Short Abstract
Over the last decades a broad collection of sophisticated fluorescent protein-based probes was engineered with the aim to specifically monitor nitric oxide (NO), one of the most important signaling molecules in biology. Here we report and discuss the characteristics and fields of applications of currently available genetically encoded fluorescent sensors for the detection of NO and its metabolites in different cell types.
Long abstract
Because of its radical nature and short half-life, real-time imaging of NO on the level of single cells is challenging. Herein we review state-of-the-art genetically encoded fluorescent sensors for NO and its by-products such as peroxynitrite, nitrite and nitrate. Such probes enable the real-time visualization of NO signals directly or indirectly on the level of single cells and cellular organelles and, hence, extend our understanding of the spatiotemporal dynamics of NO formation, diffusion and degradation. Here, we discuss the significance of NO detection in individual cells and on subcellular level with genetic biosensors. Currently available genetically encoded fluorescent probes for NO and nitrogen species are critically discussed in order to provide insights in the functionality and applicability of these promising tools. As an outlook we provide ideas for novel approaches for the design and application of improved NO probes and fluorescence imaging protocols.
Keywords: Nitric oxide, Genetically encoded fluorescent probes, FRET, Fluorescence microscopy, Single cell imaging
1. Introduction
Extensive effort has been expended in the past decades to characterize the biological function of NO in biology and medicine [1–6]. Scientists have exploited various techniques and methods including spectroscopic analysis, electrochemical sensors, fluorescent dyes, electron spin resonance, or chromatographic determinations to understand the complex NO metabolism in biological samples or even whole organisms [7–16]. On account of its major importance and relevance in physiology and pathological processes, the urgent need arose to visualize NO quantitatively, directly and with high accuracy in a dynamic manner [17,18]. However, due to its low concentration, short lifetime, and extremely high reactivity with various reactive oxygen species, the direct real-time visualization of NO represents a serious challenge [17,18]. All methods mentioned above have certain individual advantages; however, all of them have also severe disadvantages that limit their application to answer specific question related to NO signaling [17,18]. Most approaches provide only a direct or indirect read-out of NO values averaged over a population of cells. Single cell imaging techniques are quite different and enable scientists to gain important insights in cell signaling events [19,20]. Moreover, single cell imaging approaches provide high-content information about the spatiotemporal patterns of molecular processes and reveal cell-to-cell variabilities [21,22]. Such highly informative readouts can be obtained by high-resolution microscopy of fluorescent probes within cells [23–25]. Thanks to the remarkable technological progress in the field of fluorescent biosensor development during the preceding years we are now able to selectively visualize NO and NO-related species in a real-time manner on the cellular or even subcellular level [20,23,25]. By employing variously colored and targeted probes in a given cell, this approach also enables measurements of different signals such as NO and Ca2+ dynamics simultaneously and in distinct subcellular locals [26–28]. In this review we introduce and summarize features of novel genetically encoded sensors (GES) for NO imaging which have been developed recently.
2. Genetically encoded fluorescent sensors (GES), the basic principles
GES are indispensable tools that have revolutionized and accelerated biological studies [24,29]. Until now, more than 100 different GES have been developed to monitor distinct biomolecules and (sub)cellular processes including metabolites, signaling molecules, ion fluxes, and enzymatic activities with high spatial and temporal resolution [19,22,29]. Apparently, the development of novel GES and improvements of existing GES are ongoing processes [29]. Based on their operating principle, GES are usually further divided into different classes (Table 1 and Fig. 1.) [19].
Table 1.
Classes of genetically encoded fluorescent sensor.
| Class 1 GES: |
| Translocation based probes, which report cell signaling events by alterations of local fluorescence accumulations (Fig. 1A.) [30,31]. |
| Class 2 GES: |
| Intensity based single fluorescent protein (FP) probes, which sense dynamics of ion concentrations by changing their fluorescence intensity (Fig. 1B.) [32,33]. |
| Class 3 GES: |
| Ratiometric single FP probes that sense cell signaling events by distinct changes of their spectral properties (Fig. 1C.) [34–37]. |
| Class 4 GES: |
| Förster resonance energy transfer (FRET)-based probes, which are activated by conformational changes of the probe in the presence of the analyte (Fig. 1D-E.) [38,39]. |
| Class 5 GES: |
| FRET-based sensors for enzyme activity or activation. These probes are modified by enzymes which affects their conformation and, hence, the FRET signal (Fig. 1E.) [40,41]. |
| Class 6 GES: |
| FRET-based sensors, which are activated by enzyme cleavage e.g. caspase sensors (Fig. 1F.) [42,43]. |
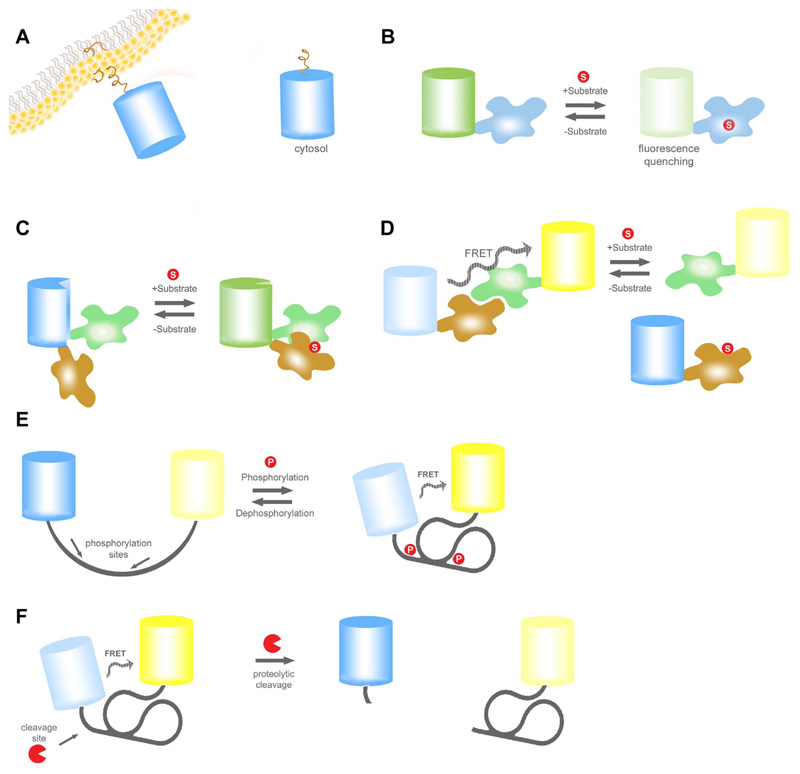
Fig. 1. Basic principle of different single and double FP-based genetically encoded probes.
(A) Basic principle of a translocation based probe. (B) A single FP based probe, where the FP is genetically fused to only one substrate specific domain. Binding of the analyte leads to a loss of fluorescence intensity. (C) A single FP-based ratiometric probe is depicted, which can be excited at two distinct wavelengths. Binding of the analyte induces the interaction of the two anayte-specificdomains. This affects the protonation status of the chromophore, which results in a shift of its spectral properties. (D) Schematic illustration of a FRET-based sensor which is activated by a conformational change upon binding of the analyte of interest to the substrate-specific domain. In the presence of the analyte FRET from CFP to YFP increases in a reversible and concentration dependent manner. Examples for commonly used FRET pairs are CFP/YFP, GFP/OFP, and BFP/GFP. (E) Illustration also represents a FRET based mechanism as shown in panel D. But here the conformational activation is triggered by a phosphorylation event. (F) Schematic overview of a protease-activated FRET sensor. The initial FRET decreases upon cleavage of the connecting domain, which consists of a cleavage site, in an irreversible manner. Examples for these kind of sensor are caspase-reporting probes.
Class 1 GES comprise a huge family of constructs, designed to visualize the subcellular distribution of signaling proteins by fusing it to a fluorescent protein (FP) variant (Fig. 1A.). Single cell imaging of cells expressing FP fusion constructs can provide a high-resolution readout of certain cell activities, if the FP-tagged protein significantly changes its subcellular location during specific cell signaling events [30,31]. In several studies, expression of eNOS fused to GFP or other FPs has been used to visualize the subcellular localization and translocation of the NO producing enzymes in single cells in response to diverse stimuli and stresses [26,44–47]. These experiments demonstrated that the subcellular localization of eNOS is dynamically altered. The subcellular localization of eNOS determines its enzymatic activity, which is known to be tightly regulated by protein-protein interactions, the local availability of (co)substrates, second messengers such as Ca2+, and diverse posttranslational modifications of the enzyme [26,44–48]. However, the visualization of dynamic NOS-FP translocation events in single cells might, if at all, only represent an indirect readout of NO formation as NOS redistributions not necessarily correlate with its enzymatic activity.
