Abstract
Introduction
Chemokines are closely related with tumor immunity, progression, and metastasis. We aimed to construct a multi-RNA classifier of chemokine family genes for predicting tumor recurrence in stage I–III patients with colorectal cancer (CRC) after operation.
Patients and methods
By analyzing microarray data, the Cox regression analysis was conducted to determine survival-related chemokine family genes and develop a multi-RNA classifier in the training set. The prognostic value of this multi-RNA classifier was further validated in the internal validation and external independent sets. Receiver operating characteristic curves were used to compare the prediction ability of the combined model of this multi-RNA classifier and stage, and this multi-RNA classifier and stage alone.
Results
Nine survival-related chemokines were identified in the training set. We identified a nine-chemokine classifier and classified the patients as high-risk or low-risk. Compared with CRC patients with high-risk scores, CRC patients with low-risk scores had longer disease-free survival in the training (HR=2.353, 95% CI=1.480–3.742, P<0.001), internal validation (HR=2.389, 95% CI=1.428–3.996, P<0.001), and external independent (HR=3.244, 95% CI=1.813–5.807, P<0.001) sets. This nine-chemokine classifier was an independent prognostic factor in these datasets (P<0.05). The combined model of this nine-chemokine classifier and tumor stage may tend to have higher accuracy than stage alone in the training (area under curve 0.727 vs 0.626, P<0.01), internal validation (0.668 vs 0.584, P=0.03), and external independent (0.704 vs 0.678, P>0.05) sets. This nine-chemokine classifier may only be applied in Marisa’s C2, C5, and C6 subtypes patients.
Conclusion
Our nine-chemokine classifier is a reliable prognostic tool for some specific biological subtypes of CRC patients. It might contribute to guide the personalized treatment for high-risk patients.
Keywords: classifier, colorectal cancer, chemokine, survival analysis, risk classification, microarray
Introduction
CRC is the third most common malignancy and the fourth leading cause of mortality worldwide.1 Risk stratification of CRC may require the combined multiple approaches, including analysis of molecular biomarkers and clinical data, and thorough experimental studies. In the past two decades, study results have confirmed that tumor is a heterogeneity disease.2 The combined model of multiple clinical and molecular parameters could be a much more objective prognostic tool for CRC patients.
Chemokines are a group of small molecular weight proteins that bind to the G protein-coupled chemokine receptors.3 Chemokines and their receptors are divided into four subfamilies (CXC, CC, CX3C, and C) based on the pattern of cysteine residues, where C represents the cysteine and X represents noncysteine amino acids.4,5 Chemokine family genes are closely related with senescence, angiogenesis, epithelial mesenchymal transition, proliferation, immune evasion, and tumor metastasis.6 The altered expression of chemokines and their receptors is found in many malignancies and may lead to aberrant chemokine receptor signaling.7–20 The chemokine ligand–receptor interactions were closely related with tumor immunity, progression, and metastasis in CRC patients, including the CXCL1– CXCR2,7 CXCL9/10–CXCR3,8,9 CXCL12–CXCR4,10–14 CCL2–CCR2,15,16 CCL5–CCR5,17 CCL15–CCR1,18 CCL20– CCR6,19 and CX3CL1–CX3CR120 axes. Although small sample studies showed that some chemokines could be used as the single biomarker for CRC, the values of these genes were not further validated in other validation sets.8,11,13,16,21–23 Until now, there is no multi-RNA classifier of chemokine family genes for predicting tumor recurrence in CRC patients. As a widely used microarray platform, Affymetrix human genome U133 plus 2.0 array included all probes of human genome U133 set and about 6,500 new genes.24 Almost all the mRNA-level data of chemokine family genes could be obtained from this microarray platform. Laetitia Marisa defined six molecular subtypes for CRC based on the main biological characteristics, including one subtype with downregulated immune pathways (C1), one deficient mismatch repair subtype (C2), one KRAS mutant subtype (C3), one cancer stem cell subtype (C4), one subtype with the upregulated Wnt pathway (C5), and one subtype with a normal-like gene expression profile (C6).25 Therefore, we performed this study to develop and validate a multi-RNA prognostic classifier of chemokine family genes for predicting recurrence in stage I–III patients with CRC by analyzing the microarray data. These multi-RNA signatures were further validated in CRC patients with different Marisa’s biological subtypes (C1–C6).25
Materials and methods
Datasets preparation
Gene expression of chemokine family genes and corresponding clinical data were obtained from the GEO database. All log2-transformed expression data of chemokines and their receptors were obtained from Affymetrix human genome U133 plus 2.0 array (GSE39582 and GSE14333). The expression values of genes with multiple probes were calculated by using the median values of multiple probes. After filtering out CRC patients without DFS and clinical data, there were a total of 718 stage I–III patients with CRC, including 492 from GSE39582 and 226 from GSE14333. The 492 patients from GSE39582 were randomly divided into training (n=246, GSE39582) and internal validation (n=246, GSE39582) sets. The 226 patients from GSE14333 were used for the external independent set.
Statistical analysis
The survival analysis was performed by the “survival” package of R software (version 3.4.3). In the training set, the association between the expression level of chemokine family genes and CRC patients’ DFS was evaluated using a univariate Cox regression analysis. Those chemokine family genes were considered to be significant if their P-values were less than 0.05. Then, the selected chemokine family genes were fitted in a multivariate Cox regression analysis in the training set. Risk scores were calculated by the selected chemokine family genes and their regression coefficients in the multivariate Cox regression analysis,26–28 as follows:
where k is the number of prognostic chemokines, Ci represents the coefficient of the ith chemokine in the multivariate Cox regression analysis, Vi is the expression value of the ith chemokine. Using the upper quartile value of risk scores in the training set as the cutoff point, CRC patients in the training, internal validation, and external independent sets were classified as low-risk or high-risk correspondingly. Survival differences between low-risk and high-risk groups were assessed by the Kaplan–Meier estimator and log-rank test. The multivariate Cox regression analysis was performed to assess whether this risk score was independent of the clinical characteristics such as stage, age, gender, and adjuvant chemotherapy. Additionally, ROC curves were used to compare the predictive value of DFS by the combined model of risk score and stage, and this risk score model and stage alone. The ROC curve analysis was performed using the “pROC” package of R software (version 3.4.3). To generate the ROC curves, those CRC patients whose durations were less than the 5-year DFS needed to be excluded, if they still did not recur at last follow-up. The remaining CRC patients were classified as having either shorter or longer than the recurrence-free survival of 60 months.28 The prognostic values of this multi-RNA prognostic classifier were further validated in different Marisa’s biological subtypes (C1–C6).25 The log-rank test, Cox regression analysis, and ROC analysis were considered to be significant if their P-values were less than 0.05.
Functional enrichment analysis
To evaluate the functional implication of these nine chemokines, functional enrichment analyses for GO and KEGG category were performed with the GeneCodis web tool (http://genecodis.cnb.csic.es/).29–31 GO and KEGG category enrichments were based on the threshold of P-value <0.05. Significant enrichment results were visualized using R software (version 3.4.3).
