Abstract
Sensing using specific and selective receptors provides two very different but complementary strategies. This Sensor Issues article will discuss the merits and challenges of specific sensors, and selective sensors based on synthetic arrays. We will examine where each has been successfully applied to a sensing challenge, and then look at how a combined approach could take elements of both to provide new sensor platforms.
Keywords: chemical sensors, array-based sensing, antibodies, specificity, selectivity
Graphical Abstract
Sensing approaches can broadly be split in to two silos: highly specific sensing, and array based selective sensing. The former is a sensor that in its ideal form would be completely specific to a single analyte, recognizing no other. This ideal is approached by antibodies, aptamers, and enzymatic lock-and-key or bio-conjugation pairs such as streptavidin/biotin.1,2 Very often, however, this ideal is not achievable, due to high similarity between analytes or a lack of tools for specific sensing of the target. Selective sensing is often the best that can be achieved, and are quite useful, as demonstrated by the use of lectin arrays for selective glycan sensing. 3
Sensors can also be engineered from the start to be selective. These systems are typically employed in an array-based format, where each sensing element interacts differentially with the analytes of interest, creating a fingerprint for that sample. The output of the array can be considered and processed as multidimensional data (multiple outputs from a single input), a feature facilitated through data analysis techniques.4 This array-based “chemical nose/tongue” approach has emerged from the world of chemometrics, gaining traction in the chemical sensor community in recent years.
Many, if not most, of the sensors and tests in widespread use today rely on specific sensor elements for individual target analytes (e.g. biomarkers) and have had excellent success in the medical and bioscience domain. With the growth and success of cross-reactive, selective sensors, we believe that sensor design would benefit from combining the best of both sensing worlds when approaching a sensing challenge.
In this Sensor Issues article we seek to compare and contrast the approaches of specific and selective array-based sensing, and show how overlap in these methodologies can be exploited to build better sensors. We will examine how to choose the best sensor type for the detection challenge at hand, and discuss where array based sensing may have a crucial role to play in an area typically dominated by specific sensors, whilst acting in tandem with the existing techniques, to provide a complete understanding of the system being examined.
The Achievements of Specific Sensing
Highly specific sensors based primarily on antibodies or enzymatic recognition, and in more recent years aptamer technology, have dominated the world of biosensing. In principle each single sensor constructed with this technology has a single target, and will bind no other, even in a complex sensing medium, such as blood serum, cell lysate or an environmental sample.5 The success story of specific sensing is well known when it comes to commercialization; examples include antibodies used in lateral flow immunochromatographic assays (LFIA) for pregnancy testing,6 and glucose-specific enzymes contained in the blood glucose meters used by diabetics.7
Specific sensing with antibodies has had a major impact on advancing the biosciences. Antibodies are the heart of the ELISA (enzyme linked immunosorbant assay)8 for proteomics screening and diagnosis of disease by sensing the up- and down-regulation of specific biomarkers, and associating them with pathologies. These assays have led to state of the art diagnostic tests for ailments such as liver fibrosis9 and cardiac disease.10 Beyond biomedicine, explosives,11 and drugs of abuse12 have likewise been successfully targeted using antibody-embedded sensors.