Class 2 GES are intensity based probes. Here the fluorescence intensity of a single FP is affected by the concentration of the analyte of interest (Fig. 1B.). In its simplest form the intrinsic sensitivity of some naturally occurring or mutated FP to e.g. H+ or chloride (Cl-) is exploited to measure the dynamics of intracellular pH or Cl- variations, respectively [32,33]. In addition, a novel class of genetically encoded NO probes, the geNOps, which will be described in detail later in this review, represent single FP quenching-based NO indicators and, hence, are rationally designed as a class 2 GES [27]. In geNOps the NO sensitivity of FPs, which naturally remain unaffected by NO, was introduced by fusing a bacteria-derived NO binding domain directly to FP variants in order to bring the radical in close vicinity to the FP chromophore [27].
Another class of single FP-based probes, referred to as class 3 GES, are ratiometric probes as their spectral properties are mutually affected by binding of the analyte of interest (Fig. 1C.). Most of these probes contain circularly permuted FPs conjugated with respective sensor domains. The Ca2+ sensitive pericams and GECOs as well as the hydrogenperoxide (H2O2) sensitive HyPer and HyPerRed are examples of class 3 GES [34–37]. Respective ratiometric single FP-based probes sensitive to NO have not been developed so far.
Many GES (class 4, 5, and 6) are based on Förster resonance energy transfer (FRET), a phenomenon that occurs when two fluorophores with overlapping excitation and emission spectra are closely aligned [38,39]. FRET-based GES naturally provide a ratiometric read-out. The FRET ratio signal may respond to the binding of an analyte (class 4, Fig. 1D-E.), posttranslational modifications (class 5, Fig. 1E.) or the cleavage of a peptide motif (class 6, Fig. 1F.). As a consequence the distance between the FRET-donor and FRET-acceptor FP and hence the FRET ratio signal is either increased or decreased [38–43]. Many FP variants are available for engineering FRET-based probes [19,24]. However, FRET-based GES most frequently consists of cyan FPs (CFPs) and yellow FP variants (YFPs) as FRET donors and FRET acceptors, respectively [38–41,43]. In addition, red-shifted FRET based probes have been developed using green FPs (GFPs) and orange (OFPs) or red FPs (RFPs), respectively [49,50]. While red-shifted FRET-based GES often have a lower dynamic range, they can be spectrally separated from UV-excitable indicators such as fura-2, a popular small chemical Ca2+ probe, to correlate the spatiotemporal patterns of certain cell signaling events with Ca2+ signals in one given cell [49,51]. For the development of direct and indirect genetic NO probes the FRET-technology has been exploited as described in chapters 3.2, 4.1 and 4.2.
3. Direct NO probes
3.1. The geNOps, differently colored FP quenching-based NO probes
We have recently developed a novel class of genetically encoded NO probes, which we named geNOps [27]. These probes consist of different FP variants directly fused to a bacteria derived NO binding domain (Fig. 2.). This domain, referred to as GAF, selectively binds NO via a non-heme iron(II) center (Fig. 2.). GAF is actually the NO sensitive domain of the transcription factor NorR, which is expressed in intestinal bacteria to protect them from toxic NO by inducing the expression of enzymes that convert NO to non-toxic laughing gas, the dinitrogen monoxide molecule (N2O) [52]. However, in geNOps the NO-binding GAF domain brings the radical in close vicinity to the chromophore of FPs, thereby probably affecting the electron density resulting in an immediate loss of fluorescence (Fig. 2.) [27]. Importantly, when NO dissociates from geNOps their fluorescence is fully recovered. Accordingly, geNOps enable the real-time visualization of NO dynamics on the level of individual cells using fluorescence microscopy [27,28].
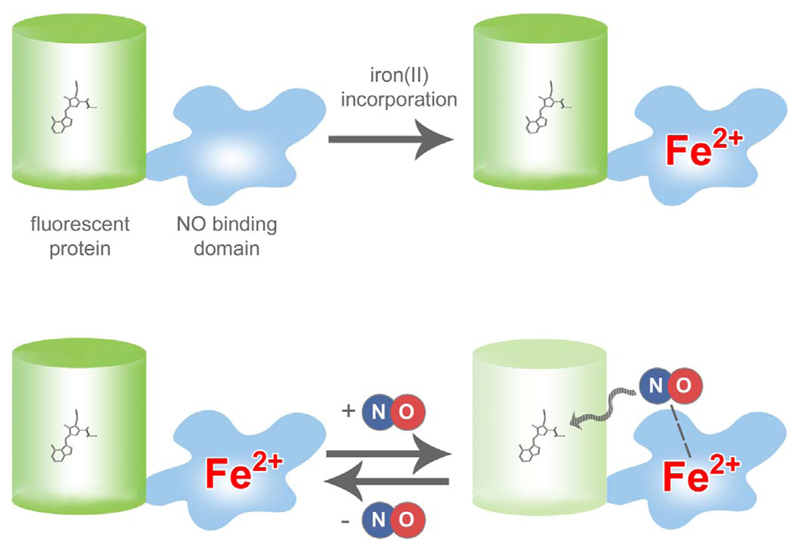
Fig. 2. Basic principle of geNOps.
The geNOps are chimeric constructs, which consist of FP variants, e.g. GFP, directly fused with a highly selective non-heme iron(II) NO binding domain. When using the probes in cells, an iron(II)-loading procedure is required (upper panel) to receive full NO responsiveness of the probes. NO binds in a reversible manner to geNOps, thereby inducing a loss of fluorescence intensity, which is fully recovered in the absence of the radical (lower panel).
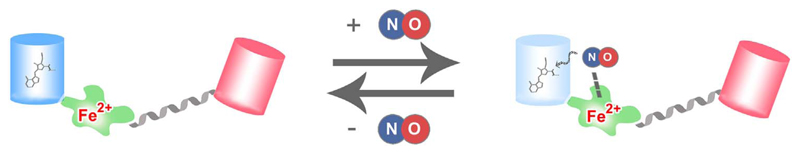
Others and we have used the differently colored geNOps to investigate NO signals in endothelial cells, vascular smooth muscle cells and HEK293 cells expressing eNOS, nNOS or iNOS under various experimental conditions [26,27,53]. The geNOps proved suitable to correlate Ca2+ and NO signals by combining fura-2 with either the green or orange geNOp, G-geNOp and O-geNOp, respectively [26,27]. In a recent video article we have introduced this powerful approach describing the respective protocol steps in detail [28]. Moreover, mitochondria targeted geNOps have been developed and used to visualize NO signals within this important organelle in endothelial cells [27]. Interestingly, we found a positive correlation between mitochondrial Ca2+ uptake and eNOS-mediated NO production exploiting the geNOps technology [26]. While the molecular mechanisms responsible for this phenomenon have not yet been discovered, these findings might lead to novel therapeutic strategies for the treatment of cardio-vascular diseases that are related to a disturbed NO homeostasis [26]. Recently, primary smooth muscle cells were infected with an AVV5 virus coding for C-geNOp in order to study the conversion of nitroglycerin to NO by the aldehydehydrogenase isoform 2 (ALDH2) and a respective ALDH2 mutant on the level of individual cells [53]. The usage of geNOps in HEK293 cells expressing different NOS isoforms unveiled that nNOS produces NO much faster and in a pulsatile manner compared to eNOS [54]. The kinetics of nNOS-mediated NO signals remain to be investigated in neurons, cardio myocytes and skeleton muscle cells, which endogenously express the NO-generating enzyme [54]. In order to estimate basal NO levels and visualize the arginine dependency of NO formation in cells expressing the constitutively active iNOS, we recently developed a novel double FP-based geNOp variant, the cyan-red geNOp (CR-geNOp, Fig. 3.). This construct consists of CFP fused to the NObinding GAF domain, which is further fused to RFP via a rigid linker (Fig. 3.). Importantly, while the fluorescence of CFP remains fully sensitive to NO, the red fluorescence of RFP remains insensitive to NO within this construct. Thus, CR-geNOp represents a (pseudo)ratiometric NO probe in which the RFP/CFP fluorescence ratio signal is a direct and real-time readout of the cellular NO concentration. The geNOps have so far not been used in vivo. Currently, we are working on the expression of G-geNOp and the ratiometric CR-geNOp in neurons of living mice in order to test the applicability of the geNOps technology for two-photon intra-vital microscopy. This approach will also unveil whether iron(II) supplementation is required in vivo. Notably, in order to obtain full NO responsiveness when using geNOps expressed in cells in situ, iron(II) supplementation was essential [27]. While a non-toxic iron (II) fumarate medium was developed for this purpose, the procedure of iron(II) supplementation might restrict the applicability of the geNOps technology. As most FP-based probes, the geNOps are more or less sensitive to pH fluctuations depending on the FP variant [27]. Hence, possible intracellular pH alterations need to be considered thoroughly when using the geNOps technology. Importantly, an acidification reduces the fluorescence intensity of the cyan, green, and yellow geNOp variants independently of cellular NO levels. Hence, any procedures and mechanisms that significantly increase the H+ concentration in cells or cellular organelles give a false positive NO signal as respective geNOps lose fluorescence intensity as if they would bind NO. In contrast, a cellular or subcellular alkalization will increase the fluorescence intensity of FPs and hence simulate a reduction of intracellular NO levels. Accordingly, both the parallel measurement of intercellular pH levels using e.g. genetically encoded pH probes and the usage of NO-insensitive mutated geNOps with unaltered pH sensitivity as negative controls are recommended to discriminate between real and false NO signals when using the geNOps technology [27].
