Abstract
Nuclear magnetic resonance (NMR)-based metabolomic approach is a high-throughput fingerprinting technique that allows a rapid snapshot of metabolites without any prior knowledge of the organism. To demonstrate the applicability of NMR-based metabolomics in the field of microalgal-based bioremediation, novel freshwater microalga Scenedesmus sp. IITRIND2 that showed hypertolerance to As(III, V) was chosen for evaluating the metabolic perturbations during arsenic stress in both its oxidation states As(III) and As(V). Using NMR spectroscopy, we were able to identify and quantify an array of ∼45 metabolites, including amino acids, sugars, organic acids, phosphagens, osmolytes, nucleotides, etc. The NMR metabolomic experiments were complemented with various biophysical techniques to establish that the microalga tolerated the arsenic stress using a complex interplay of metabolites. The two different arsenic states distinctly influenced the microalgal cellular mechanisms due to their altered physicochemical properties. Eighteen differentially identified metabolites related to bioremediation of arsenic were then correlated to the major metabolic pathways to delineate the variable stress responses of microalga in the presence of As(III, V).
1. Introduction
Metabolomics is the end point of omics cascade that represents an array of metabolites, including amino acids, carbohydrates, organic acids, nucleotides, etc.1 Metabolic profiling can be performed using nuclear magnetic resonance (NMR) spectroscopy and/or with setup of gas/liquid chromatography/capillary electrophoresis (GC/LC/CE) coupled with mass spectrometry (MS) (examples: GC–MS, LC–MS, CE–MS, etc.).2,3 The key parameters required for developing a robust metabolomics platform include reproducibility, easy and rapid quantification, and identification of large number of metabolites with minimal sample preparation steps.4 NMR spectroscopy metabolomic approach is a nondestructive and nondiscriminating technique, thus making it an ideal tool for metabolomic profiling for any chosen organism.1,5 Given the above advantages of 1H NMR metabolomics, it has been widely applied to diverse fields, including understanding of drug metabolism, disease progression, biomarker discovery, nutritional research, effects of xenobiotics on plants, photochemistry, food adulterations, etc.4,6−8
Among the above mentioned applications, environmental metabolomics is an effective tool for analyzing the changes in complex biochemical mechanisms/pathways against the stress-generating agents, such as toxic chemicals, heavy metals, and extreme pH/temperature conditions.3,9 Rapid industrialization and urbanization has led to escalation in levels of heavy metals in aquatic ecosystems, thus posing a greater threat to plant, animal, and human life.10,11 Among the heavy metals, arsenic (As) has been reported to cause high incidence of arsenicosis in more than 20 countries across the globe, thereby listing it as a category 1 and class A carcinogen by the U.S. Environmental Protection Agency.11,12 High levels of arsenic in potable water sources have been reported in various countries, including southwest Finland (17–980 mg/L), western United States (1–48 000 mg/L), and Inner Mongolia, China (1354 mg/L).13 Arsenic has a complex physicochemistry as it exists in two interchangeable forms, anoxic trivalent As(III) and oxic pentavalent As(V) in the aquatic ecosystems.14
The conventional techniques deployed for the removal of arsenic from contaminated water bodies are biased toward one form of arsenic species, pH dependent, and require high maintenance and expensive mineral adsorbents making the overall process costly and less efficient.15,16 In this regard, microalgae have emerged as budding vectors for green mitigation of arsenic(III, V) from contaminated water sources owing to their (a) inexpensive and copious availability and (b) high surface-to-volume ratio providing large contact area for the metal binding, thus increasing the binding, efficacy, as well as the removal.17 Previous studies have demonstrated the effectiveness of both marine and freshwater microalgae as phycoremediators of arsenic.14,17−22 Recently, we reported that an oleaginous microalga Scenedesmus sp. IITRIND2 was able to efficiently tolerate half-a-gram (500 mg/L) of both As(III, V) along with astonishing removal efficiency. The microalga adapted to such high arsenic levels by altering their biochemical composition, as evidenced by the observed changes in protein, carbohydrate, lipid content, and photosynthetic pigments.22
In the current investigation, 1H NMR-based metabolomic approach coupled with various biophysical techniques was used to unravel the differential metabolic profiles and morphological features of Scenedesmus sp. IITRIND2 in the presence of As(III) and As(V). Our initial goal is to identify the metabolites associated with arsenic stress and to categorize them according to biological functions of microalga. Considering the difficulties in the metabolite extraction of algal species due to their firm cell wall structure, till date only 10–12 metabolites have been reported using NMR-based approach upon heavy-metal stress (cadmium, copper, and lead) in microalgae.9,23,24 Using our improvised metabolite extraction protocol for microalgae, we were able to extract and identify a total of ∼45 metabolites. Furthermore, the current investigation unveiled 18 differential metabolites that are characteristic to mitigation and alteration of hosts signaling pathways upon uptake of As(III) and As(V). It is worth noting that no such comprehensive NMR metabolic profiling was available on As(III, V) or other heavy-metal bioremediation by microalgae.