Antibodies are quite versatile, but are prone to denaturation and cannot recognize every analyte. Aptamers – short chains of nucleic acids or peptides that are engineered to have specific binding to a target molecule – are one such strategy that is now widely being used in the specific sensing domain.13 Such sensors have been applied to detection of numerous targets including proteins of the HIV virus,14 and small molecules such as sugars.15 Another example of ‘next gen’ recognition elements are modified viruses or ‘phages’ that have ben used for sensing peptides and proteins.16
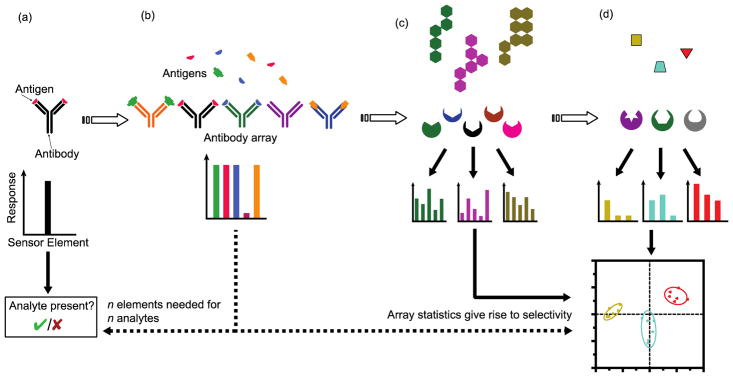
Single target specific sensing approaches are useful if one has a strong and simple hypothesis in mind related to one or two analytes, as in the above examples. However, the diagnosis of diseases can necessitate monitoring the levels of 4, up to 20+ different biomarkers,17 requiring a large array of antibodies in a sensor (Figure 1). These arrays work well to distinguish multiple components in the sample, generating a pattern that can be used to identify disease states. However, there is often incomplete knowledge of what biomarkers should even be targeted, while many diseases don’t have associated specific recognition elements currently available.
Figure 1.
The transition from specific to selective sensing, and the corresponding outputs that are generated by sensor designs: (a) an antibody gives specific information about a single analyte, due to its high specificity. (b) Multiple analytes require different antibodies, leading to the creation of an antibody array in which each array component is only specific to a single analyte with no cross-reactivity. (c) Selective sensor arrays likewise give rise to unique patterns (fingerprints) for each analyte due to the cross-reactivity of the array components. (d) Hypothesis free, cross-reactive arrays enable the differentiation of multiple analytes, even those of different types (organic, inorganic etc.), with no previous knowledge needed. In all three of the multi-sensor platforms patterns are generated that can be analyzed to match against a known pattern using statistical methods.
Selectivity by necessity
While specific and array based sensing present two distinct camps in the sensor community, there is an intermediate ground where selective sensors are employed simply because specific sensors are unavailable. One key example is the use of sugar-binding proteins, lectins, to sense carbohydrate containing biomolecules such as glycoproteins or glycolipids. Although having a high specificity for carbohydrates in general, individual lectins are not specific to individual glycosylated biomolecules. Thus, an array of lectins can be used in a single run to map the carbohydrates present.3 Another example is detection of a class of enzymes, mitogen-activated protein (MAP) kinases, in cell lysate or biological media, using an array of modified fluorescent substrates (SOX peptides).18 A single peptide substrate would give no information on the classes of kinases present, but by having an array of peptide substrates, a picture can be built up of the MAP kinases present upon analyzing the products produced. These examples of sensing arrays utilize multidimensional data analysis to leverage their cross-reactivity to give discriminatory information on the analytes present, providing output similar to both specific sensor arrays and engineered selective sensor platforms.
Selectivity by design
Selectivity and cross-reactivity are central features of olfaction, and can be used as a starting point for sensor design. Arrays can be designed using synthetic materials to target specific classes of analytes and introduce selectivity within the targets, exploiting advances in supramolecular chemistry to control analyte-sensor interactions, but without the need for the precise engineering of an antibody or aptamer (Figure 1). These sensor systems are enormously flexible; they are synthetically created from first principles, and therefore can consist of a wide variety of array types and methods to suit all sensing challenges. Through this synthetic approach, the sensor arrays can be designed to be tolerant to a wide variety of media, extremes of pH and of temperature. Sensor elements can also target a wide variety of biomolecules, small molecules or inorganic ions, all in the same test.
Array based sensing generates rich, multidimensional information from a single experimental run. Therefore, detecting multiple analytes is achievable simultaneously, in a manner similar to olfaction. This gives rise to a ‘chemical nose/tongue’ that generates a unique pattern that can be tied back to the composition of the sample. There is no need for a large number of sensor elements to detect a large number of analytes,19 if the cross-reactivity is engineered well a few sensor elements can discriminate many more analytes.
Careful selection of a suitable statistical approach allows maximal information on the sample to be gained, while avoiding biased results.20,21 In all cases, it is vitally important that separate training and test sets are generated to verify how the sensor performs in a real classification problem, rather than just being internally cross-validated. It is also beneficial to discover what interferrants or conditions may lead to sensor failure, to allow for future improvements.