Fig. 3. Basic principle of CR-geNOp.
The CR-geNOps consists of the NO sensitive CFP and the NO insensitive RFP. Due to the different chromophore and the distance to the NO binding GAF domain, the fluorescence of the RFP remains unaffected by NO. Accordingly, the probe can be used as a (pseudo) ratiometric NO sensor.
3.2. The FRET-MT, a metallothionein-based NO reporter
Pearce and colleagues developed a NO-sensitive protein-based chimeric construct by fusing CFP and YFP to the C- or N-terminal ends of human type IIa metallothionein (hMTIIa), respectively (Fig. 4.) [55]. Metallothioneins (MTs) are cysteine-rich intracellular proteins capable of binding metal ions such as cadmium, copper or zinc [56,57]. There is evidence that MTs are important for regulating metal ion homeostasis and the cellular redox status, whereas their cellular signaling functions as well as physiological and pathological roles are largely unexplored [57–60]. However, several studies showed that NO directly reacts with MTs, which might be part of a NO-sensitive downstream signaling pathway in endothelial cells and reduce NO cytotoxicity [61–64]. Based on these observations and reports that emphasized the release of metal ions from MTs in the presence of NO, the authors tested if NO directly affects the conformation and, hence, FRET ratio signal of the FRET-MT construct [55,59,61,62]. Indeed, the addition of NO to purified FRET-MT increased CFP fluorescence and decreased the respective FRET signal (Fig. 4.) [55]. This experiment proved that NO reacts with this human MT isoform, thereby affecting its structural conformation and probably also its metal ion binding capacity. Importantly, this phenomenon can be exploited to visualize NO increases in vitro as well as in biological samples [55]. Thus FRET-MT represents the first genetically encoded probe, able to directly respond to the increase of cellular NO levels. In the absence of the radical FRET-MT shows high FRET, while the reaction of NO with the central MT domain significantly reduces FRET as a consequence of a NO-induced steric rearrangement (Fig. 4.) [55]. As shown in another study, NO might affect the structure of the MT motif in a permanent manner by releasing the metal ions from the protein [59]. It is, however, unclear whether NO remains bound to the ion-free domain [59,65,66]. Furthermore it is unknown, how fast metal ions might reconstitute the MT structure in the absence of NO [65,66]. The so far published in vitro and in situ data are not conclusive on whether the probe senses NO in a fully reversible manner [67–70]. In their paper the authors mainly used FRET-MT to visualize the binding of NO to MT to determine the role of this molecular process in NO signaling in vascular tissue [55]. The characteristics of FRET-MT as a NO reporter remain widely elusive [55]. Thus, further investigations are necessary to determine the on- and off kinetics, the dynamic range, sensitivity and selectivity of FRET-MT to sense NO in vitro and living cells. Moreover, the NO-induced release of metal ions from the MT domain might impact cell fate and, hence, limit the applicability of FRET-MT [71]. Altered cell signaling events in response to the over-expression of MT domains and metal ion release should be considered as well, when interpreting results using FRET-MT to assess NO signals in living cells or animals [69,71]. Nevertheless, the FRET-based construct was successfully used to monitor NO elevations on the level of individual sheep pulmonary endothelial cells upon Ca2+ mobilization [55,69,72]. FRET-MT expressed in endothelial cells reported quite slow increases of NO upon cell treatment with fast Ca2+ mobilizing inositol 1,4,5-trisphosphate (IP3)-generating agonists such as carbachol and bradykinin [55,69]. Recently, the slow mode of eNOS-mediated NO biosynthesis despite fast cytosolic Ca2+ signals was also shown with the geNOps, in primary endothelial cells as well as in an endothelial cell line (EA.hy926) and HEK 293 cells expressing eNOS-RFP [26,54]. The comparison of all these data indicates that NO is produced slow and gradually upon activation of eNOS by Ca2+-mobilizing agonists, while the different genetically encoded NO reporter, FRET-MT as well as geNOps, are able to detect fast cellular NO levels due to their fast on kinetic. Indeed, the FRET ratio signal of FRET-MT expressed in sheep pulmonary aortic endothelial cells strongly and rapidly responded to 1 mM NO, indicating that the probe is suitable to monitor prominent cellular NO elevations in a dynamic manner when NO is applied extracellularly [55]. Overall, the usage of MT to develop FRET-based NO probes represents a promising approach. Other MT isoforms from different species might be used in future to generate novel FRET-based NO probes with probably improved characteristics. In this regard testing novel FP variants optimized for FRET-based probes also seems promising to adjust e.g. the pH sensitivity and dynamic range of future NO-sensitive FRET-MTs.
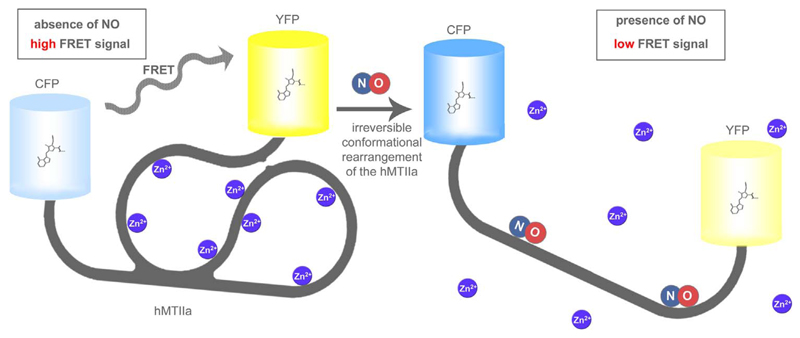
Fig. 4. Basic principle of FRET-MT.
In the absence of NO FRET-MT has a globular structure, which is stabilized by several Zn2+ ions yielding high FRET (left panel). NO binding to the MT domain yields a structural rearrangement. Thereby Zn2+ ions are released and the distance between the terminal CFP and YFP is significantly increased resulting in a decreased FRET ratio signal. The probe was mainly developed to confirm the interaction of NO with MT, but also used to measure NO in vitro and in living endothelial cells upon Ca2+ mobilization or the administration of NO and NO-donors.
