Abstract
Control of DNA methylation level is critical for gene regulation, and the factors that govern hypomethylation at CpG islands (CGIs) are still being uncovered. Here, we provide evidence that G-quadruplex (G4) DNA secondary structures are genomic features that influence methylation at CGIs. We show that the presence of G4 structure is tightly associated with CGI hypomethylation in the human genome. Surprisingly, we find that these G4 sites are enriched for DNA methyltransferase 1 (DNMT1) occupancy, which is consistent with our biophysical observations that DNMT1 exhibits higher binding affinity for G4s as compared to duplex, hemi-methylated or single-stranded DNA. The biochemical assays also show that the G4 structure itself, rather than sequence, inhibits DNMT1 enzymatic activity. Based on these data, we propose that G4 formation sequesters DNMT1 thereby protecting certain CGIs from methylation and inhibiting local methylation.
Introduction
Methylation of cytosine at C-5 is a key DNA modification in development and disease1,2. In mammals, cytosine methylation occurs predominantly at CpG dinucleotides and is installed and maintained by three DNA methyltransferase enzymes (DNMT1, 3A and 3B) that are essential for development3–5. CpGs occur less frequently than expected in the mammalian genome and show a bimodal distribution with respect to methylation6,7. Sparsely distributed CpGs (~90%), found in genic and intergenic regions, tend to be highly methylated, while CpGs found in dense GC-rich regions, so-called CpG Islands (CGIs), are largely depleted of methylation and are prevalent at the promoters of house-keeping and developmental genes8,9. Outside of embryonic development, gross methylation patterns are generally stable across different tissues10,11. Nonetheless during key cellular events, methylation can be dynamic at specific loci to modulate gene expression, such as de novo methylation of some promoter CGIs with intermediate CpG density during lineage commitment12.
General rules on maintenance of the default methylation state are being uncovered and several studies suggest that CGI hypomethylation is sequence-dependent13–18. Furthermore, DNMTs are reported to be actively and continuously excluded from CpG-poor distal regulatory regions through competitive inhibition with DNA binding proteins, such as NRF1 and CTCF/REST, thus maintaining the hypomethylated state of regulatory regions19,20. Lowly methylated regions also co-localise with DNase I hypersensitivity sites marking accessible chromatin regions21. The presence of enhancer chromatin marks, such as histone modifications, also play an important role in forming the unique chromatin structure of CGIs22,23. In mouse embryonic stem cells, the CXXC finger protein 1, Cfp1, is believed to promote CGI hypomethylation through binding unmethylated CpG and recruitment of H3K4 methyltransferases to promote H3K4me324,25. However, Cfp1 binding and/or H3K4me3 are not required for the ‘protection’ of CGI from DNA methylation since Cfp1 knockout results in a dramatic loss of H3K4me3 at CGIs without increasing DNA methylation24. This suggests that Cfp1 binding and/or H3K4me3 are not required to prevent CGIs from DNA methylation, thus there may be other factors that are fundamental to impart the hypomethylated state.
Alternative DNA secondary structures, known as G-quadruplexes (G4s) are found within certain G-rich sequences and arise through the self-association of guanine bases to form stacked tetrads (Fig. 1a)26. G4s are increasingly being recognised as important features in the genome and over 700,000 G4s have been biophysically mapped in purified human genomic DNA by high-throughput sequencing27. G4 structures have been observed in human using immunofluorescence with a G4-specific antibody (BG4)28, and linked with transcriptional regulation and are enriched in gene promoters including many oncogenes26,29. Recently, G4-chromatin immunoprecipitation sequencing (G4-ChIP-seq) has been developed to map G4 structures in human chromatin30,31. Corroborating a link with transcription, the majority of G4-ChIP-seq sites were found predominantly in regulatory, nucleosome-depleted chromatin, particularly in gene promoters31,32. As both G4s and hypomethylated CGIs are associated with actively transcribed genes9,31, this raises the question of whether there is an interplay between G4 formation and DNA methylation.
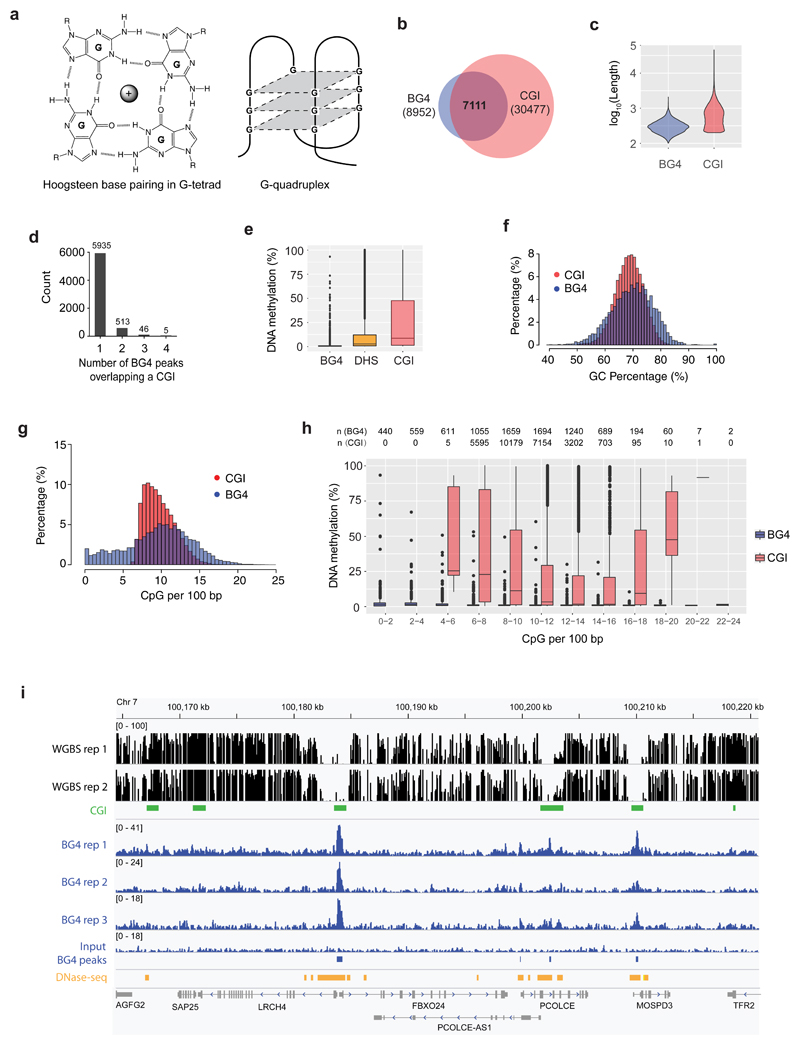
Figure 1. G4 formation is associated with hypomethylation at CGIs.