2. Results
Arsenic(III, V) is a toxic heavy metalloid whose uptake by the microalga induces stress, influencing most of its morphological, physicochemical, and biochemical characteristics. To investigate the maximum arsenic tolerance and bioremediation efficiency of Scenedesmus sp. IITRIND2, the microalga strain was cultivated at different concentrations of As(III, V), ranging from 10 to 1000 mg/L (Figure S1). Scenedesmus sp. IITRIND2 was able to tolerate up to 500 mg/L of both As(III) and As(V) with an half-maximal inhibitory concentration (IC50) value of ∼779 and 622 mg/L (Figure S1A). The microalga showed ∼98% removal of both the arsenic forms at initial metal metal concentration of 10 mg/L, which systematically reduced to ∼72% once the initial metal concentration in the growth medium was increased to 500 mg/L (Figure S1B). Further, to understand the holistic effects of arsenic on the morphological features and metabolome of microalga (Scenedesmus sp. IITRIND2), we deployed 1H NMR-based metabolomics coupled with various biophysical methods.
2.1. Interaction between As(III, V) and Microalgal Cell Surface
Arsenic in aquatic ecosystems exists mainly in two forms, anoxic trivalent As(III) and oxic pentavalent As(V).14 Both arsenic forms can further exist in different anionic and cationic species depending on the pH.22 In the present study, the pH of the growth media ranged between pH 7 and 8, causing As(V) to exist in HAsO4–2 and HsAsO4– whereas As(III) majorly (>90%) exists in its neutral form H3AsO3.22 The green microalgae cell wall possesses distinct functional groups (O-, N-, S-, and P-) that help in the binding of heavy metals on to their cell surface.25 Heavy metals are also known to bind with microalgal cell wall via phytochelatins (PCHs), which aid in detoxification of heavy metal.26 Fourier transform infrared (FT-IR) spectroscopy was performed to identify the functional groups involved in the biosorption of arsenic onto the microalgal cell surface (Figure 1). The FT-IR spectra of Scenedesmus sp. IITRIND2 loaded with As(III, V) indicated shifting of various peaks with respect to control.
Figure 1.
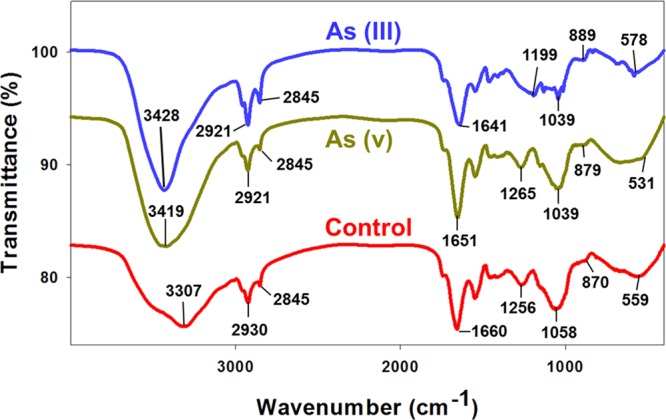
FT-IR profiles of Scenedesmus sp. IITRIND2 cultivated in the absence and presence of 500 mg/L As(III) and As(V).
On spiking with As(III), the O–H and N–H stretching at 3307 cm–1 shifted to a higher wavenumber (3428 cm–1), indicating an alteration in the H bonding. A decrease in the shift at 2930 cm–1 in control to 2921 cm–1 indicated interaction of As(III) with aliphatic C–H and aldehyde C–H stretching. Our study is in line with earlier reported studies on binding of As(III) with Ulothrix cylindrium and Chlorella pyrenoidosa.12,27 The absorption peak at 1660 cm–1 in control algal biomass shifted to 1641 cm–1 in As(III), corroborating H binding as an interaction bonding. A shift in the peak from 1058 cm–1 in nonspiked biomass to 1039 cm–1 in arsenic-spiked biomass attributing to the C–N stretching vibrations of amino groups indicates interaction between nitrogen of amino group with arsenic (Figure 1). Indeed, significant similarities existed between the spectra of algal biomass loaded with As(III) and As(V) in the prominent peaks at 2921 and 1039 cm–1, respectively. However, two notable differences existed between the FT-IR spectra of As(V); an increase in the peak at 1256 cm–1 in control to 1265 cm–1, indicating bonding of sulfide linkages, and a decrease in the peak at 559 cm–1 in control to 531 cm–1 signifying involvement of aromatic amino acids for the biosorption of As(V) (Figure 1). Such a differential characteristic peak(s) shifting of amide and sulfide groups observed in As(III)- and As(V)-spiked microalgal biomass with respect to control indicates a distinct ion-exchange interaction between the algal surface with As(III) and As(V).
