incubation, the arrays were washed on a Genechip Fluidics Station-450 (Affymetrix, Germany) and stained with an Affymetrix HWS kit as indicated in the manufacturer’s protocols. The chips were scanned with an Affymetrix Gene-Chip Scanner-3000-7G and quality control matrices were confirmed with Affymetrix GCOS software following the manufacturer’s guidelines. Background correction, summarization and normalization of all data were done with Robust Multiarray Analysis (RMA software), Affymetrix Transcriptome Analysis Console (TAC) Software and Partek Genomics Suite 6.6. Principal component analysis (PCA) plots were produced using Partek Genomics Suite 6.6. Functional toxicological analyses were performed using Ingenuity Pathways Analysis (IPA) and Partek Genomics Suite 6.6. Unless otherwise specified, the statistical thresholds for identification of significant modulated genes was set using Fisher's Exact test (p < 0.05) when using IPA and 1-way analysis of variance (ANOVA) (p < 0.05) when using Partek Genomics Suite 6.6 and TAC.
2.7. Proteomics analysis
The specification of sample preparation for proteomics analysis is outlined in detail in the supplementary materials and methods 2 (Suppl. MM2).
Cell lysis, determination of protein concentration and carbamidomethylation
HepaRG cells, whether or not treated with bosentan, were harvested and lysed with 200 µL of 50 mM Tris-HCl (pH 7.8) containing 150 mM NaCl, 1% sodium dodecyl sulfate (SDS), complete mini ethylenediaminetetraacetic acid (EDTA)-free inhibitor and phosSTOP (Roche Diagnostics, Germany). The samples were frozen immediately after collection and were kept at -80°C until further processing. Upon thawing, 6 µL of benzonase (25 U/µL) and 2 mM MgCl2 (Merck, Germany) were added to the lysates and incubated at 37°C for 30 min. Samples were clarified by centrifugation at 4°C and 18000g for 15 min. Protein concentration of the supernatant was determined by a BCA assay (Thermo Scientific, Germany) according to the manufacturer’s protocol. Cysteines were reduced with 10 mM dithiothreitol (Roche Diagnostics, Germany) at 56°C for 30 min followed by alkylation of free thiol groups with 30 mM iodoacetamide (Sigma Aldrich, Germany) at room temperature in the dark for 30 min.
Sample preparation and trypsin digestion
Sample preparation and proteolysis with trypsin were performed using filter aided sample preparation (FASP) protocol (Manza et al., 2005; Wisniewski et al., 2009) with minor changes. Thus, generated tryptic peptides were acidified (pH < 3) with trifluoroacetic acid and were quality controlled as described previously (Burkhart et al., 2012).
iTRAQ 8plex labeling, phosphopeptides enrichment and high pH reversed phase (RP) fractionation
Per sample, 50 µg of peptides were labelled using iTRAQ reagents (8plex) (Ross et al., 2004) according to the manufacturer’s instructions (AB SCIEX, Germany). After labelling and pooling of samples, the multiplexed sample (i.e. 350 µg) of each experiment was subjected to the enrichment of phosphopeptides based on the TiO2 chromatography protocol as previously described (Dickhut et al., 2014; Palmisano et al., 2012). For complete proteome analysis, the flow-through of each experimental time point from the TiO2 chromatography, which mostly consists of non-phosphorylated and unmodified peptides, was fractionated (i.e. 25 µg) by reversed phase chromatography at pH 8.0 on a Biobasic (C18; 0.5 x 150 mm; 5 µm particle size) column using an UltiMate 3000 LC system (Thermo Scientific, Germany). In total, 12 fractions were collected at 1 min intervals from min 10 to 80 in a concatenation mode.
LC-MS/MS analyses
All samples (i.e. global proteome) were dried completely, resolubilized in 15 µL of 0.1% trifluoroacetic acid (TFA) and were analysed by nano-LC-MS/MS using an Ultimate 3000 nano RSLC system coupled to a Q Exactive HF mass spectrometer (Thermo Scientific, Germany). HPLC and MS settings are described in detail in supplementary material and methods 2 (Suppl. MM2).
MS data analysis and evaluation
All iTRAQ raw data were processed with Proteome Discoverer 1.4 (Thermo Scientific, Germany). To maximize the number of peptide spectrum matches (PSMs), 3 different search algorithms were included, namely Mascot (Perkins et al., 1999), Sequest (Eng et al., 1994) and MS Amanda (Dorfer et al., 2014) using the same set of parameters, namely precursor and fragment ion tolerances of 10 ppm and 0.02 Da for MS and MS/MS, respectively; trypsin as enzyme with a maximum of 2 missed cleavages; carbamidomethylation of Cys, iTRAQ - 8plex on N-terminus and Lys as fixed modifications; oxidation of Met, phosphorylation (only for the phosphoproteome data) of Ser, Thr and Tyr as variable modifications. For the phosphoproteome data analysis, the phosphoRS (Taus et al., 2011) (version 3.1) node was used to score localization probabilities for the identified phosphorylation sites. Global data were exported with the following filter criteria: a false discovery rate (FDR) < 1% on the peptide-spectrum-match level (high confidence setting), search engine rank 1 and only proteins that were quantified with ≥ 2 unique peptides. For the phosphoproteome data, only unique PSMs filtered with Percolator (Kall et al., 2007) (FDR < 1%) and with a phosphoRS probability ≥ 90% were considered. Normalization of raw iTRAQ data and subsequent statistical evaluation are described in detail in supplementary material and methods 2 (Suppl. MM2).
2.8. Metabolomics analysis
For metabolomics analysis, 550 μL culture medium and 50 μL 11.6 mM 4,4-dimethyl-4-silapentane-[1,1,2,2,3,3-2H6]-1-ammonium trifluoroacetate (Onyx Scientific Limited, UK) in deuterium oxide as internal standard were mixed and transferred to glass 5 mm nuclear magnetic resonance (NMR) tubes. High-resolution 1H NMR spectra of cell culture media were acquired at 14.1 T (600.13 MHz 1H frequency) using a Bruker AVANCE 600 spectrometer (BrukerBiospin, Germany). All spectra were acquired using a Carr-Purcell-Meiboom-Gill (CPMG) pulse sequence with presaturation and a T2 relaxation delay of 32 ms. The sum of 64 free induction decays was collected into 64 K data-points with a spectral width of 12,019.230. After acquisition, exponential line broadening of 0.3 Hz was applied prior to Fourier transformation. 1H NMR spectra were then imported and manipulated in Matlab (Mathworks, USA) using in-house codes for automatic phasing, baseline correction and integration. Chemical shifts were referenced to the DSA resonance at δ0. The observed resonances were assigned to specific metabolites by database matching using the Chenomx NMR suite (Chenomx Incorporated, Canada) and the Human Metabolome Database.
3. Results
3.1. Selection of working concentrations of bosentan
Working concentrations of bosentan were assessed by means of a conventional MTT viability assay, whereby HepaRG cells were exposed for 24 h to a range of bosentan concentrations going from 20 to 1000 µM. The calculated values for IC10 and IC50 concentrations were 250 ± 55 µM and 744 ± 197 µM, respectively. In order to establish concentration/effect relationships, 3 concentrations of bosentan were tested in subsequent experiments, namely 250 µM (IC10), 62.5 µM (IC10/4) and 25 µM (IC10/10). Similarly, 3 exposure regimes were applied, namely 1 h exposure, 24 h exposure and 24 h exposure followed by a 72 h wash-out period. The 1 h exposure condition was based upon the previously published observation that bosentan can affect bile acid transport in primary hepatocytes already after 10 min (Kemp et al., 2005). The rationale of the wash-out step was to test the reversibility of the effects induced by bosentan.