An advantage of array-based selective sensor approach is that if a new analyte/class of sample needs to be detected, few if any changes need to be made to the array. The pattern recognition library simply needs to be updated by re-training the array on known samples to recognize new analytes. Examples of these synthetic arrays have been widely researched for detecting small molecules, explosives, drugs of abuse and also biological samples such as proteins, cells and bacteria.22,23 An important issue with these sensors is that they give ‘fingerprints’ that do not readily measure multiple individual components within the whole sample, unlike the specific analyte data provided using parallel specific sensors. Thus, many of the examples listed here can be addressed with additional information content by the application of a specific sensing regime.
When choosing a sensing approach, one must consider the particular advantages that selective array-based sensing can offer and where it might be more appropriate to use highly specific sensors. Specific sensors give direct information on biomarkers or other analytes that is often important in categorizing complex disease states or environmental samples. Selective arrays perform well when it comes to sensing the whole sample, not just its individual components. Therefore, such systems can be used in a hypothesis-less fashion. This is an enormously powerful approach in situations such as the earlier example of disease detection, where many biomarkers must be detected at once to confirm the disease, while some could possibly be unknown.
Hypothesis-free universal sensors?
Much of the work on array-based sensors has focused on detecting and differentiating single analytes in a complex sample, for example explosives in water samples, or single proteins in a serum sample.22,24 However, in these cases it may be more efficient to use a highly specific sensor, such as an antibody targeted to the analyte in question.25 A more powerful use of arrays is to target complex matrices, where all the analytes present may not be known. These applications include analyzing methods of drug action,26 atmospheric analysis to protect against toxic gasses,27 or sensing of disease for diagnosis and follow up.28 A selective array can operate in a hypothesis-less mode, where samples are discriminated based on their complete, selective interaction, rather than on the basis of any single component. A suitably cross-reactive array might be applicable to any number of these cases, for example the colorimetric arrays of Suslick et al.29 This advances the technology towards the idea of a ‘universal sensor’.
It seems foolish, however, to perpetuate the siloing of specific and selective arrays. Integrating specific and selective sensor elements has the potential to provide synergy, and hence much more effective sensor platforms. For example, by introducing class specificity – limiting the selectivity of the sensor array within a group of particularly important analytes – such as in the case of lectin arrays, we can improve stability of the array and minimize the impact of changes in the sample background on the sensing response.
A second area where specific and selective sensing are complimentary is in hypothesis-less testing for exploration of the underlying sensing mechanisms. A sample of a diseased patient will give a particular sensor response, different from a healthy patient, but understanding what components of the sample cause this difference unlocks information on the fundamentals of the disease itself and the mode of operation of the sensor array.30
Conclusions
Specific and selectivity-based strategies each have their place in the research community and in the world at large. Understanding the strengths and limitations of each provides a means of choosing the method that works best. Beyond choice of method, however, we suggest that elements of both approaches be incorporated into a new class of array-based sensors. Finally, we encourage array-based sensor researchers to consider applying specific sensors to their samples in tandem, and of course for researchers in the specific sensors camp to likewise consider selectivity-based enhancements.
Acknowledgments
WJP acknowledges funding from the EPSRC (EP/M506448/1) and the Royal Society for an International Exchange Award. VR acknowledges support form the NIH (GM077173) and the NSF (CHE-1307021).
Footnotes
Author Information
The authors declare no competing financial interest.
