Abstract
G-quadruplexes (G4s) are unique four-stranded nucleic acid secondary structures formed by G-rich nucleic acid sequences which are prevalent in gene promoter and telomere regions and deemed to play essential roles in many biological and pathological processes. Although attentions to G4s have been paid for nearly 40 years, G4 selectivity and its topology discrimination in cells is still pending. Small fluorescence molecules are emerging as a versatile tool of interrogation of cellular features in vivo. Herein, a new class of bis(4-aminobenzylidene)acetone derivatives GD1, GD2, and GD3 with excellent environment-sensitive emission properties were developed and used for fluorescent detection of G4s. Among them, compound GD3 owning four methoxy groups presented preferable capability of lighting up parallel G4s with a strong red-emission enhancement. The photophysical property of GD3 was systematically investigated to elucidate the turn-on mechanism of GD3 toward parallel G4 structures, which reveal that the binding-induced polarity change of the microenvironment around GD3 together with the fluorophore conformational confinement affected the molecular intramolecular charge-transfer state and resulted the enhanced emission. G4s staining with GD3 in fixed cells was further applied, demonstrating GD3 a promising probe with the ability to visualize the distribution of G4 structures in biological processes. In general, this study provides a new potential scaffold—bis(4-aminobenzylidene)acetone—for design of G4-selective fluorescence probes.
Introduction
G-quadruplexes (G4s) are a kind of distinctive nucleic acid secondary structures, comprising two or more G-quartet blocks bound by Hoogsteen hydrogen bonding in G-rich oligonucleotide sequences.1−3 G4 structures can be formed kinetically fast within one strand (intramolecular) or from multiple strands (intermolecular) into extensive structural morphologies depending on the characteristics of the sequences, loop geometry, and local environment, and so forth. According to the orientation of the DNA strands, the G4s are typically divided into three main topologies including parallel, antiparallel, and hybrids thereof.4,5 Recent studies reveal that the G4 forming in human genome is prevalent and responsible for a wide variety of functions including gene-expressing regulation, RNA splicing and translation, telomere maintenance, and so on.3−8 In line with their biological significance, G4-motifs are proposed as new potential therapeutic targets against cancers and other diseases.9−12 For elaborating the structure–function relationships of G4s, many analytical methods such as circular dichroism (CD) spectroscopy, gel-electrophoresis, DNA-foot printing assay, isothermal calorimetry (ITC), mass spectrometry analysis, X-ray crystallography, and nuclear magnetic resonance (NMR) spectroscopy have been employed in which fluorescent imaging as a versatile noninvasive and visualized approach has attracted special attentions.13 Under this circumstance, specific G4 fluorescent probes with good biocompatibility are essential tools for G4 imaging in vivo. During the last 20 years, various G4 probes capable of binding and reporting G4 structures have been reported.14,15 Among those, small fluorescence probes usually perform superior signal output and affinities toward G4s, such as thiazole orange and its analogues,16−20 cyanine dyes,21 heavy-metal complexes,22−24 phthalocyanines,25 squaraines,26,27 and others.28,29 Developing new probes with specific photo properties especially with red or near-infrared fluorescent emission to distinguish different G4 topologies in vitro and mark G4 structures in cells are still appealing and challenging.
Pyridodicarboxamide unit is a well-known building block for developing efficient G4 ligands and drugs, which provides G4 ligands a crescent-shape configuration and H-bonds network for G-quartet overlap.30−33 By inspiration with such structural and functional features,34 herein, we explored the possibility of bis(4-aminobenzylidene)acetone as new G4 ligands by considering the resembling structure shape and the multiple H-bond sites from the carbonyl and amino groups. Bis(4-aminobenzylidene)acetone derivatives, owing to their good biocompatibility, pharmacokinetic profile, and easy synthesis and modification properties, are widely used in biological applications from anticancer or anti-inflammatory drugs35,36 to radioactive probes.37,38 However, to the best of our knowledge, their applications as fluorescent probes, especially in cells, have not been explored. The coexistence of the central electron-withdrawing carbonyl moiety, the diene bridge, and the extended pi-conjugated structure endows these compounds with special photophysical properties39 such as large excited-state dipole moment, distinct intramolecular charge transfer (ICT) character, obvious solvatochromism effect, and microenvironmental sensitivity, which makes bis(4-aminobenzylidene)acetone derivatives becoming a kind of promising building blocks for fluorescent probes. In this work, a new series of bis(4-aminobenzylidene)acetone derivatives GD1, GD2, and GD3 with different numbers of ortho-methoxy groups (Scheme 1), which were attached to regulate hydrogen bonding and morphological fitting for G4s binding, were synthesized and characterized. The three compounds were nearly fluorescence silent in aqueous solution but showed strong red emission in organic solvents. On the basis of their obvious solvatochromism properties, fluorescent screening assays for several kinds of oligonucleotide structures were performed. The results showed that compound GD3 with four methoxy groups presented a specific affinity to parallel G4s, giving a much superior fluorescence response for G4 topology discrimination.
