Abstract
Metabolic reprogramming is a prominent feature of clear cell renal cell carcinoma (ccRCC). Here we investigated metabolic dependencies in a panel of ccRCC cell lines using nutrient depletion, functional RNAi screening and inhibitor treatment. We found that ccRCC cells are highly sensitive to the depletion of glutamine or cystine, two amino acids required for glutathione (GSH) synthesis. Moreover, silencing of enzymes of the GSH biosynthesis pathway or glutathione peroxidases, which depend on GSH for the removal of cellular hydroperoxides, selectively reduced viability of ccRCC cells but did not affect the growth of non-malignant renal epithelial cells. Inhibition of GSH synthesis triggered ferroptosis, an iron-dependent form of cell death associated with enhanced lipid peroxidation. VHL is a major tumour suppressor in ccRCC and loss of VHL leads to stabilisation of hypoxia inducible factors HIF-1α and HIF-2α. Restoration of functional VHL via exogenous expression of pVHL reverted ccRCC cells to an oxidative metabolism and rendered them insensitive to the induction of ferroptosis. VHL reconstituted cells also exhibited reduced lipid storage and higher expression of genes associated with oxidiative phosphorylation and fatty acid metabolism. Importantly, inhibition of β-oxidation or mitochondrial ATP-synthesis restored ferroptosis sensitivity in VHL reconstituted cells. We also found that inhibition of GSH synthesis blocked tumour growth in a MYC-dependent mouse model of renal cancer. Together, our data suggest that reduced fatty acid metabolism due to inhibition of β-oxidation renders renal cancer cells highly dependent on the GSH/GPX pathway to prevent lipid peroxidation and ferroptotic cell death.
Keywords: cancer metabolism, clear cell renal carcinoma, ccRCC, glutathione, ferroptosis, reactive oxygen species, ROS, glutathione peroxidases, GPX, lipid metabolism, lipid peroxidation, beta-oxidation
Introduction
Renal cell carcinoma (RCC) is a devastating disease due to frequent metastasis formation and resistance to conventional or targeted therapies 45. Clear cell renal cell carcinoma (ccRCC) is the most common type of RCC and both hereditary and sporadic ccRCC are strongly associated with mutations in the von Hippel Lindau (VHL) gene 28 that lead to the stabilisation of the hypoxia inducible factors (HIF-1α and HIF-2α) 23, 34. As metabolic reprogramming is a major feature of ccRCC tumours 8, 16, 25, targeting metabolic processes could be a successful strategy in the treatment of this disease.
One by-product of the enhanced metabolic activity of cancer cells is the increased generation of reactive oxygen species (ROS), which are generated, amongst others, by the complexes of the mitochondrial electron transport chain (ETC) 38. While high levels of ROS are detrimental to cell viability, ROS can also promote cell transformation and tumour development, for example by participating in signalling processes 46. There is now substantial evidence that oncogene activation provokes oxidative stress but also induces anti-oxidant pathways to prevent the accumulation of ROS and avoid cell damage 7, 22. Glutathione, the major non-enzymatic antioxidant in cells, is formed by the sequential condensation of glutamate, cysteine and glycine. In addition to its direct anti-oxidant function 22, GSH is a co-substrate of glutathione peroxidases (GPX), a class of selenocysteine-containing enzymes that reduce hydroperoxides, including hydrogen peroxide (H2O2) and lipid peroxides 6. While most GPX proteins reduce soluble hydroperoxides, GPX4 can also target peroxidised acyl-groups found in membrane lipids 6. Impaired GSH synthesis or inhibition of GPX4 activity has been linked to the induction of ferroptosis, an iron-mediated form of cell death caused by the accumulation of lipid peroxides 12, 53.
In this study, we investigated metabolic liabilities of ccRCC cells. Using selected amino acid depletion, RNAi screening and inhibitor treatment, we found that ccRCC cells are highly dependent on GSH biosynthesis and GPX function to prevent the induction of ferroptosis. Moreover, reconstitution of functional VHL prevented the induction of ferroptosis in response to inhibition of GSH synthesis in ccRCC cells, by reverting cells back to an oxidative metabolism and increasing fatty acid degradation through β-oxidation. We also found that inhibition of GSH biosynthesis efficiently blocked tumour growth in a mouse model of renal cancer. Our results suggest that targeting GSH biosynthesis and GPX activity could be a promising strategy for the treatment of ccRCC.
Results
Glutathione biosynthesis is essential in renal cancer cells
To establish metabolic dependencies in renal cancer, we first exposed four ccRCC cell lines (RCC4, UMRC2, A498 and 769-P) either to full medium or medium deprived of glutamine or cystine, the dipeptide precursor for intracellular cysteine. All cell lines showed severe growth inhibition when cultured in restricted media (Fig. 1A). Glutamine and cysteine are precursors for the synthesis of glutathione, a major cellular antioxidant. We therefore investigated whether glutamine and cystine deprivation could be rescued by providing cells with exogenous antioxidants. Inhibition of cell proliferation caused by glutamine starvation was not affected by the addition of N-acetylcysteine (NAC), (2,2,6,6-Tetramethylpiperidin-1-yl)oxyl (TEMPO) or cysteine (Fig. 1B). Furthermore, the reduction in cell number caused by treatment with BPTES, an allosteric inhibitor of glutaminase 1 (GLS1), was only partially rescued by addition of NAC in RCC4 or 786-O cells (Fig. S1A). In contrast, cell number reduction caused by cystine depletion was consistently rescued by addition of NAC or cysteine in several ccRCC cell lines (Fig. 1C), while TEMPO, a mimetic of mitochondrial superoxide dismutase, only had a minor effect in one of the cell lines used. Together these results suggest that renal cancer cells are highly dependent on exogenous glutamine and cystine to feed essential metabolic pathways. Moreover, cystine uptake seems to be rate-limiting for the maintenance of the intracellular cysteine pool, which is required for protein synthesis and antioxidant generation. Our results also suggest that de novo synthesis of cysteine via the transsulfuration pathway cannot substitute for cystine uptake. We next investigated whether enzymes involved in glutamine and cystine uptake and the GSH biosynthesis pathway are deregulated in renal cancer. Analysis of gene expression data showed that genes coding for the Na+-dependent glutamine transporter (SLC1A5) and a component of the cystine-glutamate antiporter XCT (SLC7A11) are upregulated in ccRCC tumours (Fig. S1B and C). Moreover, the genes coding for subunits of the γ-glutamylcysteine synthetase (GCLM and GCLC) are also highly expressed in renal tumours compared to normal renal tissue (Fig. S1D and E).
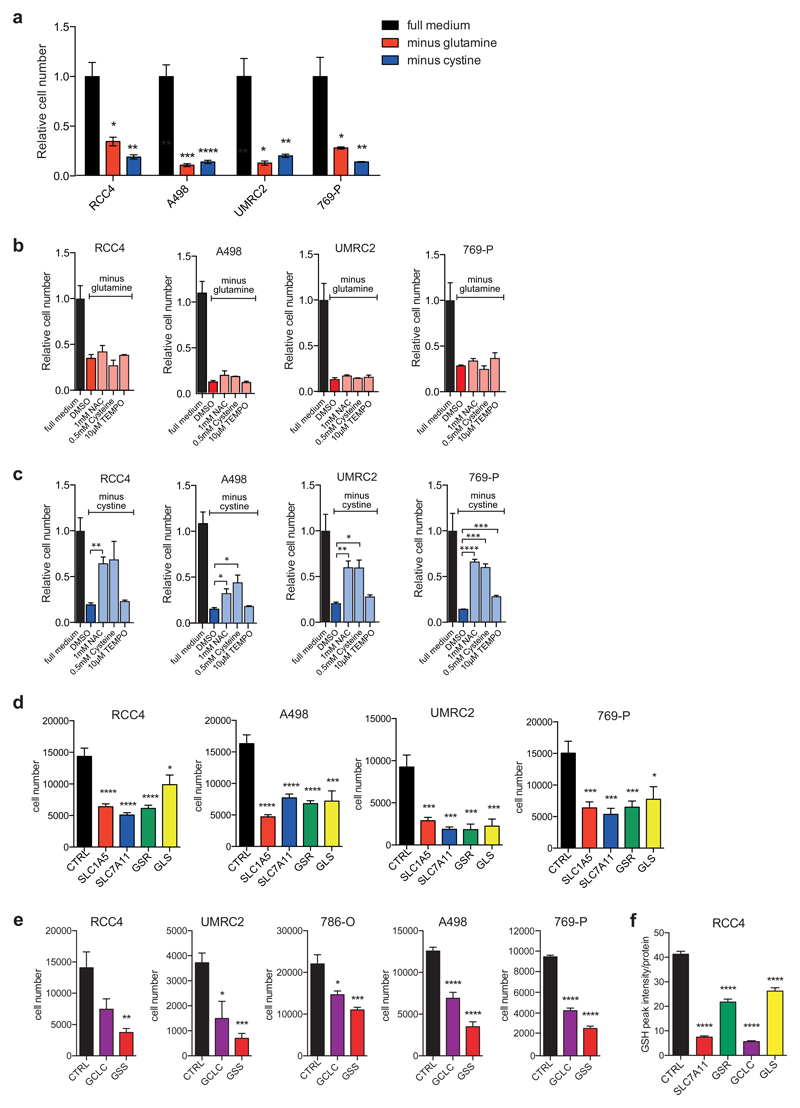
Figure 1. Renal cancer cells are sensitive to glutamine and cystine depletion.