4. Indirect NO probes
4.1. NOA-1, a bipartite amplifier-coupled fluorescent NO probe based on a cGMP indicator
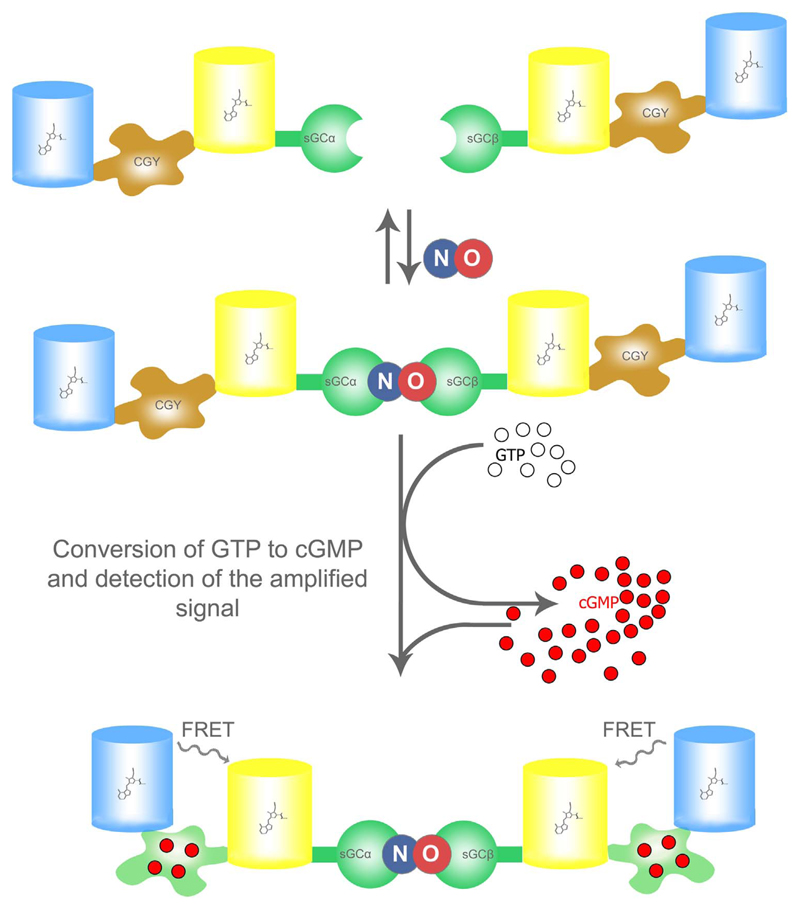
Sato and his colleagues developed a sophisticated genetically encoded fluorescent indicator system, which is able to detect cellular NO in the low nM range with a detection limit of approximately 0.1 nM [73]. The whole construct consists of the α- and β subunits (sGCα and sGCβ) of the NO-sensitive soluble guanylate cyclase (sGC). Both subunits of the enzyme are fused with a genetically encoded FRET-based cGMP indicator (Fig. 5.) [73], which is called CGY and had been developed by the same researchers [74]. Upon NO binding to the heme group of the sGCβ subunit the enzymatic conversion of GTP to cGMP by the whole heterodimeric enzyme is boosted. The FRET-based cGMP indicator CGY then senses the increase of cGMP as an amplified measure of the cellular NO concentration (Fig. 5.) [73].
Fig. 5. Basic principle of NOA-1.
NOA-1 represents a bipartite probe, which consist of the αsGC subunit fused to CGY, a FRET-based cGMP probe, and the βsGC subunit also fused to a CGY (upper panel). Upon NO binding to the βsGC-subunit the active heterodimeric sGC is formed and converts GTP into cGMP (middle panel). The NO-induced formation of cGMP is subsequently visualized with the FRET-based cGMP probe, named CGY (lower panel), which shows increased FRET in the presence of cGMP.
This bipartite probe has been named NOA-1, which stands for “a fluorescence indicator for NO with a signal amplifier” [73]. The rational design of this amplifier-coupled fluorescent NO probe is impressive and it has been convincingly demonstrated that NOA-1 is suitable to detect small intracellular NO fluctuations using CHO and endothelial cells in response to the repetitive additions of NO-liberating compounds and the activation of eNOS [73]. Although this highly sensitive single-cell NO detection approach was introduced almost 13 years ago, other researchers have not picked it up so far. What might discourage scientist applying NOA-1? One main limitation of NOA-1, is based on the fact that two bulky fusion constructs, the sGCα-CGY and sGCβ-CGY, need to be co-expressed in approximately the same amount to reconstitute the sophisticated NO-sensitive reporter system [73]. Adequate co-expression of both NOA-1 components might be well achievable in easy-to transfect cell lines, but represents a real challenge when using primary endothelial cells, cardiomyocytes, macrophages or neurons. These cell types endogenously produce NO and are therefore of special interest for NO researchers, however, they are often difficult to transfect [75–78]. Probably to overcome this limitation Sato and colleagues soon generated a cell-based indicator, referred to as Piccell, using the same NO detection principle [79]. For this purpose they selected a pig kidney-derived cells line, the PK15 cells, which endogenously and constantly express the α- and β-subunits of sGC. Using an antibiotic-containing selection medium they were then able to generate a stable fluorescent PK15 clone expressing the genetically encoded FRET-based cGMP indicator, CGY. In co-culture experiments these fluorescent NO-sensitive cGMP sensor cells could be used to visualize the release of NO from endothelial cells and neurons. In addition, the cell-based system was used to investigate the NO diffusion process by uncaging caged NO. Although this method showed exceptional sensitivity to NO allowing the detection of NO signals in the pM range, the Piccell technology has so far not been used commonly to investigate cellular NO signaling. As both NOA-1 (Fig. 5.) as well as Piccell actually sense the downstream second messenger cGMP and not NO, these methods provide only an indirect readout of cellular NO fluctuations. Pathways that influence the synthesis and breakdown of cellular cGMP influence the fluorescence signals as well and, hence, produce false results. This becomes obvious in the presence of sGC inhibitors such as NS 2028, which abolish or blunt the GCY FRET ratio signal, and inhibitors of phosphodiesterases such as zaprinast, which boost respective fluorescence signals, despite unaltered cellular NO levels [79]. Accordingly, the cellular cGMP homeostasis needs to be thoroughly considered when interpreting results obtained with the highly sensitive, but in fact indirect NO reporter systems.
4.2. The sNOOOpy, a FRET-based sensor for nitrate and nitrite
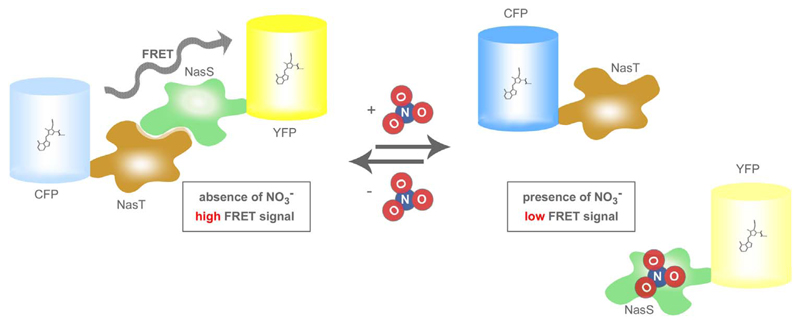
In biological systems the short-lived NO radical is rapidly metabolized by the stepwise oxidation to stable nitrite (NO2−) and nitrate (NO3−) [80]. Hence, the quantification of NO2− and NO3− levels in biological samples is frequently used as an indirect endpoint measure of cellular NO production [73,79]. For this purpose different methods for the determination of NO2− and NO3− are available. The classical Griess assay is most frequently used to indirectly estimate NO biosynthesis. It is based on an easy-to-use colorimetric detection method [8,73,79]. Interestingly, recent studies indicated that NO can be reformed from NO2− and NO3− under certain conditions in vivo [81,82]. This “NO2−- NO3−-NO” pathway most likely represents an important NO source in addition to the cellular NOS-mediated NO generation [81,82]. Accordingly, based on the close correlation between cellular NO2−/NO3- levels and the NO metabolism, the quantitative visualization of NO2−/NO3- dynamics might represent an informative indirect readout of the cellular NO homeostasis [73,79]. In 2016 the visualization of NO2−/NO3- levels in living cells became possible by a FRET-based genetically encoded fluorescent reporter, named sNOOOpy (sensor for NO2−/NO3-in physiology) [83]. As shown in Fig. 6. sNOOOpy consists of two proteins, NasT and NasS, derived from the bacterium Bradyhizobium japonicum. The bacteria-derived proteins were fused with CFP and YFP, respectively [83,84]. In the absence of NO2− and NO3− CFP-NasT and NasS-YFP interact with each other and form a heterodimer, which gives a significant FRET signal (Fig. 6.) [83]. In the presence of either NO2− or NO3− the FRET ratio signal declines due to binding of NO3−/NO2− to NasS-YFP (Fig. 6.) [83]. The reaction turned out to be fully reversible and the FRET-based sNOOOpy responds to NO3−/NO2− in a concentration dependent manner [83]. The NO3−/NO2—selective sNOOOpy also remains functional in mammalian cells, indicating that the fluorescent probe is suitable to specifically visualize intracellular NO3−/NO2− dynamics [83]. Interestingly, the inventors of sNOOOpy generated a mammalian expression plasmid in which the DNA sequence coding for a self-processing 2A peptide was inserted between the two DNA regions encoding CFP-NasT and NasS-YFP in order to express both parts in equal amounts [83].