Scheme 1. Structures of the Three Bis(4-aminobenzylidene)acetone Derivatives GD1, GD2, and GD3.
Results and Discussion
The affinity of GD1, GD2 and GD3 to G4s was first investigated through a preliminary fluorescent screening assay with 10 different oligonucleotide sequences including G4s (parallel G4s, anti-parallel G4s, and hybrid G4s), single- and double-stranded DNA, as well as triplex (the sequences and their corresponding secondary structures are shown in Table S1) in Tris-HCl (10 mM, pH = 7.4) containing 50 mM KCl. The emission enhancements of the three dyes (2 μM) after adding 0.5 equiv amount of different sequences are shown in Figures 1 and S1. All of the three dyes display very weak emissions with peaks around 600 nm initially. Addition of the 10 sequences into the three dye solutions, respectively, induces diverse fluorescence signal responses. Less than onefold enhancement of emission intensity was detected in GD1 and GD2 solutions, no matter which sequence was added. On the contrary, the emission of GD3 was enhanced significantly after being treated with c-myc, c-kit2, VEGF, and ADAM10 (RNA G4s), which had already been demonstrated to form into parallel G4 structures in buffers with K+. Whereas the addition of other oligonucleotide sequences, including hybrid G4s (22AG), anti-parallel G4s (21CTA and TBA), and other single-, double-stranded, and triplex DNA, hardly effected the emission of GD3 under the same conditions. These fluorescence results showed that compared to compounds GD1 and GD2, GD3 with four methoxy groups presented the capability of discriminating parallel G4 structures in vitro. Considering the structure differences between these dyes, it may be inferred that the additional methoxy groups played a crucial role in making GD3 an eligible ligand for parallel G4. The methoxy groups may provide more hydrogen bond acceptors to form hydrogen bond with a G-base and better morphologic fitness as well, and consequently enhance the binding affinity to parallel G4 structures. The preferable complexation of GD3 toward parallel G4s over antiparallel and hybrid G4s may result from the steric hindrance of nonparallel G4s, where dyes cannot efficiently stack on.
Figure 1.
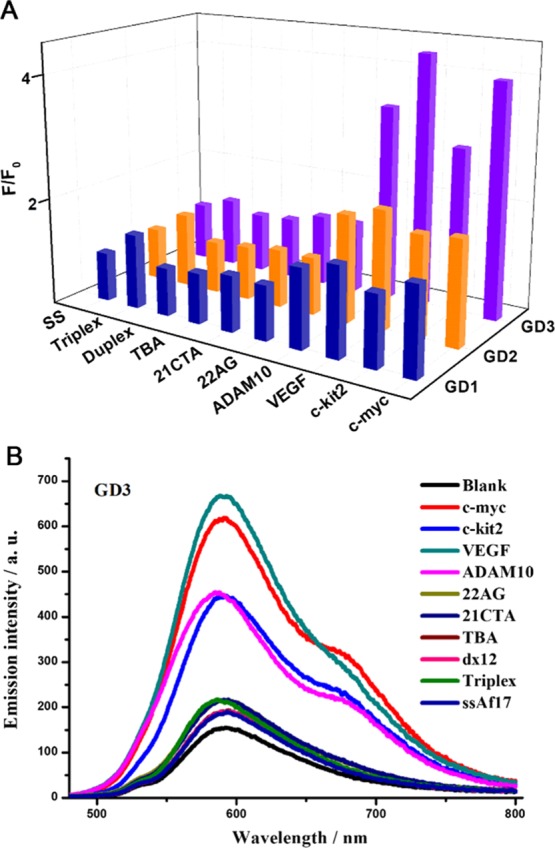
(A) Emission responses of GD1, GD2, and GD3 at 600 nm and (B) emission spectra of GD3 toward 0.5 equiv oligonucleotides of 10 different sequences (1 μM) in Tris-HCl buffer (10 mM, pH = 7.4) containing 50 mM KCl. GD1, GD2, and GD3 (2 μM) were severally incubated with DNA at room temperature for 10 min, and the solutions were excited at 450 nm.