In a first series of experiments, HepaRG cells were exposed to the 3 concentrations of bosentan in the 3 exposure regimes, followed by transcriptomics and metabolomics analyses. Based on the obtained transcriptomics results, the conditions for which the highest gene expression changes were observed was used in a second set of experiments. HepaRG cells were hereby only exposed to IC10 concentration for 1 h, 24 h and 24 h followed by the 72 h wash-out period. For these experimental conditions, proteomics analyses were performed. For analysis of BSEP functionality as well as for the determination of endogenous bile acid levels, HepaRG cells were exposed to IC10 concentration for 24 h.
3.2. Characterization and induction of the MIE
The cellular localization of BSEP in differentiated human hepatoma HepaRG cells was evaluated by immunostaining. As anticipated, BSEP was localized at the bile canalicular pole of the hepatocyte-like cells (Fig. 1A). This result confirms previously published findings showing the presence of this drug transporter in HepaRG cells and the suitability of this hepatic cell line for studying drug-induced cholestasis (Bachour-El Azzi et al., 2015; Le Vee et al., 2006; Qiu et al., 2016). The expression of ABCB11 (BSEP) was unchanged when the cells were exposed to bosentan for 1 h. However, when the exposure time was increased to 24 h, a significant downregulation was observed for the 2 highest concentrations (Fig. 1B). After the 72 h wash-out period, BSEP abundance remained significantly lower in the cells exposed to IC10 concentration compared to control samples. For cells exposed to IC10/4 concentration, gene expression returned to normal levels. Noteworthy, exposure to the lowest concentration did not influence ABCB11 expression at any time point. The downregulation of ABCB11 may have been caused by a compensatory effect in which inhibition of BSEP leads to a reduction in the expression of ABCB11. Downregulation of BSEP expression has also been observed in precision-cut human liver slices exposed to cholestatic compounds (Vatakuti et al., 2017).
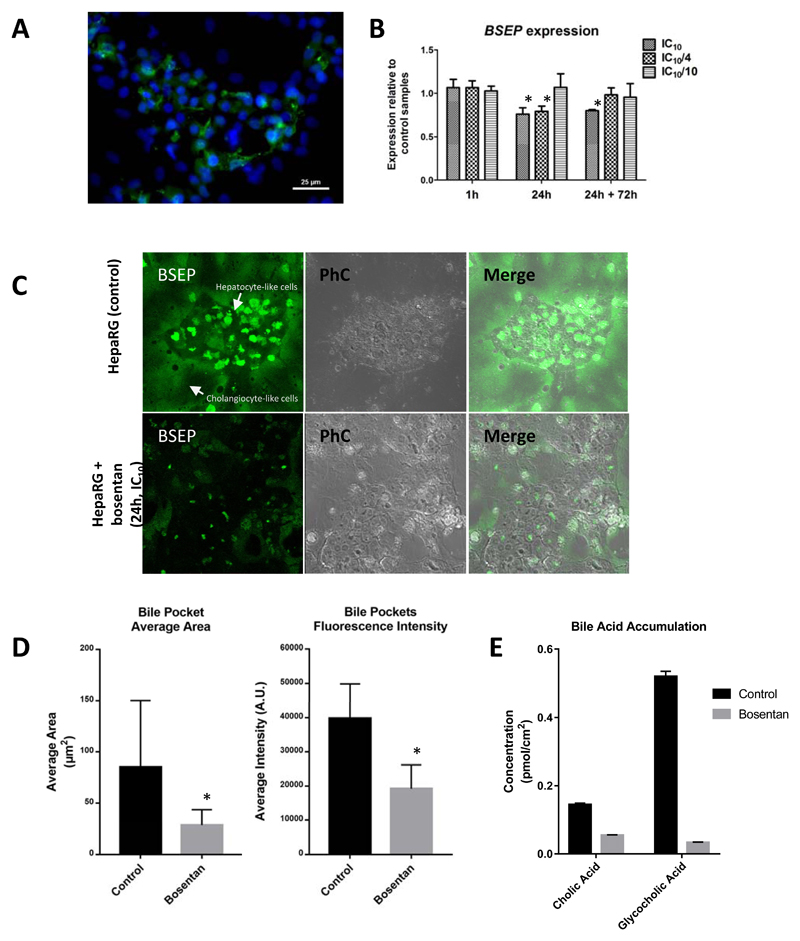
Fig. 1.
(A) Immunostaining of BSEP (green) and nuclear staining (blue) of differentiated HepaRG cells showing BSEP localization. (B) Normalized relative gene expression (microarray) of BSEP (coded by the ABCCB11) in HepaRG cells exposed to bosentan (* Student t-test p < 0.05). (C) Accumulation of fluorescent probe substrate for BSEP (green) in bile canicular structures within the hepatocyte-like cell cluster of untreated HepaRG cells and cells exposed to bosentan (24 h/IC10 concentration). Phase contrast (PhC) images show hepatocyte-like clusters of HepaRG cell culture surrounded by biliary like cells. (D) Quantification of fluorescence intensity in bile pockets of the bile pocket area in untreated cells and cells exposed to bosentan (24 h/IC10 concentration) (* Student t-test p < 0.05). (E) HPLC-MS/MS quantification of cholic acid (CA) and glycocholic acid (GCA) (pmol/cm2) in HepaRG cell pellets of control samples and samples from cells exposed to bosentan.
Functional BSEP inhibition was observed in HepaRG cells exposed for 24 h to IC10 concentration. Untreated controls showed a much higher accumulation of the BSEP probe substrate tauronor-THCA-24-DBD in the bile canaliculi compared to the cells exposed to bosentan (Fig. 1C). Although this effect was evident, the inhibition of BSEP was not absolute, as bile canalicular structures in the bosentan-treated cells still showed some accumulation of the fluorescent probe substrate. Quantification of fluorescence intensity of the BSEP probe substrate in bile pockets, and determination of the bile pocket area in untreated cells and cells exposed to bosentan (24 h/IC10 concentration), confirmed a decrease in BSEP functionality (Fig. 1D).
The accumulation of 2 bile acids, namely cholic acid (CA) and glycocholic acid (GCA), was determined in HepaRG cell pellets of control samples and samples from cells exposed to bosentan (24 h/IC10 concentration). Total CA and GCA concentrations were lower in the bosentan-treated samples compared to control samples (Fig. 1E). Overall, these results are consistent with pronounced interference of bosentan with handling of endogenous bile acids in HepaRG cells. Yet, it must be emphasized that no distinction could be made between intracellular accumulation and accumulation in bile pockets as total bile acid concentrations were determined, thus reflecting the sum of the intracellular and intracanalicular levels.