Fig. 6. Basic principle of sNOOOpy.
The sNOOOpy system consist of two FP-fusion constructs, CFP-NasT and NasS-YFP, which interact in the absence of NO3−/NO2− yielding high FRET (left panel). Upon binding of either NO3− or NO2− to the NasS domain the heterodimer falls apart resulting in reduced FRET (right panel).
So far, the probe has been used to measure NO3− and NO2− uptake of single HeLa cells upon addition of the anions [83]. The single cell experiments unveiled that sNOOOpy is less sensitive when expressed in living cells compared to experiments with the recombinant components of the protein-based probe. However, sNOOOpy represents a promising highly selective NO3−/NO2− sensor that might help to unveil missing links between the cellular NO homeostasis and NO3−/NO2− dynamics. Further experiments exploiting sNOOOpy in combination with direct NO probes such as the geNOps in NO producing cells have the potency to increase our understanding of the complex, probably cell type specific metabolism of these important nitrogen species [27].
4.3. The pnGFP, a single FP-based peroxynitrite (ONOO-) reporter
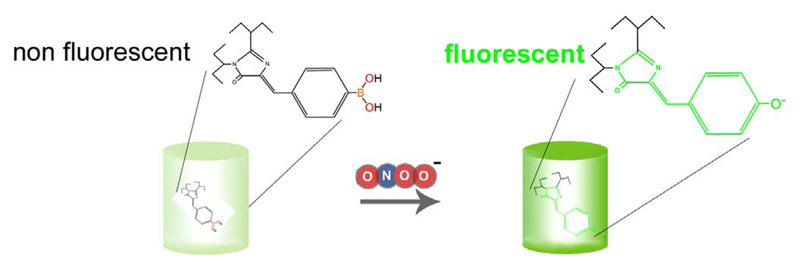
Redox signaling by nitrogen species particularly peroxynitrite (ONOO-) has been implicated in both physiological and pathophysiological conditions [85–87]. ONOO−, a product of the fast reaction between superoxide anions (O2−) and NO, is an important redox signaling molecules with strong oxidative property [88]. As most ONOO- in cells originates from this reaction, detection of ONOO- might be used as an indirect measure of cellular NO and O2− levels and dynamics [89,90]. However, due to the short half-life of ONOO- in biological samples, it is quite difficult to quantify or visualize the reactive molecule under both physiological and pathological conditions in living cells [17,18]. In 2013, a group of researchers from the University of California could go beyond this limitation by creating a novel semi-synthetic genetically-encoded probe called pnGFP, which is selectively sensitive to ONOO-[91]. It has been well-recognized that boronate organic dyes can specifically react with peroxynitrite [92]. Therefore, the scientists site-specifically introduced a boronic acid moiety into a circularly permuted green fluorescent protein by a synthetic biology approach. One mutant of this approach was named pnGFP, a sensor, which increases green fluorescence in response to ONOO− (Fig. 7.). The pnGFP is derived from a conversion of superfolder GFP to circularly permuted topology (cpsGFP). The pnGFP responded well to ONOO- but not to other reactive oxygen species such as hydrogenperoxide (H2O2), hypochlorous acid (HOCl), the hydroxyl radical (OH•), tert-butyl hydroperoxide (HOOtBu) and importantly O2− and NO [91,92]. The pnGFP was also applied in mammalian cells by constructing a mammalian expression vector, which was used to express the protein-based semi-synthetic ONOO− sensor in HEK293T cells. Addition of p-boronophenylalanine and expression of a special polyspecific synthetase is however necessary to successfully express functional pnGFP in mammalian cells. The requirement to introduce p-boronophenylalanine as a synthetic amino acid into pnGFP clearly limits the applicability of this sensor, particularly, as an expanded genetic code system is required. However, HEK293T cells expressing pnGFP showed a prominent fluorescence signal in response to SIN-1 treatment [91], a peroxynitrite-liberating compound. As expected, the selectivity of the probe to ONOO- remained high in living cells. Thus, the chemoselective and sensitive ONOO- probe seems suitable for investigating NO-related ONOO- generation under physiological and pathological conditions on the level of individual cells. However, further systematic studies are essential to evaluate the real potential of pnGFP. While pnGFP responds to ONOO- in an irreversible manner (Fig. 7), additional applications of this refined probe might have the potency to deepen our understanding of the spatiotemporal patterns of specific subcellular ROS signaling processes. Moreover, it remains to be tested if based on this principle other FP variants are also suitable to generate ONOO− probes. Such attempts might yield differently colored protein-based ONOO− sensors, which are due to their distinct spectral properties, can be combined with additional indicators to simultaneously detect diverse ROS species and signaling events in one given cell.
Fig. 7. Basic principle of pnGFP.
The semisynthetic pnGFP is a superfolder GFP variant, which consists a ONOO- sensitive boronic acid moiety. Upon ONOO- binding green fluorescence is increased.
5. Conclusion
In the last two decades, different research groups have developed several genetically encoded fluorescent probes to visualize NO and its metabolites in single cells. However, only few researchers have applied these powerful probes so far. It might take some more time until the majority of the scientific community is ready to use NO selective GES, which may allow working on so far inaccessible issues. Undoubtedly, novel FP-based probes with improved characteristics in terms of their sensitivity, selectivity, spectral properties and applicability will inspire researchers from different fields to perform insightful single cell NO imaging experiments in the near future. As in the case of other GES including different Ca2+ indicators, NO probes might soon allow investigating NO signals in vivo using informative animal models. With the development of geNOps, NOA-1, MT-FRET, pnGFP and sNOOOpy the technical basis has been set – we now need to the take the next steps in a systematic and concerted manner.
Acknowledgements
The authors acknowledge Karin Osibow for editing the manuscript. The authors further acknowledge the scientific advisory board of Next Generation Fluorescence Imaging (NGFI) GmbH (http://www.ngfi.eu/), a spin-off company of the Medical University of Graz. The authors and their research were funded by the Ph.D. program Molecular Medicine (MOLMED) of the Medical University of Graz, by Nikon Austria within the Nikon-Center of Excellence, Graz, the FWF projects P28529-B27, and the doctoral program Metabolic and Cardiovascular Disease (DK-W1226). S.C. holds an academic development scholarship from the University of Phayao (Phayao, Thailand). The Nikon Center of Excellence, Graz, is supported by the Austrian infrastructure program 2013/2014, Nikon Austria Inc., and BioTechMed, Graz.
Footnotes
FRBM: Special Issue: Current fluorescence and chemiluminescence approaches in free radical and redox biology, Edited by: Rafael Radi, Ana Denicola, Bruce Morgan and Jacek Zielonka.