The binding properties of GD3 against G4s were ascertained with the following fluorescence titration assays. When excited at 450 nm, the absorbance isosbestic point of GD3 solutions with and without equimolar G4s (Figure S2), the plots of the fluorescence intensity peaked at 600 nm as a function of oligonucleotide concentrations are recorded and depicted in Figure 2A. GD3 presents weak fluorescence responses to non-G4, anti-parallel, and hybrid G4 structures. However, the emission intensity of GD3 shows apparent dose-dependent enhancements versus the concentrations of the parallel G4 oligomers (c-myc, c-kit2, VEGF, and ADAM10). Meanwhile, the enhanced emissions induced by four parallel G4s are all accompanied with a slight hypsochromic shift of the emission maximum (Figure S3). Although GD3 can respond to all parallel G4s, the tendency of emission signal changes toward different G4 sequences is quite different. An emission plateau was first reached after adding equimolar VEGF, indicating that a GD3/VEGF complex was formed with 1:1 stoichiometry. The dissociation constant value (Kd) of GD3/VEGF complex was estimated using a nonlinear fitting equation with a 1:1 binding model to be 3.03 μM, and the Kd value provides the bis(4-aminobenzylidene)acetone derivative GD3 great potentials as a functional parallel G4 ligand. For further investigation of the application potential of GD3 in biosystems, the selectivity and anti-interference performances of GD3 to parallel G4s were examined by recording the fluorescence intensity changes of GD3/parallel G4s complexes in the absence and presence of excess ds-DNA (0−20 equiv). Figure S4 shows that the fluorescence intensity is barely affected by the added ds-DNA, indicative of excellent selectivity and anti-interference performances of GD3 to parallel G4s.
Figure 2.
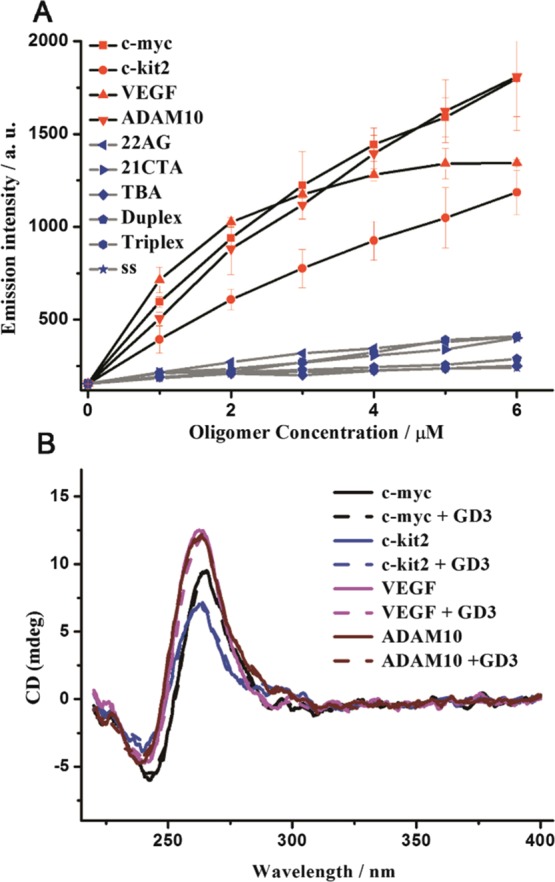
(A) Fluorescence titration of GD3 (2 μM) with the 10 preannealed oligonucleotides; (B) CD spectra of the four different parallel G4s (5 μM) and their corresponding complexes with GD3 (5 μM), in Tris-HCl buffer with 50 mM KCl (10 mM, pH = 7.4).
The topology stability of the parallel G4s with GD3 was then analyzed using CD spectroscopy. As depicted in Figure 2B, the typical CD signals of the parallel G4s with a strong positive absorption at 265 nm and a negative one at 240 nm are all conserved after addition of GD3, indicating that GD3 binds to parallel G4s without affecting their initial topologies which makes GD3 an ideal staining agent.42 To further investigate the binding mode of GD3 with parallel G4s, ITC analysis as well as molecular modeling studies were carried out, and the Kd value of GD3 to parallel G4-VEGF was reported as 3.65 μM with one binding site by ITC (Figure S5), quite agreeing with the value determined by fluorescence titration. The simulated binding site of GD3 in parallel G4s is shown in Figure S7. The result indicates that GD3 prefers to stack on one end (closing to 3′-terminal surface) of parallel G4s with a binding free energy of −36.912 kJ mol–1, which is much lower than that of GD1 and GD2 (estimated as −26.599 and −31.735 kJ mol–1, respectively), suggesting that the methoxy groups play key roles in the regulation of the binding affinity between this scaffold and parallel G4s which is mainly because of the improved H-bond forming between the methoxy groups and G-bases as well as the morphologic fitness to parallel G4 structures, and the end-stacking binding mode was further confirmed by the G4/hemin inhibition results, as shown in Figure S8.