Results
Identification of survival-related chemokines in the training set
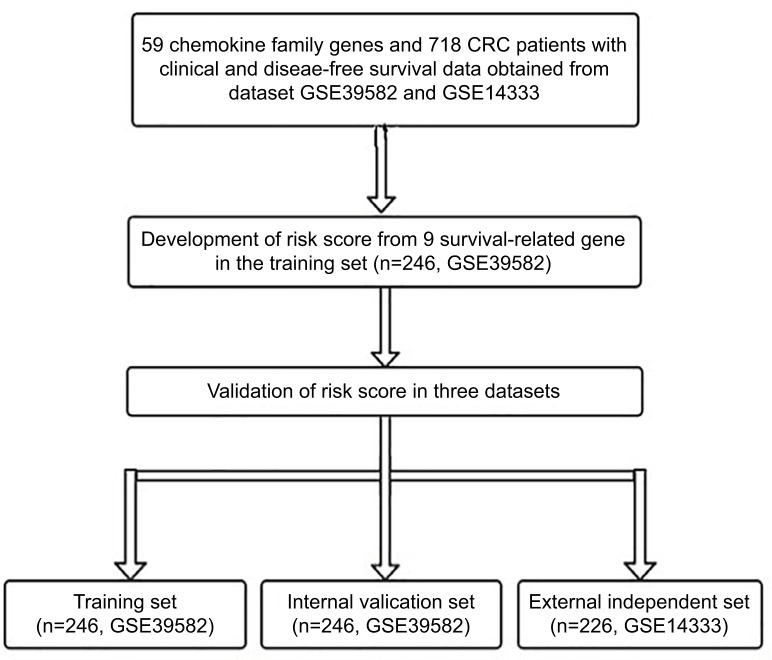
Figure 1 shows the study flow for the development and validation of the nine-chemokine classifier. By using microarray data, we identified 59 chemokine family genes from GSE39582 and GSE14333. We further analyzed these 59 genes by the univariate Cox regression analysis in the training set (n=246, GSE39582; Table S1). Consequently, we identified nine chemokines that were significantly correlated with DFS in CRC patients (shown in Table 1). The positive coefficients for three chemokines (CCL1, CCL14, and CXCL14) indicated that their higher levels of expression were associated with worse prognosis. The negative coefficients for the remaining six genes (CXCL1, CXCL3, CXCL9, CXCL10, CXCL11, and CXCL13) indicated that their higher levels of expression were associated with better prognosis.
Figure 1.
Study flow for the development and validation of the nine-chemokine classifier.
Abbreviation: CRC, colorectal cancer.
Table 1.
The characteristics of nine chemokines associated with DFS in the training set of 246 CRC patients (n=246, GSE39582)
| Gene symbol | Type | HR (95% CI) | Coefficients | P-value | Putative function |
|---|---|---|---|---|---|
| CCL1 | Ligand | 2.729 (1.118–6.662) | 1.004 | 0.027 | Risky |
| CCL14 | Ligand | 1.400 (1.043–1.879) | 0.336 | 0.025 | Risky |
| CXCL1 | Ligand | 0.824 (0.697–0.974) | −0.194 | 0.024 | Protective |
| CXCL3 | Ligand | 0.836 (0.708–0.986) | −0.179 | 0.034 | Protective |
| CXCL9 | Ligand | 0.840 (0.722–0.977) | −0.174 | 0.024 | Protective |
| CXCL10 | Ligand | 0.832 (0.712–0.972) | −0.184 | 0.021 | Protective |
| CXCL11 | Ligand | 0.878 (0.774–0.995) | −0.131 | 0.041 | Protective |
| CXCL13 | Ligand | 0.870 (0.763–0.992) | −0.140 | 0.037 | Protective |
| CXCL14 | Ligand | 1.146 (1.008–1.302) | 0.136 | 0.037 | Risky |
Abbreviation: CRC, colorectal cancer.
Survival comparisons between low-risk and high-risk groups in the training, internal validation, and external independent sets
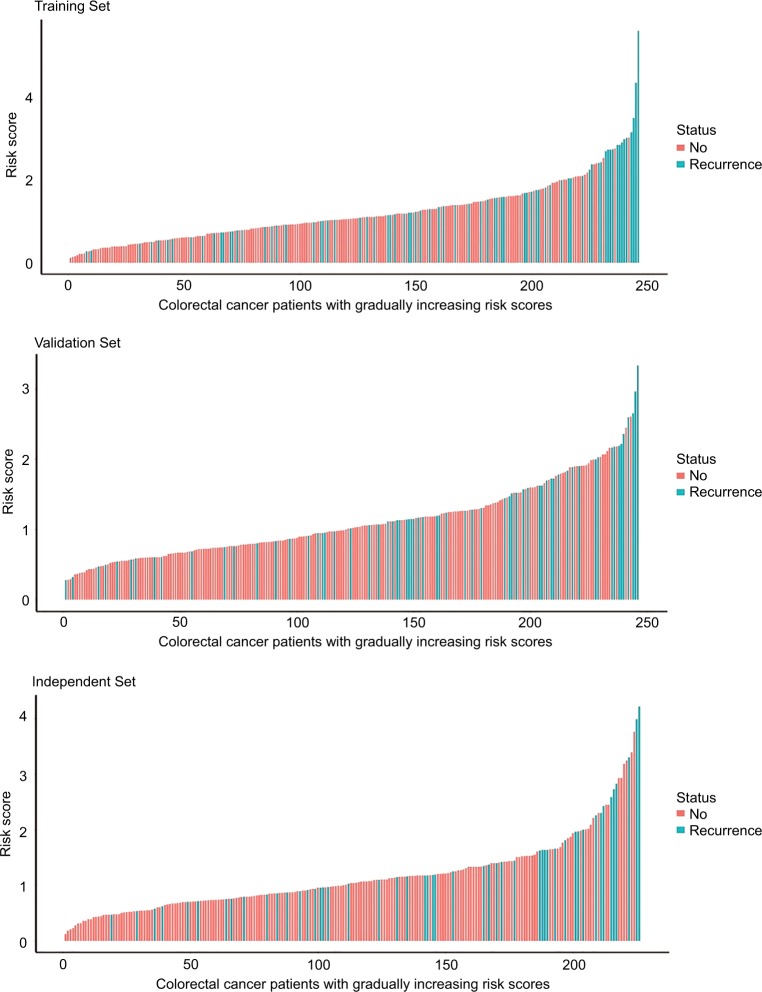
According to these chemokines and their regression coefficients in the multivariate Cox model, we calculated the risk scores for every patient in the training (n=246, GSE39582), internal validation (n=246, GSE39582), and external independent (n=226, GSE14333) sets. Using the cutoff value of risk scores (1.559), CRC patients were classified into low-risk group and high-risk group for the training (low-risk/high-risk: 185/61), internal validation (low-risk/high-risk: 196/50), and external independent (low-risk/high-risk: 185/41) sets. Figure 2 shows the distributions of this risk score and the DFS status in these three sets. As shown in Figure 2, CRC patients with high-risk scores tended to have higher risk of treatment relapse.
Figure 2.
The distributions of risk score and DFS status in the training (n=246, GSE39582), internal validation (n=246, GSE39582), and external independent (n=226, GSE14333) sets.
Abbreviation: DFS, disease-free survival.
The clinical characteristics of low-risk group and high-risk group patients with CRC in these three sets are shown in Table 2. There were no differences for clinical characteristics (age, gender, stage, and adjuvant chemotherapy) between low-risk group and high-risk group (all P>0.05).
Table 2.
Clinical characteristics of CRC patients according to the nine-chemokine classifier in the training (n=246, GSE39582), internal validation (n=246, GSE39582), and external independent (n=226, GSE14333) sets
| Characteristics | Training set (n= 246, GSE39582)
|
Internal validation set (n=246, GSE39582)
|
External independent set (n =226, GSE14333)
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| High-risk (n=61) |
Low-risk (n=185) |
P-value | High-risk (n=50) |
Low-risk (n=196) |
P-value | High-risk (n=41) |
Low-risk (n=185) |
P-value | |
| Age (years) | |||||||||
| <66 | 30 | 68 | 0.085 | 17 | 72 | 0.719 | 20 | 81 | 0.560 |
| ≥66 | 31 | 117 | 33 | 124 | 21 | 104 | |||
| Gender | |||||||||
| Male | 32 | 105 | 0.558 | 29 | 106 | 0.644 | 26 | 94 | 0.143 |
| Female | 29 | 80 | 21 | 89 | 15 | 91 | |||
| Stage | |||||||||
| I | 3 | 14 | 0.727 | 1 | 13 | 0.331 | 3 | 38 | 0.122 |
| II | 33 | 102 | 24 | 101 | 18 | 76 | |||
| III | 25 | 69 | 25 | 82 | 20 | 71 | |||
| Adjuvant chemotherapy | |||||||||
| Yes | 28 | 67 | 0.078 | 24 | 83 | 0.472 | 21 | 118 | 0.135 |
| No | 32 | 118 | 26 | 113 | 20 | 67 | |||
| Unknown | 1 | 0 | 0 | 0 | 0 | 0 | |||
Abbreviation: CRC, colorectal cancer.