3.3. Characterization and occurrence of the KEs
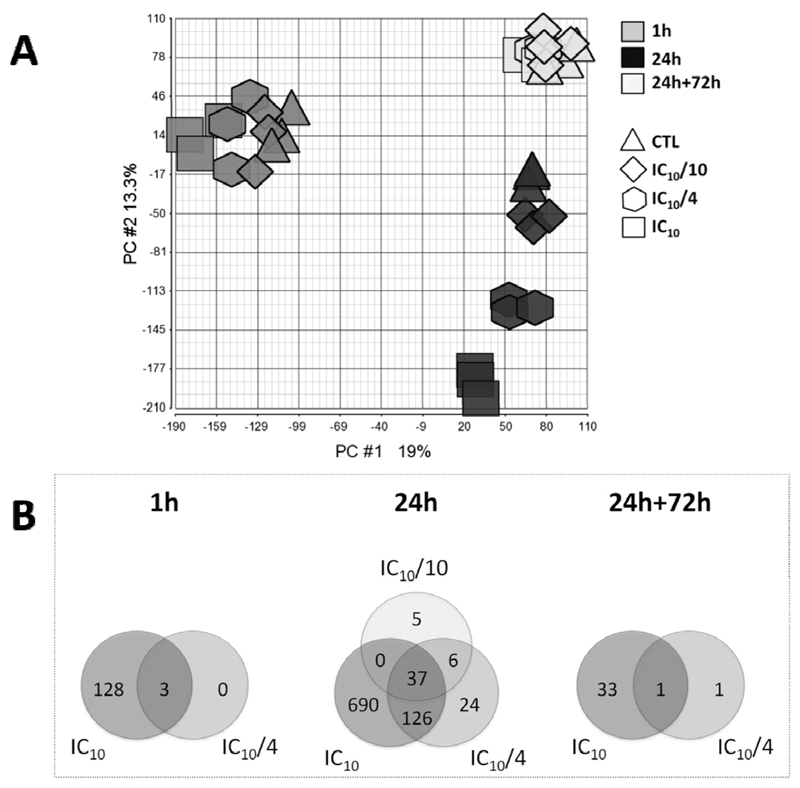
PCA evaluation of whole genome microarray data showed no differences among the different tested cell batches. The highest source of variation in the data pool was introduced by different exposure times as shown by the selective grouping of the 1 h, 24 h and 24 h+72 h wash-out samples (Fig. 2A). In addition, a clear shift with increasing bosentan concentration could be noticed in the 24 h exposure samples. This also held true for the 1 h exposure samples, albeit in a less pronounced way. On the contrary, no distinction could be made between different bosentan concentrations in the 24 h+72 h wash-out samples (Fig. 2A). The number of bosentan-modulated genes (i.e. fold change higher than 2 when compared to respective controls and Fisher’s p < 0.05) was limited after 1 h exposure and reached a maximum after 24 h of exposure. A wash-out period of 72 h reverted the effects to baseline levels (Fig. 2B). Cluster analysis, based on significantly expressed genes in the different tested conditions, showed that transcriptional modifications induced after 24 h exposure to IC10 concentration strongly differed from those observed in the other experimental conditions (Suppl. Fig. S2). Within the 24 h exposure samples, IC10 and IC10/4 concentration data showed a high pattern similarity, whereas the IC10/10 concentration data displayed only small variations compared to the control samples. Furthermore, the close proximity of the 24 h+72 h wash-out samples to the 24 h exposure control samples in the clustering dendogram suggests a return to basal gene expression levels upon wash-out (Suppl. Fig. S2).
Fig. 2.
(A) Principle component analysis (PCA) plot of microarray data sets of HepaRG cells exposed for 1 h, 24 h and 24 h+72 h to 3 concentrations of bosentan, namely IC10, IC10/4 and IC10/10, and respective vehicle controls. (B) Venn diagram of modulated genes in HepaRG cells exposed to different concentrations of bosentan (IC10, IC10/4 and IC10/10) for 1 h, 24 h and 24 h+72 h (gene selection was based on a fold change modulation > 2 when compared to respective controls and Fisher’s p < 0.05).
Functional pathway analysis of the modulated genes indicated the induction of a number of specific hepatotoxic responses in a concentration-dependent way (Table 1). In this respect, “liver cholestasis” was the most significantly enriched gene class followed by “liver necrosis” and “liver damage”. Exposure to the highest concentration of bosentan consistently triggered the highest number of transcriptional changes in each of these hepatotoxicity classes (Table 1). Particularly, HepaRG cells exposed for 24 h to IC10 concentration showed a high significant enrichment of genes of the “liver cholestasis” gene class (Table 1).
Table 1.
Enriched toxicological gene classes, identified by Ingenuity Pathway Analysis, in HepaRG cells exposed for 24 h to bosentan at 3 different concentrations, namely IC10, IC10/4 and IC10/10.
| Ratio (number of modulated genes/ total number genes) |
Fisher’s
Exact p-value |
||
|---|---|---|---|
| Liver cholestasis | 24 h IC10 versus control |
10.4 % (23/221) |
1.58E-10 |
| 24 h IC10/4 versus control |
5.0 % (11/221) |
1.41E-08 | |
| 24 h IC10/10 versus control |
2.7% (6/221) |
6.34E-06 | |
| Liver necrosis | 24 h IC10 versus control |
4.8 % (28/583) |
8.66E-05 |
| 24 h IC10/4 versus control |
1.5 % (9/583) |
3.33E-03 | |
| 24 h IC10/10 versus control |
0.7 % (4/583) |
6.79E-03 | |
| Liver damage | 24 h IC10 versus control |
4.3 % (28/656) |
3.64E-04 |
| 24 h IC10/4 versus control |
1.7 % (28/656) |
7.88E-04 | |
| 24 h IC10/10 versus control |
0.8 % (28/656) |
8.41E-04 |
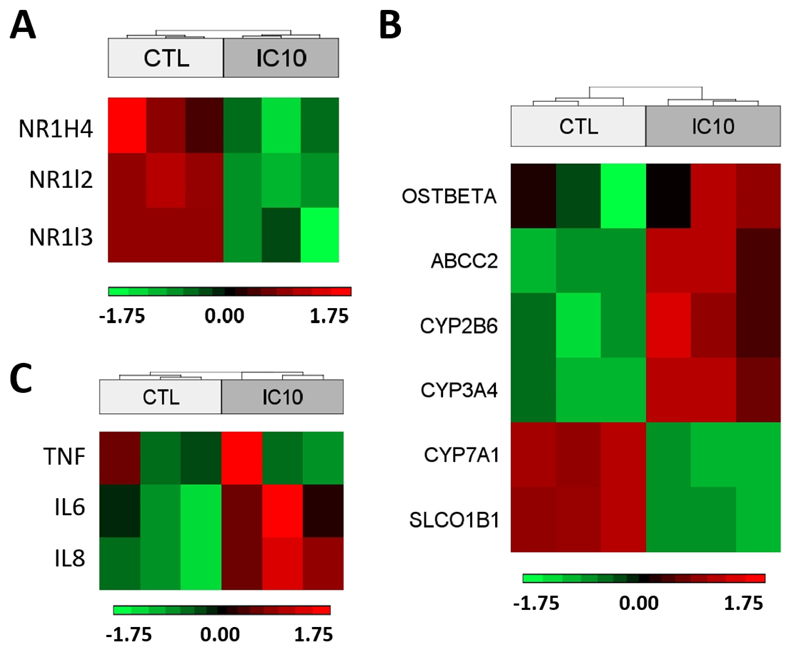
As predicted by the AOP, the inhibition of BSEP in casu provoked by bosentan, leads to activation of the nuclear receptors FXR, PXR and CAR (Vinken et al., 2013). This activation induces a number of transcriptional changes that are linked to the removal of the excess of bile acids and their metabolites. When activated, FXR translocates to the nucleus, where it forms a dimer with RXR, which binds to the hormone response elements in specific gene promoters leading to modulation in transcriptional activity (Beuers, 2015; Forman et al., 1995). In this context, Table 2 shows genes regulated by active FXR-RXR dimers in HepaRG cells exposed for 24 h to IC10 concentration. Six transcriptional changes induced by the activation of the nuclear receptors were reproduced in compliance with AOP prediction (Fig. 3B). More specifically, 4 genes (OSTBETA, ABCC2 (MRP2), CYP2B6 and CYP3A4) were upregulated and 2 genes (CYP7A1 and SLCO1B1 (SLCO1B1)) were downregulated (Suppl. Table S2). These changes are part of the adaptive cellular response that aims at counteracting the primary cholestatic insults induced by bosentan. It should be mentioned that murine CYP2B10 included in the AOP was replaced during this analysis by its human counterpart CYP2B6. Modulation of OSTALPHA, UGT2B4, SULT2A1 and ABCC3, which are also under control of the activation of the same nuclear receptors, was inconsistent with the AOP (Suppl. Table S2). The reason for this discrepancy is unknown and deserves further scrutiny.
Table 2.