The “light up” mechanism of GD3 toward parallel G4s was further investigated by fluorescence spectroscopy. There are possibly three main reasons accounting for the emission changes of GD3 induced by complexation with G4s, including changes of fluorophore aggregation or dispersion state, influence on ICT process, and molecular conformation planarity or confinement. These factors may exist lonely or synergistically in different scenarios. The aggregation or dispersion state change of GD3 was first excluded by the concentration- and temperature-dependent absorption experiments (Figure S9). The unchanged absorption spectrum of GD3 under heating and the linear decrease of absorbance with dilution suggested that GD3 was molecularly dispersed and chemically stable under the experimental condition as well as during the binding procedure, indicative of the unchanged physical state of GD3. The solvatochromism effect is a typical phenomenon shown by a fluorophore with the ICT property; thus, GD3 was dissolved into six solvents with different polarities, and the fluorescence spectra of the solutions are recorded and shown in Figure 3A. The remarkable variation of emission maximum in the six solvents with a positive solvatochromism response identified GD3 as a typical ICT compound. The fluorescence quantum yields (ΦF) of GD3 in organic solvents were above 0.18, whereas the value sharply dropped to 0.003 in Tris-HCl (10 mM, pH = 7.4). The mixture of methanol–Tris-HCl was chosen for monitoring the emission changes of GD3 induced by subtle variations of the solvent environment. Emission spectra in Figure 3B show that with increasing the proportion of methanol in mixed solvent from 0 to 1, the emission of GD3 gradually enhanced accompanied with a hypsochromic shift of 15 nm. The variation tendency of the emission spectra of GD3 treated with increasing parallel G4s resembles that with environmental polarity decreasing, indicating that the complexation of GD3 with parallel G4s decreases the microenvironmental polarity around GD3 and then enhances the emission. In addition, complexation with G4s may introduce conformational confinement that will enhance the chromophore emission. A viscosity-dependent emission assay was carried out to evaluate the emission enhancement induced by molecular conformation confinement. Glycerol–methanol mixtures were used as the dispersant media of different viscosities, and the emission spectra of GD3 therein are recorded and shown in Figure S10. With increasing the fraction of glycerol and the consequent medium viscosity, the emission of GD3 was gradually enhanced, which is attributed to the restrained intramolecular rotation and nonradiative decay in high-viscosity medium. Therefore, taking account of these photophysical results, the lighting-up mechanism of GD3 toward parallel G4s can be mainly ascribed to the binding-induced polarity change of the microenvironment around GD3, which enhances the emission of GD3 ICT state. Meanwhile, the conformational confinement of the fluorophore also synergistically contributes to the sensing signal.
Figure 3.
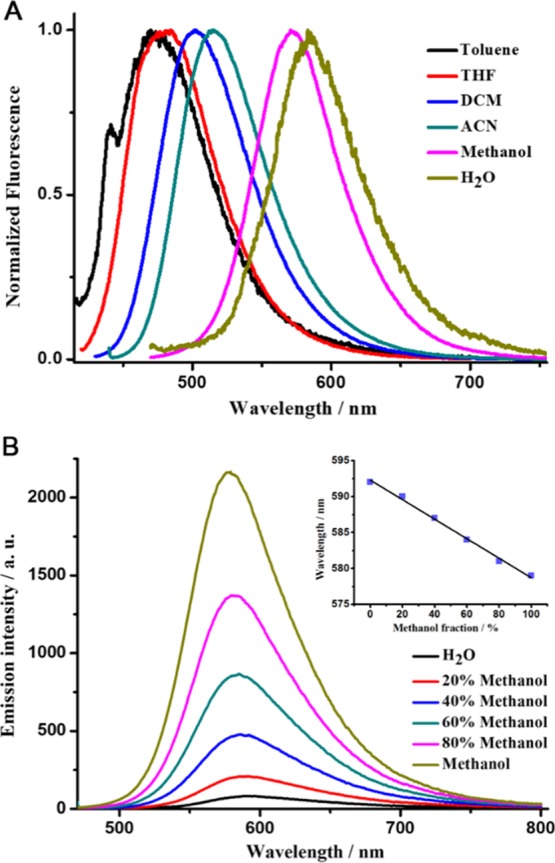
(A) Normalized fluorescence spectra of GD3 (2 μM) in six kinds of solvents with different polarities; (B) fluorescence spectra of GD3 (2 μM) in the mixtures of methanol and Tris-HCl with different ratios (Inset: the emission maximum wavelength of GD3 at different methanol fractions).
The intracellular application of GD3 as a selective parallel G4 staining ligand in ARPE-19 cell lines was investigated as follows. The confocal fluorescence images of methanol-fixed ARPE-19 cells costained with GD3 and nuclear-localized dye Hoechst 33258 (Figure 4A–C) showed that bright-red emission spots appeared in certain regions of nucleus, coinciding with the areas of nucleoli.43 Besides, red emission signals also distributed in some cytoplasm regions near the nucleus where parallel G4s might exist as well. To preclude the background interferences and the unspecific binding in cells, compounds GD1 and GD2 with similar scaffold and photophysical properties of GD3 were used to stain ARPE-19 cells as negative controls, and their imaging results are recorded in Figure S12. Compared to the strong red emission in cells stained with GD3, no signal could be detected in cells with GD1 and only weak red emission could be detected from cells with GD2, which were highly consistent with the results of G4 detection in vitro, demonstrating that GD3 would be a new kind of G4 fluorescent probes with low interferences in cells. To confirm the sensing targets of GD3 in nucleus and in cytoplasm regions, respectively, deoxyribonuclease (DNase) and ribonuclease (RNase) digestion experiments were performed, and the results are shown in Figure 4D,F. Predigesting cells with DNase made the emission spots in nucleus dramatically weakened or even disappeared, but reserved in cytoplasm. In contrast, the RNase predigested only decreased emission signals in cytoplasm regions, but kept the signal in nucleus. When predigesting cells with both DNase and RNase, no emission signal was observed with GD3 staining (Figure 4F) which results that GD3 is capable of lighting up nucleic acid in cells. In combination with the in vitro discrimination results, we presume that the signal is mainly located at DNA G4s foci and RNA G4s foci in cells, demonstrating that GD3 might be a competent G4s staining agent in cells with a clear background.