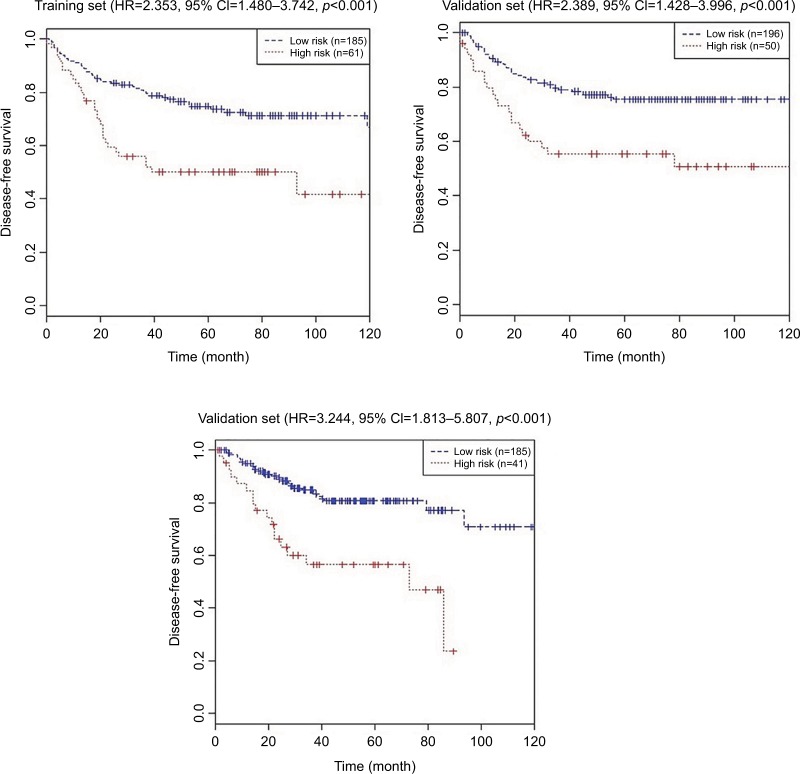
Figure 3 and Table 3 show the DFS differences between high-risk and low risk groups in these three sets. The log-rank test showed that CRC patients with low-risk scores had significantly longer DFS than those with high-risk scores in the training set (HR=2.353, 95% CI=1.480–3.742, P<0.001) and internal validation (HR=2.389, 95% CI=1.428–3.996, P<0.001), and external independent (HR=3.244, 95% CI=1.813–5.807, P<0.001) sets.
Figure 3.
Kaplan–Meier curves of disease-free survival according to the nine-chemokine classifier in the training (n=246, GSE39582), internal validation (n=246, GSE39582), and external independent (n=226, GSE14333) sets.
Table 3.
Log-rank test of disease-free survival according to the nine-chemokine classifier in the training (n=246, GSE39582), internal validation (n=246, GSE39582), and external independent (n=226, GSE14333) sets
| Datasets | Risk group (n) | Disease-free survival
|
||||
|---|---|---|---|---|---|---|
| 1-year | 3-year | 5-year | HR (95% CI) | P-value | ||
| Training set (n=246) | High-risk (n=61) | 91.0% | 55.9% | 50.1% | 2.353 (1.480–3.742) | <0.001 |
| Low-risk (n=185) | 92.6% | 80.2% | 74.8% | |||
| Validation set (n=246) | High-risk (n=50) | 77.5% | 55.4% | 55.4% | 2.389 (1.428–3.996) | <0.001 |
| Low-risk (n=196) | 90.4% | 79.0% | 75.5% | |||
| Independent set (n=226) | High-risk (n=41) | 84.7% | 56.5% | 56.5% | 3.244 (1.813–5.807) | <0.001 |
| Low-risk (n=185) | 95.0% | 84.9% | 80.7% | |||
Multivariate Cox regression analysis in the training, internal validation, and external independent sets
Table 4 shows the multivariate Cox regression analysis results of the nine-chemokine classifier, gender, age, stage, adjuvant chemotherapy, and DFS in the training, internal validation, and external independent sets. Both the nine-chemokine classifier and stages were significantly associated with CRC patients’ DFS in these datasets (all P<0.05).
Table 4.
Multivariate Cox regression analysis of the nine-chemokine classifier, gender, age, stage, adjuvant chemotherapy, and disease-free survival in the training (n=246, GSE39582), internal validation (n=246, GSE39582), and external independent (n=226, GSE14333) sets
| Datasets | Variable | Disease-free survival
|
|
|---|---|---|---|
| HR (95% CI) | P-value | ||
| Training set (n=246) | Nine-chemokine classifier (high- vs low-risk) | 2.107 (1.313–3.382) | 0.002 |
| Age (≥66 years vs <66 years) | 0.944 (0.567–1.571) | 0.824 | |
| Gender (female vs male) | 1.023 (0.644–1.626) | 0.923 | |
| Tumor stage (III vs II vs I) | 1.763 (1.083–2.869) | 0.023 | |
| Adjuvant chemotherapy (unknown/no vs yes) | 0.761 (1.425–1.362) | 0.358 | |
| Validation set (n=246) | Nine-chemokine classifier (high- vs low-risk) | 2.298 (1.370–3.855) | 0.002 |
| Age (≥66 years vs <66 years) | 1.181 (0.695–2.004) | 0.538 | |
| Gender (female vs male) | 0.624 (0.377–1.035) | 0.068 | |
| Tumor stage (III vs II vs I) | 2.001 (1.186–3.378) | 0.009 | |
| Adjuvant chemotherapy (unknown/no vs yes) | 1.296 (0.726–2.317) | 0.380 | |
| Independent set (n=226) | Nine-chemokine classifier (high- vs low-risk) | 2.914 (1.599–5.311) | <0.001 |
| Age (≥66 years vs <66 years) | 0.666 (0.374–1.186) | 0.167 | |
| Gender (female vs male) | 1.102 (0.621–1.954) | 0.740 | |
| Tumor stage (III vs II vs I) | 3.235 (1.845–5.673) | <0.001 | |
| Adjuvant chemotherapy (no vs yes) | 0.672 (0.350–1.290) | 0.232 | |
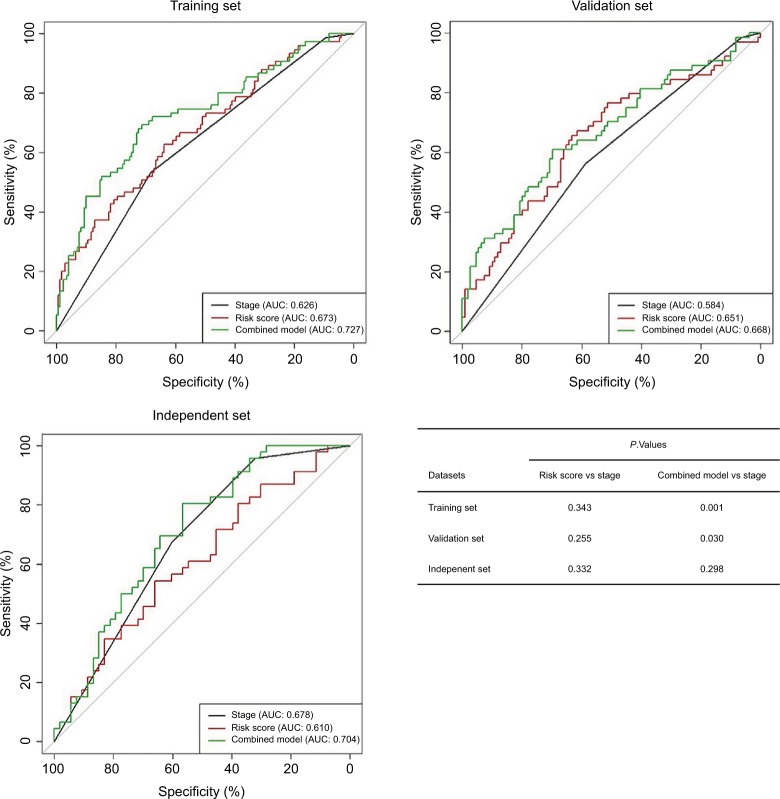
ROC analysis in the training, internal validation, and external independent sets
Figure 4 shows the ROC curves for predicting DFS in the training, internal validation, and external independent sets. AUC of the nine-chemokine classifier is similar with that of stage alone in the training (0.673 vs 0.626, P=0.343), internal validation (0.651 vs 0.584, P=0.332), and external independent (0.609 vs 0.678, P=0.888) sets. AUC of the combined model (nine-chemokine classifier and tumor stage) may tend to be higher than that of stage alone in the training (0.727 vs 0.626, P=0.001), internal validation (0.668 vs 0.584, P=0.030), and external independent (0.704 vs 0.678, P=0.298) sets.