a) A G-tetrad stabilized by Hoogsteen hydrogen bonding and a central monovalent cation (left). Schematic representations of a three-tetrad G4 structure (Right). b) Venn diagram illustrating the overlap of G4 structure formation (BG4 peaks) and CGIs. c) Violin plot showing size distribution of BG4 peaks and CGIs. d) Count of BG4 peaks overlapping a CGI. e) Box and whisker plot showing the average methylation for BG4 peaks (n = 8,210), DHSs (n = 142,115) and CGIs (n = 27,073). Centre line represents the median value separating upper and lower quartiles in the box, whiskers indicate 1.5× interquartile range (IQR), points are actual values of outliers. Note that methylation level at CpG sites with less than 5x coverage is considered unreliable and discarded. f) Histogram showing the distribution of BG4 peaks and CGIs relative to percentage of GC. g) Histogram showing the distribution of BG4 peaks and CGIs relative to percentage of CpGs per 100 bp. h) Box and whisker plot showing the methylation levels for BG4 peaks and CGIs at different CpG densities. Note that by definition there are no CGIs at a CpG density < 5 CpGs/100bp and that at > 20 CpGs/100bp there are few CGIs (1) and BG4 (36) peaks to consider. The number of CGI regions and BG4 peaks in each category are presented on top of the plot. i) An IGV screen shot illustrating the co-incidence of BG4 peaks (blue) with hypomethylated promoter CGIs (green) and DHSs (orange) for a representative genome region from Chr 7. Shown are normalised signal. Whole genome bisulfite sequence tracks are in black (top). RefGene tracks are in grey (bottom).
Herein we present evidence that most G4 structures, as detected by G4-ChIP-seq, are formed in regions comprising unmethylated CGIs in the human genome. We also uncover a striking co-localisation of G4 structures and DNMT1 docked at CGIs, and we demonstrate that DNMT1 methylation activity is inhibited by DNA G4 structures. Our data suggest a mechanism for the ‘protection’ of CGIs from methylation by G-quadruplex structures that locally sequester and inhibit DNMT1.
Results
G4 structures in active chromatin are found within hypomethylated CGIs
To explore any potential relationship between G4 structures in chromatin and methylation levels, we employed human K562 chronic myelogenous leukaemia cells in which methylation has been comprehensively characterised at single base resolution using whole genome bisulfite sequencing (WGBS) by the ENCODE project33. We generated a genome-wide dataset for G4 structures by G4 ChIP-Seq31 using the G4-specific antibody BG428 and compared the BG4 peak overlap with CGIs34. Strikingly, we found that the majority of BG4 peaks (79%, 7111/8952) overlapped with a CGI (covering 23% of all CGIs) (Fig. 1b). The majority of CpG island regions span 200 to 1000 bp (median/mean, 569/775 bp), while BG4 peak regions span 100 to 400 bp (median/mean, 205/226 bp) (Fig. 1c). 83% (5935/7111) of these CGIs overlap with one BG4 peak (Fig. 1d). Furthermore, when the level of methylation at BG4 peaks was considered, we noted that there was a dramatic absence of methylation at BG4 peaks (mean 1%, median 0.5%), compared with average genome methylation (28.4%) (Fig. 1e). To rule out any effect of the cytosine methylation state on the ability of the BG4 antibody to recognise a G4 structure, an ELISA binding assay was used to show that BG4 can bind to G4 structured DNA with equal affinity irrespective of the presence of cytosine methylation (SI Fig. 1). DNase I hypersensitive sites (DHS), which mark open chromatin, are also mainly hypomethylated (mean 11%, median 2.5%, Fig. 1e). Confirming our previous observations31, the majority of BG4 peaks are found in open chromatin (DHSs, 97%, 8655/8952), and it is notable that these sites have the lowest methylation levels (Fig. 1e). Overall CGI methylation (mean 27%, median 8%, Fig. 1e) shows a broader distribution than BG4 regions, since some CGIs are associated with active hypomethylated promoters while others with inactive genes or gene bodies and thus are more heavily methylated9,35. This prominent association between BG4 peaks, hypomethylation and particular CGIs is suggestive of a functional link between G4 secondary structures and the establishment and/or maintenance of low methylation status at these CGIs in active chromatin.
It has recently been concluded from work in mouse embryonic stem cells, that both high CpG-density and high GC-richness are required to establish the hypomethylated state at CGIs15. It is therefore notable that BG4 peaks have a similar level of GC richness to CGIs (Fig. 1f) with most (79%) being located in regions of CpG density comparable to that seen in CGIs (Fig. 1g). It has been suggested that CpG density alone is only a minor determinant of the unmethylated state, as dense CpG sequences embedded within an AT-rich context are invariably highly methylated when inserted into the mouse genome15,16. Indeed, when we compare the average methylation of CGIs (Fig. 1h) to that of BG4 peaks at different CpG dinucleotide densities, it is noteworthy that across a wide range of CpG densities, BG4 peak regions are always largely devoid of methylation (Fig. 1h). We also confirmed these observations using an alternative CGI definition set generated by CpGCluster algorithm36 (SI Fig. 2a, 2b). Furthermore, when methylation at CGIs is considered with respect to the presence or absence of a BG4 peak, it is noteworthy that there is an almost a total lack of methylation at CGIs with BG4 than CGIs without (SI Fig. 2c). This strongly suggests that CGIs associated with the physical presence of a G4 structure generally have particularly low methylation.
To explore low methylation in different CGI contexts, we calculated methylation levels relative to BG4 presence in CGIs containing i) no promoter or DHS site, ii) a promoter alone, iii) a DHS site alone and iv) both a promoter and DHS site. It is apparent that CGIs containing a BG4 peak always have lower methylation in open (DHS +) or closed (DHS −) chromatin, or in the presence or absence of a promoter. (SI Fig. 2d). CGIs with a DHS site and promoter but without a BG4 peak (4500 sites) have higher methylation (mean 2%, median 2%) than those CGIs (5567 sites) with a BG4 peak plus promoter and DHSs (mean 1%, median 0.5%) (SI Fig. 2d, right two panels). The lowest observed methylation states are found therefore at sites carrying a G4 structure, suggesting that the physical presence of a G4 structure within CGI is an important feature with respect to the hypomethylation state. This is illustrated in Fig. 1i which shows the co-incidence of BG4 peaks with hypomethylated promoter CGIs for a representative genome region.