A) RCC4, A498, UMRC2 and 769-P cells were seeded in full medium. After 24h, medium was replaced to normal medium or medium deprived of glutamine or cystine for 72h and cell numbers were determined. Values represent mean cell number ±SEM relative to full medium (n=3).
B) RCC4, A498, UMRC2 and 769-P cells were seeded in full medium. After 24h, cells were transferred to glutamine-free medium containing the different supplements and antioxidants. Cells were fixed after 72h and cell numbers were determined. Values represent mean cell number ±SEM relative to full medium (n=3).
C) RCC4, A498, UMRC2 and 769-P cells were seeded in full medium. After 24h, cells were transferred to cystine-free medium containing the different supplements and antioxidants. Cells were fixed after 72h and cell numbers were determined. Values represent mean cell number ±SEM relative to full medium (n=3).
D) RCC4, A498, UMRC2 and 769-P cells were transfected with siRNA oligonucleotides targeting SLC7A11, SLC1A5, GSR or GLS or a non-targeting control (CTRL). Cell numbers were determined 96h post transfection. Values represent mean cell number ±SEM (n=3).
E) RCC4, A498, UMRC2, 786-O and 769-P cells were transfected with siRNA oligonucleotides targeting GCLC, GSS or a non-targeting control (CTRL). Cell number was determined 96h post transfection. Values represent mean cell number ±SEM (n=3).
F) RCC4 cells were transfected with siRNA oligonucleotides targeting SLC7A11, GSR, GCLC, GLS or a non-targeting control (CTRL). After 96 hours, cells were lysed and levels of reduced glutathione (GSH) were determined by mass spectrometry. Values represent mean peak intensity normalised to protein ±SEM (n=4).
*p≤0.05; **p≤0.01; ***p≤0.005; ****p≤0.001 unpaired two-tailed Student’s t-test.
We next investigated whether siRNA-mediated silencing of transporters required for the uptake of glutamine and cystine or components of the GSH synthesis and regeneration pathway had an effect on the viability of renal cancer cells. Interestingly, silencing of SLC1A5, SLC7A11, glutathione reductase (GSR), glutaminase (GLS), GCLC and glutathione synthase (GSS) resulted in a strong reduction in cell number in several ccRCC cell lines (Fig. 1D and E). This was accompanied by a substantial reduction in cellular GSH levels (Fig. 1F), supporting the importance of GSH maintenance in these cells.
Functional screen identifies glutathione peroxidases as essential genes in renal cancer
To obtain a more comprehensive insight into metabolic dependencies of renal cancer cells, we performed an siRNA screen targeting 230 metabolic enzymes, nutrient transporters and regulators of metabolism in a subset of renal cancer cell lines 17. Cells were cultured for 96 hours after transfection and numbers of remaining cells were used to calculate z-scores for each gene (supplementary Table 1). Ranking of averaged z-scores indicated that all cell lines are highly sensitive towards depletion of the MYC proto-oncogene (Fig. 2A and B), which has been shown repeatedly to be essential for the proliferation of ccRCC cells 20, 47. Moreover, depletion of the glycolytic enzyme ALDOA also caused a strong reduction in cell number in all cell lines tested (Fig. 2A and B). This is not surprising as enhanced glycolysis is an important feature of ccRCC 43. In addition, we found that silencing of DHCR24, the gene coding for an enzyme in the cholesterol biosynthesis pathway, also reduced cell numbers in renal cancer cells (Fig. 2A and B). While increased levels of compartmentalised cholesterol and cholesterol esters have been found in renal tumours compared to normal kidney 18, a clear connection between cholesterol metabolism and renal cancer development has not yet been established 14. Among the top ranking genes were also two genes coding for glutathione peroxidases, GPX3 and GPX4 (Fig. 2A and B).
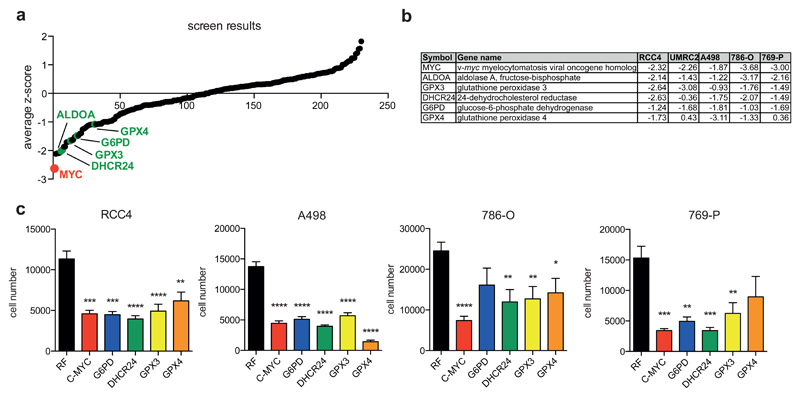
Figure 2. Functional siRNA screen identifies glutathione peroxidases as essential enzymes in renal cancer cells.
A) RCC4, A498, UMRC2, 786-O and 769-P cells were transfected with siRNA oligonucleotides targeting 230 different metabolic enzymes, nutrient transporters and metabolic regulators. 96h post transfection, number of remaining cells was determined and used to calculate z-scores. Graph displays mean z-scores across all cell lines and selected genes are highlighted.
B) Table showing z-scores of selected genes in each of the five cell lines.
C) RCC4, A498, 786-O and 769-P cells were transfected with siRNA oligonucleotides targeting MYC, G6PD, DHCR24, GPX3 or GPX4 or a non-targeting control (RF). Cell numbers were determined 96h post transfection. Values represent mean cell number ±SEM (n=3).
*p≤0.05; **p≤0.01; ***p≤0.005; ****p≤0.001 unpaired two-tailed Student’s t-test.
GPX4 targets phospholipid hydroperoxides within membranes and lipoproteins 6. GPX3 is a secreted protein that is produced by epithelial cells of the proximal renal tubules 1. It has been shown that GPX3 is induced by hypoxia 4 but its role in renal cancer has not been explored in detail. Finally, silencing of G6PD, the gene coding for the first NADPH producing enzyme within the pentose phosphate pathway (PPP), was also detrimental to most ccRCC cell lines (Fig. 2A and B). This is consistent with previous findings demonstrating that fructose-1,6-bisphosphatase 1 (FBP1) restrains ccRCC cell proliferation by interfering with PPP activity 32. As NADPH is an essential cofactor for the synthesis and regeneration of GSH, this finding also further supports the importance of GSH for ccRCC cell survival.
We also confirmed the screen results by showing that individual silencing of MYC, G6PD, DHCR24, GPX3 and GPX4 significantly reduces cell numbers in several renal cancer cell lines (Fig. 2C). Moreover, we found that MYC, GPX4 and G6PD are upregulated in ccRCC tumours compared to normal kidney (Fig. S2A-C), further supporting the importance of these genes in ccRCC development.
Inhibition of GSH synthesis induces ferroptosis in renal cancer cells
The results obtained so far indicate that ccRCC cells are highly sensitive to the disruption of the GSH biosynthesis and regeneration pathway or depletion of specific GPX enzymes. Inhibition of cystine uptake or inhibition of GPX4 has been linked to ferroptosis, caused by the accumulation of lipid hydroperoxides (13 and Fig. 3A). We therefore investigated whether inhibition of cystine uptake and GSH synthesis could cause ferroptosis in ccRCC cells. Inhibition of cystine import using the XCT-inhibitor Erastin resulted in a robust cell number reduction in RCC4 and 786-O cells in a dose dependent manner (Fig. 3B). This was alleviated by treatment with ferrostatin-1 (fer-1), a lipophilic small molecule antioxidant that was identified as a selective inhibitor of ferroptosis 12. Similarly, reduction in cell number in response to treatment with L-buthionine-S,R-sulfoximine (BSO), an irreversible inhibitor of γ-cysteine synthetase, was partially rescued by fer-1 treatment (Fig. 3C). We also confirmed that loss of cell viability caused by silencing of SLC7A11 could be restored by treatment with fer-1 (Fig. 3D).
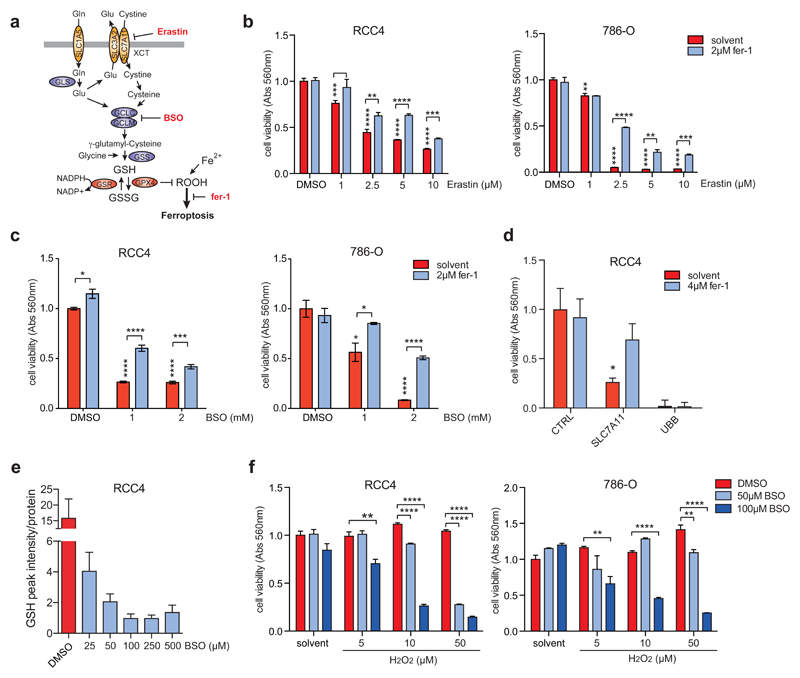
Figure 3. Inhibition of GSH synthesis induces ferroptosis in ccRCC cells.