Figure 4.
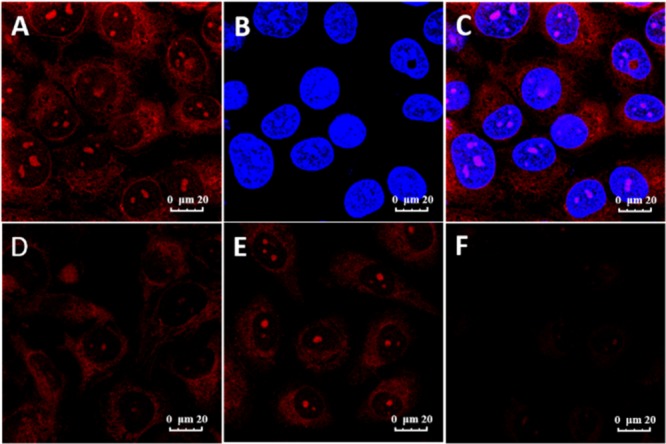
Fluorescence images of ARPE-19 cells costained with 1 μM GD3 and 1 μg/mL Hoechst 33258. (A) GD3 red channel (580–620 nm, λex = 488 nm); (B) Hoechst 33258 blue channel (420–450 nm, λex = 405 nm); (C) merged image of (A,B); (D) cell lines pretreated with DNase, (E) cell lines pretreated with RNase, and (F) pretreated with both.
Conclusions
In summary, three bis(4-aminobenzylidene)acetone derivatives with different numbers of methoxy substituents were synthesized, and their spectral responses to several oligonucleotides with different secondary structures were investigated and compared. Among them, compound GD3 with four methoxy groups exhibited a preferable binding property to parallel G4 structures in vitro, accompanied with an evident red emission enhancement. The lighting-up of parallel G4s is attributed to the microenvironmental polarity changes and the conformational confinement induced by the complexation of GD3 with parallel G4s. Therefore, GD3 was applied to stain G4 structures in cells and exhibited preferential localization of DNA G4 and RNA G4 structures. In general, this study provided a new building block, bis(4-aminobenzylidene)acetone derivative, for developing fluorescence probes to detect and investigate G4s in biological processes.
Experimental Section
General Information
All reagents were obtained from Beijing Innochem Co. and used without further purification. 1H NMR and 13C NMR spectra were recorded on Bruker AVANCE 400 spectrometer. Ultrapure water (18 MΩ/cm) was used for the preparation of all aqueous solutions. Matrix-assisted laser desorption/ionization–time of flight–mass spectrum was measured by a Bruker BIFLEX III spectrometer. UV–vis absorption spectra were recorded with a Hitachi U-3010 spectrophotometer. Photoluminescence spectra were carried out on a Hitachi F-7000 spectrophotometer. Fluorescence quantum yields were determined by using Nile Red (0.38 in methanol) as standards.
Molecular Docking Method
Molecular docking was carried out using the CDOCKER module in Discovery Studio (BIOVIA), and the G4 structures (PDB codes: 2L7V(40) and 3EM2(41)) were retrieved from the RCSB PDB databank. All water molecules were deleted. Explicit hydrogen atoms of the G4 were added. The active site center was set as the mean coordinates of the atoms from the original ligand. The radius of the binding site was set to 15 Å. Then, GD1, GD2, and GD3 were prepared for Docking by using “prepare ligands” module in Discovery Studio (BIOVIA). All other parameters were set as default. The Docking results were displayed by Discovery Studio Visualizer.
Cell Culture and Imaging
ARPE-19 cells were grown at 37 °C under 5% CO2 in Dulbecco’s modified Eagle medium (Gibco; Invitrogen), supplemented with 10% fetal bovine serum. Before staining experiments, cultured cells grown on special confocal microscope dishes were fixed by precooled methanol (−20 °C) for 1 min and washed with phosphate-buffered saline (PBS) three times. The cell membrane was permeabilized by immersing 0.5% Triton X-100 for 1 min and then washed with PBS three times. For the DNase and RNase digestion experiments, the cells were incubated with DNase or RNase (100 μg/mL) at 37 °C for 2 h first and then stained with GD3 at 37 °C for 15 min. Cells were rinsed by PBS three times before imaging. The fluorescence imaging pictures in the experiments were obtained with an equal parameter.