Figure 4.
Receiver operating characteristics curves of the combined model of the nine-chemokine classifier and stage, the nine-chemokine classifier and stage alone for predicting disease-free survival in the training (n=246, GSE39582), internal validation (n=246, GSE39582), and external independent (n=226, GSE14333) sets.
Abbreviation: AUC, area under the curve.
Survival analysis between low-risk and high-risk groups in the combined training and validation set
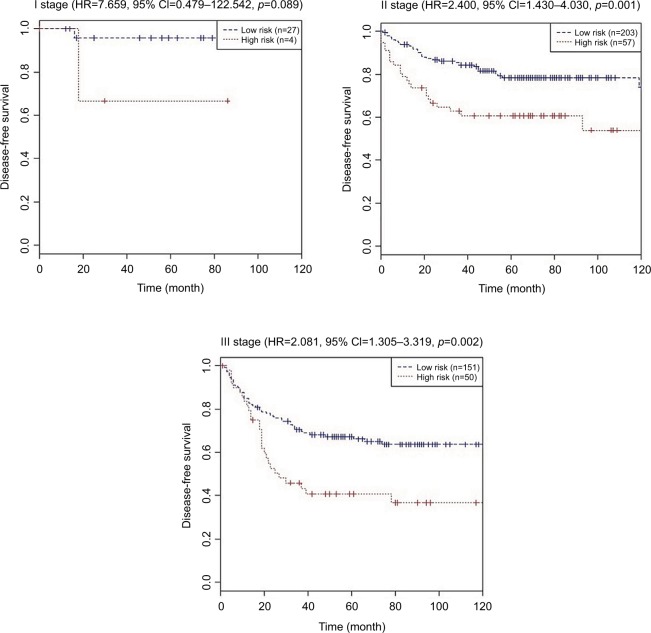
Using the same cutoff point (1.559), CCR patients were categorized into low-risk group (n=381) and high-risk group (n=111) in the combined training and validation set. CRC patients with low-risk scores had significantly longer DFS (HR=2.383, 95% CI=1.690–3.359, P<0.001) and OS (HR=1.603, 95% CI=1.115–2.305, P=0.010) than those with high-risk scores (shown in Table 5). Table 6 shows the multivariate Cox regression analysis results of the nine-chemokine classifier, gender, age, stage, adjuvant chemotherapy, and survival in the combined training and validation set (n=492, GSE39582). The multivariate Cox analysis showed that the nine-chemokine classifier was significantly associated with patients’ DFS (HR=2.292, 95% CI=1.622–3.239, P<0.001) and OS (HR=1.640, 95% CI=1.139–2.362, P=0.008; Table 6). Subgroup analysis showed that stage II–III patients with low-risk scores had significantly longer DFS (P<0.05) than those with high-risk scores (shown in Figure 5).
Table 5.
Comparison of the survival of colorectal cancer patients according to the nine-chemokine classifier in the combined training and validation set (n=492, GSE39582)
| Set | Risk group (n) | Disease-free survival
|
Overall survival
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1-year | 3-year | 5-year | HR (95% CI) | P-value | 1-year | 3-year | 5-year | HR (95% CI) | P-value | ||
| The combined set (n=492) | High-risk (n=111) | 79.8% | 55.6% | 52.3% | 2.383 | <0.001 | 93.6% | 77.1% | 63.5% | 1.603 | 0.010 |
| Low-risk (n=381) | 90.7% | 79.6% | 75.2% | (1.690–3.359) | 96.5% | 85.2% | 76.1% | (1.115–2.305) | |||
Table 6.
Multivariate Cox regression analysis of the nine-chemokine classifier, gender, age, stage, adjuvant chemotherapy, and survival in the combined training and validation set (n=492, GSE39582)
| Datasets | Variable | Disease-free survival
|
Overall survival
|
||
|---|---|---|---|---|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value | ||
| The combined set (n=492) | Nine-chemokine classifier (high- vs low-risk) | 2.292 (1.622–3.239) | <0.001 | 1.640 (1.139–2.362) | 0.008 |
| Age (≥66 years vs <66 years) | 1.019 (0.714–1.454) | 0.919 | 1.395 (0.967–2.031) | 0.075 | |
| Gender (female vs male) | 0.806 (0.573–1.132) | 0.213 | 0.768 (0.549–1.074) | 0.122 | |
| Tumor stage (III vs II vs I) | 1.867 (1.303–2.674) | 0.001 | 1.122 (0.804–1.565) | 0.497 | |
| Adjuvant chemotherapy (unknown/no vs yes) | 0.995 (0.662–1.495) | 0.980 | 1.198 (0.798–1.796) | 0.384 | |
Figure 5.
Kaplan–Meier curves of disease-free survival according to the nine-chemokine classifier for different stage patients with CRC in the combined training and validation set (n=492, GSE39582).
Abbreviation: CRC, colorectal cancer.
The prediction values of the nine-chemokine classifier for different biological subtypes in the combined training and validation set
To verify the value of the nine-chemokine classifier, we further validated our findings in different molecular subtypes of Laetitia Marisa in the combined training and validation set (n=460, GSE39582).
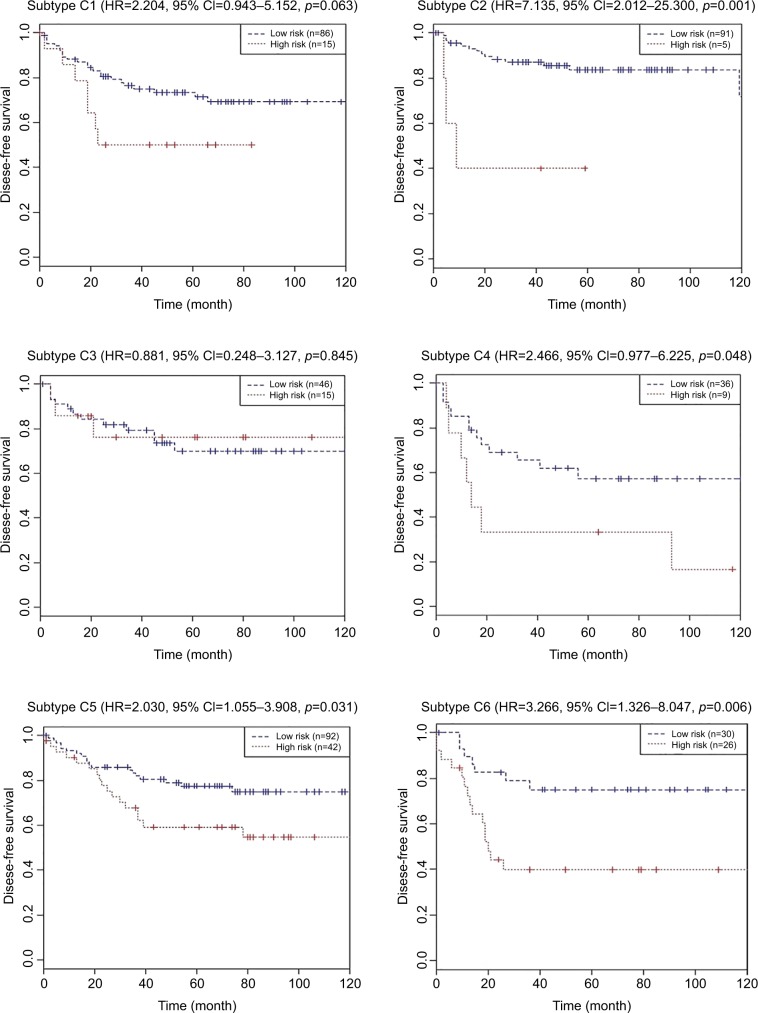
We further validated the values of the nine-chemokine classifier for different biological subtypes in the combined training and validation set (n=492, GSE39582). Patients with high-risk scores had similar DFS (all P>0.05) with those with low-risk scores in the subtypes C1 (one subtype with downregulated immune pathways) and C3 (one KRAS mutant subtype) (shown in Figure 6). Patients with high-risk scores had significantly shorter DFS (all P>0.05) than those with low-risk scores in the subtypes C2 (one deficient mismatch repair subtype), C4 (one cancer stem cell subtype), C5 (one subtype with the upregulated Wnt pathway), and C6 (one subtype with a normal-like gene expression profile) (shown in Figure 6). But the Cox regression analysis showed that the integrated lncRNA-mRNA classifier was not significantly associated with the subtypes C1, C3, and C4 patients’ DFS (all P≥0.05). The integrated lncRNA-mRNA classifier was an independent prognostic factor for the subtypes C2, C5, and C6 patients’ DFS (all P<0.05).