Normalized expression of a selection of genes regulated by active FXR-RXR dimers in HepaRG cells exposed for 24 h to bosentan in IC10 concentration.
| Gene | Fold change (bosentan versus control) |
p-value (Fisher’s Exact) |
|---|---|---|
| ABCB4 | -2.9 | 5.93E-05 |
| HNF4A | -2.0 | 1.15E-03 |
| MTTP | -4.7 | 8.88E-03 |
| NR1H4 | -2.4 | 2.06E-03 |
| PCK2 | -2.5 | 9.33E-03 |
| PON1 | -4.4 | 5.79E-03 |
| PPARG | -2.1 | 2.90E-02 |
| PPARGC1A | -2.6 | 6.84E-05 |
| SLC51A | 3.0 | 2.29E-02 |
| SLCO1B1 | -2.6 | 7.90E-06 |
| UGT2B4 | -5.1 | 7.50E-04 |
Fig. 3.
Heat map of NR1H4, NR1I2 and NR1I2 genes representing the downregulation of FXR, PXR and CAR in HepaRG cells exposed to bosentan (24 h/IC10 concentration) (A). Heat map of a selection of modulated genes in HepaRG cells exposed to bosentan (24 h/IC10 concentration) that correctly represent the corresponding KE of the AOP for cholestatic liver injury (Vinken et al., 2013) (B). Heat map of tumour necrosis factor (TNF), interleukin 6 (IL6) and interleukin 8 (IL8) in HepaRG cells exposed to bosentan (24 h/IC10 concentration) (C).
Importantly, the microarray analysis equally revealed a number of transcriptional changes that have not yet been included in the AOP. As such, the expression of the genes coding for PXR, FXR and CAR, namely NRIH4 (1.2-fold), NR1I2 (2.4-fold) and NR1I3 (2.4-fold), respectively, was significantly downregulated (Fig. 3A). Suppression of FXR expression has also been observed in precision-cut human liver slices exposed to cholestatic compounds (Vatakuti et al., 2017). This probably reflects a compensatory response to the their functional activation. Downregulation of FXR production was previously observed in human cholestasis (Alvarez et al., 2004; Chen et al., 2004; Demeilliers et al., 2006) as well as in murine cholestasis models (Szalowska et al., 2013). FXR is a mediator of bile acid synthesis, but also controls the expression of alcohol dehydrogenases (ADH). Its downregulation in cells exposed to bosentan is further propagated and can indeed be observed by the downregulation of several isoforms of ADH. These enzymes are known to play a direct role in bile acid biosynthesis (Langhi et al., 2013). In parallel, several isoforms of ADH have been identified by selecting only those genes that are consistently modulated in cells exposed to different concentrations of bosentan. Hereby, a set of 37 genes was identified that might be specific for drug-induced hepatotoxicity (Fig. 2B and Suppl. Table S3). Several of these transcriptional changes (8 out of the 37 genes) were parallelled by similar modifications at the protein level (Table 3). This list also contains fatty acid binding protein, which was downregulated. A downregulation of this gene has been previously reported to contribute to hepatic oxidative stress and consequently to the pathogenesis of biliary disease (Wang et al., 2007).
Table 3.
Molecules consistently modulated at gene and protein expression levels in HepaRG cells exposed to decreasing concentrations of bosentan.
| Gene expression | Protein expression | |||||
|---|---|---|---|---|---|---|
| Gene name | Family | Fold change | p-value | Fold change | p-value | |
| ADH1A | alcohol dehydrogenase 1A | enzyme | -12.39 | 0.0050 | -1.52 | 0.0007 |
| ADH1B | alcohol dehydrogenase 1B | enzyme | -11.59 | 0.0025 | -1.31 | 0.0213 |
| ADH1C | alcohol dehydrogenase 1C | enzyme | -10.67 | 0.0041 | -1.24 | 0.0049 |
| ADH4 | alcohol dehydrogenase 4 | enzyme | -12.78 | 0.0061 | -1.39 | 0.0021 |
| ALDOB | aldolase, fructose-bisphosphate B | enzyme | -46.97 | 0.0033 | -1.52 | 0.0001 |
| CPS1 | carbamoyl-phosphate synthase 1 | enzyme | -27.40 | 0.0013 | -1.22 | 0.0034 |
| FABP1 | fatty acid binding protein 1 | transporter | -6.42 | 0.0001 | -1.50 | 0.0005 |
| SLC3A2 | solute carrier family 3 member 2 | transporter | 5.85 | 0.0000 | 2.25 | 0.0000 |
The enhanced transcription of genes coding for pro-inflammatory tumour necrosis factor (TNF), interleukin 6 (IL6) and interleukin 8 (IL8) were observed in HepaRG cells treated with bosentan, thereby suggesting induction of oxidative stress and inflammation (Fig. 3C). Simultaneously, several genes related to the production of reactive oxygen species (ROS) were also found to be upregulated, while those that counteract oxidative stress were downregulated (Suppl. Fig. S4). As such, activation of production of ROS was mainly modulated by an upregulation of thioredoxin reductase 1 (TXNRD1), glutamate-cysteine ligase catalytic subunit (GCLC), metallothionein 1X (MT1X), heme oxygenase 1 (HMOX1), NAD(P)H quinone dehydrogenase 1 (NQO1) and superoxide dismutase 2 (SOD2) (Suppl. Fig. S4). In parallel, the repression of ROS producing systems is supported by the downregulation of nuclear factor I X (NFIX) and xanthine dehydrogenase (XDH), which contribute to the inhibition of oxidases (Morel et al., 1999).
Several transcriptomics findings were confirmed by proteomics analysis. By using iTRAQ technology, 3578, 3698 and 3500 proteins (with ≥ 2 unique peptides and 1% FDR) could be relatively quantified in HepaRG cells exposed to IC10 concentration for 1 h, 24 h and 24 h+72 h wash-out, respectively. Of these quantified proteins, 3 (~0.08%), 145 (~4%) and 109 (~3%) proteins were differentially altered at the respective time points. The correlation between the obtained proteomics and transcriptomics data was determined using log2-fold change data to calculate Pearson’s r coefficient. The 24 h exposure and the 24 h+72 h wash-out samples showed the highest correlation, with Pearson’s r coefficient values of 0.4534 and 0.2937, respectively (Suppl. Fig. S3). On the contrary, for the 1 h exposure samples, a low correlation coefficient of 0.0399 between transcriptomics and proteomics data was found. The latter can be due to insufficient time between evaluation of gene modulation and the corresponding translational changes.
Functional pathway analysis of the identified proteins showed the enrichment of toxicological classes “cholestasis” and “intrahepatic cholestasis” in HepaRG cells exposed for 24 h to the highest concentration of bosentan. Hereby, 22.8% and 40.9%, respectively, of the molecules in each toxicological gene class were modulated (Suppl. Table S1). These proteomics data support the transcriptomics findings and clearly show the occurrence of the cholestatic cellular response in HepaRG cells exposed to bosentan.