A) Diagram illustrating the mechanisms of GSH synthesis and induction of ferroptosis.
B) RCC4 and 786-O cells were treated with the indicated concentrations of Erastin or solvent (DMSO) either in the presence or absence of ferrostatin (fer-1) for 72h. Cell viability was determined by crystal violet staining. Values represent mean ±SEM relative to solvent-treated controls (n=3).
C) RCC4 and 786-O cells were treated with the indicated concentrations of BSO or solvent (DMSO) either in the presence or absence of ferrostatin (fer-1) for 72h. Cell viability was determined by crystal violet staining. Values represent mean ±SEM relative to solvent-treated controls (n=3).
D) RCC4 cells were transfected with siRNA oligonucleotides targeting SLC7A11, UBB or non-targeting controls (CTRL) and treated with 4 μM ferrostatin (fer-1) or solvent for 96 h. Cell viability was determined by crystal violet staining. Values represent mean ±SEM relative to solvent-treated controls (n=3).
E) RCC4 cells were treated with the indicated concentrations of BSO or solvent (DMSO) for 24h. GSH levels were determined by mass spectrometry. Values represent mean ±SEM (n=2).
F) RCC4 and 786-O cells were treated with the indicated concentrations of BSO or solvent (DMSO) with or without different concentrations of hydrogen peroxide (H2O2) for 72 hours. Cell viability was determined by crystal violet staining. Values represent mean ±SEM relative to solvent-treated controls (n=3).
*p≤0.05; **p≤0.01; ***p≤0.005; ****p≤0.001 unpaired two-tailed Student’s t-test.
We next investigated the effect of BSO treatment on cellular GSH levels. This analysis showed that even low concentrations of this drug (25-50 μM) already cause a substantial reduction in GSH after 24 hours of treatment. A further reduction of GSH levels was achieved by lethal BSO concentrations (100 μM and higher) (Fig. 3E). Nevertheless, the low levels of residual GSH detected after treatment with 50 μM of BSO were sufficient to maintain cell viability in the presence of an oxidative insult, as demonstrated by the treatment with H2O2 (Fig. 3F). This suggests that renal cancer cells maintain a large spare capacity in their GSH production and that GSH levels have to be reduced below a certain threshold before cell viability is compromised.
Restoration of pVHL function induces resistance to ferroptosis in renal cancer cells
To investigate the molecular basis for the high sensitivity of ccRCC cells towards induction of ferroptosis, we used derivatives of the VHL-deficient RCC4 cell line that were engineered to re-express wild type pVHL or an empty vector (RCC4-VHL and RCC4-EV) 28, 33. In ccRCC cells, restoration of pVHL function blocks tumour formation but does not alter cell proliferation in vitro 28, 33.
Immunoblotting showed that VHL-reconstituted cells express lower levels of HIF-1α and HIF-2α proteins (Fig. 4A) and display reduced mRNA levels of the HIF-target genes VEGFA and PDHK1 (Fig. 4B), confirming the functional restoration of pVHL-dependent regulation of HIF activity in these cells. Moreover, VHL reconstitution also reduced serine 473 phosphorylation of AKT and serine 235/236 phosphorylation of pS6 (Fig. 4C). We also observed a strong upregulation of basal and maximal respiration as well as enhanced spare capacity and mitochondrial ATP production, which is consistent with a reversal of the glycolytic phenotype that is established by HIF and AKT activation in VHL mutant renal cancer cells (Fig. 4D).
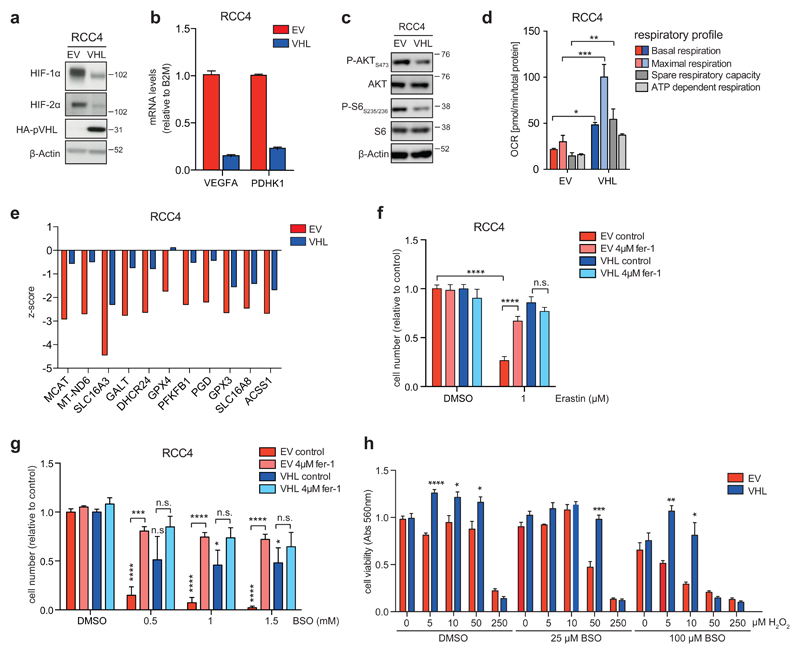
Figure 4. Restoration of pVHL function leads to ferroptosis resistance in ccRCC cells.
A) Isogenic RCC4 cells were generated by expression of functional VHL (RCC4-VHL) or empty vector (RCC4-EV) 33. Analysis of protein expression shows reduced expression of HIF-1α and HIF-2α in VHL-restored cells. Expression of HA-tagged pVHL was also confirmed. β-Actin was used as loading control.
B) mRNA levels of VEGFA and PDHK1 in isogenic RCC4 cell lines. Values represent mean ±SD relative to RCC4-EV and are normalised to B2M (n=2).
C) Phosphorylation of AKT (serine 473) and pS6 (serines 235 and 236) was determined by immunoblotting using phospho-specific antibodies.
D) Oxygen consumption rates (OCR) were determined in RCC4-EV and RCC4-VHL cells using a Seahorse Bioanalyzer and used to calculate basal respiration, maximal respiration in the presence of FCCP, spare respiratory capacity and ATP-dependent respiration. Values represent mean ±SEM of 6 replicates.
E) Genes showing differential essentiality in RCC4-EV and RCC4-VHL cells (Δz-score≥1).
F) RCC4-EV and RCC4-VHL cells were treated with 1μM Erastin or solvent (DMSO) in the presence or absence of 4μM ferrostatin (fer-1) for 72h. Cell viability was determined by crystal violet staining. Values represent mean ±SEM relative to solvent-treated controls (n=3).
G) RCC4-EV and RCC4-VHL cells were treated with the indicated concentrations of BSO or solvent (DMSO) in the presence or absence of ferrostatin (fer-1) for 72h. Cell viability was determined by crystal violet staining. Values represent mean ±SEM relative to solvent treated controls (n=3).
H) RCC4-EV and RCC4-VHL cells were treated with the indicated concentrations of BSO or solvent (DMSO) together with increasing concentrations of H2O2 for 72 hours. Cell viability was determined by crystal violet staining. Values represent mean ±SEM relative to solvent-treated controls (n=3).
*p≤0.05; **p≤0.01; ***p≤0.005; ****p≤0.001 unpaired two-tailed Student’s t-test.
VHL-isogenic cells were also subjected to siRNA screening and differential z-score analysis identified those genes that are selectively required in EV cells. Among the VHL-mutant specific targets was SLC16A3, the gene coding for the lactate transporter MCT4, which has been shown previously to be essential for ccRCC cell survival 19. Moreover, DHCR24, GPX4 and GPX3 were also among the genes for which silencing had a greater effect in VHL-deficient cells (Fig. 4E).
We next asked whether VHL reconstitution alters the sensitivity of cells towards inhibition of GSH synthesis. While treatment with Erastin caused a robust reduction in cell number in VHL-deficient cells, which was almost completely blocked by fer-1 treatment, VHL-restored cells were largely insensitive towards treatment with this compound (Fig. 4F). This effect was not due to differential expression of the XCT-transporter, as expression of SLC7A11 mRNA was similar in both cell lines (Fig. S3A). Similarly, treatment with BSO efficiently reduced cell numbers in empty vector cells, while leaving VHL-restored cells largely unaffected. The effect of BSO on empty vector cells was also completely blocked by addition of fer-1 (Fig. 4G).
We also confirmed that VHL-deficient and restored cells contain similar levels of intracellular GSH and that Erastin and BSO efficiently depleted GSH levels in both cell lines (Fig. S3B). In addition, while sub-lethal concentrations of BSO increased the sensitivity of VHL-deficient cells towards oxidative stress induced by H2O2 treatment, VHL-restored cells showed higher resistance towards oxidative stress both in the absence and presence of BSO (Fig. 4H). Together, these results demonstrate that VHL reconstitution decreases the sensitivity of renal cancer cells towards oxidative stress and the induction of ferroptosis in response to inhibition of GSH synthesis.