References
- [1].Godo S, Shimokawa H. Divergent roles of endothelial nitric oxide synthases system in maintaining cardiovascular homeostasis. Free Radic Biol Med. 2017;109:4–10. doi: 10.1016/j.freeradbiomed.2016.12.019. [DOI] [PubMed] [Google Scholar]
- [2].Garland CJ, Dora KA. EDH: endothelium-dependent hyperpolarization and microvascular signalling. Acta Physiol. 2017;219(1):152–161. doi: 10.1111/apha.12649. [DOI] [PubMed] [Google Scholar]
- [3].Umbrello M, Dyson A, Feelisch M, Singer M. The key role of nitric oxide in hypoxia: hypoxic vasodilation and energy supply-demand matching. Antioxid Redox Signal. 2013;19(14):1690–1710. doi: 10.1089/ars.2012.4979. [DOI] [PubMed] [Google Scholar]
- [4].San Martin A, Arce-Molina R, Galaz A, Perez-Guerra G, Barros LF. Nanomolar nitric oxide concentrations quickly and reversibly modulate astrocytic energy metabolism. J Biol Chem. 2017;292(22):9432–9438. doi: 10.1074/jbc.M117.777243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hagan G, Pepke-Zaba J. Pulmonary hypertension, nitric oxide and nitric oxide-releasing compounds. Expert Rev Respir Med. 2011;5(2):163–171. doi: 10.1586/ers.11.5. [DOI] [PubMed] [Google Scholar]
- [6].Vanhoutte PM, Zhao Y, Xu A, Leung SW. Thirty years of saying NO: sources, fate, actions, and misfortunes of the endothelium-derived vasodilator mediator. Circ Res. 2016;119(2):375–396. doi: 10.1161/CIRCRESAHA.116.306531. [DOI] [PubMed] [Google Scholar]
- [7].Saldanha C, Lopes de Almeida JP, Silva-Herdade AS. Application of a nitric oxide sensor in biomedicine. Biosensors. 2014;4(1):1–17. doi: 10.3390/bios4010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Green LC, Wagner DA, Glogowski J, Skipper PL, Wishnok JS, Tannenbaum SR. Analysis of nitrate, nitrite, and [15N]nitrate in biological fluids. Anal Biochem. 1982;126(1):131–138. doi: 10.1016/0003-2697(82)90118-x. [DOI] [PubMed] [Google Scholar]
- [9].Shibuki K. An electrochemical microprobe for detecting nitric oxide release in brain tissue. Neurosci Res. 1990;9(1):69–76. doi: 10.1016/0168-0102(90)90048-j. [DOI] [PubMed] [Google Scholar]
- [10].Privett BJ, Shin JH, Schoenfisch MH. Electrochemical nitric oxide sensors for physiological measurements. Chem Soc Rev. 2010;39(6):1925–1935. doi: 10.1039/b701906h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kojima H, Nakatsubo N, Kikuchi K, Kawahara S, Kirino Y, Nagoshi H, Hirata Y, Nagano T. Detection and imaging of nitric oxide with novel fluorescent indicators: diaminofluoresceins. Anal Chem. 1998;70(13):2446–2453. doi: 10.1021/ac9801723. [DOI] [PubMed] [Google Scholar]
- [12].Feelisch M, Noack EA. Correlation between nitric oxide formation during degradation of organic nitrates and activation of guanylate cyclase. Eur J Pharmacol. 1987;139(1):19–30. doi: 10.1016/0014-2999(87)90493-6. [DOI] [PubMed] [Google Scholar]
- [13].Ghebremariam YT, Huang NF, Kambhampati S, Volz KS, Joshi GG, Anslyn EV, Cooke JP. Characterization of a fluorescent probe for imaging nitric oxide. J Vasc Res. 2014;51(1):68–79. doi: 10.1159/000356445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Coneski PN, Schoenfisch MH. Nitric oxide release: part III. Measurement and reporting. Chem Soc Rev. 2012;41(10):3753–3758. doi: 10.1039/c2cs15271a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Gopalakrishnan B, Nash KM, Velayutham M, Villamena FA. Detection of nitric oxide and superoxide radical anion by electron paramagnetic resonance spectroscopy from cells using spin traps. J Vis Exp. 2012;66:e2810. doi: 10.3791/2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Funazo K, Tanaka M, Shono T. Gas chromatographic determination of nitric oxide at sub-ppm levels. Anal Chim Acta. 1980;119(2):291–297. [Google Scholar]
- [17].Hunter RA, Storm WL, Coneski PN, Schoenfisch MH. Inaccuracies of nitric oxide measurement methods in biological media. Anal Chem. 2013;85(3):1957–1963. doi: 10.1021/ac303787p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bryan NS, Grisham MB. Methods to detect nitric oxide and its metabolites in biological samples. Free Radic Biol Med. 2007;43(5):645–657. doi: 10.1016/j.freeradbiomed.2007.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Newman RH, Fosbrink MD, Zhang J. Genetically encodable fluorescent biosensors for tracking signaling dynamics in living cells. Chem Rev. 2011;111(5):3614–3666. doi: 10.1021/cr100002u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Mullassery D, Horton CA, Wood CD, White MR. Single live-cell imaging for systems biology. Essays Biochem. 2008;45:121–133. doi: 10.1042/BSE0450121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ettinger A, Wittmann T. Fluorescence live cell imaging. Methods Cell Biol. 2014;123:77–94. doi: 10.1016/B978-0-12-420138-5.00005-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Correa IR., Jr Live-cell reporters for fluorescence imaging. Curr Opin Chem Biol. 2014;20:36–45. doi: 10.1016/j.cbpa.2014.04.007. [DOI] [PubMed] [Google Scholar]
- [23].Adams SR, Mackey MR, Ramachandra R, Palida Lemieux SF, Steinbach P, Bushong EA, Butko MT, Giepmans BN, Ellisman MH, Tsien RY. Multicolor electron microscopy for simultaneous visualization of multiple molecular species. Cell Chem Biol. 2016;23(11):1417–1427. doi: 10.1016/j.chembiol.2016.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Stepanenko OV, Stepanenko OV, Shcherbakova DM, Kuznetsova IM, Turoverov KK, Verkhusha VV. Modern fluorescent proteins: from chromophore formation to novel intracellular applications. Biotechniques. 2011;51(5):313–314. doi: 10.2144/000113765. (316, 318 passim) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Galdeen SA, North AJ. Live cell fluorescence microscopy techniques. Methods Mol Biol. 2011;769:205–222. doi: 10.1007/978-1-61779-207-6_14. [DOI] [PubMed] [Google Scholar]
- [26].Charoensin S, Eroglu E, Opelt M, Bischof H, Madreiter-Sokolowski CT, Kirsch A, Depaoli MR, Frank S, Schrammel A, Mayer B, Waldeck-Weiermair M, et al. Intact mitochondrial Ca2+ uniport is essential for agonist-induced activation of endothelial nitric oxide synthase (eNOS) Free Radic Biol Med. 2017;102:248–259. doi: 10.1016/j.freeradbiomed.2016.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Eroglu E, Gottschalk B, Charoensin S, Blass S, Bischof H, Rost R, Madreiter-Sokolowski CT, Pelzmann B, Bernhart E, Sattler W, Hallstrom S, et al. Development of novel FP-based probes for live-cell imaging of nitric oxide dynamics. Nat Commun. 2016;7:10623. doi: 10.1038/ncomms10623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Eroglu E, Rost R, Bischof H, Blass S, Schreilechner A, Gottschalk B, Depaoli MR, Klec C, Charoensin S, Madreiter-Sokolowski CT, Ramadani J, et al. Application of genetically encoded fluorescent nitric oxide (NO•) probes, the geNOps, for real-time imaging of NO• signals in single cells. J Vis Exp. 2017;(121) doi: 10.3791/55486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Rodriguez EA, Campbell RE, Lin JY, Lin MZ, Miyawaki A, Palmer AE, Shu X, Zhang J, Tsien RY. The growing and glowing toolbox of fluorescent and photoactive proteins. Trends Biochem Sci. 2017;42(2):111–129. doi: 10.1016/j.tibs.2016.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Downes CP, Gray A, Lucocq JM. Probing phosphoinositide functions in signaling and membrane trafficking. Trends Cell Biol. 2005;15(5):259–268. doi: 10.1016/j.tcb.2005.03.008. [DOI] [PubMed] [Google Scholar]
- [31].Varnai P, Balla T. Live cell imaging of phosphoinositide dynamics with fluorescent protein domains. Biochim Biophys Acta. 2006;1761(8):957–967. doi: 10.1016/j.bbalip.2006.03.019. [DOI] [PubMed] [Google Scholar]