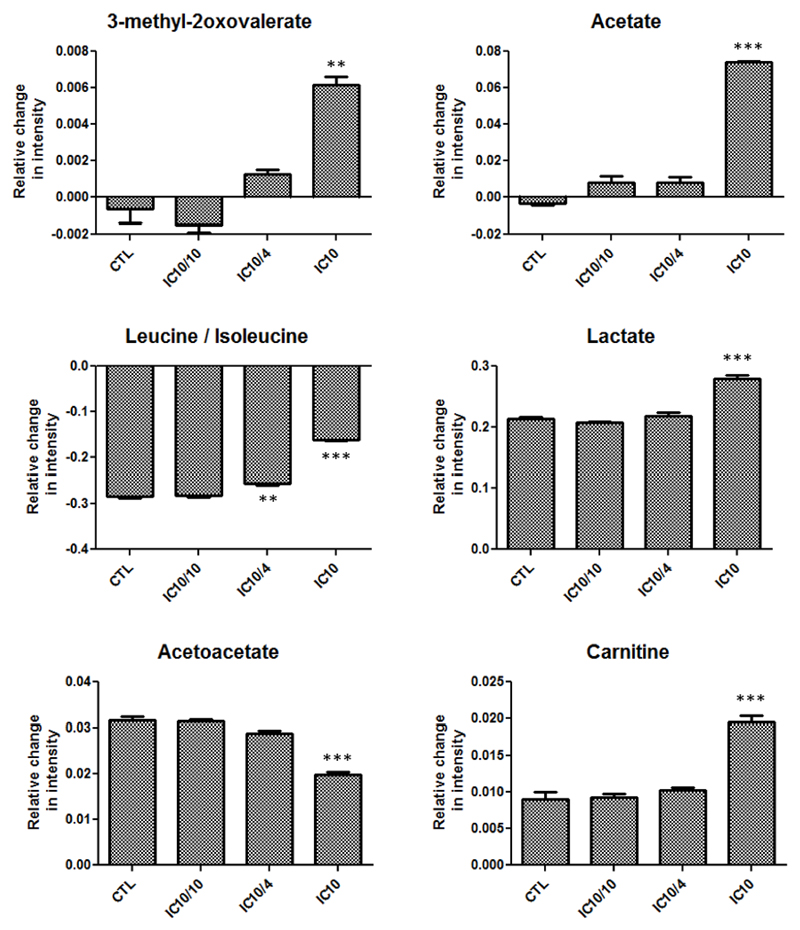
Compared to the transcriptomics and proteomics data, the number of the identified endogenous metabolites by metabolomics analysis of the cell supernatants was relatively low. Nevertheless, some of the identified metabolites were significantly modulated in HepaRG cells exposed to the highest concentration of bosentan. Thus, a concentration-dependent increase was noticed for 3-methyl-2-oxovalerate, carnitine, acetate, acetoacetate and lactate. Additionally, reduced amino acid uptake, in particular for leucine and isoleucine quantification, was observed (Fig. 4). These findings collectively point to mitochondrial impairment caused by bosentan and therefore corroborate with mitochondrial permeability pore transition, which is a KE of the AOP (Labbe et al., 2008; Lynch et al., 2014; Vinken et al., 2013). 3-Methyl-2-oxovalerate is a metabolite of isoleucine that belongs to the branched chain amino acid metabolites (BCAA). BCAA metabolites are often associated with mitochondrial dysfunction, stress signalling and apoptosis (Lynch et al., 2014). The reduced amino acids leucine/isoleucine quantities further suggest their catabolism towards BCAA metabolites, hereby also pointing to mitochondrial dysfunction. Carnitine, which is a non-essential amino acid, is responsible for carrying fatty acids across the inner mitochondrial membrane for β-oxidation (Bremer, 1983). A correlation exists between plasma concentrations of carnitine and mitochondrial dysfunction (Adeva-Andany et al., 2017). The observed increased levels of acetate and lactate indicate a higher rate of glycolysis and hence impairment of fatty acid oxidation in mitochondria. The increase of acetoacetate levels again suggests a decrease in mitochondrial function (Begriche et al., 2011).
Fig. 4.
Alterations in metabolite production detected by NMR spectroscopy of media from HepaRG cells exposed to bosentan (24 h/IC10 concentration) compared to control samples (** Student t-test p < 0.01; *** Student t-test p < 0.001).
4. Discussion
The goal of the present study was to characterize the hepatotoxic effects of the drug bosentan at the transcriptomic, proteomic and metabolomic level in vitro and to anchor this in the previously introduced AOP on cholestatic liver injury (Vinken et al., 2013). A number of in vitro models are currently in use to study cholestatic liver injury, among which human hepatoma HepaRG cells. Cholestatic responses can be triggered in human hepatoma HepaRG cells by exposure to cholestatic drugs alone, in casu bosentan, without exogenous addition of high concentrations of bile acids (Burbank et al., 2017; Sharanek et al., 2016). Therapeutic oral doses of bosentan typically vary around 125 mg twice a day, corresponding with clinical Cmax/total values up to 7.5 µM (Chatterjee et al., 2014; Dingemanse and van Giersbergen, 2004). Although this value is 10 times lower than the concentrations necessary to inhibit BSEP, cholestasis may still occur during therapy (Dawson et al., 2011; Lepist et al., 2014). Most likely, this is due to active uptake and accumulation of bosentan in hepatocytes, thereby locally increasing its concentration in the liver (Muller and Milton, 2012). Therefore, the actual concentration in hepatocytes that causes cholestatic injury in vivo is thought to be considerably higher than Cmax/total and might be within the concentration range tested in the present in vitro study, namely 250 µM (IC10), 62.5 µM (IC10/4) and 25 µM (IC10/10).
Several studies have been conducted to demonstrate that HepaRG cells express the transporters and enzymes involved in bile acid disposition in hepatocytes in vivo, although differences in expression and activity levels exist (Bachour-El Azzi et al., 2015). Moreover, in a recently conducted in vitro study (Sharanek et al., 2015), HepaRG cells were shown to support endogenous bile acid synthesis and bile acid disposition processes. These findings justified the use of HepaRG cells in the present study to explore the expected interference of bosentan with bile acid homeostasis. It was found that accumulation of the marker bile acids CA and GCA in the combined intracellular and intracanalicular compartments of HepaRG cells was very low. This result is in agreement with the data by Sharanek and colleagues (Sharanek et al., 2015), reporting undetectable accumulation in HepaRG cells at the 24 h time point. The higher sensitivity of the bioanalytical method used in the present study can explain this difference. Interestingly, treatment of the HepaRG cells with bosentan induced a very pronounced reduction in the accumulated amounts of both CA and GCA. Recognizing the known effects of bosentan on both sodium-taurocholate cotransporting polypeptide (NTCP) and BSEP as well as on OATPs provides a good starting point for interpretation of the observed effect of bosentan. Lepist and group (Lepist et al., 2014) reported IC50 values for inhibition of hepatic transporters by bosentan, namely organic anion transporter 1B1 OATP1B1 (5.0 µM), OATP1B3 (5.2 µM), NTCP (36 µM) and BSEP (42 µM). This non-selective profile of bosentan in terms of inhibition of transporters involved in hepatic bile acid handling implies that exposure of HepaRG cells to bosentan will eventually result in reduced cellular uptake of bile acids as well as reduced canalicular efflux of bile acids. Although the IC50 value for BSEP inhibition by bosentan is relatively high, it should be noted that intracellular accumulation of bosentan will most likely result in intrahepatic concentrations that are sufficiently high to cause BSEP inhibition even at therapeutically relevant concentrations. This concept is also in line with in vitro data obtained in sandwich-cultured human hepatocytes, indicating an intracellular to extracellular bosentan concentration ratio of more than 10-fold for an extracellular dose concentration of 10 µM (Lepist et al., 2014). Importantly, the experimental set-up used in the present study likely did not provide the most sensitive approach to indirectly detect BSEP inhibition, as bile acids levels were determined on cell lysates, including canaliculi. In other words, shifts in bile acid levels between canaliculi and cells are not expected to be observed. On the other hand, as bosentan also influences bile acid uptake, intracellular bile acid levels are likely to gradually shift to the extracellular medium at the expense of intracellular levels. Indeed, as bile acids are effluxed to the medium by sinusoidal efflux transporters, including MRP3 and organic solute transporter α and β (OSTα/β), re-uptake will be blocked by bosentan due to NTCP and OATP1B1/3 inhibition. Furthermore, bile acids that had accumulated in bile pockets prior to addition of bosentan will be gradually released in the extracellular medium via bile canalicular contraction (Reif et al., 2015). Again, NTCP and OATP1B1/3 inhibition by bosentan will result in decreased re-uptake of these bile acids. Taken together, the inhibition of hepatic bile acid uptake by bosentan is expected to result in a shift of the total bile acid pool in the cell cultures from intracellular to extracellular. This scenario is fully consistent with the observations in the present study. Moreover, the present observation of reduced CA and GCA levels in HepaRG cells exposed to bosentan is completely in line with previously reported findings in sandwich-cultured human hepatocytes, namely that total GCA accumulation at 24 h was reduced by 3- to 6-fold in the presence of 10-100 µM bosentan (Lepist et al., 2014). In the same study, similar effects were seen for glycochenodeoxycholic acid. In a more recent study (Burbank et al., 2017), exposure of HepaRG cells to 10-100 µM bosentan also resulted in a pronounced decrease in cellular levels of total bile acids, including CA and GCA, both at the 4 h and 24 h time points. It is noteworthy that a reduction in bile acid levels has also been observed previously in HepaRG cells exposed to the cholestatic compound cyclosporin A (Sharanek et al., 2015). In view of the known effects of bosentan and cyclosporin A on multiple hepatic transporters, the findings of the present study as well as previous results confirm the importance of evaluating the possible influence of cholestatic drugs on all bile acid transporters, not just BSEP. Further experiments, at multiple earlier time points, and including determination of additional bile acids, if detectable in the cell cultures, also in the extracellular medium, will be required to generate evidence for this hypothesis regarding the in vitro interaction of bosentan, and other cholestatic compounds, with bile acids.