In earlier work, we found that treatment of human epidermal keratinocytes (HaCaT) cells with the HDAC inhibitor entinostat led to increased BG4 binding signal primarily located in open chromatin promoter regions37. We therefore generated WGBS datasets to examine DNA methylation changes with respect to BG4 signal. Consistent with our observation in K562 cells, BG4 peaks in HaCaT cells have lower methylation compared with open chromatin and CGI regions (SI Fig. 2e). In open-chromatin promoter CGI regions, 307 had a significant increase in BG4 signal (BG4 increase, > 1.5-fold change in signal and FDR < 0.05, see Online Methods). No change in BG4 signal was seen in 3261 CGI promoter regions before and after treatment (BG4 constant), or for 1504 G-rich CGI promoter regions that do not have a BG4 peak (BG4 negative) but have the potential to form a G4 in vitro27. Despite open-chromatin promoter CGI regions already being predisposed to low methylation, we see a statistically significant additional drop in methylation levels at CGIs where BG4 peak size increases after HDAC inhibition (SI Fig. 2f). Overall, these data support that formation of G-quadruplex structures in CGIs is linked to lower methylation.
DNMT1 is sequestered at G4 structures associated with low methylation
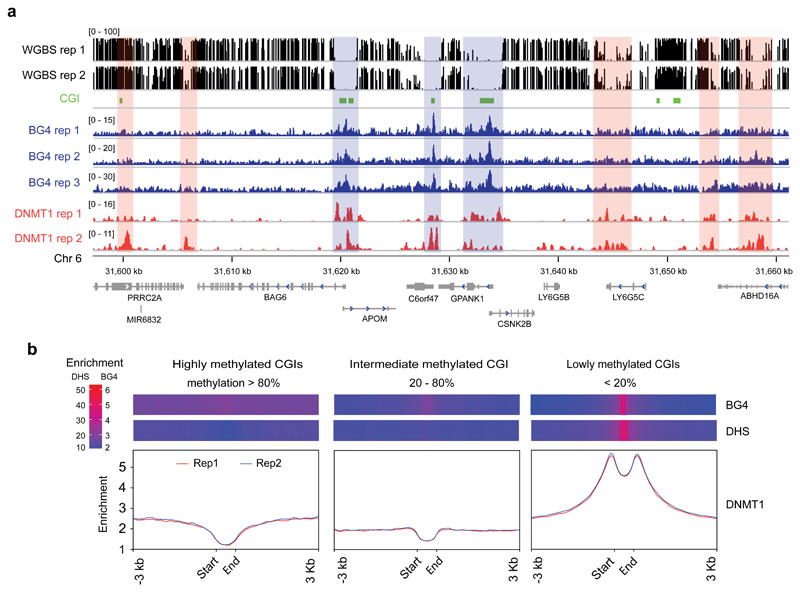
Given that regions where G4 structures marked by BG4 peaks are generally observed to be hypomethylated, we considered that the DNA methyltransferases might have some form of physical interaction with G4 structures in the chromatin context. We focused on DNMT1 since DNMT1 knockout is lethal causing global DNA methylation loss in all dividing somatic cells and human embryonic stem cells (ESCs)3,5,38,39, whereas DNMT3A/B knockouts mainly affect non-CpG methylation in human ESCs5. When we considered the distribution of DNMT1 binding sites in K562 cells, downloaded from ENCODE33 (516,483 peaks in total across both biological replicates), we found that 52% (4611/8952, Monte Carlo simulation’s P-value 1.25e-04) of the G4 structures mapped by G4-ChIP (BG4 peaks) overlapped with at least one DNMT1 binding site. Of the remaining 4341 BG4 peaks, 4003 were within 1 Kb of a DNMT1 binding site. The proximity of BG4 peaks to DNMT1 recruitment sites is illustrated graphically in Fig. 2a for a representative genome region. Intriguingly, when the distribution of DNMT1 binding is plotted relative to high, intermediate or low methylated CGIs, we observe a prominent enrichment of DNMT1 binding at lowly methylated CGIs which overlaps with those regions with the highest BG4 peak density as well as DHS sites (Fig. 2b). A similar profile is also seen using alternative CGI definition set generated by CpGCluster36 (SI Fig. 3). The observation that DNMT1 enrichment at G4 regions that lack methylation is, at first glance, somewhat unexpected and counter-intuitive, given that DNMT1 installs methylation. We therefore considered the possibility of a mechanism whereby DNMT1 protein is sequestered at these sites in active chromatin but prevented from methylating CpGs in that locality.
Figure 2. DNMT1 is recruited to BG4 peaks associated with low methylation.
a) An IGV screen shot showing the co-incidence (blue-masked) of BG4 peaks (blue) with DNMT1 ChIP-seq peaks (red) and CGIs (green) at hypomethylated region from Chr 6. Orange-masked regions are hypermethylated and enriched with DNMT1 presence, but not BG4 signal. Whole genome bisulfite sequence tracks in black (top). b) Binding profile of DNMT1 shown across CGIs with low (less than 20%, n = 16,523), intermediate (between 20% and 80%, n = 6,042) and high (more than 80%, n = 4,266) methylation. Y-axis shows the number of reads in the ChIP normalised to 1 of sequencing depth (also known as Reads Per Genomic Content (RPGC), more details in computational methods). Replicate 1 and 2 are indicated in red and blue respectively. Above each plot is a heat map showing the enrichment of BG4 peaks and DHSs across the respective regions. The heat maps show RPGC of active chromatin marks (DHSs) and BG4 peaks on these three classes of CGIs.