- [32].Zhong S, Navaratnam D, Santos-Sacchi J. A genetically-encoded YFP sensor with enhanced chloride sensitivity, photostability and reduced ph interference demonstrates augmented transmembrane chloride movement by gerbil prestin (SLC26a5) PLoS One. 2014;9(6):e99095. doi: 10.1371/journal.pone.0099095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Jayaraman S, Haggie P, Wachter RM, Remington SJ, Verkman AS. Mechanism and cellular applications of a green fluorescent protein-based halide sensor. J Biol Chem. 2000;275(9):6047–6050. doi: 10.1074/jbc.275.9.6047. [DOI] [PubMed] [Google Scholar]
- [34].Nagai T, Sawano A, Park ES, Miyawaki A. Circularly permuted green fluorescent proteins engineered to sense Ca2+ Proc Natl Acad Sci USA. 2001;98(6):3197–3202. doi: 10.1073/pnas.051636098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zhao Y, Araki S, Wu J, Teramoto T, Chang YF, Nakano M, Abdelfattah AS, Fujiwara M, Ishihara T, Nagai T, Campbell RE. An expanded palette of genetically encoded Ca2+ indicators. Science. 2011;333(6051):1888–1891. doi: 10.1126/science.1208592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ermakova YG, Bilan DS, Matlashov ME, Mishina NM, Markvicheva KN, Subach OM, Subach FV, Bogeski I, Hoth M, Enikolopov G, Belousov VV. Red fluorescent genetically encoded indicator for intracellular hydrogen peroxide. Nat Commun. 2014;5:5222. doi: 10.1038/ncomms6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Markvicheva KN, Bilan DS, Mishina NM, Gorokhovatsky AY, Vinokurov LM, Lukyanov S, Belousov VV. A genetically encoded sensor for H2O2 with expanded dynamic range. Bioorg Med Chem. 2011;19(3):1079–1084. doi: 10.1016/j.bmc.2010.07.014. [DOI] [PubMed] [Google Scholar]
- [38].Dooley CT, Dore TM, Hanson GT, Jackson WC, Remington SJ, Tsien RY. Imaging dynamic redox changes in mammalian cells with green fluorescent protein indicators. J Biol Chem. 2004;279(21):22284–22293. doi: 10.1074/jbc.M312847200. [DOI] [PubMed] [Google Scholar]
- [39].Lohman JR, Remington SJ. Development of a family of redox-sensitive green fluorescent protein indicators for use in relatively oxidizing subcellular environments. Biochemistry. 2008;47(33):8678–8688. doi: 10.1021/bi800498g. [DOI] [PubMed] [Google Scholar]
- [40].Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, Ikura M, Tsien RY. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature. 1997;388(6645):882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- [41].Ohkura M, Matsuzaki M, Kasai H, Imoto K, Nakai J. Genetically encoded bright Ca2+ probe applicable for dynamic Ca2+ imaging of dendritic spines. Anal Chem. 2005;77(18):5861–5869. doi: 10.1021/ac0506837. [DOI] [PubMed] [Google Scholar]
- [42].Edgington LE, Berger AB, Blum G, Albrow VE, Paulick MG, Lineberry N, Bogyo M. Noninvasive optical imaging of apoptosis by caspase-targeted activity-based probes. Nat Med. 2009;15(8):967–973. doi: 10.1038/nm.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Takemoto K, Nagai T, Miyawaki A, Miura M. Spatio-temporal activation of caspase revealed by indicator that is insensitive to environmental effects. J Cell Biol. 2003;160(2):235–243. doi: 10.1083/jcb.200207111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Sowa G, Liu J, Papapetropoulos A, Rex-Haffner M, Hughes TE, Sessa WC. Trafficking of endothelial nitric-oxide synthase in living cells. Quantitative evidence supporting the role of palmitoylation as a kinetic trapping mechanism limiting membrane diffusion. J Biol Chem. 1999;274(32):22524–22531. doi: 10.1074/jbc.274.32.22524. [DOI] [PubMed] [Google Scholar]
- [45].Iwakiri Y, Satoh A, Chatterjee S, Toomre DK, Chalouni CM, Fulton D, Groszmann RJ, Shah VH, Sessa WC. Nitric oxide synthase generates nitric oxide locally to regulate compartmentalized protein S-nitrosylation and protein trafficking. Proc Natl Acad Sci USA. 2006;103(52):19777–19782. doi: 10.1073/pnas.0605907103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].van Haperen R, Cheng C, Mees BM, van Deel E, de Waard M, van Damme LC, van Gent T, van Aken T, Krams R, Duncker DJ, de Crom R. Functional expression of endothelial nitric oxide synthase fused to green fluorescent protein in transgenic mice. Am J Pathol. 2003;163(4):1677–1686. doi: 10.1016/S0002-9440(10)63524-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Church JE, Fulton D. Differences in eNOS activity because of subcellular localization are dictated by phosphorylation state rather than the local calcium environment. J Biol Chem. 2006;281(3):1477–1488. doi: 10.1074/jbc.M505968200. [DOI] [PubMed] [Google Scholar]
- [48].Boo YC, Kim HJ, Song H, Fulton D, Sessa W, Jo H. Coordinated regulation of endothelial nitric oxide synthase activity by phosphorylation and subcellular localization. Free Radic Biol Med. 2006;41(1):144–153. doi: 10.1016/j.freeradbiomed.2006.03.024. [DOI] [PubMed] [Google Scholar]
- [49].Waldeck-Weiermair M, Bischof H, Blass S, Deak AT, Klec C, Graier T, Roller C, Rost R, Eroglu E, Gottschalk B, Hofmann NA, et al. Generation of red-shifted cameleons for imaging Ca2+ dynamics of the endoplasmic reticulum. Sensors. 2015;15(6):13052–13068. doi: 10.3390/s150613052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Bajar BT, Wang ES, Zhang S, Lin MZ, Chu J. A guide to fluorescent protein FRET pairs. Sensors. 2016;16(9) doi: 10.3390/s16091488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Grynkiewicz G, Poenie M, Tsien RY. A new generation of Ca2+ indicators with greatly improved fluorescence properties. J Biol Chem. 1985;260(6):3440–3450. [PubMed] [Google Scholar]
- [52].D'Autreaux B, Tucker NP, Dixon R, Spiro S. A non-haem iron centre in the transcription factor NorR senses nitric oxide. Nature. 2005;437(7059):769–772. doi: 10.1038/nature03953. [DOI] [PubMed] [Google Scholar]
- [53].Opelt M, Eroglu E, Waldeck-Weiermair M, Russwurm M, Koesling D, Malli R, Graier WF, Fassett JT, Schrammel A, Mayer B. Formation of nitric oxide by aldehyde dehydrogenase-2 is necessary and sufficient for vascular bioactivation of nitroglycerin. J Biol Chem. 2016;291(46):24076–24084. doi: 10.1074/jbc.M116.752071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Eroglu E, Hallstrom S, Bischof H, Opelt M, Schmidt K, Mayer B, Waldeck-Weiermair M, Graier WF, Malli R. Real-time visualization of distinct nitric oxide generation of nitric oxide synthase isoforms in single cells. Nitric Oxide. 2017;70:59–67. doi: 10.1016/j.niox.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Pearce LL, Gandley RE, Han W, Wasserloos K, Stitt M, Kanai AJ, McLaughlin MK, Pitt BR, Levitan ES. Role of metallothionein in nitric oxide signaling as revealed by a green fluorescent fusion protein. Proc Natl Acad Sci USA. 2000;97(1):477–482. doi: 10.1073/pnas.97.1.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Nielson KB, Atkin CL, Winge DR. Distinct metal-binding configurations in metallothionein. J Biol Chem. 1985;260(9):5342–5350. [PubMed] [Google Scholar]
- [57].Rigby Duncan KE, Stillman MJ. Metal-dependent protein folding: metallation of metallothionein. J Inorg Biochem. 2006;100(12):2101–2107. doi: 10.1016/j.jinorgbio.2006.09.005. [DOI] [PubMed] [Google Scholar]
- [58].Carpene E, Andreani G, Isani G. Metallothionein functions and structural characteristics. J Trace Elem Med Biol. 2007;21(Suppl 1):S35–S39. doi: 10.1016/j.jtemb.2007.09.011. [DOI] [PubMed] [Google Scholar]
- [59].Zangger K, Oz G, Haslinger E, Kunert O, Armitage IM. Nitric oxide selectively releases metals from the amino-terminal domain of metallothioneins: potential role at inflammatory sites. FASEB J. 2001;15(7):1303–1305. doi: 10.1096/fj.00-0641fje. [DOI] [PubMed] [Google Scholar]
- [60].Meloni G, Faller P, Vasak M. Redox silencing of copper in metal-linked neurodegenerative disorders: reaction of Zn7metallothionein-3 with Cu2+ ions. J Biol Chem. 2007;282(22):16068–16078. doi: 10.1074/jbc.M701357200. [DOI] [PubMed] [Google Scholar]
- [61].Sato M, Bremner I. Oxygen free radicals and metallothionein. Free Radic Biol Med. 1993;14(3):325–337. doi: 10.1016/0891-5849(93)90029-t. [DOI] [PubMed] [Google Scholar]