2.2. Effect of Arsenic on Morphology and Cell Surface of Scenedesmus sp. IITRIND2
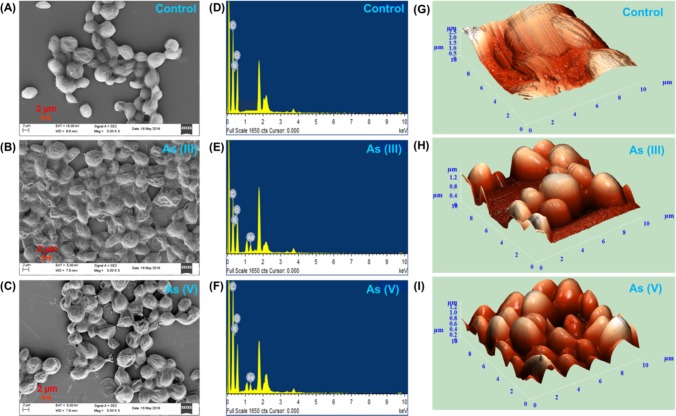
Interaction of As(III, V) with the surface markers of the microalga and their presence inside the algal cell can significantly influence its shape/size and morphology. To elucidate the physical characteristics (cell size, cell shape, surface texture, etc.) of As(III, V)-spiked algal cells, field emission scanning electron microscopy (FE-SEM) and atomic force microscopy (AFM) studies were performed. FE-SEM micrographs of As(III)- and As(V)-spiked Scenedesmus sp. IITRIND2 suggested that the microalgal cells retained their ellipsoidal shape (Figure 2A–C). However, the surface of the cells appeared more ruptured, rough with ridged textures, as compared to control cells which were smooth (Figure 2A–C). The resultant rigid texture of algal cell spiked with arsenic can be attributed to the adsorption of arsenic onto the cell surface, which was confirmed by energy-dispersive X-ray (EDX) spectrum (Figure 2D–F). The surface characteristics of the algal cells visualized by three-dimensional AFM image suggested that the microalgal cell surface of control was smooth without drops, whereas on treatment with arsenic(III, V), the surface of the cell became rough and irregular with frequent drops throughout (Figure 2G–I). Thus, AFM analysis validated the aberrations caused by As(III) and As(V) to the surface of microalgal cells.
Figure 2.
Scanning electron micrographs (SEMs) (A–C), energy-dispersive X-ray (EDX) (D–F) analysis, and (G–I) atomic force microscopy of Scenedesmus sp. IITRIND2. Control: (A, D, G); As(III): (B, E, H); and As(V): (C, F, I). The images were collected at a concentration of 500 mg/L of As(III, V).
2.3. Metabolic Changes Observed in Scenedesmus sp. IITRIND2 upon Exposure of As(III) and As(V)
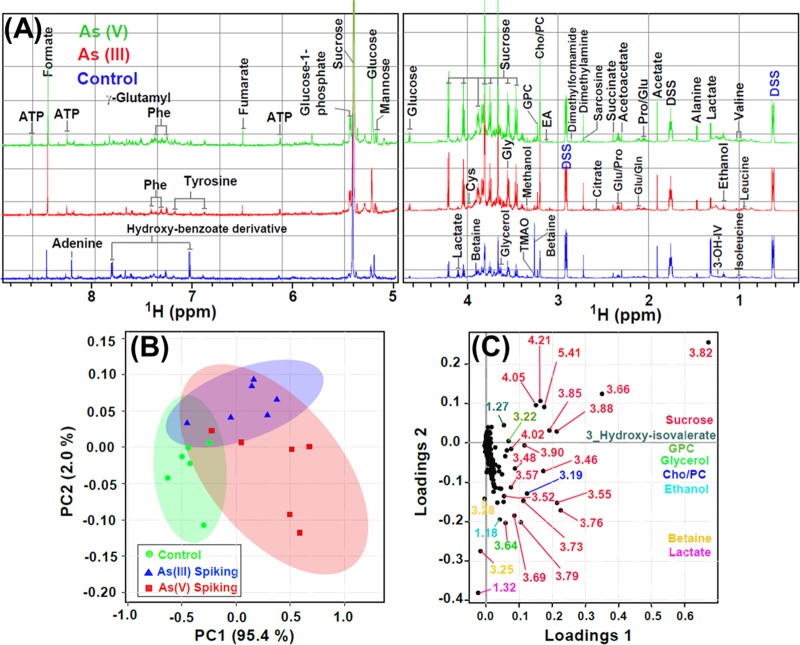
To gain deep insights on the variable effects of As(III) and As(V) on Scenedesmus sp. IITRIND2, the metabolites obtained from aqueous methanolic extract of microalga were analyzed using NMR spectroscopy. The cumulative 1H NMR spectra (n = 6 replicates) of Scenedesmus sp. IITRIND2 control polar extracts stacked up with those of cultures spiked with As(III) and As(V) metal systems were recorded (Figure 3A). A total of 45 metabolites were identified composed of carbohydrates/sugar (5), amino acids (17), organic acids (7), phosphagen (2), nucleotides (2), osmolytes (3), and others (9) (Table S1). Several of the assigned metabolites were validated using the two-dimensional (2D) NMR experiments, such as 1H–1H total correlation spectroscopy (TOCSY), 1H–13C single-quantum correlation spectroscopy (HSQC), and 2D J-resolved spectroscopy (JRES). Representative plots for the metabolite assignments using these experiments were presented in Figures S2–S4.