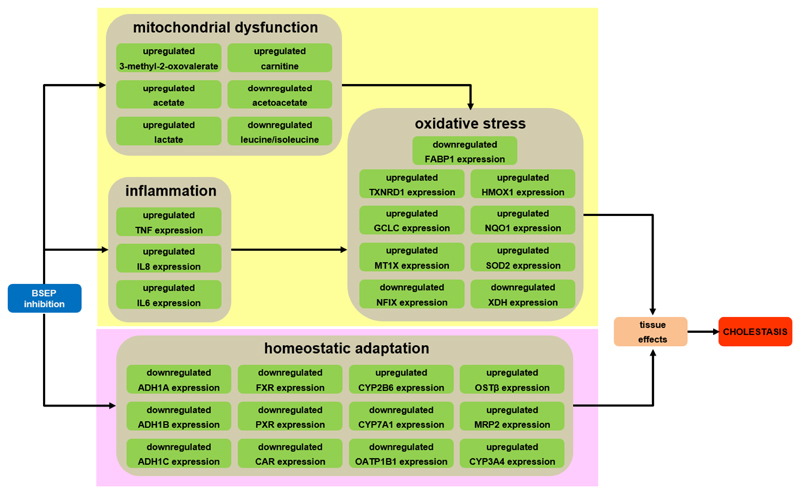
In analogy with the established AOP from BSEP inhibition to cholestatic liver injury (Vinken et al., 2013), the KEs picked up in the present study reflect a deteriorative cellular response and an adaptive cellular response (Fig. 5). The main MIE is the inhibition of BSEP, which mediates efflux of bile acids from the hepatocyte cytosol into the bile canaliculi (Vinken et al., 2013). BSEP inhibition is indeed frequently linked to the clinical manifestation of cholestasis (Dawson et al., 2011; Kis et al., 2012). As expected, exposure to bosentan inhibited BSEP functionality in the established human hepatoma-derived HepaRG cell culture system, as evidenced by lower accumulation of a fluorescent probe substrate tauro-nor-THCA-24-DBD, which is specifically transported by BSEP. This is in line with data published by other groups that tested bosentan in sandwich-cultured primary hepatocytes (Chatterjee et al., 2014; Hartman et al., 2010; Lepist et al., 2014). It was also observed that the expression of the BSEP gene ABCB11 was downregulated in HepaRG cells exposed to the highest concentrations of bosentan. The altered expression of ABCB11 by BSEP inhibitors has been previously associated with drug-induced cholestatic liver toxicity (Garzel et al., 2014). Furthermore, it should be stressed that adverse effects are rarely induced by merely 1 trigger. In this respect, BSEP inhibition does not necessarily lead to cholestasis and, vice versa, cholestasis is not always associated with BSEP modulation. Rather, drugs tend to simultaneously inhibit a number of transporters in addition to BSEP, which synergistically triggers cholestatic injury (Köck et al., 2014; Mita et al., 2006). Bosentan was indeed found to negatively affect the activity of multidrug resistance-associated protein 2 (MRP2) in HepaRG cells (Suppl. MM3; Suppl. Fig. S5). In fact, in follow-up initiatives of the present study, the proposed AOP will be further elaborated in order to establish a so-called AOP network, which includes multiple MIEs. Unlike individual AOP constructs, such AOP networks provide a better reflection of the in vivo complexity of toxicological effects (Vinken et al., 2017). Also, next series of experiments will include several additional cholestatic drugs as well as alternative BSEP inhibitory strategies, such as RNA interference technology. This will allow to test the overall validity of the results of the present study, using only bosentan, which may also cause toxicity through mechanisms not related to BSEP inhibition or cholestasis as such, as well as to challenge the robustness of the AOP on cholestatic liver injury.
Fig. 5.
In vitro specifications of KE (green) related to the deteriorative cellular response (yellow background) and adaptive cellular response (pink background) of the AOP of cholestasis (Vinken et al., 2013).
Supplementary Material
Acknowledgements
This work was financially supported by the grants of European Union (FP7)/Cosmetics Europe (SEURAT-1 projects DETECTIVE (HEALTH-F5-2010-266838) and HeMiBio (HEALTH-F5-2010-266777)), the European Research Council (Starting Grant 335476), the Fund for Scientific Research-Flanders (FWO grants G009514N, G010214N, G012318N, G020018N and 12H2216N), the University Hospital of the Vrije Universiteit Brussel-Belgium (“Willy Gepts Fonds” UZ-VUB) and the Center for Alternatives to Animal Testing (CAAT) at Johns Hopkins University Baltimore-USA. The authors also gratefully acknowledge the financial support from the Ministerium für Innovation, Wissenschaft und Forschung des Landes Nordrhein-Westfalen, Senatsverwaltung für Wirtschaft, Technologie und Forschung des Landes Berlin, and the Bundesministerium für Bildung und Forschung. The authors like to thank Dr. Christophe Chesné (Biopredic) for making HepaRG cells available, Dr. Stefan Vinckier for assistance with confocal microscopy and Miss Tineke Vanhalewyn for technical assistance.