Increased lipid peroxidation causes ferroptosis in VHL-deficient renal cancer cells
A central mechanism of ferroptosis is the accumulation of oxidised lipids produced by the cytoplasmic labile iron pool (Fe2+) via the Fenton reaction 13 and through the function of Fe-dependent lipoxygenases 54. In agreement with this hypothesis, we found that VHL-deficient renal cancer cells showed enhanced lipid peroxidation in response to Erastin treatment (Fig. 5A).
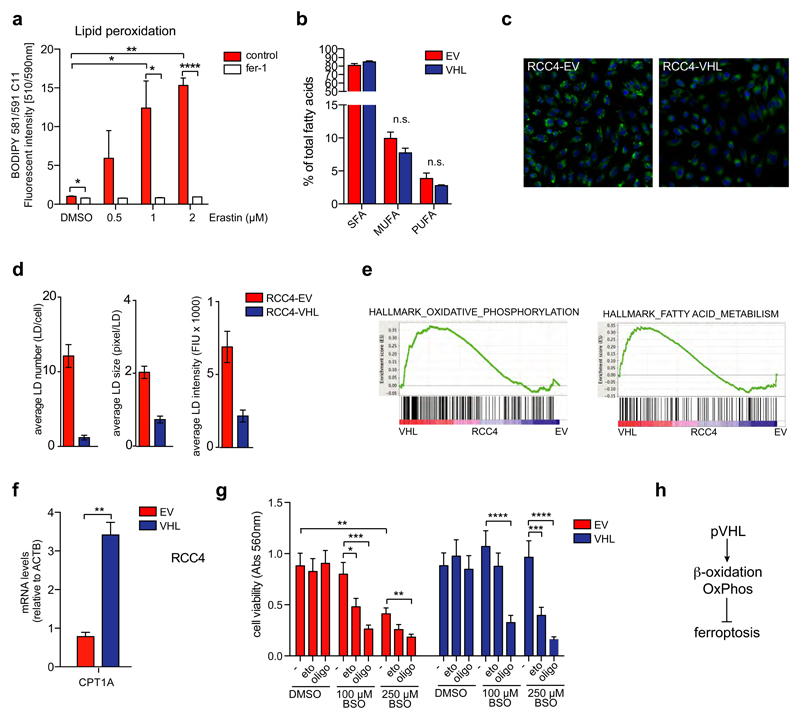
Figure 5. Induction of ferroptosis in ccRCC cells is mediated by lipid peroxidation.
A) RCC4-EV cells were treated with the indicated concentrations of Erastin for 24h. Lipid peroxidation was determined using the BODIPY 581/591 C11 reagent. Values represent mean ±SEM relative to solvent-treated controls (n=3).
B) Relative amounts of saturated (SFA), mono-unsaturated (MUFA) and poly-unsaturated (PUFA) fatty acids in RCC4-EV and RCC4-VHL cells. Values represent mean percentages ±SEM of the total fatty acid content (n=3).
C) RCC4-EV and RCC4-VHL cells were cultured for 48 hours in full medium before fixation and staining with Nile Red to detect lipid droplets (LD). Cells were imaged using a Cellomics high content microscope.
D) Average numbers of LDs per cell, average LD size (pixel per LD) and average LD staining intensity (fluorescence intensity units, FIU). Values represent mean ± SEM (n=3).
E) GSEA results of expression data from RCC4-EV and RCC4-VHL cells from 48. Enrichment plots for two gene sets from the hallmark database associated with oxidative phosphorylation and fatty acid metabolism are shown.
F) Expression of CPT1A in RCC4-EV and RCC4-VHL cells. Values represent mean ±SEM relative to RCC4-EV and are normalised to ACTB (n=3).
G) RCC4-EV and RCC4-VHL cells were treated with the indicated concentrations of BSO or solvent (DMSO) together with 100 μM of Etomoxir or 2 μM of Oligomycin for 72h. Cell viability was determined by crystal violet staining. Values represent mean ±SEM relative to solvent-treated controls (n=7).
H) Diagram illustrating the link between induction of β-oxidation and oxidative phosphorylation (OxPhos) and protection from ferroptosis by pVHL.
*p≤0.05; **p≤0.01; ***p≤0.005; ****p≤0.001 unpaired two-tailed Student’s t-test.
To address the mechanism underlying the differential sensitivity of VHL-isogenic cells towards ferroptosis, we first determined total fatty acid composition derived from all lipid species in RCC4-EV and RCC4-VHL cells. We found that the relative proportion of saturated fatty acids (SFA), mono-unsaturated fatty acids (MUFA) and poly-unsaturated fatty acids (PUFA) (the latter being more susceptible to peroxidation) in the total lipid pool were similar in both cell lines (Fig. 5B). Nevertheless, when we stained for neutral lipids using Nile Red, we found that numbers, size and intensity of lipid droplets (LD) are strongly reduced in VHL-restored cells compared to their VHL deficient counterparts (Fig. 5C and D). Extensive lipid storage is an important feature of ccRCC and high levels of LD are found in renal tumours compared to normal kidney 40. Moreover, hypoxia induces lipid uptake and the formation of LD via a HIF-1α-dependent mechanism 2. This suggests that the high abundance of LD found in RCC4-EV cells could be indicative of higher levels of lipid accumulation and storage compared to RCC4-VHL cells.
We next investigated the expression of proteins controlling the cellular labile iron pool in the isogenic cell line pair. The transferrin receptor (TFRC), which mediates iron uptake by receptor-mediated endocytosis, was more strongly expressed in VHL-restored cells (Fig. S3C), arguing against the possibility that differences in iron uptake mediate differential ferroptosis sensitivity in these cells. Furthermore, the gene coding for heme oxygenase HMOX1, the enzyme releasing iron during heme degradation, which has been shown to accelerate Erastin induced ferroptosis 31, did not show differential expression between the two cell lines (Fig. S3C). In contrast, expression of the lipoxygenase ALOX5, which catalyses the conversion of arachidonic acid to 5(S)-hydroperoxy-6-trans-8,11,14-cis-eicosatetraenoic acid (HPETE) as part of the leukotriene pathway 41, was significantly lower in VHL-reconstituted cells (Fig. S3D). Moreover, ALOX5 mRNA also showed strong upregulation in ccRCC tumours compared to normal tissue (Fig. S3E), while TFRC mRNA was not deregulated (data not shown).
To further explore differences between the cell lines that could be involved in determining their differential sensitivity towards induction of ferroptosis, we compared gene expression profiles from both cell lines 48. Gene set enrichment analysis (GSEA) revealed an upregulation of hallmark gene sets linked to oxidative phosphorylation and fatty acid metabolism in VHL-restored cells (Fig. S3F and 5E). Moreover, expression of carnitine palmitoyl-transferase 1A (CPT1A), the enzyme responsible for the transfer of acyl-groups from coenzyme A to carnitine for import into the mitochondrial matrix and subsequent degradation via β-oxidation, was strongly increased in VHL-reconstituted cells (Fig. 5F), further indicating that VHL-deficient and reconstituted cells differ in their ability to perform fatty acid degradation.
We therefore investigated whether inhibition of fatty acid metabolism altered ferroptosis sensitivity in ccRCC cells. Treatment with Etomoxir, an inhibitor of CPT1, or Oligomycin, an inhibitor of mitochondrial ATP-synthase, had no effect on cell number but fully restored the sensitivity of VHL-reconstituted cells towards inhibition of GSH synthesis by BSO (Fig. 5G). In addition, exposure to these compounds together with low concentrations of BSO caused a synergistic reduction in cell number in VHL-deficient cells (Fig. 5G). Together, these results suggest that increased lipid storage and impaired fatty acid degradation as a consequence of inhibition of β-oxidation renders ccRCC cells highly dependent on cystine uptake and GSH synthesis to prevent the accumulation of lipid peroxides and maintain cell viability (Fig. 5H).
Inhibition of GSH synthesis blocks growth of renal tumours
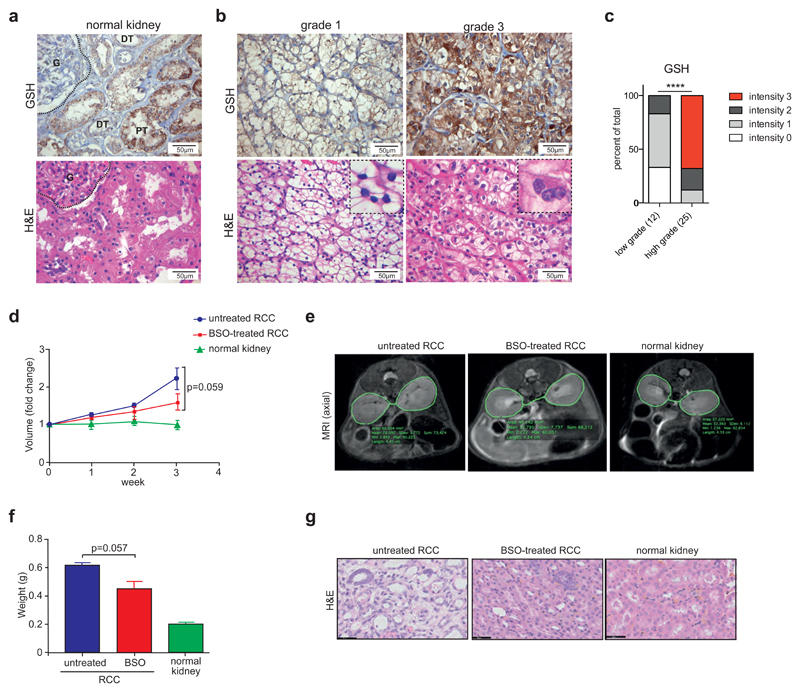
As our results indicate that GSH biosynthesis and GPX function are important in ccRCC cells, we investigated GSH levels in human ccRCC tissue by histological staining. In normal kidney tissue, protein-bound GSH was mainly localised to the cytoplasm of epithelial cells within the proximal renal tubuli, while glomeruli showed low levels of staining (Fig. 6A). Furthermore, a clear distinction in GSH levels between high and low grade tumours was observed. While low-grade tumours (grade 1) showed lower levels of GSH staining compared to normal renal tissue, high-grade tumours (grade 3) displayed strongly elevated staining (Fig. 6B). Analysis of multiple tumour specimens confirmed that a larger proportion of high-grade tumours (grade 3 and 4) exhibit strong GSH staining compared to low-grade tumours (grade 1 and 2) (Fig. 6C), suggesting that the GSH pathway is highly important in advanced cancers.