The detailed synthesis procedures of compounds GD1, GD2, and GD3 are provided in the Supporting Information.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (21233011) and the Youth Innovation Promotion Association (2017032) of Chinese Academy of Sciences.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.8b01190.
Synthesis processes of GD1–GD3; oligonucleotides used in this study; fluorescence spectra of 2 μM GD1 and GD2; fluorescence titration of GD3; selectivity of GD3 to G4s; ITC analysis of GD3 binding affinity to G4-structure VEGF; CD spectra of oligonucleotides; molecular docking results of GD1, GD2, and GD3 with parallel G4 structure; Inhibition activity of GD3 on the peroxidase activity; and absorption spectra of GD3 in experimental buffer at different temperatures and concentrations (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Sen D.; Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature 1988, 334, 364–366. 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- Davis J. T. G-Quartets 40 Years Later: From 5′-GMP to Molecular Biology and Supramolecular Chemistry. Angew. Chem., Int. Ed. 2004, 43, 668–698. 10.1002/anie.200300589. [DOI] [PubMed] [Google Scholar]
- Lipps H. J.; Rhodes D. G-quadruplex structures: in vivo evidence and function. Trends Cell Biol. 2009, 19, 414–422. 10.1016/j.tcb.2009.05.002. [DOI] [PubMed] [Google Scholar]
- Hu M.-H.; Chen S.-B.; Wang Y.-Q.; Zeng Y.-M.; Ou T.-M.; Li D.; Gu L.-Q.; Huang Z.-S.; Tan J.-H. Accurate high-throughput identification of parallel G-quadruplex topology by a new tetraaryl-substituted imidazole. Biosens. Bioelectron. 2016, 83, 77–84. 10.1016/j.bios.2016.04.029. [DOI] [PubMed] [Google Scholar]
- Bochman M. L.; Paeschke K.; Zakian V. A. DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012, 13, 770–780. 10.1038/nrg3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maizels N. Dynamic roles for G4 DNA in the biology of eukaryotic cells. Nat. Struct. Mol. Biol. 2006, 13, 1055–1059. 10.1038/nsmb1171. [DOI] [PubMed] [Google Scholar]
- Schaffitzel C.; Berger I.; Postberg J.; Hanes J.; Lipps H. J.; Pluckthun A. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc. Natl. Acad. Sci. U.S.A. 2001, 98, 8572–8577. 10.1073/pnas.141229498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paeschke K.; Simonsson T.; Postberg J.; Rhodes D.; Lipps H. J. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat. Struct. Mol. Biol. 2005, 12, 847–854. 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- Collie G. W.; Parkinson G. N. The application of DNA and RNA G-quadruplexes to therapeutic medicines. Chem. Soc. Rev. 2011, 40, 5867–5892. 10.1039/c1cs15067g. [DOI] [PubMed] [Google Scholar]
- Ou T.-m.; Lu Y.-j.; Tan J.-h.; Huang Z.-s.; Wong K.-Y.; Gu L.-q. G-Quadruplexes: Targets in Anticancer Drug Design. ChemMedChem 2008, 3, 690–713. 10.1002/cmdc.200700300. [DOI] [PubMed] [Google Scholar]
- Maizels N. G4-associated human diseases. EMBO Rep. 2015, 16, 910–922. 10.15252/embr.201540607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S.; Hurley L. H.; Neidle S. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy?. Nat. Rev. Drug Discovery 2011, 10, 261–275. 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suseela Y. V.; Narayanaswamy N.; Pratihar S.; Govindaraju T. Far-red fluorescent probes for canonical and non-canonical nucleic acid structures: current progress and future implications. Chem. Soc. Rev. 2018, 47, 1098–1131. 10.1039/c7cs00774d. [DOI] [PubMed] [Google Scholar]
- Bhasikuttan A. C.; Mohanty J. Targeting G-quadruplex structures with extrinsic fluorogenic dyes: promising fluorescence sensors. Chem. Commun. 2015, 51, 7581–7597. 10.1039/c4cc10030a. [DOI] [PubMed] [Google Scholar]