List of abbreviations
- ABCB11
ATP binding cassette subfamily B member 11
- ABCC2
ATP binding cassette subfamily C member 2
- ADH
alcohol dehydrogenase
- AOP
adverse outcome pathway
- BCA
bicinchoninic acid assay
- BCAA
branched chain amino acid metabolites
- BSEP
bile salt export pump
- CA
cholic acid
- CAR
constitutive androstane receptor
- CPMG
Carr-Purcell-Meiboom-Gill
- CYP2B6
cytochrome P450 2B6
- CYP3A4
cytochrome P450 3A4
- DAPI
4’,6-diamidino-2-phenylindole
- DMSO
dimethyl sulfoxide
- FASP
filter aided sample preparation
- FDR
false discovery rate
- FXR
farnesoid X receptor
- GCA
glycocholic acid
- HBSS
Hank’s balanced salt solution
- IPA
ingenuity pathways analysis
- IL6
interleukin 6
- IC10
inhibitory concentration of 10% (cell viability)
- IC50
inhibitory concentration of 50% (cell viability)
- IL8
interleukin 8
- iTRAQ
isobaric tags for relative and absolute quantification
- KE(s)
key event(s)
- LC-MS/MS
liquid chromatography-mass spectrometry/mass spectrometry
- LFC
log2 fold change
- MIE
molecular initiating event
- MRP2
multidrug resistance-associated protein 2
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- NMR
nuclear magnetic resonance
- NTCP
sodium-taurocholate cotransporting polypeptide
- OATP1B1/3
organic anion transporter 1B1/3
- OSTα/β
organic solute transporter α and β
- PCA
principal component analysis
- phosSTOP
phosphatase inhibitor
- PSMs
peptide spectrum matches
- PSW-ANOVA
probe sliding window-analysis of variance
- PTX
proteomics
- PXR
pregnane X receptor
- RT
room temperature
- ROS
reactive oxygen species
- RXR
retinoid X receptor
- SDS
sodium dodecyl sulphate
- SLCO1B1
solute carrier organic anion transporter family member 1B1
- TAC
Affymetrix transcriptome analysis console
- TCX
transcriptomics
- TFA
trifluoroacetic acid
- TNF
tumor necrosis factor
References
- Alvarez L, Jara P, Sánchez-Sabaté E, Hierro L, Larrauri J, Díaz MC, Camarena C, De la Vega A, Frauca E, López-Collazo E, Lapunzina P. Reduced hepatic expression of farnesoid X receptor in hereditary cholestasis associated to mutation in ATP8B1. Hum Mol Genet. 2004;13:2451–2460. doi: 10.1093/hmg/ddh261. [DOI] [PubMed] [Google Scholar]
- Bachour-El Azzi P, Sharanek A, Burban A, Li R, Guével RL, Abdel-Razzak Z, Stieger B, Guguen-Guillouzo C, Guillouzo A. Comparative localization and functional activity of the main hepatobiliary transporters in HepaRG cells and primary human hepatocytes. Toxicol Sci. 2015;145:157–168. doi: 10.1093/toxsci/kfv041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begriche K, Massart J, Robin MA, Borgne-Sanchez A, Fromenty B. Drug-induced toxicity on mitochondria and lipid metabolism: mechanistic diversity and deleterious consequences for the liver. J Hepatol. 2011;54:773–794. doi: 10.1016/j.jhep.2010.11.006. [DOI] [PubMed] [Google Scholar]
- Beuers U, Trauner M, Jansen P, Poupon R. New paradigms in the treatment of hepatic cholestasis: from UDCA to FXR, PXR and beyond. J Hepatol. 2015;62:S25–S37. doi: 10.1016/j.jhep.2015.02.023. [DOI] [PubMed] [Google Scholar]
- Bremer J. Carnitine-metabolism and functions. Physiol Rev. 1983;63:1420–1480. doi: 10.1152/physrev.1983.63.4.1420. [DOI] [PubMed] [Google Scholar]
- Burbank MG, Sharanek A, Burban A, Mialanne H, Aerts H, Guguen-Guillouzo C, Weaver RJ, Guillouzo A. From the cover: mechanistic insights in cytotoxic and cholestatic potential of the endothelial receptor antagonists using HepaRG Cells. Toxicol Sci. 2017;157:451–464. doi: 10.1093/toxsci/kfx062. [DOI] [PubMed] [Google Scholar]
- Burkhart JM, Schumbrutzki C, Wortelkamp S, Sickmann A, Zahedi RP. Systematic and quantitative comparison of digest efficiency and specificity reveals the impact of trypsin quality on MS-based proteomics. J Proteomics. 2012;75:1454–1462. doi: 10.1016/j.jprot.2011.11.016. [DOI] [PubMed] [Google Scholar]
- Chatterjee S, Richert L, Augustijns P, Annaert P. Hepatocyte-based in vitro model for assessment of drug-induced cholestasis. Toxicol Appl Pharmacol. 2014;274:124–136. doi: 10.1016/j.taap.2013.10.032. [DOI] [PubMed] [Google Scholar]
- Chen F, Ananthanarayanan M, Emre S, Neimark E, Bull LN, Knisely AS, Strautnieks SS, Thompson RJ, Magid MS, Gordon R, Balasubramanian N, et al. Progressive familial intrahepatic cholestasis, type 1, is associated with decreased farnesoid X receptor activity. Gastroenterology. 2004;126:756–764. doi: 10.1053/j.gastro.2003.12.013. [DOI] [PubMed] [Google Scholar]
- Dawson S, Stahl S, Paul N, Barber J, Kenna JG. In vitro inhibition of the bile salt export pump correlates with risk of cholestatic drug-induced liver injury in humans. Drug Metab Dispos. 2011;40:130–138. doi: 10.1124/dmd.111.040758. [DOI] [PubMed] [Google Scholar]
- Demeilliers C, Jacquemin E, Barbu V, Mergey M, Paye F, Fouassier L, Chignard N, Housset C, Lomri NE. Altered hepatobiliary gene expressions in PFIC1: ATP8B1 gene defect is associated with CFTR downregulation. Hepatology. 2006;43:1125–1134. doi: 10.1002/hep.21160. [DOI] [PubMed] [Google Scholar]
- Dickhut C, Radau S, Zahedi R. Fast, efficient, and quality-controlled phosphopeptide enrichment from minute sample amounts using ditanium Dioxide. In: Martins-de-Souza D, editor. Shotgun Proteomics. Springer; New York: 2014. pp. 417–430. [DOI] [PubMed] [Google Scholar]
- Dingemanse J, van Giersbergen PL. Clinical pharmacology of bosentan, a dual endothelin receptor antagonist. Clin Pharmacokinet. 2004;43:1089–1115. doi: 10.2165/00003088-200443150-00003. [DOI] [PubMed] [Google Scholar]
- Fattinger K, Funk C, Pantze M, Weber C, Reichen J, Stieger B, Meier PJ. The endothelin antagonist bosentan inhibits the canalicular bile salt export pump: a potential mechanism for hepatic adverse reactions. Clin Pharmacol Ther. 2001;69:223–231. doi: 10.1067/mcp.2001.114667. [DOI] [PubMed] [Google Scholar]
- Forman BM, Goode E, Chen J, Oro AE, Bradley DJ, Perlmann T, Noonan DJ, Burka LT, McMorris T, Lamph WW, Evans RM. Identification of a nuclear receptor that is activated by farnesol metabolites. Cell. 1995;81:687–693. doi: 10.1016/0092-8674(95)90530-8. [DOI] [PubMed] [Google Scholar]
- Garzel B, Yang H, Zhang L, Huang SM, Polli JE, Wang H. The role of bile salt export pump gene repression in drug-induced cholestatic liver toxicity. Drug Metab Dispos. 2014;42:318–322. doi: 10.1124/dmd.113.054189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman JC, Brouwer K, Mandagere A, Melvin L, Gorczynski R. Evaluation of the endothelin receptor antagonists ambrisentan, darusentan, bosentan, and sitaxsentan as substrates and inhibitors of hepatobiliary transporters in sandwich-cultured human hepatocytes. Can J Physiol Pharmacol. 2010;88:682–691. doi: 10.1139/Y10-060. [DOI] [PubMed] [Google Scholar]
- Kall L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat Meth. 2007;4:923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- Kemp DC, Zamek-Gliszczynski MJ, Brouwer KL. Xenobiotics inhibit hepatic uptake and biliary excretion of taurocholate in rat hepatocytes. Toxicol Sci. 2005;83:207–214. doi: 10.1093/toxsci/kfi020. [DOI] [PubMed] [Google Scholar]
- Kis E, Ioja E, Rajnai Z, Jani M, Méhn D, Herédi-Szabó K, Krajcsi P. BSEP inhibition: in vitro screens to assess cholestatic potential of drugs. Toxicol In Vitro. 2012;26:1294–1299. doi: 10.1016/j.tiv.2011.11.002. [DOI] [PubMed] [Google Scholar]