Figure 6. Inhibition of GSH synthesis blocks renal tumour growth.
A) Detection of GSH in normal kidney tissue. Corresponding sections of normal renal cortex were stained for in situ protein-bound GSH. H&E staining is shown for comparison. G=glomerulus, PT=proximal tubulus, DT=distal tubulus.
B) Detection of in situ protein-bound GSH in tissue from ccRCC patients. Representative sections of grade 1 and grade 3 tumours are shown with corresponding H&E staining for comparison. Insets indicate nuclear morphology of grade 1 and grade 3 tumours.
C) Quantification of GSH staining in 12 low- and 25 high-grade ccRCC tumours. Relative staining intensities were scored in a blinded fashion. Statistical comparison was performed using a two-tailed Chi-square test with 95% confidence intervals.
D) Transgenic mice with the γ-glutamyl transferase (GGT) promoter driving the tetracycline transactivator protein (tTA) and MYC under the control of the tetracycline-responsive element (MYC-GGT-tTA) were treated with 100 μg/ml doxycycline in the drinking water. To induce the formation of renal cell carcinomas (RCC), mice were shifted to normal drinking water to induce expression of MYC. After 2 weeks of MYC activation, one cohort of mice was treated with 20 mM BSO in the drinking water. Kidney volumes were monitored by magnetic resonance imaging (MRI) over a period of 3 weeks (BSO-treated) and compared to untreated MYC-expressing mice (untreated RCC) and kidneys from normal controls (normal kidney). Values show mean kidney volume ±SEM (n=3).
E) Representative MRI images indicating differences in kidney volume in BSO treated animals.
F) Kidney weight at endpoint of D in the 3 cohorts.
G) Representative histological sections of kidneys at endpoint of D illustrating the restoration of normal tissue morphology by BSO treatment.
****p≤0.001 unpaired two-tailed Student’s t-test.
Inactivation of VHL is a crucial event in the development of sporadic ccRCC. However, between 20% and 50% of human ccRCC tumours retain a functional VHL allele. We therefore expanded our panel of renal cancer cell lines to include three additional VHL mutant cell lines as well as four lines that retain functional VHL. In addition, a non-malignant renal epithelial cell line (HK-2) was used. As expected, VHL mutant cell lines showed high normoxic levels of either HIF-2α alone (786-O and 769-P) or both HIF-1α and HIF-2α, while VHL wild type cells display low to undetectable levels of these proteins (Fig. S4A). VHL mutant cells also showed higher levels of AKT phosphorylation compared to wild type cells. In contrast, VHL wild type cells expressed higher levels of the MYC oncoprotein compared to VHL mutant lines while non-malignant HK-2 cells showed high levels of HIF-1α, MYC and phospho-AKT, most likely due to the culture medium containing added growth factors (Fig. S4A). Nevertheless, all cell lines were highly sensitive towards depletion of glutamine or cystine (Fig. S4B). Moreover, no difference in the sensitivity between VHL wild type and mutant cell lines towards inhibitors of glutaminolysis (BPTES) or GSH synthesis (BSO) was detected (Fig. S4C). In addition, silencing of components of the GSH synthesis pathway as well as MYC, DHCR24, GPX3 or GPX4 also reduced the viability of most ccRCC cells (Fig. S4D). In all cases, ccRCC cells showed a higher sensitivity towards gene depletion compared to non-malignant HK-2 cells, indicating that the ccRCC cells have a higher dependency on GSH synthesis. These results also suggest that this dependency represents a general metabolic liability in this tumour type and can be induced not only through inactivation of VHL but also as a consequence of other oncogenic pathways, such as MYC activation.
Finally, we examined whether inhibition of GSH synthesis can interfere with renal tumourigenesis. We used a previously described model of renal cancer in which the human MYC protein is expressed in the mouse kidney following induction by the removal of doxycycline 44. Upon induction of tumour growth, GSH synthesis was inhibited by addition of BSO to the drinking water and tumour volume was assessed by magnetic resonance imaging (MRI) for three weeks. While BSO treatment did not affect the volume of normal kidneys in control mice, the volume of the cancer-bearing kidneys was reduced by at least 30% (Fig. 6D and E). The reduction in tumour growth by BSO treatment was even more obvious when the weight of the tumour was assessed after 2 weeks of treatment (Fig. 6F). Moreover, histological sections of BSO treated tumours also showed a clear reversal of dysplasia and restoration of a more normal renal appearance (Fig. 6G).
Taken together, our results demonstrate that ccRCC cells are highly dependent on GSH synthesis in order to counteract lipid peroxidation and ferroptotic cell death. Moreover, we found that inhibition of GSH synthesis interferes with renal tumour growth and restores normal renal tissue morphology.
Discussion
Altered metabolic activity is a central feature of ccRCC, the most common form of kidney cancer. Here we show that ccRCC cells are highly susceptible to disruption of the GSH/GPX pathway, which prevents the accumulation of ROS, including lipid peroxides. We found that renal cancer cells are exquisitely sensitive to depletion of glutamine or cystine, two metabolites required for GSH biosynthesis. Glutamine supports multiple cellular metabolic processes, for example as a substrate for TCA cycle anaplerosis, a nitrogen source for nucleotide biosynthesis or as a substrate for reductive carboxylation to form citrate for lipogenesis 36, 37, 50. The dipeptide cystine is a major source of cellular cysteine, which is used for protein synthesis and GSH production.
We also used functional screening to identify metabolic genes that are essential for the survival of ccRCC cells. Unsurprisingly, several of these genes mapped to the glycolytic pathway as the metabolic changes driven by HIF1α activation are reminiscent of the Warburg effect 43 and deregulated expression of glycolytic enzymes are frequently found in kidney cancer 39. Moreover, molecular characterisation of genetic and transcriptional alterations in ccRCC confirmed the strong association between deregulated glycolysis and patient survival in this disease 8. We also found an enzyme within the cholesterol biosynthesis pathway, namely DHCR24, to be essential for ccRCC cells. Cholesterol synthesis requires molecular oxygen and hypoxia-dependent regulation of this pathway has been described in yeast 27. Cholesterol and cholesterylesters are also required to form lipid droplets, lipid storage organelles that are a prominent feature in ccRCC 18, 40. Interestingly, isopentenyl pyrophosphate, an intermediate of the mevalonate pathway, is required for the maturation of the selenocysteine tRNA, which is essential for the production of selenocysteine-containing proteins, including GPX3 and GPX4 49, suggesting that high cholesterol synthesis may be a requirement in our system to maintain GPX3 and GPX4 protein levels.
Inhibition of GSH biosynthesis in ccRCC cells, either by blocking cystine uptake or through inhibition of γ-glutamylcysteine synthetase by the small molecule inhibitors Erastin and BSO, respectively, triggered ferroptosis, an iron-dependent form of cell death 12. This was confirmed by our finding that the effect of Erastin or BSO on cell viability was blocked by treatment with fer-1, a free radical scavenger that can remove lipid peroxides from lipid membranes 12. Interestingly, we found that restoration of VHL function protected renal cancer cells from induction of cell death caused by depletion of GPX3 or GPX4 and also from the detrimental effect of Erastin and BSO. A previous study linked cystine deprivation in VHL-deficient renal cancers to induction of programmed necrosis via the tumour necrosis factor alpha (TNFα) 48. This study showed that depletion of components of the TNFα signalling pathway rendered VHL-deficient cancer cells insensitive to low doses of Erastin. We found that the lipid-antioxidant ferrostatin protected VHL-deficient renal cancer cells over a range of different doses of Erastin or BSO treatment, confirming that cell death induced by inhibition of GSH biosynthesis involves a ferroptotic mechanism.
Ferroptosis is linked to the accumulation of peroxidised lipids produced from PUFAs through enzymatic and non-enzymatic processes 52, 55. It has been shown that p53 induces ferroptosis by downregulating the expression of SLC7A11 29. However, we observed that VHL-restoration did not alter SLC7A11 expression levels, making it unlikely that the differential sensitivity of VHL-deficient and restored cells towards inhibition of GSH biosynthesis is caused by different levels of this transporter. Moreover, GSH levels were similar between both cell types but strongly reduced in response to Erastin or BSO treatment, indicating that the differential sensitivity is not caused by altered activity of the GSH synthesis pathway. Instead, it is likely that activation of oxidative metabolism in VHL-restored cells, including increased metabolism of fatty acid by β-oxidation, results in a larger turnover of damaged lipids, thereby reducing the requirement for the GSH pathway for the detoxification of lipid peroxides. Indeed, inhibition of β-oxidation by treatment with the CPT1 inhibitor Etomoxir or Oligomycine, an inhibitor of mitochondrial ATP-synthase (complex V) completely restored ferroptosis induction in VHL-restored cells.