Figure 6.
Kaplan–Meier curves of disease-free survival according to the nine-chemokine classifier for different biological subtypes in the combined training and validation set (n=492, GSE39582).
Functional enrichment analysis
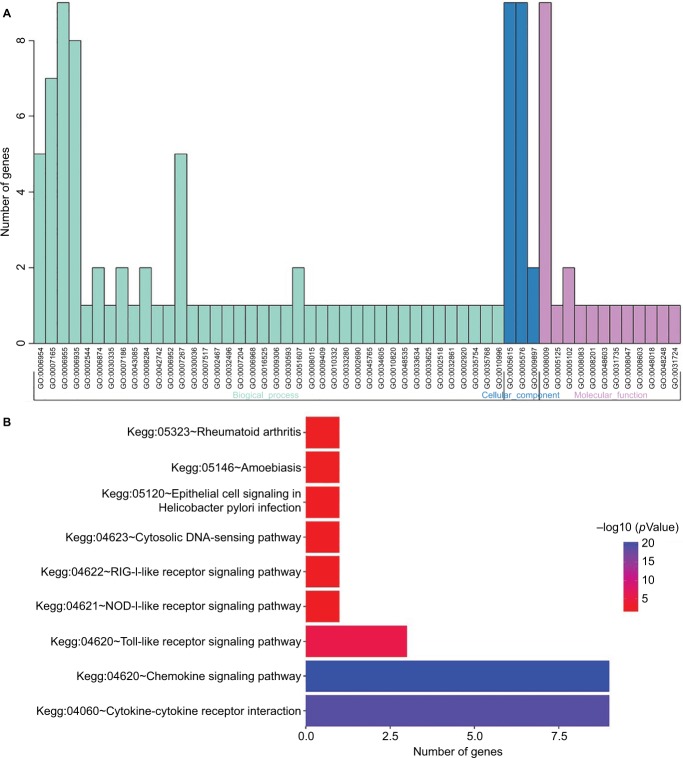
To explore the functional implication of nine chemokines, we performed functional category enrichment analysis. Functional enrichment analysis showed that the nine chemokine family genes were significantly enriched in 55 GO terms and 9 KEGG pathways (shown in Figure 7). The functional categories are mainly involved in eight GO terms, including immune response (GO:0006955), inflammatory response (GO:0006954), signal transduction (GO:0007165), chemotaxis (GO:0006935), cell–cell signaling (GO:0007267), extracellular space (GO:0005615), extracellular region (GO:0005576), and chemokine activity (GO:0008009). The mainly involved KEGG pathways included cytokine–cytokine receptor interaction (KEGG:04060), chemokine signaling pathway (KEGG:04062), and toll-like receptor signaling pathway (KEGG:04620).
Figure 7.
Results of functional enrichment analyses for GO (A) and KEGG (B) category.
Discussion
Although the TNM staging system is widely used as the risk stratification of CRC patients, it is insufficient in the prediction of prognosis and estimation for some patients.32,33 Conflict clinical outcomes may exist among some CRC patients with the same stage.31,32 To date, there are no clinically utilized prognostic biomarkers in CRC patients. An increasing amount of evidence demonstrates that chemokines and their receptors play an important role in tumor immunity, progression, and metastasis of CRC patients.7–20 The discovery and application of a multiple-chemokine biomarker will promote the evaluation and identification of potential high-risky recurrence in CRC patients. To identify the prognostic chemokines, we profiled chemokines by mining the existing microarray data of Affymetrix human genome U133 plus 2.0 array. We applied a univariate Cox regression analysis to select DFS-related chemokines. Based on this data-mining method, we have developed and validated a nine-chemokine classifier. The utility of this nine-chemokine classifier may add to the prognostic value of the TNM stage system. Furthermore, the clinical application of this nine-chemokine classifier might stratify CRC patients with the same stage into low-risk and high-risk groups of recurrence after operation. CRC patients of high-risk group had shorter DFS than those of low-risk group in stage II and III patients. The nine-chemokine classifier may provide an additional biomarker for identifying potential candidates for aggressive treatment strategies. The molecular subtype should be considered before the clinical application of the prognostic signatures. Subgroup showed that the nine-chemokine classifier was not an independent prognostic factor for DFS in Marisa’s C1, C3, and C4 subtypes patients. Therefore, the nine-chemokine classifier may only be applied in Marisa’s C2, C5, and C6 subtypes patients.
The nine-chemokine classifier included three risky genes (CCL1, CCL14, and CXCL14) and six protective genes (CXCL1, CXCL3, CXCL9, CXCL10, CXCL11, and CXCL13). The previous study showed that high tissue levels of CXCL14 was associated with increased risk of recurrence and mortality among CRC patients.34 However, high expressions of CXCL1,CXCL9, CXCL10, and CXCL13 may be correlated with better prognosis of CRC patients.23,35–37 Moreover, the prognostic values of these five chemokines were not further confirmed in another validation sets. The relationship of the remaining four chemokines (CCL1, CCL14, CXCL3, CXCL11) and CRC patients’ prognosis should be further studied.
Although our study possessed the larger sample size for developing and validating this nine-chemokine classifier, we should acknowledge certain potential limitations. First, preliminary functional enrichment analysis indicated that the nine-chemokine classifier was mainly involved in immune response, inflammatory response, signal transduction, chemotaxis, and cell–cell signaling. But the mechanism of these nine chemokines has not been confirmed through experimental studies. Further experimental studies may provide potential therapeutic targets for CRC patients. Second, Affymetrix human genome U133 plus 2.0 array was used for obtaining the mRNA level data of chemokine family genes in this study. But the mRNA level data of some chemokines (CCL6, CCL12, CXCL7, and CXCL15) were not available in this microarray platform. The link between the mRNA levels of these chemokines and survival of CRC patients should be further investigated by experimental studies. Third, this multi-RNA classifier was only developed and validated by the mRNA-level data. The value of this multi-RNA classifier was not further confirmed by the protein-level data. Moreover, future analysis of protein level on additional independent datasets would contribute to determine the potential importance of population and geographical differences.