- Köck K, Ferslew BC, Netterberg I, Yang K, Urban TJ, Swaan PW, Stewart PW, Brouwer KL. Risk factors for development of cholestatic drug-induced liver injury: inhibition of hepatic basolateral bile acid transporters multidrug resistance-associated proteins 3 and 4. Drug Metab Dispos. 2014;42:665–674. doi: 10.1124/dmd.113.054304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollipara L, Zahedi RP. Protein carbamylation: in vivo modification or in vitro artefact? Proteomics. 2013;13:941–944. doi: 10.1002/pmic.201200452. [DOI] [PubMed] [Google Scholar]
- Labbe G, Pessayre D, Fromenty B. Drug-induced liver injury through mitochondrial dysfunction: mechanisms and detection during preclinical safety studies. Fundam Clin Pharmacol. 2008;22:335–353. doi: 10.1111/j.1472-8206.2008.00608.x. [DOI] [PubMed] [Google Scholar]
- Langhi C, Pedraz-Cuesta E, Haro D, Marrero PF, Rodríguez JC. Regulation of human class I alcohol dehydrogenases by bile acids. J Lipid Res. 2013;54:2475–2484. doi: 10.1194/jlr.M039404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Vee M, Jigorel E, Glaise D, Gripon P, Guguen-Guillouzo C, Fardel O. Functional expression of sinusoidal and canalicular hepatic drug transporters in the differentiated human hepatoma HepaRG cell line. Eur J Pharm Sci. 2006;28:109–117. doi: 10.1016/j.ejps.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Lepist EI, Gillies H, Smith W, Hao J, Hubert C, St Claire RL, 3rd, Brouwer KR, Ray AS. Evaluation of the endothelin receptor antagonists ambrisentan, bosentan, macitentan, and sitaxsentan as hepatobiliary transporter inhibitors and substrates in sandwich-cultured human hepatocytes. PLoS One. 2014;9:e87548. doi: 10.1371/journal.pone.0087548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch CJ, Adams SH. Branched-chain amino acids in metabolic signalling and insulin resistance. Nat Rev Endocrinol. 2014;10:723–736. doi: 10.1038/nrendo.2014.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mano Y, Usui T, Kamimura H. Effects of bosentan, an endothelin receptor antagonist, on bile salt export pump and multidrug resistance-associated protein 2. Biopharm Drug Dispos. 2007;28:13–18. doi: 10.1002/bdd.527. [DOI] [PubMed] [Google Scholar]
- Manza LL, Stamer SL, Ham AJ, Codreanu SG, Liebler DC. Sample preparation and digestion for proteomic analyses using spin filters. Proteomics. 2005;5:1742–1745. doi: 10.1002/pmic.200401063. [DOI] [PubMed] [Google Scholar]
- Mita S, Suzuki H, Akita H, Hayashi H, Onuki R, Hofmann AF, Sugiyama Y. Inhibition of bile acid transport across Na+/taurocholate cotransporting polypeptide (SLC10A1) and bile salt export pump (ABCB 11)-coexpressing LLC-PK1 cells by cholestasis-inducing drugs. Drug Metab Dispos. 2006;34:1575–1581. doi: 10.1124/dmd.105.008748. [DOI] [PubMed] [Google Scholar]
- Morel Y, Barouki R. Repression of gene expression by oxidative stress. Biochem J. 1999;342:481–496. [PMC free article] [PubMed] [Google Scholar]
- Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Meth. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- Muller PY, Milton MN. The determination and interpretation of the therapeutic index in drug development. Nat Rev Drug Discov. 2012;11:751–761. doi: 10.1038/nrd3801. [DOI] [PubMed] [Google Scholar]
- Oorts M, Richert L, Annaert P. Drug-induced cholestasis detection in cryopreserved rat hepatocytes in sandwich culture. J Pharmacol Toxicol Methods. 2015;73:63–71. doi: 10.1016/j.vascn.2015.03.002. [DOI] [PubMed] [Google Scholar]
- Padda MS, Sanchez M, Akhtar AJ, Boyer JL. Drug-induced cholestasis. Hepatology. 2011;53:1377–1387. doi: 10.1002/hep.24229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmisano G, Parker BL, Engholm-Keller K, Lendal SE, Kulej K, Schulz M, Schwämmle V, Graham ME, Saxtorph H, Cordwell SJ, Larsen MR. A novel method for the simultaneous enrichment, identification, and quantification of phosphopeptides and sialylated glycopeptides applied to a temporal profile of mouse brain development. Mol Cell Proteomics. 2012;11:1191–1202. doi: 10.1074/mcp.M112.017509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Zhang Y, Liu T, Shen H, Xiao Y, Bourner MJ, Pratt JR, Thompson DC, Marathe P, Humphreys WG, Lai Y. Disruption of BSEP function in HepaRG cells alters bile acid disposition and is a susceptive factor to drug-induced cholestatic injury. Mol Pharm. 2016;13:1206–1216. doi: 10.1021/acs.molpharmaceut.5b00659. [DOI] [PubMed] [Google Scholar]
- Reif R, Karlsson J, Günther G, Beattie L, Wrangborg D, Hammad S, Begher-Tibbe B, Vartak A, Melega S, Kaye PM, Hengstler JG, et al. Bile canalicular dynamics in hepatocyte sandwich cultures. Arch Toxicol. 2015;89:1861–1870. doi: 10.1007/s00204-015-1575-9. [DOI] [PubMed] [Google Scholar]
- Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, et al. Multiplexed protein quantitation in saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- Sharanek A, Burban A, Humbert L, Bachour-El Azzi P, Felix-Gomes N, Rainteau D, Guillouzo A. Cellular accumulation and toxic effects of bile acids in cyclosporine A-treated HepaRG hepatocytes. Toxicol Sci. 2015;147:573–587. doi: 10.1093/toxsci/kfv155. [DOI] [PubMed] [Google Scholar]
- Sharanek A, Burban A, Burbank M, Le Guevel R, Li R, Guillouzo A, Guguen-Guillouzo C. Rho-kinase/myosin light chain kinase pathway plays a key role in the impairment of bile canaliculi dynamics induced by cholestatic drugs. Sci Rep. 2016;6 doi: 10.1038/srep24709. 24709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szalowska E, Stoopen G, Groot MJ, Hendriksen PJ, Peijnenburg AA. Treatment of mouse liver slices with cholestatic hepatotoxicants results in down-regulation of Fxr and its target genes. BMC Med Genomics. 2013;10:6–39. doi: 10.1186/1755-8794-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taus T, Köcher T, Pichler P, Paschke C, Schmidt A, Henrich C, Mechtler K. Universal and confident phosphorylation site localization using phosphoRS. J Proteome Res. 2011;10:5354–5362. doi: 10.1021/pr200611n. [DOI] [PubMed] [Google Scholar]
- Tein I. Carnitine transport: pathophysiology and metabolism of known molecular defects. J Inherit Metab Dis. 2003;26:147–169. doi: 10.1023/a:1024481016187. [DOI] [PubMed] [Google Scholar]
- Vatakuti S, Olinga P, Pennings JL, Groothuis GM. Validation of precision-cut liver slices to study drug-induced cholestasis: a transcriptomics approach. Arch Toxicol. 2017;91:1401–1412. doi: 10.1007/s00204-016-1778-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinken M, Landesmann B, Goumenou M, Vinken S, Shah I, Jaeschke H, Willett C, Whelan M, Rogiers V. Development of an adverse outcome pathway from drug-mediated bile salt export pump inhibition to cholestatic liver injury. Toxicol Sci. 2013;136:97–106. doi: 10.1093/toxsci/kft177. [DOI] [PubMed] [Google Scholar]
- Vinken M, Knapen D, Vergauwen L, Hengstler JG, Angrish M, Whelan M. Adverse outcome pathways: a concise introduction for toxicologists. Arch Toxicol. 2017;91:3697–3707. doi: 10.1007/s00204-017-2020-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Shen H, Rajaraman G, Roberts MS, Gong Y, Jiang P, Burczynski F. Expression and antioxidant function of liver fatty acid binding protein in normal and bile-duct ligated rats. Eur J Pharmacol. 2007;560:61–68. doi: 10.1016/j.ejphar.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Wang G, Bonkovsky HL, de Lemos A, Burczynski FJ. Recent insights into the biological functions of liver fatty acid binding protein 1. J Lipid Res. 2015;56:2238–2247. doi: 10.1194/jlr.R056705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Meth. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- Zollner G, Trauner M. Mechanisms of cholestasis. Clin Liver Dis. 2008;12:1–26. doi: 10.1016/j.cld.2007.11.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.