A recent lipidomic analysis revealed a large increase in triacylglycerides and cholesterol-esters in ccRCC tumour tissue 42, suggesting that renal cancer cells may be particularly sensitive to ferroptosis due to their high lipid content. Although we did not observe major differences in fatty acid composition between VHL-mutant and VHL-restored cells, we found that pVHL expression reduced the amount of LDs, the major storage organelle for neutral lipids, including triacylglycerides and cholesterol. Moreover, expression of ALOX5, the enzyme that controls the production of lipid peroxides from arachidonic acid, was repressed by pVHL. Consistent with a previous study linking increased expression of ALOX5 in ccRCC with enhanced expression of VEGF and high-grade disease 15, we found that ALOX5 mRNA is strongly overexpressed in ccRCC tumours compared to normal renal tissue. Induction of ALOX5 in renal cancer would increase the production of pro-inflammatory leukotrienes 51. However, enhanced lipid peroxidation could also increase the dependency of cancer cells on the GSH/GPX system to prevent excess oxidative damage.
Somewhat surprisingly, we did not observe major differences in the sensitivity towards nutrient restriction or inhibition of GSH synthesis between VHL wild type and mutant ccRCC cell lines. One explanation for this finding could be the elevated levels of MYC expression that we observed in cell lines expressing functional pVHL. MYC is frequently overexpressed in cancer and promotes cell growth and proliferation by acting as a global modulator of gene expression. MYC and HIF have partially overlapping target genes and both transcription factors enhance the expression of glycolytic enzymes 21. MYC also induces widespread metabolic reprogramming in cancer cells, including activation of glucose and glutamine metabolism 11 and increased lipid synthesis 9. Experiments in transgenic mice have shown that MYC induces a profound lipogenic programme during renal cancer development 44. It is therefore likely that enhanced dependence on GSH biosynthesis is a general feature of renal cancer cells and linked to their highly glycolytic metabolism coupled with reduced fatty acid degradation through β-oxidation. Accordingly, we found that ccRCC cells showed an overall higher dependency on GSH biosynthesis enzymes compared to non-malignant cells, confirming the importance of this pathway in renal cancer.
We also observed that levels of GSH bound to proteins were enhanced in high-grade renal tumour tissue. This is in agreement with a previous study showing strong elevation of GSH in ccRCC tumours compared to normal tissue 32. Coupling of GSH is involved in the removal of damaged cellular components, including toxic protein-lipid adducts formed as a result of lipid peroxidation 10, by the biotransformation pathway.
Finally, our study demonstrates that inhibition of GSH biosynthesis by the γ-cysteine synthetase inhibitor BSO attenuates tumour formation in a MYC-dependent mouse model of renal carcinogenesis. In this system, enhanced proliferation of renal epithelial cells can already be seen after one week of MYC induction 44. Interestingly, we found that BSO caused a reduction in tumour growth when delivered before tumour onset. This is consistent with the previously reported data from a mouse model of breast cancer (MMTV-PyMT), where BSO alone could prevent carcinogenesis only when delivered before tumour onset, while combined inhibition of GSH and thioredoxin was required to reduce tumour burden at later stages 26.
Overall, the results presented here demonstrate that renal cancer cells are highly dependent on the GSH pathway for the detoxification of reactive oxygen radicals, including lipid peroxides, and suggest that targeting components of this pathway could be an effective strategy for the treatment of this disease.
Material and Methods
Cell culture and reagents
RCC4, UMRC2, A498, 786-O, 769-P, CAKI1, CAKI2, TK10, A704, UMRC3, ACHN and UO31 cells as well as the non-tumorigenic proximal tubular HK-2 cell line were obtained from Cell Services (CRUK-LRI). VHL-isogenic ccRCC cell line pairs RCC4-EV and RCC4-VHL were obtained from William Kaelin Jr. (Dana Farber Cancer Institute, Boston, MA, USA) and used at low passage numbers. ccRCC cells were cultured in DMEM (Life Technologies) with 10% foetal bovine serum (FBS, Gibco), 4mM L-glutamine and 100U/ml Penicillin / 100μg/ml Streptomycin. HK-2 cells were cultured on collagen-coated cell culture dishes (BD Bioscience) in Keratinocyte-Serum-free medium (KSFM, Gibco) supplemented with 5ng/ml human recombinant EGF and 50ng/ml Bovine Pituitary Extract (BPE). All cells were cultured at 37°C in a humidified incubator at 5% CO2 and frequently tested to confirm absence of mycoplasma contamination. BPTES, BSO, H2O2, NAC, GSH, L-Cysteine hydrochloride, TEMPO, Erastin, BCNU and ferrostatin were obtained from Sigma-Aldrich.
Cell viability and cell number assays
Cells were seeded at low density and treated as indicated. After incubation, cells were fixed with 70% ethanol, stained with 0.01% crystal violet, washed and dried. For quantification, dye was extracted with 10% acetic acid and OD was measured at 550nm. For the determination of cell number, cells were stained with DAPI to visualize nuclei and analysed using an Acumen X3 laser scanning imaging cytometer (TTP Labtech) or an Operetta high content microscope (Perkin Elmer).
Nutrient depletion experiments
2000 cells/well were plated in 96-well plates in full growth medium. The next day, cells were washed with PBS before adding full medium or medium lacking glutamine or cystine, containing either solvent or GSH, NAC, L-cysteine or TEMPO, as indicated. Cell numbers were analysed 72h post depletion/supplementation.
Analysis of cellular respiration and extracellular acidification
10-35 x 103 cells/well were plated in sextuplets in regular culture medium in a 96-well XF culture plate. 16 hours later, cells were washed twice in PBS before changing to assay medium (Seahorse Biosciences) supplemented with 10mM sodium pyruvate and 25mM glucose with the pH adjusted to 7.4. Cells were incubated at 37°C in a CO2-free atmosphere for 1 hour and oxygen consumption rate (OCR) was determined using an XF96 Extracellular Flux Analyser (Software Version 1.4, Seahorse Biosciences, North Billerica, USA). During the experiment, 1.3μM oligomycin was injected, followed by 0.4 μM FCCP (Sigma) to measure the respiratory profile. OCRs were normalised to total protein content determined by sulforhodamine B staining.
Protein analysis
Cells were lysed in NP-40 lysis buffer (1% NP-40, 20mM Tris pH 7.5, 137mM NaCl, 1mM EGTA, 1mM DTT, 10% Glycerol, Protease-Inhibitor-Cocktail (Roche) and Phosphatase-Inhibitor-Cocktail (Roche)). Proteins were separated on SDS-PAGE and blotted onto PVDF membrane (Immobilon). Membranes were blocked with 3% BSA, incubated with antibody solutions and signals were detected using ECL-reagent (Amersham). Antibodies for MYC (9E10, Sigma), phospho-AKT (S473, ), pan-AKT (both Cell Signaling Technology, #9271), HIF-1α (BD Biosciences, #610959), HIF-2α (Novus, #NB100-122), HA-tag (Covance, #MMS-101P) and HRP-conjugated anti-β-Actin (Sigma-Aldrich, #A3854) were used.
Analysis of patient data and survival analysis
Comparative gene expression analyses between ccRCC and normal kidney tissue were performed with Oncomine™ v4.5. Datasets used were from 56, 30, 24 and 3.
siRNA transfection and determination of cell number
The siRNA screen targeting 230 metabolic genes in renal cancer cells has been described before 17. Cell numbers were used to calculate z-scores based on the median absolute deviation (MAD) of the population 35.
For individual gene silencing, 2000 cells/well were reverse transfected with 37.5nM siRNA (Dharmacon siGENOME, SMARTpools) on 96-well plates. 96h post transfection, cells were fixed with 80% ice-cold ethanol. For the determination of cell number, cells were stained with DAPI to visualise nuclei and analysed using an Acumen X3 laser scanning imaging cytometer (TTP Labtech) or an Operetta high content microscope (Perkin Elmer). RNAi effects are represented as fold-change relative to scrambled-control transfection (CTRL). siUBB was used as positive control.
Detection of GSH using mass spectrometry
For the determination of GSH in cell extracts, adherent cells were washed with PBS and snap frozen in liquid N2. After addition of 250μl cold methanol/water (4/1, v/v), cells were scraped off and transferred into a tube. Water-soluble metabolites were extracted in a 530μl of a final mixture of methanol/water/chloroform (2/1.8/2, v/v/v) 5. After centrifugation, the Bligh/Dyer-upper phase was transferred into a new tube, evaporated for 30min at 35°C under a stream of nitrogen gas and taken to dryness in SpeedVac at room temperature. The dry residue was dissolved in CH3CN/5 mM NH4OAc (25/75, v/v) and measured by direct injection with a flow rate of 10 μl/min in negative ion mode using a Thermo Scientific™ Q-Exactive™ quadrupole-Orbitrap mass spectrometer. Full mass scan (m/z 67-1000) at a resolution of 70 000 with automatic gain control target of 1x106 ions and a maximum ion injection time of 100 ms was used. Source ionization parameters were optimized with the spray voltage of 3.5 kV; transfer temperature of 320 °C; S-Lens level of 50; heater temperature of 325°C and Sheath gas 7.