- Vummidi B. R.; Alzeer J.; Luedtke N. W. Fluorescent probes for G-quadruplex structures. ChemBioChem 2013, 14, 540–558. 10.1002/cbic.201200612. [DOI] [PubMed] [Google Scholar]
- Lubitz I.; Zikich D.; Kotlyar A. Specific high-affinity binding of thiazole orange to triplex and G-quadruplex DNA. Biochemistry 2010, 49, 3567–3574. 10.1021/bi1000849. [DOI] [PubMed] [Google Scholar]
- Yang P.; De Cian A.; Teulade-Fichou M.-P.; Mergny J.-L.; Monchaud D. Engineering bisquinolinium/thiazole orange conjugates for fluorescent sensing of G-quadruplex DNA. Angew. Chem., Int. Ed. 2009, 48, 2188–2191. 10.1002/anie.200805613. [DOI] [PubMed] [Google Scholar]
- Guan A.-j.; Zhang X.-F.; Sun X.; Li Q.; Xiang J.-F.; Wang L.-X.; Lan L.; Yang F.-M.; Xu S.-J.; Guo X.-M.; Tang Y.-L. Ethyl-substitutive Thioflavin T as a highly-specific fluorescence probe for detecting G-quadruplex structure. Sci. Rep. 2018, 8, 2666. 10.1038/s41598-018-20960-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S.; Li Q.; Xiang J.; Yang Q.; Sun H.; Guan A.; Wang L.; Liu Y.; Yu L.; Shi Y.; Chen H.; Tang Y. Thioflavin T as an efficient fluorescence sensor for selective recognition of RNA G-quadruplexes. Sci. Rep. 2016, 6, 24793. 10.1038/srep24793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J.-W.; Ye W.-J.; Chen S.-B.; Wu W.-B.; Hou J.-Q.; Ou T.-M.; Tan J.-H.; Li D.; Gu L.-Q.; Huang Z.-S. Development of a Universal Colorimetric Indicator for G-Quadruplex Structures by the Fusion of Thiazole Orange and Isaindigotone Skeleton. Anal. Chem. 2012, 84, 6288–6292. 10.1021/ac300207r. [DOI] [PubMed] [Google Scholar]
- Xu S.; Li Q.; Xiang J.; Yang Q.; Sun H.; Guan A.; Wang L.; Liu Y.; Yu L.; Shi Y.; Chen H.; Tang Y. Directly lighting up RNA G-quadruplexes from test tubes to living human cells. Nucleic Acids Res. 2015, 43, 9575–9586. 10.1093/nar/gkv1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D.-L.; Che C.-M.; Yan S.-C. Platinum(II) Complexes with Dipyridophenazine Ligands as Human Telomerase Inhibitors and Luminescent Probes for G-Quadruplex DNA. J. Am. Chem. Soc. 2009, 131, 1835–1846. 10.1021/ja806045x. [DOI] [PubMed] [Google Scholar]
- Gill M. R.; Garcia-Lara J.; Foster S. J.; Smythe C.; Battaglia G.; Thomas J. A. A ruthenium(II) polypyridyl complex for direct imaging of DNA structure in living cells. Nat. Chem. 2009, 1, 662–667. 10.1038/nchem.406. [DOI] [PubMed] [Google Scholar]
- Leung K.-H.; He H.-Z.; He B.; Zhong H.-J.; Lin S.; Wang Y.-T.; Ma D.-L.; Leung C.-H. Label-free luminescence switch-on detection of hepatitis C virus NS3 helicase activity using a G-quadruplex-selective probe. Chem. Sci. 2015, 6, 2166–2171. 10.1039/c4sc03319a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alzeer J.; Vummidi B. R.; Roth P. J. C.; Luedtke N. W. Guanidinium-modified phthalocyanines as high-affinity G-quadruplex fluorescent probes and transcriptional regulators. Angew. Chem., Int. Ed. 2009, 48, 9362–9365. 10.1002/anie.200903685. [DOI] [PubMed] [Google Scholar]
- Jin B.; Zhang X.; Zheng W.; Liu X.; Zhou J.; Zhang N.; Wang F.; Shangguan D. Dicyanomethylene-Functionalized Squaraine as a Highly Selective Probe for Parallel G-Quadruplexes. Anal. Chem. 2014, 86, 7063–7070. 10.1021/ac501619v. [DOI] [PubMed] [Google Scholar]
- Grande V.; Doria F.; Freccero M.; Würthner F. An Aggregating Amphiphilic Squaraine: A Light-up Probe That Discriminates Parallel G-Quadruplexes. Angew. Chem., Int. Ed. 2017, 56, 7520–7524. 10.1002/anie.201702096. [DOI] [PubMed] [Google Scholar]
- Doria F.; Oppi A.; Manoli F.; Botti S.; Kandoth N.; Grande V.; Manet I.; Freccero M. A naphthalene diimide dyad for fluorescence switch-on detection of G-quadruplexes. Chem. Commun. 2015, 51, 9105–9108. 10.1039/c5cc01536g. [DOI] [PubMed] [Google Scholar]