Figure 3.
(A) Cumulative one-dimensional (1D) 1H NMR spectra (n = 6) of Scenedesmus sp. IITRIND2 control polar extracts (blue) stacked up with those of cultures spiked with As(III) (red) and As(V) (green) metal systems. The spectral peaks were assigned for particular small-molecule metabolites. The water region at δ 4.6–4.9 was removed for clarity. The abbreviations used are: 3-OH-IV: 3-hydroxyisovalerate; Gln: glutamine; Glu: glutamate; EA: ethanolamine; Cys: cysteine; Cho: choline; PC: phosphocholine; GPC: glycerophosphocholine; DSS: 4,4-dimethyl-4-silapentane-1-sulfonic acid; TMAO: trimenthylamine-N-oxide; DMG: N,N-dimethylglycine; Phe: phenylalanine; and ATP: adenosine triphosphate. (B) The combined principal component analysis (PCA) 2D score plot resulted from the analysis of 1D 1H Carr–Purcell–Meiboom–Gill (CPMG) spectra of Scenedesmus sp. IITRIND2 exposed to different metal treatments over a period of 10 days (green = control; blue = As(III) spiking; red = As(V) spiking). The semitransparent red and blue ovals represent the 95% confidence interval. (C) PCA loading plot revealing the metabolites responsible for the discrimination pattern; the more the metabolite is away from the origin (0, 0), the more it contributes in the group discrimination.
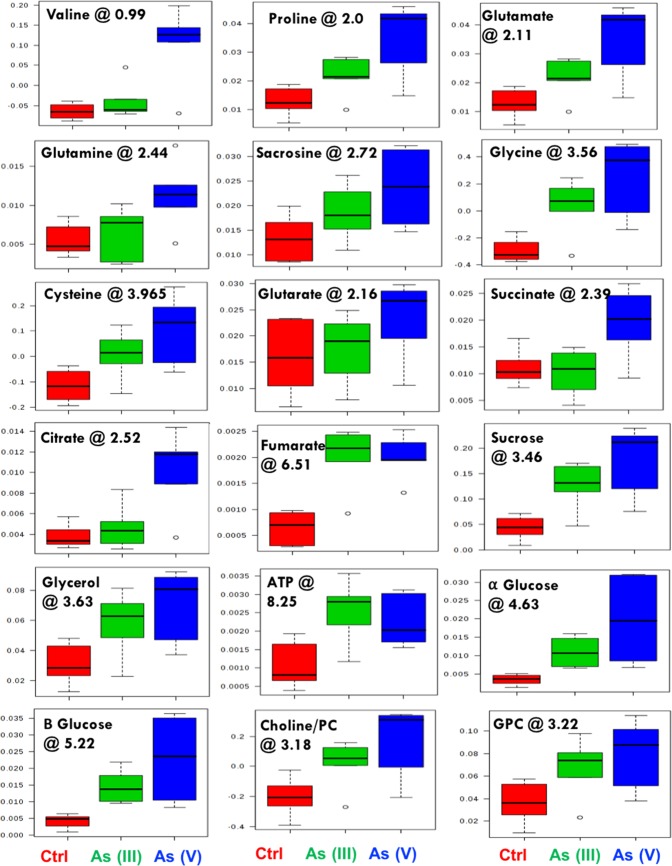
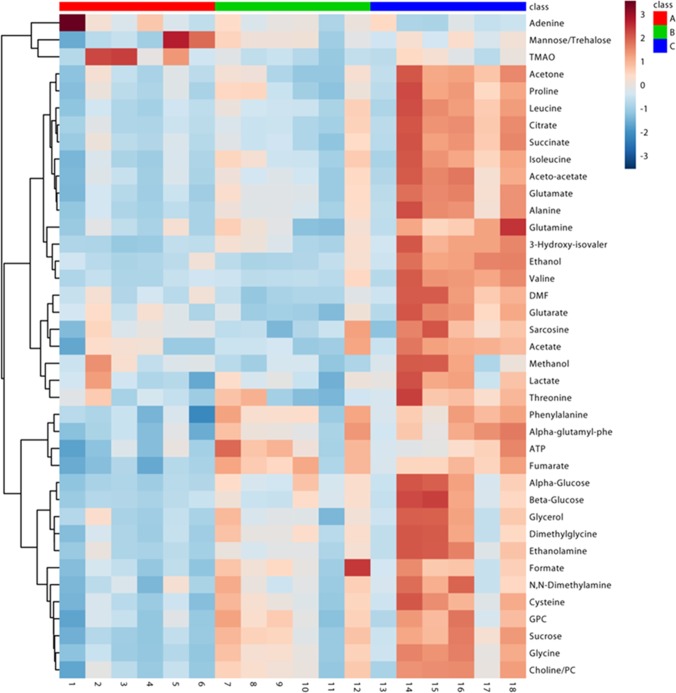
The assignment of metabolites using NMR data showed clear differences in the peaks of carbohydrates (sucrose, glucose, and mannose), amino acids (leucine, alanine, valine, serine, and cysteine), ATP, organic acids (fumarate, succinate, citrate, and acetate), and nucleotides in control as compared to those in As(III) and As(V) algal extracts and also between the two arsenic species (Figure 3A). To validate these variabilities across the three treatments, multivariate analysis was performed using principal component analysis (PCA). The PCA analysis showed statistically significant clustering of the six biological replicates and distinct differences between the control and arsenic-treated algal samples, as well as between As(III) and As(V), assuring a differential metabolic profiling in all three cases (Figure 3B). The PCA loading plot revealing metabolites responsible for the discrimination pattern along with the assignment of few relevant metabolites is shown in Figure 3C. Univariate analysis was further performed to identify the relative change in the metabolite levels. Representative box-cum-whisker plots derived from the univariate analysis shown in Figure 4, clearly revealed the quantitative variations of relative signal integrals for algal metabolites in response to As(III) and As(V) treatment. The quantitative data of significantly altered algal metabolites were visualized using quantitative NMR analysis and hierarchically clustered heat maps to discern the dissimilarity between the three experimental groups (Figure 5). The results clearly established that the As(V)-treated group was remarkably