DNMT1 selectively binds to and is inhibited by G4 structures
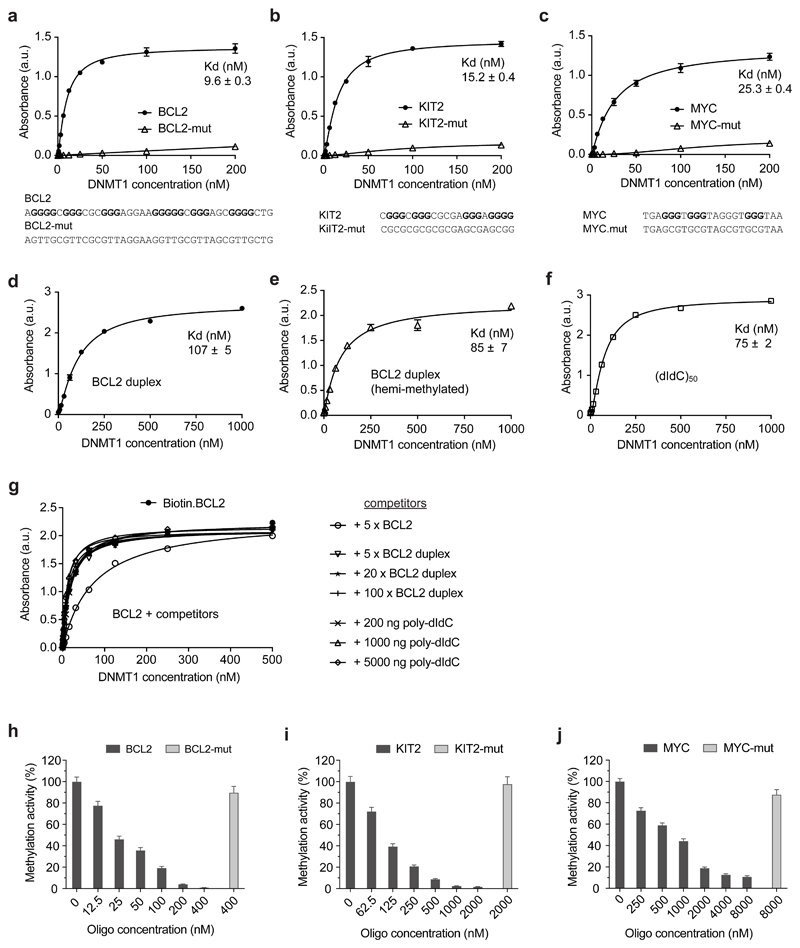
To address whether DNMT1 binds G4 structures directly, we carried out biophysical measurements using an enzyme-linked immunosorbent assay (ELISA) to measure the binding of recombinant human FLAG-tagged full-length DNMT1 protein to immobilized target DNA structures (see Online Methods). Biotinylated single-stranded oligonucleotides of sequence based on the promoters of BCL2, KIT2 and MYC were chosen as these fold into well-characterised G4 structures40–42. Mutated versions (BCL2-mut, KIT2-mut and MYC-mut) that are unable to form a G4 structure were also used as controls. The presence or absence of G4 folded structure with G4 oligonucleotides and mutated controls was confirmed by circular dichroism (CD) spectroscopy and ultraviolet (UV) thermal melting spectroscopic analysis (SI Fig. 4a-f). We found that DNMT1 binds to all three G4 structures with low nanomolar affinity (Kd[BCL2] = 9.6 ± 0.3 nM; Kd [KIT2] = 15.2 ± 0.4 nM, Kd [MYC] = 25.3 ± 0.4 nM, n=3; Fig. 3a-c). DNMT1 showed a lower binding affinity for unmethylated duplex DNA (BCL2, 107±5 nM; Fig. 3d) and there was no specific binding observed for the single stranded mutated oligonucleotide controls. Notably, DNMT1 generally showed a greater affinity for G4 structures than known DNMT1 substrates such as a hemi-methylated duplex DNA (BCL2, 85 ± 7 nM, Fig. 3e), or a synthetic poly(dI-dC)50 substrate (75 ± 2 nM, Fig. 3f). DNMT1 binding to G4 structures does not appear to depend on CpG dinucleotides, since the absence of CpG in the MYC G4 did not preclude DNMT1 binding (Fig. 3c). To begin to dissect the binding mode of DNMT1 to G4s, we used a competition ELISA assay. 50 nM immobilized BCL2 G4 (Kd for DNMT1 = 9.6 nM, Fig. 3a) was incubated with DNMT1 protein in the presence of increasing concentrations of competitors. Even with 100-fold excess (5 μM) of DNA duplex (Kd for DNMT1 = 107 nM, Fig. 3d) or poly-dIdC (Kd for DNMT1 = 75 nM, Fig. 3f), there was no inhibition (Fig. 3g). This suggests that G4s and duplex DNA occupy different binding sites and that the catalytic domain is not involved in G4 binding. A similar, non-overlapping G4 and duplex binding has also been observed in other proteins such as TRF243 and Rap144.
Figure 3. DNMT1 selectively binds and is inhibited by G4 structures.
a-f) ELISA assays testing the binding of recombinant DNMT1 to G4 structure and control oligonucleotides. Binding curves for: a) BCL2 G4 and non-G4-forming control (BCL2-mut); b) KIT2 G4 and non-G4-forming control (KIT2-mut); c) MYC G4 and non-G4-forming control (MYC-mut); d) BCL2 duplex DNA; e) BCL2 hemi-methylated duplex DNA; f) poly(dI-dC), 100 nt. Absorbance was measured at 450 nm. a.u., arbitrary unit. Sequences of oligonucleotides are given below the graphs. g) Binding curve of BCL2 G4 in presence of different concentration of BCL2 duplex or poly(dIdC)n. h-j) Relative methylation activity of recombinant DNMT1 in presence of G4 structure and control oligonucleotides: h) BCL2 G4 and BCL2-mut; i) KIT2 G4 and KIT2-mut; j) MYC G4 and MYC-mut. Shown are mean ± s.d., n = 3 independent experiments in all plots but g (n = 2).
The relatively high binding affinity and selectivity of DNMT1 for DNA G4 structures is consistent with the observation that DNMT1 shows some localisation to G4 structures in K562 cells (Fig. 2a, b). To validate the association with low methylation in the locality G4 sites in the genome, we evaluated whether G4 DNA could actually inhibit DNA methylation on a standard assay using poly(dI-dC)n as substrate45 of DNMT1 using a fluorometric biochemical assay (Abcam, see Online Methods). Specifically, we evaluated different concentrations of folded G4-structured oligonucleotides or mutated non-G4 controls, where the presence or absence of G4 structure had been confirmed by CD spectroscopy (SI Fig. 4g-i). We indeed found that each of three G4 structures resulted in significant inhibition of DNMT1 methyltransferase activity whereas the mutated control oligonucleotides did not (Fig 3h-j). Gratifyingly, the potency of inhibition by each G4 was related to the binding affinity for DNMT1, as determined by ELISA with BCL2 being the most potent inhibitor (50% inhibition at ~25 nM), MYC being least potent (50% inhibition at ~1 μM) and KIT2 being intermediate (50% inhibition at 90 nM). No inhibition of activity was seen with mutated controls ranging from 400 nM-8 μM concentration. C-rich oligonucleotides complementary to the G4 sequences (BCL2-CCC, KIT2-CCC and MYC-CCC) or corresponding duplex DNA also had no effect on DNMT1 activity (SI Fig. 4j-l). We also tested G4 oligonucleotides that were able to fold into a G4 structure but carried a reduced number CpGs (BCL2, KIT) or had a number of artificially introduced CpGs (MYC). In all cases, changes in the number of CpG sites only had minor effects on DNMT1 inhibition (SI Fig. 4j-l). Taken together, these results indicate a novel and unexpected feature of G4 structures as potential genomic regions that promote the unmethylated state through recruitment and inhibition of DNMT1 activity.