- Bhasikuttan A. C.; Mohanty J.; Pal H. Interaction of malachite green with guanine-rich single-stranded DNA: preferential binding to a G-quadruplex. Angew. Chem., Int. Ed. 2007, 46, 9305–9307. 10.1002/anie.200703251. [DOI] [PubMed] [Google Scholar]
- Granotier C.; Pennarun G.; Riou L.; Hoffschir F.; Gauthier L. R.; De Cian A.; Gomez D.; Mandine E.; Riou J.-F.; Mergny J.-L.; Mailliet P.; Dutrillaux B.; Boussin F. D. Preferential binding of a G-quadruplex ligand to human chromosome ends. Nucleic Acids Res. 2005, 33, 4182–4190. 10.1093/nar/gki722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cian A.; DeLemos E.; Mergny J.-L.; Teulade-Fichou M.-P.; Monchaud D. Highly efficient G-quadruplex recognition by bisquinolinium compounds. J. Am. Chem. Soc. 2007, 129, 1856–1857. 10.1021/ja067352b. [DOI] [PubMed] [Google Scholar]
- Müller S.; Kumari S.; Rodriguez R.; Balasubramanian S. Small-molecule-mediated G-quadruplex isolation from human cells. Nat. Chem. 2010, 2, 1095–1098. 10.1038/nchem.842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T.; Li W.; Ding Z.; Cheng S.; Liang K.; Liu X.; Bing T.; Shangguan D. Detection of G-Quadruplex Structures Formed by G-Rich Sequences from Rice Genome and Transcriptome Using Combined Probes. Anal. Chem. 2017, 89, 8162–8169. 10.1021/acs.analchem.7b01992. [DOI] [PubMed] [Google Scholar]
- Jin B.; Zhang X.; Zheng W.; Liu X.; Qi C.; Wang F.; Shangguan D. Fluorescence Light-Up Probe for Parallel G-Quadruplexes. Anal. Chem. 2014, 86, 943–952. 10.1021/ac403676x. [DOI] [PubMed] [Google Scholar]
- Modzelewska A.; Pettit C.; Achanta G.; Davidson N. E.; Huang P.; Khan S. R. Anticancer activities of novel chalcone and bis-chalcone derivatives. Bioorg. Med. Chem. 2006, 14, 3491–3495. 10.1016/j.bmc.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Yin S.; Zheng X.; Yao X.; Wang Y.; Liao D. Synthesis and Anticancer Activity of Mono-Carbonyl Analogues of Curcumin. J. Cancer Ther. 2013, 04, 113–123. 10.4236/jct.2013.41016. [DOI] [Google Scholar]
- Cui M.; Ono M.; Kimura H.; Liu B.; Saji H. Synthesis and Structure–Affinity Relationships of Novel Dibenzylideneacetone Derivatives as Probes for β-Amyloid Plaques. J. Med. Chem. 2011, 54, 2225–2240. 10.1021/jm101404k. [DOI] [PubMed] [Google Scholar]
- Yang Y.; Cui M.; Jin B.; Wang X.; Li Z.; Yu P.; Jia J.; Fu H.; Jia H.; Liu B. 99mTc-labeled dibenzylideneacetone derivatives as potential SPECT probes for in vivo imaging of β-amyloid plaque. Eur. J. Med. Chem. 2013, 64, 90–98. 10.1016/j.ejmech.2013.03.057. [DOI] [PubMed] [Google Scholar]
- DeVoe R. J.; Sahyun M. R. V.; Schmidt E.; Sadrai M.; Serpone N.; Sharma D. K. Photophysics of bis(p-N,N-dimethylamino)benzylidene acetone: Does photoinduced electron transfer occur from a twisted excited state?. Can. J. Chem. 1989, 67, 1565–1575. 10.1139/v89-239. [DOI] [Google Scholar]
- Dai J.; Carver M.; Hurley L. H.; Yang D. Solution Structure of a 2:1 Quindoline-c-MYC G-Quadruplex: Insights into G-Quadruplex-Interactive Small Molecule Drug Design. J. Am. Chem. Soc. 2011, 133, 17673–17680. 10.1021/ja205646q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell N. H.; Patel M.; Tofa A. B.; Ghosh R.; Parkinson G. N.; Neidle S. Selectivity in Ligand Recognition of G-Quadruplex Loops†. Biochemistry 2009, 48, 1675–1680. 10.1021/bi802233v. [DOI] [PubMed] [Google Scholar]
- Chen S.-B.; Wu W.-B.; Hu M.-H.; Ou T.-M.; Gu L.-Q.; Tan J.-H.; Huang Z.-S. Discovery of a new fluorescent light-up probe specific to parallel G-quadruplexes. Chem. Commun. 2014, 50, 12173–12176. 10.1039/c4cc05394j. [DOI] [PubMed] [Google Scholar]
- Lu Y.-J.; Hu D.-P.; Zhang K.; Wong W.-L.; Chow C.-F. New pyridinium-based fluorescent dyes: A comparison of symmetry and side-group effects on G-Quadruplex DNA binding selectivity and application in live cell imaging. Biosens. Bioelectron. 2016, 81, 373–381. 10.1016/j.bios.2016.03.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.