different in terms of the expression levels of metabolites compared with control and As(III)-treated groups (Figure 5 and Table S1). These metabolites, including amino acids, organic acids, sugars, and osmolytes, are distinct between As(III) and As(V) species. Among the identified metabolites, 28 metabolites were overexpressed in both As(III, V) -spiked microalgal cells, whereas only two metabolites (betaine and TMAO) showed reduced levels as compared with control (Table S1). Apart from 28 mutually overexpressed metabolites of As(III, V), 12 metabolites were exclusively enhanced in As(V)-treated microalgal cells (Table S1).
Figure 4.
Box plots showing relative abundance of some of the metabolites showing significant variation after As(III) and As(V) spiking compared with normal algal culture. In the box plots, the boxes denote interquartile ranges, horizontal line inside the box denotes the median, and bottom and top boundaries of boxes are 25th and 75th percentiles, respectively. Lower and upper whiskers are 5th and 95th percentiles, respectively. The corresponding chemical shift (in ppm) for each of the metabolites was also presented.
Figure 5.
Heat maps showing z-scores of discriminatory metabolite entities altered in either As(III) or As(V) spiking compared with control algal culture. X axis represents the six replicates of the culture (A) control: lane (1–6), red bar; (B) As(III): lane (7–12), green bar; (C) As(V): lane (13–18), blue bar. The color scheme through signifies the elevation and reduction in metabolite concentration in As(III) or As(V) spiking compared with normal algal culture: dark blue, lowest; dark red, highest.
Among the identified differential metabolites, a total of 18 metabolites was then correlated for their role in As(III, V) bioremediation by the microalga. Out of these 18 metabolites, 13 were differentially regulated for both As(III, V). A 2-fold increase in the levels of free amino acids involved in bioremediation, such as glutamate, valine, glycine, proline, and cysteine, was recorded in As(V)-treated microalgal cells in comparison with As(III) treatment (Table 1). Analogous to the results of the free amino acids, the levels of carbohydrates (sucrose, glycerol, and glucose) and ATP were higher in As(V)-spiked cells as compared to those in As(III). The levels of fumarate, choline/phosphocholine (PC), and glycerophosphocholine (GPC) were also high in As(V) as compared to that in As(III)-spiked microalgal cells. Thus, the above metabolic responses established that As(V) was more toxic to the microalgal cells as compared with As(III). Such an enhanced toxicity indeed resulted in overexpression of five specific metabolites (sarcosine, succinate, citrate, glutarate, and glutamine) exclusively under As(V) stress conditions (Table 1). Differential regulation of these extra four metabolites suggests that Scenedesmus sp. IITRIND2 has activated additional defense mechanism to cope with the enhanced toxicity of As(V).