In conclusion, we performed a comprehensive analysis of chemokine expression levels and corresponding survival information of CRC patients. We have successfully developed and validated a nine-chemokine classifier that may be a useful prognostic biomarker for the personalized treatment. It was the first study demonstrating a link between a multiple-chemokine classifier and tumor recurrence in CRC patients. This nine-chemokine classifier may provide an effective risk stratification of disease-free survival in CRC patients with the same stage, especially for stage II–III patients. But we should acknowledge that the protein-level data of this nine-chemokine classifier should be further validated before its clinical application.
Supplementary material
Table S1.
Univariate Cox regression analysis of chemokine family genes associated with disease-free survival in the training set (n=246, GSE39582)
| Gene symbol | Type | Sub-family | HR (95% CI) | Coefficient | P-value |
|---|---|---|---|---|---|
| CCL1 | Ligand | CC | 2.729 (1.118–6.662) | 1.004 | 0.027 |
| CCL2 | Ligand | CC | 1.169 (0.970–1.410) | 0.156 | 0.102 |
| CCL3 | Ligand | CC | 1.004 (0.844–1.194) | 0.004 | 0.962 |
| CCL4 | Ligand | CC | 1.019 (0.823–1.261) | 0.019 | 0.863 |
| CCL5 | Ligand | CC | 0.918 (0.757–1.114) | −0.085 | 0.387 |
| CCL7 | Ligand | CC | 1.118 (0.728–1.719) | 0.112 | 0.610 |
| CCL8 | Ligand | CC | 0.993 (0.830–1.190) | −0.007 | 0.943 |
| CCL11 | Ligand | CC | 1.019 (0.868–1.196) | 0.018 | 0.822 |
| CCL13 | Ligand | CC | 1.152 (0.743–1.784) | 0.141 | 0.527 |
| CCL14 | Ligand | CC | 1.400 (1.043–1.879) | 0.336 | 0.025 |
| CCL15 | Ligand | CC | 1.051 (0.823–1.344) | 0.050 | 0.688 |
| CCL16 | Ligand | CC | 1.556 (0.916–2.643) | 0.442 | 0.102 |
| CCL17 | Ligand | CC | 1.240 (0.699–2.203) | 0.215 | 0.462 |
| CCL18 | Ligand | CC | 0.987 (0.853–1.142) | −0.013 | 0.861 |
| CCL19 | Ligand | CC | 1.063 (0.902–1.252) | 0.061 | 0.467 |
| CCL20 | Ligand | CC | 0.885 (0.771–1.015) | −0.122 | 0.082 |
| CCL21 | Ligand | CC | 1.124 (0.924–1.366) | 0.117 | 0.242 |
| CCL22 | Ligand | CC | 0.877 (0.538–1.430) | −0.131 | 0.600 |
| CCL23 | Ligand | CC | 1.211 (0.701–2.090) | 0.191 | 0.492 |
| CCL24 | Ligand | CC | 1.132 (0.919–1.395) | 0.124 | 0.243 |
| CCL25 | Ligand | CC | 0.739 (0.466–1.172) | −0.302 | 0.199 |
| CCL26 | Ligand | CC | 0.938 (0.705–1.250) | −0.064 | 0.664 |
| CCL27 | Ligand | CC | 1.507 (0.673–3.373) | 0.410 | 0.319 |
| CCL28 | Ligand | CC | 1.058 (0.821–1.363) | 0.056 | 0.664 |
| CXCL1 | Ligand | CXC | 0.824 (0.697–0.974) | −0.194 | 0.024 |
| CXCL2 | Ligand | CXC | 0.870 (0.608–1.243) | −0.140 | 0.444 |
| CXCL3 | Ligand | CXC | 0.836 (0.708–0.986) | −0.179 | 0.034 |
| CXCL5 | Ligand | CXC | 0.939 (0.824–1.071) | −0.062 | 0.349 |
| CXCL6 | Ligand | CXC | 0.951 (0.810–1.115) | −0.051 | 0.534 |
| CXCL8 | Ligand | CXC | 0.933 (0.810–1.073) | −0.070 | 0.329 |
| CXCL9 | Ligand | CXC | 0.840 (0.722–0.977) | −0.174 | 0.024 |
| CXCL10 | Ligand | CXC | 0.832 (0.712–0.972) | −0.184 | 0.021 |
| CXCL11 | Ligand | CXC | 0.878 (0.774–0.995) | −0.131 | 0.041 |
| CXCL12 | Ligand | CXC | 1.107 (0.907–1.352) | 0.102 | 0.319 |
| CXCL13 | Ligand | CXC | 0.870 (0.763–0.992) | −0.140 | 0.037 |
| CXCL14 | Ligand | CXC | 1.146 (1.008–1.302) | 0.136 | 0.037 |
| CXCL16 | Ligand | CXC | 0.901 (0.650–1.249) | −0.104 | 0.531 |
| CXCL17 | Ligand | CXC | 0.814 (0.525–1.262) | −0.206 | 0.357 |
| XCL1 | Ligand | XC | 0.923 (0.524–1.627) | −0.080 | 0.782 |
| XCL2 | Ligand | XC | 1.004 (0.744–1.355) | 0.004 | 0.978 |
| CX3CL1 | Ligand | CX3C | 0.896 (0.613–1.312) | −0.109 | 0.574 |
| CCR1 | Receptor | CC | 1.079 (0.786–1.481) | 0.076 | 0.638 |
| CCR2 | Receptor | CC | 0.965 (0.632–1.475) | −0.035 | 0.870 |
| CCR3 | Receptor | CC | 1.001 (0.553–1.815) | 0.001 | 0.996 |
| CCR4 | Receptor | CC | 1.356 (0.600–3.062) | 0.304 | 0.464 |
| CCR5 | Receptor | CC | 0.861 (0.595–1.244) | −0.15 | 0.425 |
| CCR6 | Receptor | CC | 0.937 (0.622–1.411) | −0.065 | 0.756 |
| CCR7 | Receptor | CC | 0.880 (0.667–1.161) | −0.128 | 0.364 |
| CCR8 | Receptor | CC | 1.127 (0.429–2.960) | 0.119 | 0.808 |
| CCR9 | Receptor | CC | 1.190 (0.391–3.620) | 0.174 | 0.759 |
| CCR10 | Receptor | CXC | 1.239 (0.880–1.745) | 0.214 | 0.220 |
| CXCR1 | Receptor | CXC | 0.511 (0.211–1.240) | −0.671 | 0.138 |
| CXCR2 | Receptor | CXC | 1.005 (0.746–1.355) | 0.005 | 0.973 |
| CXCR3 | Receptor | CXC | 0.732 (0.373–1.438) | −0.312 | 0.365 |
| CXCR4 | Receptor | CXC | 0.979 (0.793–1.208) | −0.021 | 0.842 |
| CXCR5 | Receptor | CXC | 0.966 (0.412–2.267) | −0.034 | 0.937 |
| CXCR6 | Receptor | CXC | 0.803 (0.490–1.317) | −0.219 | 0.385 |
| XCR1 | Receptor | XC | 1.648 (0.515–5.274) | 0.499 | 0.400 |
| CX3CR1 | Receptor | CX3C | 1.569 (0.864–2.849) | 0.45 | 0.139 |
Acknowledgments
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations
- AUC
area under the curve
- CRC
colorectal cancer
- DFS
disease-free survival
- GEO
Gene Expression Omnibus
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes OS, overall survival
- ROC
receiver operating characteristics
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Alizadeh AA, Aranda V, Bardelli A, et al. Toward understanding and exploiting tumor heterogeneity. Nat Med. 2015;21(8):846–853. doi: 10.1038/nm.3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raman D, Sobolik-Delmaire T, Richmond A. Chemokines in health and disease. Exp Cell Res. 2011;317(5):575–589. doi: 10.1016/j.yexcr.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zlotnik A, Yoshie O. Chemokines: a new classification system and their role in immunity. Immunity. 2000;12(2):121–127. doi: 10.1016/s1074-7613(00)80165-x. [DOI] [PubMed] [Google Scholar]
- 5.Murphy PM, Baggiolini M, Charo IF, et al. International union of pharmacology. XXII. Nomenclature for chemokine receptors. Pharmacol Rev. 2000;52(1):145176. [PubMed] [Google Scholar]