RNA extraction, reverse transcription and RT-qPCR
Total RNA was isolated using an RNeasy kit (Qiagen). 1-5μg of total RNA was used for first strand cDNA synthesis with SuperScript II Reverse Transcriptase and oligo-dT primers (Invitrogen). Real time PCR was performed with SYBR® Green PCR Master Mix (Applied Biosystems) using Quantitect primers (Qiagen) in an ABI PRISM 7900 Sequence Detection System (Applied Biosystems). All reactions were performed at least in duplicate. The relative amount of all mRNAs was calculated using the comparative CT method after normalization to ACTB or B2M.
Lipid peroxidation assay
For detection of lipid peroxidation, the Image-iT Lipid peroxidation kit (Life Technologies, Cat.no. C10445) was used. Cells were seeded in black 96-well plates with clear bottom and treated with Erastin for 24 hours. During the last 30min of incubation, 10μM of sensor was added to cultures. Cells were then washed 3 times with PBS and imaged immediately with a high throughput fluorescence microscope (Perkin Elmer, Operetta) using filters for FITC (Alexa488) and Texas Red (Alexa594). Signal intensities at 510nm and 590nm were used to quantify lipid peroxidation.
Lipid droplet staining
4000 cells/well were plated in 96-well plates and cultured for 48 hours before fixation with 4% paraformaldehyde for 15 min, washed with PBS and stained for 30 minutes with 0.1μg/ml Nile Red (Life Technologies) and 1μg/ml DAPI (Sigma) in PBS. Cells were washed three times in PBS and analysed using a Cellomics ArrayScan VT high content microscope (Thermo Fisher) with associated HCS studio:Cellomics Navigator Version 6.4.4 software.
Histology
Paraffin-embedded 4μm thick tissue sections were deparaffinized and rehydrated. Antigen retrieval was performed with citrate buffer (pH 6.0) in a microwave oven for 30min. Endogenous peroxidase activity was blocked by incubating sections in 3% (v/v) hydrogen peroxide for 10min. Sections were blocked with 3% (w/v) BSA for 30min and incubated with an anti-GSH antibody (Abcam ab-19534), diluted 1:200 overnight at 4°C in a humidified chamber. Sections were washed in PBS and incubated with biotinylated secondary antibody followed by streptavidin-horseradish-peroxidase assay (Vector Labs, Dako). Reactions were developed using 3,3'-diaminobenzidine (Cell Signaling). Slides were counterstained with Gilmore 3 hematoxylin, dehydrated, cleared, and mounted with coverslips.
In vivo experiments
Transgenic mice GGT-TTA in S129 background and Tet-O-MYC in FVB/N background were bred to create a double-transgenic mouse line in which human MYC protein is expressed in the mouse kidney. MYC inactivation was achieved by supplementing the drinking water with 100 μg/mL doxycycline since birth. MYC activation at 4 weeks of age for both male and female mice was achieved by removal of doxycline from the drinking water as previously described 44. All animal experiments were performed under the guidelines established by the Administrative Panel on Laboratory Animal Care at Stanford University. Sizes of animal cohorts were based on previous experiments 44. Animals were randomly assigned to the groups and no animals were excluded from the analysis. MYC inactivation was achieved by adding 100 μg/mL doxycycline to the drinking water and replacing it weekly. MYC activation was achieved by administering regular drinking water at 4 weeks of age. Glutathione pathway inhibition was examined for antitumor activity in vivo in the transgenic mouse model by adding 20mM of BSO (L-buthionine (S,R)-sulfoximine); Sigma (Cat No B2515)) to the drinking water for treatment after 2 weeks of MYC activation.
Mice were visualised by magnetic resonance imaging (MRI) at 0, 7, and 14 days following treatment. Mice were sequentially imaged using a 7 Tesla MRI system at Stanford Small Animal Imaging Center. Kidney measurements were performed using T2-weighted axial image stacks using Osirix software to obtain the tumour volumes. Tissues were fixed in 10% (v/v) buffered formalin (Starplex Scientific) for 24 h, followed by transfer to 70% (v/v) ethanol. Samples were processed, embedded, and 5-μm paraffin sections were cut by Stanford Histology Services. H&E staining was performed using standard procedures. Kidneys are weighed when mice are sacrificed at the end of the experiment.
Statistical analysis
Graphs were generated using GraphPad Prism 6.0 (GraphPad software). All experiments have been performed independently at least twice with at least three biological replicates. Unless otherwise indicated, statistical significance of magnitude of changes between different conditions was calculated using the parametric two-tailed unpaired Student t-test. Statistical significance was defined as a p-value of less than 0.05. P-values are depicted as follows: *p≤0.05, **p≤0.01, ***p≤0.001 and ****p≤0.0001. n.s. = not significant.
Supplementary Material
Acknowledgements
We thank the LRI research services for technical support. We also thank Prof. William Kaelin (Dana Faber Cancer Institute, Boston) for providing isogenic renal cancer cell lines. This study was funded by Cancer Research UK, the German Research Foundation (FOR2314) and the German Cancer Aid (111917).
Footnotes
Author Contribution
H.M. and A.S. conceived the project and wrote the manuscript. H.M. performed the starvation and rescue experiments, the RNAi screen and most of the silencing experiments and cell analyses. B.D. performed ferroptosis analyses including rescue experiments and RNA analyses. W.S. performed metabolite analyses. B.P. contributed to the study design and manuscript preparation. M.J., B.S. and M.H. were involved in optimisation, execution and analysis of the RNAi screen. M.R. performed histological analysis and interpretation. A.G. and D.F. provided the mouse model and performed the in vivo experiments. All authors commented on the manuscript.
Author Information
The authors declare no other competing financial interests.
References
- 1.Avissar N, Ornt DB, Yagil Y, Horowitz S, Watkins RH, Kerl EA, et al. Human kidney proximal tubules are the main source of plasma glutathione peroxidase. Am J Physiol. 1994;266:C367–375. doi: 10.1152/ajpcell.1994.266.2.C367. [DOI] [PubMed] [Google Scholar]
- 2.Bensaad K, Favaro E, Lewis CA, Peck B, Lord S, Collins JM, et al. Fatty acid uptake and lipid storage induced by HIF-1alpha contribute to cell growth and survival after hypoxia-reoxygenation. Cell reports. 2014;9:349–365. doi: 10.1016/j.celrep.2014.08.056. [DOI] [PubMed] [Google Scholar]
- 3.Beroukhim R, Brunet JP, Di Napoli A, Mertz KD, Seeley A, Pires MM, et al. Patterns of gene expression and copy-number alterations in von-hippel lindau disease-associated and sporadic clear cell carcinoma of the kidney. Cancer research. 2009;69:4674–4681. doi: 10.1158/0008-5472.CAN-09-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bierl C, Voetsch B, Jin RC, Handy DE, Loscalzo J. Determinants of human plasma glutathione peroxidase (GPx-3) expression. The Journal of biological chemistry. 2004;279:26839–26845. doi: 10.1074/jbc.M401907200. [DOI] [PubMed] [Google Scholar]
- 5.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 6.Brigelius-Flohe R, Kipp A. Glutathione peroxidases in different stages of carcinogenesis. Biochimica et biophysica acta. 2009;1790:1555–1568. doi: 10.1016/j.bbagen.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 7.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 8.Cancer Genome Atlas Research N. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carroll PA, Diolaiti D, McFerrin L, Gu H, Djukovic D, Du J, et al. Deregulated Myc Requires MondoA/Mlx for Metabolic Reprogramming and Tumorigenesis. Cancer Cell. 2015;27:271–285. doi: 10.1016/j.ccell.2014.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dalleau S, Baradat M, Gueraud F, Huc L. Cell death and diseases related to oxidative stress: 4-hydroxynonenal (HNE) in the balance. Cell Death Differ. 2013;20:1615–1630. doi: 10.1038/cdd.2013.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dixon SJ, Stockwell BR. The role of iron and reactive oxygen species in cell death. Nat Chem Biol. 2014;10:9–17. doi: 10.1038/nchembio.1416. [DOI] [PubMed] [Google Scholar]
- 14.Drabkin HA, Gemmill RM. Cholesterol and the development of clear-cell renal carcinoma. Current opinion in pharmacology. 2012;12:742–750. doi: 10.1016/j.coph.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 15.Faronato M, Muzzonigro G, Milanese G, Menna C, Bonfigli AR, Catalano A, et al. Increased expression of 5-lipoxygenase is common in clear cell renal cell carcinoma. Histol Histopathol. 2007;22:1109–1118. doi: 10.14670/HH-22.1109. [DOI] [PubMed] [Google Scholar]
- 16.Gatto F, Nookaew I, Nielsen J. Chromosome 3p loss of heterozygosity is associated with a unique metabolic network in clear cell renal carcinoma. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E866–875. doi: 10.1073/pnas.1319196111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gatto F, Miess H, Schulze A, Nielsen J. Flux balance analysis predicts essential genes in clear cell renal cell carcinoma metabolism. Scientific reports. 2015;5 doi: 10.1038/srep10738. 10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gebhard RL, Clayman RV, Prigge WF, Figenshau R, Staley NA, Reesey C, et al. Abnormal cholesterol metabolism in renal clear cell carcinoma. Journal of lipid research. 1987;28:1177–1184. [PubMed] [Google Scholar]