Table 1. List of Metabolites Involved in Bioremediation along with Their Respective Chemical Shifts and Their Respective Fold Change as a Consequence of Uptake of Arsenic(III, V) Metal Uptakea,b.
| relative
fold change |
||||
|---|---|---|---|---|
| metabolite name | assignment | chemical shifts (δ) in ppm | As(III) spiking vs control | As(V) spiking vs control |
| Amino Acids | ||||
| valine | γ-CH3 | 0.98 (d) | 4.1 | |
| γ-CH3 | 1.03 (d)c | |||
| proline | γ-CH2 | 2.00 (m)c | 1.3 | 2.3 |
| 1/2 β-CH2 | 2.06 (m) | |||
| 1/2 β-CH2 | 2.34 (m) | |||
| glutamate | β-CH2 | 2.11 (m)c | 1.7 | 2.8 |
| γ-CH2 | 2.34 (m) | |||
| glutamine | β-CH2 | 2.12 (m) | 2.1 | |
| γ-CH2 | 2.44 (m)c | |||
| sacrosine | N–CH3 | 2.72 (s) | 1.3 | |
| glycine | α-CH2 | 3.56 (s) | 2.2 | 3.0 |
| cysteine | β-CH2 | 3.07 (m) | 1.8 | 2.5 |
| α-CH | 3.97 (dd)c | |||
| Organic Acids | ||||
| glutarate | β,δ-CH2 | 2.16 (t) | 1.4 | |
| succinate | α,β-CH2 | 2.39 (s) | 2.2 | |
| citrate | 1/2 γ-CH2 | 2.52 (d)c | 2.7 | |
| 1/2 γ-CH2 | 2.69 (d) | |||
| fumarate | CH | 6.51 (s) | 3.3 | 3.3 |
| Carbohydrates/Sugar | ||||
| sucrose | C10H | 3.46 (t) | 3.2 | 4.2 |
| C12H | 3.55 (dd) | |||
| C13H | 3.66 (s) | |||
| C11H | 3.75 (m) | |||
| C17H and C19H | 3.77 (m) | |||
| C5H and C9H | 3.81 (dd) | |||
| C4H | 4.04 (t) | |||
| C3H | 4.21(d)c | |||
| C7H | 5.40(d) | |||
| α-glucose | C1H | 4.63 (d) | 2.7 | 5.1 |
| β-glucose | C1H | 5.22 (d) | 3.4 | 5.3 |
| Phosphagen | ||||
| choline/PC | N–(CH3)3 | 3.20 (s) | 1.9 | 2.4 |
| GPC | N–(CH3)3 | 3.22 (s) | 1.8 | 2.2 |
| Osmolytes | ||||
| glycerol | 1/2 γ-CH2 | 3.63 (d)c | 1.3 | 1.6 |
| 1/2 γ-CH2 | 3.65 (d) | |||
| Nucleotides | ||||
| ATP | C7H | 8.61 (s)c | 2.2 | 2.0 |
| C12H | 8.25 (s) | |||
| C2H | 6.13 (d) | |||
One-way analysis of variance (ANOVA) was conducted to determine significant (p < 0.001) metabolic changes. All differentially expressed metabolites involved in bioremediation pathway of As(III, V) are represented in bold. Metabolites that are specific to As(V) mitigation are marked with bold and italics.
All values of the metabolites were statistically significant with p value 0.001, respectively.
Metabolite peak used for evaluating the quantitative difference, as represented here by fold changes.
3. Discussion
3.1. Mechanistic Insights into Bioremediation of As(III, V) by Scenedesmus sp. IITRIND2 Using NMR-Based Metabolite Data
The current study is an attempt to deploy a comprehensive NMR-based metabolomic approach to quantify an array of metabolites responsible for efficient bioremediation of arsenic by the hypertolerant microalga. The first step toward the bioremediation of any metal by microalgae is binding and adsorption to the cell wall (Figure 6). The elevated levels of glycine, valine, and glutamine under arsenic stress signified complexation of arsenic to the microalga biomass.28 Further, a distinct peak of choline/PC and GPC was observed in the NMR spectra of arsenic-spiked microalgal cells. Phytochelatins (PCHs) play all three essential roles in metal bioremediation; (a) help in metal binding, (b) act as antioxidant, and (c) assist in signaling, thereby protecting the algal cell from the deleterious effect of heavy metal.26 The differential shifting of functional groups in FT-IR spectra between As(III) and As(V) combined with distinct levels of choline/PC observed through NMR evidenced the distinct binding pathways of these two arsenic species by microalgae. Post internalization, an increase in the levels of proline content in As(V)-spiked algal cultures followed by that in As(III) as compared with control suggested activation of arsenic detoxification process by the microalga (Figure 6). Proline has been reported to have diverse roles during stress, such as it acts as a metal chelator, osmoprotectant, inhibitor of lipid peroxidation, reactive oxygen species (ROS) scavenger, and has antioxidant properties.29 Under oxidative stress, the microalgal cell starts generating components such as ascorbate, glutathione, and pyridine nucleotides (NAD+/NADP+) to combat this stress.26 The metabolomic profile also showed an increase in the levels of cysteine and glutamate in arsenic-spiked cultures as compared with control, with more elevation in the As(V) cultures. Indeed, these metabolites also protect the cell by scavenging ROS, thereby aiding the cell survival.30 In a previous study, two different microalgae Chlorella sp. and Monoraphidium arcuatum were evaluated for their arsenic toxicity mechanism.30 The authors reported that Macrostemum arcuatum was more sensitive to arsenic and actively excreted both As(III, V) into the medium, whereas Chlorella sp. was more tolerant and mitigated arsenic by binding to thiols, undergoing complexation in intracellular vacuoles, followed by reduction to methylated forms.30 As Scenedesmus sp. IITRIND2 and Chlorella belong to Chlorophyceae algal class, we presume a similar arsenic tolerance mechanism, which was also evident by increase in PC, glutamate, glutamine, and cysteine responsible for thiol oxidation.