References
- 1.Peng HP, Lee KH, Jian JW, Yang AS. Origins of Specificity and Affinity in Antibody-Protein Interactions. Proc Natl Acad Sci US A. 2014;111:E2656–E2665. doi: 10.1073/pnas.1401131111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Livnah O, Bayer EA, Wilchek M, Sussman JL. Three-Dimensional Structures of Avidin and the Avidin-Biotin Complex. Proc Natl Acad Sci US A. 1993;90:5076–5080. doi: 10.1073/pnas.90.11.5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hirabayashi J, Yamada M, Kuno A, Tateno H. Lectin Microarrays: Concept, Principle and Applications. Chem Soc Rev. 2013;42:4443–4458. doi: 10.1039/c3cs35419a. [DOI] [PubMed] [Google Scholar]
- 4.Anzenbacher P, Jr, Palacios MA. Array-Based Sensors. In: Anslyn EV, Wang B, editors. Chemosensors: Principles, Strategies, and Applications. John Wiley & Sons; New Jersey: 2011. pp. 345–368. [Google Scholar]
- 5.Saper CB. A Guide to the Perplexed on the Specificity of Antibodies. J Histochem Cytochem. 2009;57:1–5. doi: 10.1369/jhc.2008.952770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wide L, Gemzell CA. An Immunological Pregnancy Test. Acta Endocrinol. 1960;35:261–267. doi: 10.1530/acta.0.xxxv0261. [DOI] [PubMed] [Google Scholar]
- 7.Heller A, Feldman B. Electrochemical Glucose Sensors and Their Applications in Diabetes Management. Chem Rev. 2008;108:2482–2505. doi: 10.1021/cr068069y. [DOI] [PubMed] [Google Scholar]
- 8.Engvall E, Perlmann P. Enzyme-Linked Immunosorbent Assay (ELISA) Quantitative Assay of Immunoglobulin G. Immunochemistry. 1971;8:871–874. doi: 10.1016/0019-2791(71)90454-x. [DOI] [PubMed] [Google Scholar]
- 9.Rosenberg WMC, Voelker M, Thiel R, Becka M, Burt A, Schuppan D, Hubscher S, Roskams T, Pinzani M, Arthur MJP. Serum Markers Detect the Presence of Liver Fibrosis: a Cohort Study. Gastroenterology. 2004;127:1704–1713. doi: 10.1053/j.gastro.2004.08.052. [DOI] [PubMed] [Google Scholar]
- 10.Kaneko N, Matsuda R, Hosoda S, Kajita T, Ohta Y. Measurement of Plasma Annexin v by ELISA in the Early Detection of Acute Myocardial Infarction. Clin Chim Acta. 1996;251:65–80. doi: 10.1016/0009-8981(96)06294-8. [DOI] [PubMed] [Google Scholar]
- 11.Smith RG, D’Souza N, Nicklin S. A Review of Biosensors and Biologically-Inspired Systems for Explosives Detection. Analyst. 2008;133:571–584. doi: 10.1039/b717933m. [DOI] [PubMed] [Google Scholar]
- 12.Hazarika P, Jickells SM, Wolff K, Russell DA. Multiplexed Detection of Metabolites of Narcotic Drugs From a Single Latent Fingermark. Anal Chem. 2010;82:9150–9154. doi: 10.1021/ac1023205. [DOI] [PubMed] [Google Scholar]
- 13.Blind M, Blank M. Aptamer Selection Technology and Recent Advances. Mol Ther Nucleic Acids. 2015;4:e223. doi: 10.1038/mtna.2014.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamamoto R, Kumar PKR. Molecular Beacon Aptamer Fluoresces in the Presence of Tat Protein of HIV-1. Genes to Cells. 2000;5:389–396. doi: 10.1046/j.1365-2443.2000.00331.x. [DOI] [PubMed] [Google Scholar]