Recruitment of DNMT to G4 structures shapes the methylome
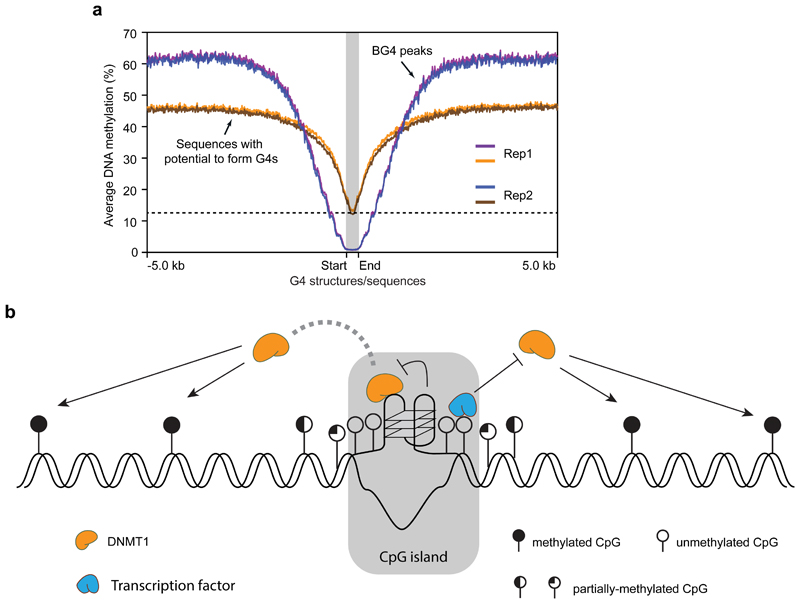
The above data suggest that there is a striking lack of methylation (Fig. 1e, h, SI Fig. 2a-d) in chromatin regions where G4 structure formation is observed. To rigorously question whether this observation was related to the detectable formation of a G4 structure in chromatin (i. e. a BG4 peak), or merely the G-rich sequences per se with potential to form a G4 structure, the methylation profile for BG4 peak regions was compared to those of G-rich sequences that can physically form a G4 structure in an in vitro sequencing assay27 (here called Sequences with potential to form G4s, Fig. 4a). As the majority of BG4 peaks (8,210) are found in open chromatin, only sequences with potential to form G4s located in open chromatin (36,015) were considered. The mean and median length is 226/205 bp for BG4 peaks, and 383/285 bp for the latter. G-rich sequences with the potential to form a G4 are largely hypomethylated (12%), with methylation levels rising in the flanking regions (45%), whereas BG4 peaks have substantially lower methylation (down to 1%) and flanking regions being more methylated (60%). The contrast between lowest methylation at BG4 sites with highest methylation at distal flanking regions is also exemplified in the genome browser view in Fig. 1i. While G-richness as defined by G4 sequence without structure is a feature that correlates with the lack of methylation, there is a further dramatic loss of methylation due to the physical presence of a G4 structure with these regions also being marked by a greater methylation flanking the G4 structure. Regions with a G4 structure also correspond to CGIs that mark particular active genes, and is in keeping with our previous data showing that G4s are associated with particular chromatin states to promote elevated transcription31. R-loops (three-stranded DNA-RNA hybrids) have also been linked to reduced methylation in transcribed CpG island promoters46,47. As R-loops form in a similar genomic context to G4s, we tested the correlation of R-loops, BG4 peaks and methylation. Using the K562 R-loop dataset46, we found that 5685 BG4 peaks overlap with a R-loop, while 3267 BG4 peaks do not and that BG4 peaks are depleted of methylation independent of R-loop presence (SI Fig. 5). This suggests that G4 structure is strongly linked to hypomethylation, irrespective of the presence of R-loop.
Figure 4. Recruitment of DNMT1 by G4 structures shapes the methylome in G-rich regions.
a) Plot showing the average methylation profile centred around G4 forming regions (red and blue are replicates 1 and 2 respectively, n = 7,491) or G4 sequences without structure (orange and green are replicates 1 and 2 respectively, n = 36,015). The plot extends ± 5 Kb from the centre. The dotted line denotes the lowest methylation level of G4 sequence without structure. b) Proposed model for potential involvement of G4 structures and methylation control at CGIs: i) G4 structures sequester DNMT1 due to high affinity binding; ii) G4 structures inhibit the methylation activity of DNMT1. Together with the binding of transcription factors, G4 structures contribute to protection of CGIs from methylation.
Discussion
Here we have provided evidence for a link between a DNA secondary structure formation and epigenetic status. We have uncovered a unique chromatin context whereby certain CGIs in active chromatin are depleted in methylation but carry a G4 structure and also the surrounding flanking regions display higher than average methylation. This suggests that G4s may impart a previously unknown and important function in the establishment of epigenome.
We propose a model (Fig. 4b) in which G4 formation, together with transcription factor binding19,20, contributes to loss of methylation at key genomic loci by sequestering DNMT1, via G4 recognition, and locally inhibiting DNMT1 function at CpG islands. It is noteworthy that this mechanism resembles a recently proposed model for recruitment and inhibition of PRC2 complex by a RNA G-quadruplex present in the HOTAIR lncRNA48,49. This suggests there may be other mechanisms for epigenetic regulation that operate by the sequestration and inhibition of epigenetic modifiers mediated by high affinity interactions with nucleic acid secondary structures.
Online Methods
Cell culture
Mycoplasma-free human chronic myelogenous leukaemia K-562 cells (CCL-243) were purchased from ATCC and grown in RPMI1640 (Glutamine plus, Life Technologies) supplemented with 10% of fetal bovine serum and 100 U/ml penicillin-streptomycin (Life Technologies). All cell stocks were regularly tested for mycoplasma contamination.
G-quadruplex ChIP-seq
ChIP-seq for G-quadruplex structures (G4-ChIP-seq) was performed using the G4-specific antibody BG4 essentially as described previously31.