- [62].Thornalley PJ, Vasak M. Possible role for metallothionein in protection against radiation-induced oxidative stress. Kinetics and mechanism of its reaction with superoxide and hydroxyl radicals. Biochim Biophys Acta. 1985;827(1):36–44. doi: 10.1016/0167-4838(85)90098-6. [DOI] [PubMed] [Google Scholar]
- [63].Wright J, George S, Martinez-Lara E, Carpene E, Kindt M. Levels of cellular glutathione and metallothionein affect the toxicity of oxidative stressors in an established carp cell line. Mar Environ Res. 2000;50(1–5):503–508. doi: 10.1016/s0141-1136(00)00125-2. [DOI] [PubMed] [Google Scholar]
- [64].Schwarz MA, Lazo JS, Yalowich JC, Allen WP, Whitmore M, Bergonia HA, Tzeng E, Billiar TR, Robbins PD, Lancaster JR, Jr, et al. Metallothionein protects against the cytotoxic and DNA-damaging effects of nitric oxide. Proc Natl Acad Sci USA. 1995;92(10):4452–4456. doi: 10.1073/pnas.92.10.4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Palmiter RD. The elusive function of metallothioneins. Proc Natl Acad Sci USA. 1998;95(15):8428–8430. doi: 10.1073/pnas.95.15.8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Kennedy MC, Gan T, Antholine WE, Petering DH. Metallothionein reacts with Fe2+ and NO to form products with A g = 2.039 ESR signal. Biochem Biophys Res Commun. 1993;196(2):632–635. doi: 10.1006/bbrc.1993.2296. [DOI] [PubMed] [Google Scholar]
- [67].Zhang LM, Croix C, Cao R, Wasserloos K, Watkins SC, Stevens T, Li S, Tyurin V, Kagan VE, Pitt BR. Cell-surface protein disulfide isomerase is required for transnitrosation of metallothionein by S-nitroso-albumin in intact rat pulmonary vascular endothelial cells. Exp Biol Med. 2006;231(9):1507–1515. doi: 10.1177/153537020623100909. [DOI] [PubMed] [Google Scholar]
- [68].Croix CM, Leelavaninchkul K, Watkins SC, Kagan VE, Pitt BR. Nitric oxide and zinc homeostasis in acute lung injury. Proc Am Thorac Soc. 2005;2(3):236–242. doi: 10.1513/pats.200501-007AC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Croix CM, Wasserloos KJ, Dineley KE, Reynolds IJ, Levitan ES, Pitt BR. Nitric oxide-induced changes in intracellular zinc homeostasis are mediated by metallothionein/thionein. Am J Physiol Lung Cell Mol Physiol. 2002;282(2):L185–L192. doi: 10.1152/ajplung.00267.2001. [DOI] [PubMed] [Google Scholar]
- [70].Croix CM, Stitt MS, Leelavanichkul K, Wasserloos KJ, Pitt BR, Watkins SC. Nitric oxide-induced modification of protein thiolate clusters as determined by spectral fluorescence resonance energy transfer in live endothelial cells. Free Radic Biol Med. 2004;37(6):785–792. doi: 10.1016/j.freeradbiomed.2004.06.004. [DOI] [PubMed] [Google Scholar]
- [71].Cherian MG, Apostolova MD. Nuclear localization of metallothionein during cell proliferation and differentiation. Cell Mol Biol (Noisy-le-Grand) 2000;46(2):347–356. [PubMed] [Google Scholar]
- [72].Pearce LL, Wasserloos K, Croix CM, Gandley R, Levitan ES, Pitt BR. Metallothionein, nitric oxide and zinc homeostasis in vascular endothelial cells. J Nutr. 2000;130(5S Suppl):S1467S–S1470SS. doi: 10.1093/jn/130.5.1467S. [DOI] [PubMed] [Google Scholar]
- [73].Sato M, Hida N, Umezawa Y. Imaging the nanomolar range of nitric oxide with an amplifier-coupled fluorescent indicator in living cells. Proc Natl Acad Sci USA. 2005;102(41):14515–14520. doi: 10.1073/pnas.0505136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Sato M, Hida N, Ozawa T, Umezawa Y. Fluorescent indicators for cyclic GMP based on cyclic GMP-dependent protein kinase Ialpha and green fluorescent proteins. Anal Chem. 2000;72(24):5918–5924. doi: 10.1021/ac0006167. [DOI] [PubMed] [Google Scholar]
- [75].Tricoire L, Vitalis T. Neuronal nitric oxide synthase expressing neurons: a journey from birth to neuronal circuits. Front Neural Circuits. 2012;6:82. doi: 10.3389/fncir.2012.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].McNeill E, Crabtree MJ, Sahgal N, Patel J, Chuaiphichai S, Iqbal AJ, Hale AB, Greaves DR, Channon KM. Regulation of iNOS function and cellular redox state by macrophage Gch1 reveals specific requirements for tetrahydrobiopterin in NRF2 activation. Free Radic Biol Med. 2015;79:206–216. doi: 10.1016/j.freeradbiomed.2014.10.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Sessa WC. eNOS at a glance. J Cell Sci. 2004;117(Pt 12):2427–2429. doi: 10.1242/jcs.01165. [DOI] [PubMed] [Google Scholar]
- [78].Seddon M, Shah AM, Casadei B. Cardiomyocytes as effectors of nitric oxide signalling. Cardiovasc Res. 2007;75(2):315–326. doi: 10.1016/j.cardiores.2007.04.031. [DOI] [PubMed] [Google Scholar]
- [79].Sato M, Nakajima T, Goto M, Umezawa Y. Cell-based indicator to visualize picomolar dynamics of nitric oxide release from living cells. Anal Chem. 2006;78(24):8175–8182. doi: 10.1021/ac061791b. [DOI] [PubMed] [Google Scholar]
- [80].Thomas DD, Liu X, Kantrow SP, Lancaster JR., Jr The biological lifetime of nitric oxide: implications for the perivascular dynamics of NO and O2. Proc Natl Acad Sci USA. 2001;98(1):355–360. doi: 10.1073/pnas.011379598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Shiva S. Nitrite: a physiological store of nitric oxide and modulator of mitochondrial function. Redox Biol. 2013;1(1):40–44. doi: 10.1016/j.redox.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Lundberg JO, Weitzberg E. NO-synthase independent NO generation in mammals. Biochem Biophys Res Commun. 2010;396(1):39–45. doi: 10.1016/j.bbrc.2010.02.136. [DOI] [PubMed] [Google Scholar]
- [83].Hidaka M, Gotoh A, Shimizu T, Minamisawa K, Imamura H, Uchida T. Visualization of NO3(-)/NO2(-) dynamics in living cells by Fluorescence Resonance Energy Transfer (FRET) imaging employing a rhizobial two-component regulatory system. J Biol Chem. 2016;291(5):2260–2269. doi: 10.1074/jbc.M115.687632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Sanchez C, Itakura M, Okubo T, Matsumoto T, Yoshikawa H, Gotoh A, Hidaka M, Uchida T, Minamisawa K. The nitrate-sensing NasST system regulates nitrous oxide reductase and periplasmic nitrate reductase in Bradyrhizobium japonicum. Environ Microbiol. 2014;16(10):3263–3274. doi: 10.1111/1462-2920.12546. [DOI] [PubMed] [Google Scholar]
- [85].Liaudet L, Vassalli G, Pacher P. Role of peroxynitrite in the redox regulation of cell signal transduction pathways. Front Biosci (Landmark Ed.) 2009;14:4809–4814. doi: 10.2741/3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Klotz LO, Schroeder P, Sies H. Peroxynitrite signaling: receptor tyrosine kinases and activation of stress-responsive pathways. Free Radic Biol Med. 2002;33(6):737–743. doi: 10.1016/s0891-5849(02)00892-4. [DOI] [PubMed] [Google Scholar]
- [87].Pacher P, Beckman JS, Liaudet L. Nitric oxide and peroxynitrite in health and disease. Physiol Rev. 2007;87(1):315–424. doi: 10.1152/physrev.00029.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Szabo C, Ischiropoulos H, Radi R. Peroxynitrite: biochemistry, pathophysiology and development of therapeutics. Nat Rev Drug Discov. 2007;6(8):662–680. doi: 10.1038/nrd2222. [DOI] [PubMed] [Google Scholar]
- [89].Kiechle FL, Malinski T. Indirect detection of nitric oxide effects: a review. Ann Clin Lab Sci. 1996;26(6):501–511. [PubMed] [Google Scholar]
- [90].Tarpey MM, Fridovich I. Methods of detection of vascular reactive species: nitric oxide, superoxide, hydrogen peroxide, and peroxynitrite. Circ Res. 2001;89(3):224–236. doi: 10.1161/hh1501.094365. [DOI] [PubMed] [Google Scholar]
- [91].Chen ZJ, Ren W, Wright QE, Ai HW. Genetically encoded fluorescent probe for the selective detection of peroxynitrite. J Am Chem Soc. 2013;135(40):14940–14943. doi: 10.1021/ja408011q. [DOI] [PubMed] [Google Scholar]
- [92].Sikora A, Zielonka J, Lopez M, Joseph J, Kalyanaraman B. Direct oxidation of boronates by peroxynitrite: mechanism and implications in fluorescence imaging of peroxynitrite. Free Radic Biol Med. 2009;47(10):1401–1407. doi: 10.1016/j.freeradbiomed.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]