- 15.Yang KA, Barbu M, Halim M, Pallavi P, Kim B, Kolpashchikov DM, Pecic S, Taylor S, Worgall TS, Stojanovic MN. Recognition and Sensing of Low-Epitope Targets via Ternary Complexes with Oligonucleotides and Synthetic Receptors. Nat Chem. 2014;6:1003–1008. doi: 10.1038/nchem.2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mao C, Liu A, Cao B. Virus-Based Chemical and Biological Sensing. Angew Chem Int Ed. 2009;48:6790–6810. doi: 10.1002/anie.200900231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin D, Cohen Freue G, Hollander Z, John Mancini GB, Sasaki M, Mui A, Wilson-McManus J, Ignaszewski A, Imai C, Meredith A, et al. Plasma Protein Biosignatures for Detection of Cardiac Allograft Vasculopathy. J Heart Lung Transplant. 2013;32:723–733. doi: 10.1016/j.healun.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 18.Zamora-Olivares D, Kaoud TS, Jose J, Ellington A, Dalby KN, Anslyn EV. Differential Sensing of MAP Kinases Using SOX−Peptides. Angew Chem Int Ed. 2014;53:14064–14068. doi: 10.1002/anie.201408256. [DOI] [PubMed] [Google Scholar]
- 19.Liu Y, Minami T, Nishiyabu R, Wang Z, Anzenbacher P. Sensing of Carboxylate Drugs in Urine by a Supramolecular Sensor Array. J Am Chem Soc. 2013;135:7705–7712. doi: 10.1021/ja4015748. [DOI] [PubMed] [Google Scholar]
- 20.Askim JR, Mahmoudi M, Suslick KS. Optical Sensor Arrays for Chemical Sensing: the Optoelectronic Nose. Chem Soc Rev. 2013;42:8649–8682. doi: 10.1039/c3cs60179j. [DOI] [PubMed] [Google Scholar]
- 21.Stewart S, Ivy MA, Anslyn EV. The Use of Principal Component Analysis and Discriminant Analysis in Differential Sensing Routines. Chem Soc Rev. 2014;43:70–84. doi: 10.1039/c3cs60183h. [DOI] [PubMed] [Google Scholar]
- 22.De M, Rana S, Akpinar H, Miranda OR, Arvizo RR, Bunz UHF, Rotello VM. Sensing of Proteins in Human Serum Using Conjugates of Nanoparticles and Green Fluorescent Protein. Nat Chem. 2009;1:461–465. doi: 10.1038/nchem.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peveler WJ, Roldan A, Hollingsworth N, Porter MJ, Parkin IP. Multichannel Detection and Differentiation of Explosives with a Quantum Dot Array. ACS Nano. 2016;10:1139–1146. doi: 10.1021/acsnano.5b06433. [DOI] [PubMed] [Google Scholar]
- 24.Zhu W, Li W, Wang C, Cui J, Yang H, Jiang Y, Li G. CB[8]-Based Rotaxane as a Useful Platform for Sensitive Detection and Discrimination of Explosives. Chem Sci. 2013;4:3583–3590. [Google Scholar]
- 25.Anderson GP, Moreira SC, Charles PT, Medintz IL, Goldman ER, Zeinali M, Taitt CR. TNT Detection Using Multiplexed Liquid Array Displacement Immunoassays. Anal Chem. 2013;78:2279–2285. doi: 10.1021/ac051995c. [DOI] [PubMed] [Google Scholar]
- 26.Rana S, Le NDB, Mout R, Saha K, Tonga GY, Bain RES, Miranda OR, Rotello CM, Rotello VM. A Multichannel Nanosensor for Instantaneous Readout of Cancer Drug Mechanisms. Nat Nanotechnol. 2015;10:65–69. doi: 10.1038/nnano.2014.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lim SH, Feng L, Kemling JW, Musto CJ, Suslick KS. An Optoelectronic Nose for the Detection of Toxic Gases. Nat Chem. 2009;1:562–567. doi: 10.1038/nchem.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mazzone PJ, Wang XF, Xu Y, Mekhail T, Beukemann MC, Na J, Kemling JW, Suslick KS, Sasidhar M. Exhaled Breath Analysis with a Colorimetric Sensor Array for the Identification and Characterization of Lung Cancer. J Thorac Oncol. 2012;7:137–142. doi: 10.1097/JTO.0b013e318233d80f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rakow NA, Suslick KS. A Colorimetric Sensor Array for Odour Visualization. Nature. 2000;406:710–713. doi: 10.1038/35021028. [DOI] [PubMed] [Google Scholar]
- 30.Peng G, Tisch U, Adams O, Hakim M, Shehada N, Broza YY, Billan S, Abdah-Bortnyak R, Kuten A, Haick H. Diagnosing Lung Cancer in Exhaled Breath Using Gold Nanoparticles. Nat Nanotechnol. 2009;4:669–673. doi: 10.1038/nnano.2009.235. [DOI] [PubMed] [Google Scholar]