Oligonucleotide annealing
All oligonucleotides were PAGE purification quality (Sigma). For G4 formation, 10 µM DNA oligonucleotide was annealed in 10 mM Tris HCl, pH 7.4, 100 mM KCl by heating at 95 °C for 5 min followed by gradually cooling to 21 °C. For double-stranded DNA, 10 µM forward and reverse strand oligonucleotides were mixed and annealed in 10 mM Tris HCl, pH 7.4, 100 mM NaCl in the same manner. 20 µM poly(dI-dC)50 was annealed as for double-stranded DNA.
Enzyme-linked immunosorbent assay (ELISA)
ELISAs for binding affinity and specificity were performed as described previously28 with minor modifications. Briefly, biotinylated oligonucleotides were bound to Pierce™ Streptavidin Coated High Capacity Plates (ThermoFisher) followed by blocking with 1.5% BSA and incubation with recombinant full-length human FLAG-tagged DNMT1 protein (Active Motif, Cat. No: 31404) in ELISA buffer (100 mM KCl, 50 mM KH2PO4, pH7.4). After three washes with ELISA buffer, detection was achieved with an anti-FLAG horseradish peroxidase (HRP)-conjugated antibody (ab1238, Abcam) and TMB (3,3′,5,5′-tetramethylbenzidine) ELISA Substrate (Fast Kinetic Rate, ab171524, Abcam). Signal intensity was measured at 450 nm on a PHERAstar microplate reader (BMG Labtech). Dissociation constants (Kd) were calculated from saturation binding curves assuming one-site binding using Prism (GraphPad Software Inc.). Standard error of mean (s.e.m.) values were calculated from three replicates.
In vitro DNA methylation assay
DNA methylation assays were performed using a DNMT Activity Assay Kit (Fluorometric, ab113468, Abcam) as per manufacturer's instructions. Briefly, 100ng recombinant DNMT1 was incubated with substrate assay wells in presence of different concentrations of G4 or non-G4 oligonucleotides at 37 °C for 90 min. Methylation levels were quantified from the binding of an anti-5-methylcytosine antibody detected by fluorescent secondary antibody. Fluorescence signal was measured using a PHERAstar microplate reader (530 nm excitation, 590 nm emission). DNMT enzyme activity is proportional to the fluorescence intensity (RFU, relative fluorescence unit) measured. Relative methylation activity is then calculated against mock control.
Circular dichroism spectroscopy
CD spectra were recorded on an Applied Photo-physics Chirascan circular dichroism spectropolarimeter using a 1 mm path length quartz cuvette. CD measurements were performed at 298 K over a range of 220-300 nm using a response time of 0.5 s, 1 nm pitch and 0.5 nm bandwidth. The recorded spectra represent a smoothed average of three scans, zero-corrected at 300 nm (Molar ellipticity θ is quoted in 105 deg cm2 dmol-1). The absorbance of the buffer was subtracted from the recorded spectra. Oligonucleotides were dissolved in lithium cacodylate buffer (100 mM, pH 7.2) containing 100 mM of KCl and 1 mM EDTA to the concentration of 10 μM. 200 μL of the oligonucleotides were annealed prior measurement by warming up to 90 °C and slowly cooling down at room temperature.
UV Melting
For UV melting experiments, measurements were collected using a Varian Cary 100-Bio UV–visible spectrophotometer by following absorbance at 295 nm. Samples (200 μl) with final concentration of 2 μM were measured in black, small window, 1 cm path-length quartz cuvettes, covered with a layer of mineral oil (50 μl). Samples were equilibrated at 5 °C for 10 min, heated to 95 °C and cooled back to 5 °C at a rate of 0.5 °C/min. The samples were held for a further 10 min and then the 5 °C to 95 °C ramp was repeated. Data were recorded every 1 °C during both the melting and cooling steps. 200 μL of oligonucleotides were annealed prior measurement by warming up to 90 °C and slowly cooling down at room temperature.
Bioinformatics Software and Scripts
Bioinformatic data analyses and processing were performed using Perl, Bash, Python and R programming languages. The following tools were also used: cutadapt (1.15)50, BWA (0.7.15)51, Picard (2.8.3), (http://broadinstitute.github.io/picard), MACS (2.1.1)52, Bedtools (2.26.0), (http://bedtools.readthedocs.io/en/latest/content/overview.html), Deeptools (2.5.1)53 and Bismark (v0.19.0)54.
All scripts and software developed are released in the following GitHub page: https://github.com/sblab-bioinformatics/dna-g4-methylation-dnmt1
G4-ChIP-seq analysis
Raw fastq reads from G4-ChIP-seq in K562 cells were trimmed with cutadapt50 to remove adapter sequences and low-quality reads (mapping quality < 10). Reads were aligned to the human genome (version hg19) with BWA51 and duplicates were removed using Picard. Peaks were called by MACS252 (p < 10-5) following our previous work31: https://github.com/sblab-bioinformatics/dna-secondary-struct-chrom-lands/blob/master/Methods.md
Peaks were merged from different replicates with bedtools multiIntersect. Only peaks overlapping in 3 out 5 replicates were considered high-confidence. K562 datasets for DHSs (ENCSR000EPC), DNMT1 ChIP-seq (ENCSR987PBI), whole genome bisulfite sequencing (ENCSR765JPC) were downloaded from ENCODE. Promoters were defined as 1 kb (+/−) from the transcription start sites of 31,239 hg19 transcripts. Methylation levels at CpG sites with less than 5x coverage were discarded. If not otherwise specified, CGI34 were downloaded using the UCSC’s table browser and then ported to human genome release hg38 using the batch coordinate conversion (liftover) tool of the UCSC. The alternative CGI sets were generated using CpGCluster36.
Enrichment analysis
ENCODE DHS and ChIP-seq data sets were normalised to sequencing depth of 1 (i.e. RPGC, Reads Per Genomic Content). Sequencing depth is defined as: (total number of mapped reads * fragment length) / effective genome size. The effective genome size was set to be 3,209,286,105 and enrichment values for DHSs and BG4 peaks over CGIs and their flanking sequences were visualised in R using ggplot2 library. Enrichment values for DNMT1 over CGIs and their flanks were visualised with DeepTools53.