Figure 6.
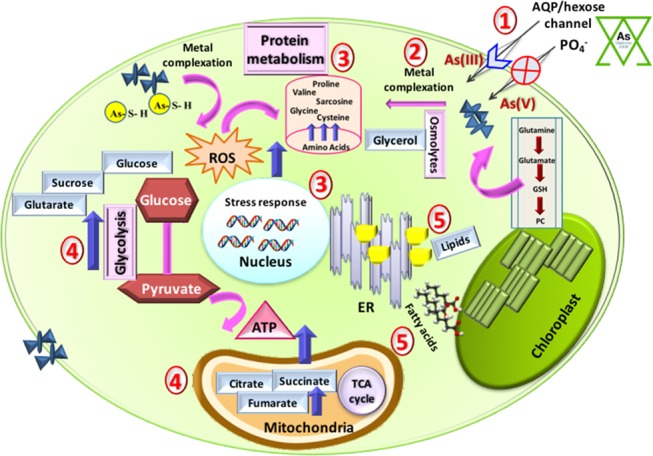
Schematic showing the hierarchy of arsenic(III, V)-induced metabolic changes in the microalgal cells: (1) Arsenic intake via AQP/hexose and phosphate channels, (2) metal complexation by PC, (3) ROS generation in response to arsenic stress, (4) mitigating ROS stress by modulating protein metabolism, glycolysis, and tricarboxylic acid, and (5) endoplasmic reticulum stress resulting in increase in lipid content.
Parallel to changes in the amino acids and PC/GPC, an increase in the soluble sugars was recorded in arsenic-spiked microalgal cells (Figure 6). High levels of sucrose and α/β-glucose were observed in As(V) cultures compared to those in As(III), which help in maintaining osmotic balance during the bioremediation by microalga. Moreover, an elevation in the levels of glycerol (osmolytic polyol) also suggested maintenance of carbon pool during stress conditions to protect the photosystem.31 Carbohydrates and polyols also act as osmoprotectants, which helps to stabilize the cell membrane.32,33 Further an increase in the levels of fumarate in both As(III, V)-spiked cultures indicated limitation of the deleterious effects of the heavy metal.29 Interestingly, the levels of several of the organic acids, such as succinate, citrate, and glutarate, were unaltered upon As(III) spiking whereas they were enhanced substantially upon As(V) spiking. Citrate acts as a detoxifying molecule by quenching the metal ions, whereas glutarate is involved in PC synthesis. All above results concisely deduce the hierarchy of arsenic tolerance/bioremediation (Figure 6). To gain detailed insights into the hierarchical activation of these multiple pathways of metabolites and mitigation of arsenic, a future time course metabolomic analysis integrated with proteomics and transcriptomics studies is quintessential.
4. Concluding Remarks
In a nutshell, we demonstrated the effectiveness of using proton NMR-based metabolomic approach in answering environmental toxicological responses. The current study identified an array of metabolites, which provided information on the arsenic mitigation mechanism by a hypertolerant microalgae. In a recent study, 1H NMR, in conjunction with high-resolution mass spectroscopy (HRMS), was deployed to identify and quantitate metabolites in bacteria on exposure to metal nanoparticles.34 The authors initially utilized 1H NMR to tentatively assign metabolites by matching with an open-access Escherichia coli metabolome database (ECMDB) and structurally validated via HRMS. In the current study, we have developed an exclusive NMR (1D and 2D) metabolomic workflow to efficaciously assign algal metabolites that can serve as a starting framework for algal metabolome database. Such a database can be enriched with more metabolites using similarity search algorithms on plant, yeast, and bacterial metabolite databases and metabolites assigned using NMR/MS techniques. The workflow developed could be utilized as a centralized source for robustly assigning algal metabolites, which can shed light on the different biochemical pathways instrumental in exploiting algae on biotechnological platforms for flux analysis, and also to study the effect of environmental factors or any stress stimulus.