- 6.Sarvaiya PJ, Guo D, Ulasov I, Gabikian P, Lesniak MS. Chemokines in tumor progression and metastasis. Oncotarget. 2013;4(12):2171–2185. doi: 10.18632/oncotarget.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamamoto M, Kikuchi H, Ohta M, et al. TSU68 prevents liver metastasis of colon cancer xenografts by modulating the premetastatic niche. Cancer Res. 2008;68(23):9754–9762. doi: 10.1158/0008-5472.CAN-08-1748. [DOI] [PubMed] [Google Scholar]
- 8.Kawada K, Hosogi H, Sonoshita M, et al. Chemokine receptor CXCR3 promotes colon cancer metastasis to lymph nodes. Oncogene. 2007;26(32):4679–4688. doi: 10.1038/sj.onc.1210267. [DOI] [PubMed] [Google Scholar]
- 9.Murakami T, Kawada K, Iwamoto M, et al. The role of CXCR3 and CXCR4 in colorectal cancer metastasis. Int J Cancer. 2013;132(2):276–287. doi: 10.1002/ijc.27670. [DOI] [PubMed] [Google Scholar]
- 10.Kim J, Takeuchi H, Lam ST, et al. Chemokine receptor CXCR4 expression in colorectal cancer patients increases the risk for recurrence and for poor survival. J Clin Oncol. 2005;23(12):2744–2753. doi: 10.1200/JCO.2005.07.078. [DOI] [PubMed] [Google Scholar]
- 11.Ottaiano A, Franco R, Aiello Talamanca A, et al. Overexpression of both CXC chemokine receptor 4 and vascular endothelial growth factor proteins predicts early distant relapse in stage II-III colorectal cancer patients. Clin Cancer Res. 2006;12(9):2795–2803. doi: 10.1158/1078-0432.CCR-05-2142. [DOI] [PubMed] [Google Scholar]
- 12.Schimanski CC, Schwald S, Simiantonaki N, et al. Effect of chemokine receptors CXCR4 and CCR7 on the metastatic behavior of human colorectal cancer. Clin Cancer Res. 2005;11(5):1743–1750. doi: 10.1158/1078-0432.CCR-04-1195. [DOI] [PubMed] [Google Scholar]
- 13.Yopp AC, Shia J, Butte JM, et al. CXCR4 expression predicts patient outcome and recurrence patterns after hepatic resection for colorectal liver metastases. Ann Surg Oncol. 2012;19(Suppl 3):339–346. doi: 10.1245/s10434-011-1774-4. [DOI] [PubMed] [Google Scholar]
- 14.Matsusue R, Kubo H, Hisamori S, et al. Hepatic stellate cells promote liver metastasis of colon cancer cells by the action of SDF-1/CXCR4 axis. Ann Surg Oncol. 2009;16(9):2645–2653. doi: 10.1245/s10434-009-0599-x. [DOI] [PubMed] [Google Scholar]
- 15.Bailey C, Negus R, Morris A, et al. Chemokine expression is associated with the accumulation of tumour associated macrophages (TAMs) and progression in human colorectal cancer. Clin Exp Metastasis. 2007;24(2):121–130. doi: 10.1007/s10585-007-9060-3. [DOI] [PubMed] [Google Scholar]
- 16.Hu H, Sun L, Guo C, et al. Tumor cell-microenvironment interaction models coupled with clinical validation reveal CCL2 and SNCG as two predictors of colorectal cancer hepatic metastasis. Clin Cancer Res. 2009;15(17):5485–5493. doi: 10.1158/1078-0432.CCR-08-2491. [DOI] [PubMed] [Google Scholar]
- 17.Chang LY, Lin YC, Mahalingam J, et al. Tumor-derived chemokine CCL5 enhances TGF-β-mediated killing of CD8(+) T cells in colon cancer by T-regulatory cells. Cancer Res. 2012;72(5):1092–1102. doi: 10.1158/0008-5472.CAN-11-2493. [DOI] [PubMed] [Google Scholar]
- 18.Itatani Y, Kawada K, Fujishita T, et al. Loss of SMAD4 from colorectal cancer cells promotes CCL15 expression to recruit CCR1+ myeloid cells and facilitate liver metastasis. Gastroenterology. 2013;145(5):1064–1075. doi: 10.1053/j.gastro.2013.07.033. [DOI] [PubMed] [Google Scholar]
- 19.Liu J, Zhang N, Li Q, et al. Tumor-associated macrophages recruit CCR6+ regulatory T cells and promote the development of colorectal cancer via enhancing CCL20 production in mice. PLoS One. 2011;6(4):e19495. doi: 10.1371/journal.pone.0019495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng J, Yang M, Shao J, et al. Chemokine receptor CX3CR1 contributes to macrophage survival in tumor metastasis. Mol Cancer. 2013;12(1):141. doi: 10.1186/1476-4598-12-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeng YJ, Lai W, Wu H, et al. Neuroendocrine-like cells -derived CXCL10 and CXCL11 induce the infiltration of tumor-associated macrophage leading to the poor prognosis of colorectal cancer. Oncotarget. 2016;7(19):27394–27407. doi: 10.18632/oncotarget.8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stanisavljević L, Aßmus J, Storli KE, et al. CXCR4, CXCL12 and the relative CXCL12-CXCR4 expression as prognostic factors in colon cancer. Tumour Biol. 2016;37(6):7441–7452. doi: 10.1007/s13277-015-4591-8. [DOI] [PubMed] [Google Scholar]
- 23.Jiang Z, Xu Y, Cai S. CXCL10 expression and prognostic significance in stage II and III colorectal cancer. Mol Biol Rep. 2010;37(6):3029–3036. doi: 10.1007/s11033-009-9873-z. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Szustakowski J, Schinke M. Bioinformatics analysis of micro-array data. Methods Mol Biol. 2009;573:259–284. doi: 10.1007/978-1-60761-247-6_15. [DOI] [PubMed] [Google Scholar]
- 25.Marisa L, de Reyniès A, Duval A, et al. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med. 2013;10(5):e1001453. doi: 10.1371/journal.pmed.1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lossos IS, Czerwinski DK, Alizadeh AA, et al. Prediction of survival in diffuse large-B-cell lymphoma based on the expression of six genes. N Engl J Med. 2004;350(18):1828–1837. doi: 10.1056/NEJMoa032520. [DOI] [PubMed] [Google Scholar]
- 27.Hu Z, Chen X, Zhao Y, et al. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J Clin Oncol. 2010;28(10):1721–1726. doi: 10.1200/JCO.2009.24.9342. [DOI] [PubMed] [Google Scholar]