Monte Carlo Simulation
Monte Carlo simulation was used to calculate the significance of overlap between BG4 peaks and high confidence DNMT1 peaks, defined by Irreproducible Discovery Rate (IDR) in ENCODE’s ChIP pipeline. We first counted how many BG4 peaks overlapped with OQSs in open chromatin (defined as all OQSs seen potassium and/or PDS conditions (749,339 sequences)27, which overlap at least one DHS region (43,506 sequences)). We then randomly selected the same number of OQSs from all OQSs in open chromatin and counted how many overlapped with at least one high confidence DNMT1 peak. The Monte Carlo P-value was then calculated as (N+1)/(M+1), where M is the number of iterations and N is the number of times the same or more overlaps were observed between randomised OQSs and high confidence DNMT1 peaks (compared to the number of overlaps observed between BG4 peaks and high confidence DNMT1 peaks). Randomisation was repeated for 8000 times and on average the number of overlaps between the shuffled OQSs and DNMT1 were two-fold less than those observed between BG4 and DNMT1 peaks.
Differential methylation and BG4 binding analysis of entinostat treated HaCaT cells
HaCaT cells were treated with 10 μM entinostat for 48 hours as we previously described31.
Genomic DNA from untreated and treated cells were extracted with phenol/chloroform. 50 ng DNA were used to generate whole genome bisulfite sequencing libraries using Pico Methyl-Seq Library Prep Kit from Zymo research. Libraries were sequenced using the pair-end 150 bp high-output kit on Illumina Next-seq platform. Data from 4 runs were pooled together. After quality assessment using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/), reads were processed to remove adaptors and low-quality bases using cutadapt3. Options –u 6 –u -1 –U 6 –U -1 were used to trim the initial six and last nucleotide bases. High-quality reads were aligned using Bismark in paired-end mode with options --non_directional, --unmapped and -N 0 to hg19 reference genome. Reads were then de-duplicated and methylation was extracted. To increase mapping efficiency and following previous work55, unmapped reads resulting from the paired-end alignments were then re-aligned in single-end mode with options --non_directional and -N 0, and then deduplicated. Methylation was extracted for paired-end and single-end alignments separately and then aggregated. Technical replicates for each condition (before and after entinostat treatment) were merged and methylation counts were aggregated by CpG site. A threshold was then applied to keep CpG sites with more than 5X bisulfite sequencing depth both before and after treatment. This resulted in 21,106,307 CpG sites, 75% of all CpG sites in hg19.
Differential BG4 binding analysis was done as previously reported31. Analysis focused on open chromatin promoter regions (5351) which have at least one G-rich sequence27 and ATAC-seq peak unaltered in size (log2 fold change = −0.6 to 0.6, FDR < 0.05) between untreated and entinostat-treated HaCaT cells. Depending on differential BG4 signal, these regions were categorized into BG4 gain (> 1.5-fold change in signal and FDR < 0.05) and BG4 constant and BG4 negative. 95% (5072/5351) of these regions are overlapping with CGIs. Difference of the percentage methylation of the overlapping CGIs in each of the categories were calculated. Statistic test was done with Mann–Whitney U test. Plotting of methylation data were performed in the R programming language.
Supplementary Material
Acknowledgements
This work is supported by a core CRUK award (C14303/A17197). S.B. is a Senior Investigator of the Wellcome Trust (grant no. 099232/z/12/z). JS is a Marie Curie Fellow of the European Union (747297-QAPs-H2020-MSCA-IF-2016).
Footnotes
Data availability. K562 datasets for DHS (ENCSR000EPC), DNMT1 ChIP-seq (ENCSR987PBI) and whole genome bisulfate sequencing (ENCSR765JPC) were downloaded from ENCODE. G4-ChIP-seq data sets for K562 and WGBS datasets for entinostat-treated and untreated HaCaT cells are available at the NCBI GEO repository under accession number GSE107690. G4-ChIP-seq data in entinostat-treated and untreated HaCaT cells were taken from GSE76688. Source data for figure 1d, e, h and Figure 3 are available with the paper online.
Accession Codes
K562 datasets for DHS (ENCSR000EPC), DNMT1 ChIP-seq (ENCSR987PBI) and whole genome bisulfate sequencing (ENCSR765JPC) were downloaded from ENCODE. G4-ChIP-seq data sets for K562 and WGBS datasets for entinostat-treated and untreated HaCaT cells are available at the NCBI GEO repository under accession number GSE107690. G4-ChIP-seq data in entinostat-treated and untreated HaCaT cells were taken from GSE76688.
Author Contributions
The project was conceived by SM and SB. SM designed and carried out all the experiments with discussions from DB, JS, RHH, DT & SB. SM designed the analysis strategies with discussions from AG, DT & SB. JS performed G4-ChIP-seq experiments. AG & SMC carried out all computational analysis with discussions from SM, DT, DB, RHH & GM. MD carried out the CD spectroscopy and UV melting experiments. All authors interpreted the results. SM, DT & SB wrote the paper with input from all authors.
Competing interests
SB is an advisor and shareholder of Cambridge Epigenetix limited.