- 19.Gerlinger M, Santos CR, Spencer-Dene B, Martinez P, Endesfelder D, Burrell RA, et al. Genome-wide RNA interference analysis of renal carcinoma survival regulators identifies MCT4 as a Warburg effect metabolic target. J Pathol. 2012;227:146–156. doi: 10.1002/path.4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gordan JD, Bertout JA, Hu CJ, Diehl JA, Simon MC. HIF-2alpha promotes hypoxic cell proliferation by enhancing c-myc transcriptional activity. Cancer cell. 2007;11:335–347. doi: 10.1016/j.ccr.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gordan JD, Thompson CB, Simon MC. HIF and c-Myc: sibling rivals for control of cancer cell metabolism and proliferation. Cancer cell. 2007;12:108–113. doi: 10.1016/j.ccr.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorrini C, Harris IS, Mak TW. Modulation of oxidative stress as an anticancer strategy. Nature reviews Drug discovery. 2013;12:931–947. doi: 10.1038/nrd4002. [DOI] [PubMed] [Google Scholar]
- 23.Gossage L, Eisen T, Maher ER. VHL, the story of a tumour suppressor gene. Nat Rev Cancer. 2015;15:55–64. doi: 10.1038/nrc3844. [DOI] [PubMed] [Google Scholar]
- 24.Gumz ML, Zou H, Kreinest PA, Childs AC, Belmonte LS, LeGrand SN, et al. Secreted frizzled-related protein 1 loss contributes to tumor phenotype of clear cell renal cell carcinoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2007;13:4740–4749. doi: 10.1158/1078-0432.CCR-07-0143. [DOI] [PubMed] [Google Scholar]
- 25.Hakimi AA, Reznik E, Lee CH, Creighton CJ, Brannon AR, Luna A, et al. An Integrated Metabolic Atlas of Clear Cell Renal Cell Carcinoma. Cancer cell. 2016;29:104–116. doi: 10.1016/j.ccell.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harris IS, Treloar AE, Inoue S, Sasaki M, Gorrini C, Lee KC, et al. Glutathione and thioredoxin antioxidant pathways synergize to drive cancer initiation and progression. Cancer cell. 2015;27:211–222. doi: 10.1016/j.ccell.2014.11.019. [DOI] [PubMed] [Google Scholar]
- 27.Hughes AL, Todd BL, Espenshade PJ. SREBP pathway responds to sterols and functions as an oxygen sensor in fission yeast. Cell. 2005;120:831–842. doi: 10.1016/j.cell.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 28.Iliopoulos O, Kibel A, Gray S, Kaelin WG., Jr Tumour suppression by the human von Hippel-Lindau gene product. Nature medicine. 1995;1:822–826. doi: 10.1038/nm0895-822. [DOI] [PubMed] [Google Scholar]
- 29.Jiang L, Kon N, Li T, Wang SJ, Su T, Hibshoosh H, et al. Ferroptosis as a p53-mediated activity during tumour suppression. Nature. 2015;520:57–62. doi: 10.1038/nature14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones J, Otu H, Spentzos D, Kolia S, Inan M, Beecken WD, et al. Gene signatures of progression and metastasis in renal cell cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2005;11:5730–5739. doi: 10.1158/1078-0432.CCR-04-2225. [DOI] [PubMed] [Google Scholar]
- 31.Kwon MY, Park E, Lee SJ, Chung SW. Heme oxygenase-1 accelerates erastin-induced ferroptotic cell death. Oncotarget. 2015;6:24393–24403. doi: 10.18632/oncotarget.5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li B, Qiu B, Lee DS, Walton ZE, Ochocki JD, Mathew LK, et al. Fructose-1,6-bisphosphatase opposes renal carcinoma progression. Nature. 2014;513:251–255. doi: 10.1038/nature13557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li L, Zhang L, Zhang X, Yan Q, Minamishima YA, Olumi AF, et al. Hypoxia-inducible factor linked to differential kidney cancer risk seen with type 2A and type 2B VHL mutations. Molecular and cellular biology. 2007;27:5381–5392. doi: 10.1128/MCB.00282-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Molecular cell. 2010;40:294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malo N, Hanley JA, Cerquozzi S, Pelletier J, Nadon R. Statistical practice in high-throughput screening data analysis. Nature biotechnology. 2006;24:167–175. doi: 10.1038/nbt1186. [DOI] [PubMed] [Google Scholar]
- 36.Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2011;481:380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mullen AR, Wheaton WW, Jin ES, Chen PH, Sullivan LB, Cheng T, et al. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2011;481:385–388. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murphy MP. How mitochondria produce reactive oxygen species. Biochem J. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perroud B, Lee J, Valkova N, Dhirapong A, Lin PY, Fiehn O, et al. Pathway analysis of kidney cancer using proteomics and metabolic profiling. Mol Cancer. 2006;5:64. doi: 10.1186/1476-4598-5-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qiu B, Ackerman D, Sanchez DJ, Li B, Ochocki JD, Grazioli A, et al. HIF2alpha-Dependent Lipid Storage Promotes Endoplasmic Reticulum Homeostasis in Clear-Cell Renal Cell Carcinoma. Cancer discovery. 2015;5:652–667. doi: 10.1158/2159-8290.CD-14-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Radmark O, Werz O, Steinhilber D, Samuelsson B. 5-Lipoxygenase, a key enzyme for leukotriene biosynthesis in health and disease. Biochimica et biophysica acta. 2015;1851:331–339. doi: 10.1016/j.bbalip.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 42.Saito K, Arai E, Maekawa K, Ishikawa M, Fujimoto H, Taguchi R, et al. Lipidomic Signatures and Associated Transcriptomic Profiles of Clear Cell Renal Cell Carcinoma. Scientific reports. 2016;6 doi: 10.1038/srep28932. 28932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Semenza GL. HIF-1 mediates the Warburg effect in clear cell renal carcinoma. Journal of bioenergetics and biomembranes. 2007;39:231–234. doi: 10.1007/s10863-007-9081-2. [DOI] [PubMed] [Google Scholar]
- 44.Shroff EH, Eberlin LS, Dang VM, Gouw AM, Gabay M, Adam SJ, et al. MYC oncogene overexpression drives renal cell carcinoma in a mouse model through glutamine metabolism. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:6539–6544. doi: 10.1073/pnas.1507228112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singer EA, Gupta GN, Marchalik D, Srinivasan R. Evolving therapeutic targets in renal cell carcinoma. Current opinion in oncology. 2013;25:273–280. doi: 10.1097/CCO.0b013e32835fc857. [DOI] [PubMed] [Google Scholar]
- 46.Sullivan LB, Chandel NS. Mitochondrial reactive oxygen species and cancer. Cancer Metab. 2014;2:17. doi: 10.1186/2049-3002-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang SW, Chang WH, Su YC, Chen YC, Lai YH, Wu PT, et al. MYC pathway is activated in clear cell renal cell carcinoma and essential for proliferation of clear cell renal cell carcinoma cells. Cancer letters. 2009;273:35–43. doi: 10.1016/j.canlet.2008.07.038. [DOI] [PubMed] [Google Scholar]
- 48.Tang X, Wu J, Ding CK, Lu M, Keenan MM, Lin CC, et al. Cystine Deprivation Triggers Programmed Necrosis in VHL-Deficient Renal Cell Carcinomas. Cancer research. 2016;76:1892–1903. doi: 10.1158/0008-5472.CAN-15-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Warner GJ, Berry MJ, Moustafa ME, Carlson BA, Hatfield DL, Faust JR. Inhibition of selenoprotein synthesis by selenocysteine tRNA[Ser]Sec lacking isopentenyladenosine. The Journal of biological chemistry. 2000;275:28110–28119. doi: 10.1074/jbc.M001280200. [DOI] [PubMed] [Google Scholar]
- 50.Wise DR, Ward PS, Shay JE, Cross JR, Gruber JJ, Sachdeva UM, et al. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of alpha-ketoglutarate to citrate to support cell growth and viability. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:19611–19616. doi: 10.1073/pnas.1117773108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wymann MP, Schneiter R. Lipid signalling in disease. Nature reviews Molecular cell biology. 2008;9:162–176. doi: 10.1038/nrm2335. [DOI] [PubMed] [Google Scholar]
- 52.Xie Y, Hou W, Song X, Yu Y, Huang J, Sun X, et al. Ferroptosis: process and function. Cell Death Differ. 2016;23:369–379. doi: 10.1038/cdd.2015.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang WS, SriRamaratnam R, Welsch ME, Shimada K, Skouta R, Viswanathan VS, et al. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156:317–331. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang WS, Kim KJ, Gaschler MM, Patel M, Shchepinov MS, Stockwell BR. Peroxidation of polyunsaturated fatty acids by lipoxygenases drives ferroptosis. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:E4966–4975. doi: 10.1073/pnas.1603244113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang WS, Stockwell BR. Ferroptosis: Death by Lipid Peroxidation. Trends Cell Biol. 2016;26:165–176. doi: 10.1016/j.tcb.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yusenko MV, Kuiper RP, Boethe T, Ljungberg B, van Kessel AG, Kovacs G. High-resolution DNA copy number and gene expression analyses distinguish chromophobe renal cell carcinomas and renal oncocytomas. BMC Cancer. 2009;9:152. doi: 10.1186/1471-2407-9-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.