- 28.Kang J, D’Andrea AD, Kozono D. A DNA repair pathway-focused score for prediction of outcomes in ovarian cancer treated with platinum-based chemotherapy. J Natl Cancer Inst. 2012;104(9):670–681. doi: 10.1093/jnci/djs177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tabas-Madrid D, Nogales-Cadenas R, Pascual-Montano A. Gene-Codis3: a non-redundant and modular enrichment analysis tool for functional genomics. Nucleic Acids Res. 2012;40(Web Server issue):W478–W483. doi: 10.1093/nar/gks402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nogales-Cadenas R, Carmona-Saez P, Vazquez M, et al. GeneCodis: interpreting gene lists through enrichment analysis and integration of diverse biological information. Nucleic Acids Res. 2009;37(Web Server issue):W317–W322. doi: 10.1093/nar/gkp416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carmona-Saez P, Chagoyen M, Tirado F, Carazo JM, Pascual-Montano A. GENECODIS: a web-based tool for finding significant concurrent annotations in gene lists. Genome Biol. 2007;8(1):R3. doi: 10.1186/gb-2007-8-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zinicola R, Pedrazzi G, Haboubi N, Nicholls RJ. The degree of extramural spread of T3 rectal cancer: a plea to the UICC and AJCC. Colorectal Dis. 2017;19(3):310. doi: 10.1111/codi.13456. [DOI] [PubMed] [Google Scholar]
- 33.Li J, Yang S, Hu J, et al. Tumor deposits counted as positive lymph nodes in TNM staging for advanced colorectal cancer: a retrospective multicenter study. Oncotarget. 2016;7(14):18269–18279. doi: 10.18632/oncotarget.7756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeng J, Yang X, Cheng L, et al. Chemokine CXCL14 is associated with prognosis in patients with colorectal carcinoma after curative resection. J Transl Med. 2013;11:6. doi: 10.1186/1479-5876-11-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Le Rolle AF, Chiu TK, Fara M, et al. The prognostic significance of CXCL1 hypersecretion by human colorectal cancer epithelia and myofibroblasts. J Transl Med. 2015;13:199. doi: 10.1186/s12967-015-0555-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu Z, Huang X, Han X, et al. The chemokine CXCL9 expression is associated with better prognosis for colorectal carcinoma patients. Biomed Pharmacother. 2016;78:8–13. doi: 10.1016/j.biopha.2015.12.021. [DOI] [PubMed] [Google Scholar]
- 37.Agesen TH, Sveen A, Merok MA, et al. ColoGuideEx: a robust gene classifier specific for stage II colorectal cancer prognosis. Gut. 2012;61(11):1560–1567. doi: 10.1136/gutjnl-2011-301179. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
Univariate Cox regression analysis of chemokine family genes associated with disease-free survival in the training set (n=246, GSE39582)
| Gene symbol | Type | Sub-family | HR (95% CI) | Coefficient | P-value |
|---|---|---|---|---|---|
| CCL1 | Ligand | CC | 2.729 (1.118–6.662) | 1.004 | 0.027 |
| CCL2 | Ligand | CC | 1.169 (0.970–1.410) | 0.156 | 0.102 |
| CCL3 | Ligand | CC | 1.004 (0.844–1.194) | 0.004 | 0.962 |
| CCL4 | Ligand | CC | 1.019 (0.823–1.261) | 0.019 | 0.863 |
| CCL5 | Ligand | CC | 0.918 (0.757–1.114) | −0.085 | 0.387 |
| CCL7 | Ligand | CC | 1.118 (0.728–1.719) | 0.112 | 0.610 |
| CCL8 | Ligand | CC | 0.993 (0.830–1.190) | −0.007 | 0.943 |
| CCL11 | Ligand | CC | 1.019 (0.868–1.196) | 0.018 | 0.822 |
| CCL13 | Ligand | CC | 1.152 (0.743–1.784) | 0.141 | 0.527 |
| CCL14 | Ligand | CC | 1.400 (1.043–1.879) | 0.336 | 0.025 |
| CCL15 | Ligand | CC | 1.051 (0.823–1.344) | 0.050 | 0.688 |
| CCL16 | Ligand | CC | 1.556 (0.916–2.643) | 0.442 | 0.102 |
| CCL17 | Ligand | CC | 1.240 (0.699–2.203) | 0.215 | 0.462 |
| CCL18 | Ligand | CC | 0.987 (0.853–1.142) | −0.013 | 0.861 |
| CCL19 | Ligand | CC | 1.063 (0.902–1.252) | 0.061 | 0.467 |
| CCL20 | Ligand | CC | 0.885 (0.771–1.015) | −0.122 | 0.082 |
| CCL21 | Ligand | CC | 1.124 (0.924–1.366) | 0.117 | 0.242 |
| CCL22 | Ligand | CC | 0.877 (0.538–1.430) | −0.131 | 0.600 |
| CCL23 | Ligand | CC | 1.211 (0.701–2.090) | 0.191 | 0.492 |
| CCL24 | Ligand | CC | 1.132 (0.919–1.395) | 0.124 | 0.243 |
| CCL25 | Ligand | CC | 0.739 (0.466–1.172) | −0.302 | 0.199 |
| CCL26 | Ligand | CC | 0.938 (0.705–1.250) | −0.064 | 0.664 |
| CCL27 | Ligand | CC | 1.507 (0.673–3.373) | 0.410 | 0.319 |
| CCL28 | Ligand | CC | 1.058 (0.821–1.363) | 0.056 | 0.664 |
| CXCL1 | Ligand | CXC | 0.824 (0.697–0.974) | −0.194 | 0.024 |
| CXCL2 | Ligand | CXC | 0.870 (0.608–1.243) | −0.140 | 0.444 |
| CXCL3 | Ligand | CXC | 0.836 (0.708–0.986) | −0.179 | 0.034 |
| CXCL5 | Ligand | CXC | 0.939 (0.824–1.071) | −0.062 | 0.349 |
| CXCL6 | Ligand | CXC | 0.951 (0.810–1.115) | −0.051 | 0.534 |
| CXCL8 | Ligand | CXC | 0.933 (0.810–1.073) | −0.070 | 0.329 |
| CXCL9 | Ligand | CXC | 0.840 (0.722–0.977) | −0.174 | 0.024 |
| CXCL10 | Ligand | CXC | 0.832 (0.712–0.972) | −0.184 | 0.021 |
| CXCL11 | Ligand | CXC | 0.878 (0.774–0.995) | −0.131 | 0.041 |
| CXCL12 | Ligand | CXC | 1.107 (0.907–1.352) | 0.102 | 0.319 |
| CXCL13 | Ligand | CXC | 0.870 (0.763–0.992) | −0.140 | 0.037 |
| CXCL14 | Ligand | CXC | 1.146 (1.008–1.302) | 0.136 | 0.037 |
| CXCL16 | Ligand | CXC | 0.901 (0.650–1.249) | −0.104 | 0.531 |
| CXCL17 | Ligand | CXC | 0.814 (0.525–1.262) | −0.206 | 0.357 |
| XCL1 | Ligand | XC | 0.923 (0.524–1.627) | −0.080 | 0.782 |
| XCL2 | Ligand | XC | 1.004 (0.744–1.355) | 0.004 | 0.978 |
| CX3CL1 | Ligand | CX3C | 0.896 (0.613–1.312) | −0.109 | 0.574 |
| CCR1 | Receptor | CC | 1.079 (0.786–1.481) | 0.076 | 0.638 |
| CCR2 | Receptor | CC | 0.965 (0.632–1.475) | −0.035 | 0.870 |
| CCR3 | Receptor | CC | 1.001 (0.553–1.815) | 0.001 | 0.996 |
| CCR4 | Receptor | CC | 1.356 (0.600–3.062) | 0.304 | 0.464 |
| CCR5 | Receptor | CC | 0.861 (0.595–1.244) | −0.15 | 0.425 |
| CCR6 | Receptor | CC | 0.937 (0.622–1.411) | −0.065 | 0.756 |
| CCR7 | Receptor | CC | 0.880 (0.667–1.161) | −0.128 | 0.364 |
| CCR8 | Receptor | CC | 1.127 (0.429–2.960) | 0.119 | 0.808 |
| CCR9 | Receptor | CC | 1.190 (0.391–3.620) | 0.174 | 0.759 |
| CCR10 | Receptor | CXC | 1.239 (0.880–1.745) | 0.214 | 0.220 |
| CXCR1 | Receptor | CXC | 0.511 (0.211–1.240) | −0.671 | 0.138 |
| CXCR2 | Receptor | CXC | 1.005 (0.746–1.355) | 0.005 | 0.973 |
| CXCR3 | Receptor | CXC | 0.732 (0.373–1.438) | −0.312 | 0.365 |
| CXCR4 | Receptor | CXC | 0.979 (0.793–1.208) | −0.021 | 0.842 |
| CXCR5 | Receptor | CXC | 0.966 (0.412–2.267) | −0.034 | 0.937 |
| CXCR6 | Receptor | CXC | 0.803 (0.490–1.317) | −0.219 | 0.385 |
| XCR1 | Receptor | XC | 1.648 (0.515–5.274) | 0.499 | 0.400 |
| CX3CR1 | Receptor | CX3C | 1.569 (0.864–2.849) | 0.45 | 0.139 |