References
- 1.Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet. 2013;14:204–20. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 2.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4 doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 3.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 4.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 5.Liao J, et al. Targeted disruption of DNMT1, DNMT3A and DNMT3B in human embryonic stem cells. Nat Genet. 2015;47:469–478. doi: 10.1038/ng.3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 7.Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Illingworth RS, Bird AP. CpG islands - ‘A rough guide’. FEBS Lett. 2009;583:1713–1720. doi: 10.1016/j.febslet.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 9.Deaton A, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–93. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 11.Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet. 2002;3:662–673. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- 12.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 13.Long HK, King HW, Patient RK, Odom DT, Klose RJ. Protection of CpG islands from DNA methylation is DNA-encoded and evolutionarily conserved. Nucleic Acids Res. 2016;44:6693–6706. doi: 10.1093/nar/gkw258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lienert F, et al. Identification of genetic elements that autonomously determine DNA methylation states. Nat Genet. 2011;43:1091–1097. doi: 10.1038/ng.946. [DOI] [PubMed] [Google Scholar]
- 15.Wachter E, et al. Synthetic CpG islands reveal DNA sequence determinants of chromatin structure. Elife. 2014;3:1–16. doi: 10.7554/eLife.03397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krebs AR, Dessus-Babus S, Burger L, Schübeler D. High-throughput engineering of a mammalian genome reveals building principles of methylation states at CG rich regions. Elife. 2014;3:e04094. doi: 10.7554/eLife.04094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takahashi Y, et al. Integration of CpG-free DNA induces de novo methylation of CpG islands in pluripotent stem cells. Science. 2017;356:503–508. doi: 10.1126/science.aag3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quante T, Bird A. Do short, frequent DNA sequence motifs mould the epigenome? Nat Rev Mol Cell Biol. 2016;17:257–62. doi: 10.1038/nrm.2015.31. [DOI] [PubMed] [Google Scholar]
- 19.Domcke S, et al. Competition between DNA methylation and transcription factors determines binding of NRF1. Nature. 2015;528:575–579. doi: 10.1038/nature16462. [DOI] [PubMed] [Google Scholar]
- 20.Stadler MB, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–5. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 21.Thurman RE, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ooi SKT, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du J, Johnson LM, Jacobsen SE, Patel DJ. DNA methylation pathways and their crosstalk with histone methylation. Nat Rev Mol Cell Biol. 2015;16:519–532. doi: 10.1038/nrm4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clouaire T, et al. Cfp1 integrates both CpG content and gene activity for accurate H3K4me3 deposition in embryonic stem cells. Genes Dev. 2012;26:1714–1728. doi: 10.1101/gad.194209.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomson JP, et al. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature. 2010;464:1082–1086. doi: 10.1038/nature08924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hänsel-Hertsch R, Di Antonio M, Balasubramanian S. DNA G-quadruplexes in the human genome: Detection, functions and therapeutic potential. Nat Rev Mol Cell Biol. 2017;18:279–284. doi: 10.1038/nrm.2017.3. [DOI] [PubMed] [Google Scholar]
- 27.Chambers VS, et al. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat Biotechnol. 2015;33:1–7. doi: 10.1038/nbt.3295. [DOI] [PubMed] [Google Scholar]
- 28.Biffi G, Tannahill D, McCafferty J, Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat Chem. 2013;5:182–6. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rhodes D, Lipps HJ. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015;43:8627–8637. doi: 10.1093/nar/gkv862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hänsel-Hertsch R, Spiegel J, Marsico G, Tannahill D, Balasubramanian S. Genome-wide mapping of endogenous G-quadruplex DNA structures by chromatin immunoprecipitation and high-throughput sequencing. Nat Protoc. 2018;13:551–564. doi: 10.1038/nprot.2017.150. [DOI] [PubMed] [Google Scholar]
- 31.Hänsel-Hertsch R, et al. G-quadruplex structures mark human regulatory chromatin. Nat Genet. 2016;48:1267–1272. doi: 10.1038/ng.3662. [DOI] [PubMed] [Google Scholar]
- 32.De S, Michor F. DNA secondary structures and epigenetic determinants of cancer genome evolution. Nat Struct Mol Biol. 2011;18:950–5. doi: 10.1038/nsmb.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dunham I, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gardiner-Garden M, Frommer M. CpG Islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 35.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–92. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 36.Hackenberg M, et al. CpGcluster: A distance-based algorithm for CpG-island detection. BMC Bioinformatics. 2006;7:1–13. doi: 10.1186/1471-2105-7-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen L, et al. R-ChIP Using Inactive RNase H Reveals Dynamic Coupling of R-loops with Transcriptional Pausing at Gene Promoters. Mol Cell. 2017;68:745–757.e5. doi: 10.1016/j.molcel.2017.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fan G, et al. DNA hypomethylation perturbs the function and survival of CNS neurons in postnatal animals. J Neurosci. 2001;21:788–797. doi: 10.1523/JNEUROSCI.21-03-00788.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jackson-Grusby L, et al. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat Genet. 2001;27:31–39. doi: 10.1038/83730. [DOI] [PubMed] [Google Scholar]
- 40.Dai J, et al. An intramolecular G-quadruplex structure with mixed parallel/antiparallel G-strands formed in the human BCL-2 promoter region in solution. J Am Chem Soc. 2006;128:1096–1098. doi: 10.1021/ja055636a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuryavyi V, Phan AT, Patel DJ. Solution structures of all parallel-stranded monomeric and dimeric G-quadruplex scaffolds of the human c-kit2 promoter. Nucleic Acids Res. 2010;38:6757–6773. doi: 10.1093/nar/gkq558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ambrus A, Chen D, Dai J, Jones RA, Yang D. Solution structure of the biologically relevant G-quadruplex element in the human c-MYC promoter. Implications for G-quadruplex stabilization. Biochemistry. 2005;44:2048–2058. doi: 10.1021/bi048242p. [DOI] [PubMed] [Google Scholar]
- 43.Biffi G, Tannahill D, Balasubramanian S. An intramolecular G-quadruplex structure is required for binding of telomeric repeat-containing RNA to the telomeric protein TRF2. J Am Chem Soc. 2012;134:11974–11976. doi: 10.1021/ja305734x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giraldo R, Rhodes D. The yeast telomere-binding protein RAP1 binds to and promotes the formation of DNA quadruplexes in telomeric DNA. EMBO J. 1994;13:2411–2420. doi: 10.1002/j.1460-2075.1994.tb06526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bacolla A, Pradhan S, Roberts RJ, Wells RD. Recombinant Human DNA (Cytosine-5) Methyltransferase. J Biol Chem. 1999;274:33002–33010. doi: 10.1074/jbc.274.46.33002. [DOI] [PubMed] [Google Scholar]
- 46.Sanz LA, et al. Prevalent, Dynamic, and Conserved R-Loop Structures Associate with Specific Epigenomic Signatures in Mammals. Mol Cell. 2016;63:167–178. doi: 10.1016/j.molcel.2016.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ginno PA, Lott PL, Christensen HC, Korf I, Chédin F. R-Loop Formation Is a Distinctive Characteristic of Unmethylated Human CpG Island Promoters. Mol Cell. 2012;45:814–825. doi: 10.1016/j.molcel.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang X, et al. Targeting of Polycomb Repressive Complex 2 to RNA by Short Repeats of Consecutive Guanines. Mol Cell. 2017;65:1056–1067.e5. doi: 10.1016/j.molcel.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 49.Wang X, et al. Molecular analysis of PRC2 recruitment to DNA in chromatin and its inhibition by RNA. Nat Struct Mol Biol. 2017 doi: 10.1038/nsmb.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet journal. 2011;17:10. [Google Scholar]
- 51.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. 2013;00:1–3. Prepr. https://arxiv.org/abs/1303.3997. [Google Scholar]
- 52.Zhang Y, et al. Model-based Analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramírez F, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krueger F, Andrews SR. Bismark: A flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peat JR, et al. Genome-wide Bisulfite Sequencing in Zygotes Identifies Demethylation Targets and Maps the Contribution of TET3 Oxidation. Cell Rep. 2014;9:1990–2000. doi: 10.1016/j.celrep.2014.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.