5. Materials and Methods
5.1. Microalgae Cultivation and Experimental Design
Scenedesmus sp. IITRIND2 (Genebank Accession number KT932960) was isolated from a freshwater lake in India and maintained in modified Bold’s Basal medium.35 Arsenic stock solutions (10 g/L) were prepared by dissolving salts of NaAsO2 (As(III)) and Na2HAsO4.7H2O (As(V)) in sterilized distilled water. To perform the experiments, the microalga were adapted and cultivated in synthetic soft water (SSW).36 The microalga were cultivated in Erlenmeyer flasks (250 mL) containing 75 mL of culture for 96 h (log phase) in SSW at 27 °C, with a photoperiod of 16 h [8 h light–dark cycle irradiated with six white fluorescent lights (300 μmol/(m2 s))]. The cells were then centrifuged at 6000g for 10 min, and the cell pellet (1 × 105 cells/mL) obtained was washed thrice with autoclaved distilled water and then resuspended for inoculation in SSW, SSW with 500 mg/L of As(III), and SSW with 500 mg/L of As(V), respectively. The number of cells were counted using a haemocytometer cell counter by staining cells with trypan blue. Briefly, 100 μL of microalgal cells were mixed with 100 μL of trypan blue (0.4% in phosphate-buffered saline; pH 7.2) and incubated for 5 min. The cell suspension (10 μL) was then used to count the cells using a compound microscope. The cell density was then calculated according to the formula
The toxicity of As(III) and As(V) to the microalga was determined using a 96 h growth inhibition bioassay.37 Inductive coupled plasma mass spectroscopy (PerkinElmer, ELAN DRC-e) was used to estimate the amount of arsenic(III, V) left in the SSW after 10 days of algal growth. The concentration of arsenic was calculated by plotting a standard curve from different concentrations (0, 10, 20, 50, 100, 250, and 500 mg/L) of As(III) and As(V) using a mixture of Rh, Ge, and Ir (100 mg/L) as internal standard. The metal uptake capacity was evaluated using the following equation
 |
5.2. Characterization of Arsenic Interaction and Morphological Changes in Scenedesmus sp. IITRIND2
The adsorption of As(III, V) on to the microalga was analyzed by Fourier transform infrared (FT-IR) spectroscopy (Thermo Nicolet NEXUS, Maryland) at 400–4000 cm–1 wavenumber range, with field emission scanning electron microscopy (FE-SEM) coupled to energy-dispersive X-ray (EDX) spectroscopy (FE-SEM Quanta 200 FEG).22,38 Atomic force microscopy (AFM) analysis (NT-MDT-INTEGRA) was performed to visualize the changes in the surface morphology of arsenic-spiked microalgal cells. Briefly, the microalgal cells (2 × 106) were fixed on poly-l-lysine-coated glass slides using 2.5% glutaraldehyde (24 h in dark at 4 °C) followed by dehydration (10–100% ethanol) and then visualized under the microscope.
5.3. NMR-Based Metabolomics and Multivariate Analysis
For the extraction of the metabolites, 40 mg of lyophilized microalgal biomass (harvested on the 10th day) from the control, As(III), and As(V) cultures was ground with liquid N2 using 1 mL of 20% methanol, as mixture of methanol/water allows a better separation of polar components from the nonpolar components by lowering down the surface tension and polarity of water and increasing the density.23 The process was repeated twice; the supernatant was pooled together and lyophilized overnight. Lyophilized samples were reconstituted in phosphate buffer (0.1 M, pH 7.4), and the samples were relyophilized to maintain a uniform pH across all samples to have a spectral overlap of chemical shifts across the samples and also to facilitate the NMR acquisition in deuterium oxide (D2O). The lyophilized samples were dissolved in 550 μL of D2O containing a chemical shift indicator (4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS), 0.5 mM). All proton NMR spectra were acquired on an 800 MHz NMR spectrometer equipped with a cryoprobe. Each NMR spectrum consisted of 128 scans of 16 384 data points in the frequency domain, and the spectra were collected using a Carr–Purcell–Meiboom–Gill (CPMG) pulse sequence consisting of water presaturation (during relaxation delay of 4.00 s).
The 1D spectra were Fourier transformed using an exponential window with a line broadening value of 0.5 Hz, phased and baseline-corrected using Chenomx NMR suite 8.1 (Chenomx Inc., AB, Canada) prior to chemical shift and intensity measurements. 1H NMR chemical shifts in all spectra [control and arsenic(III, V)-spiked] were referenced with respect to the methyl peak of DSS at 0.00 ppm. Chemical shifts in the 1D 1H NMR spectra were identified and assigned using the 800 MHz chemical shift database in Chenomx Profiler and were validated by comparing them with other databases and the literature report.23,39
The resonance assignments obtained for the microalga samples were further validated using two-dimensional NMR experiments such as homonuclear 1H–1H total correlation spectroscopy (TOCSY), 1H–13C single-quantum correlation spectroscopy (HSQC), and 2D J-resolved (JRES) NMR spectra, on the basis of their specific patterns of HH/CH correlations and 1H–1H scalar coupling constants, respectively. The complete details of experimental procedures for 2D NMR experiments are provided in Appendix S1 (Supporting Information). HSQC and TOCSY analysis was validated using MetaboMiner with tolerances of 0.02 ppm (1H) and 0.5 ppm (13C).40
For multivariate data analysis, the NMR spectra were integrated and normalized against internal standard area of DSS and the data were reduced into spectral bins (0.03 ppm width) using Pathomx.39 The resultant data was imported into Metaboanalyst (v3.0) software for multivariate data analysis, performed using the principal component analysis (PCA).41,42 Metabolites that influenced the differentiation pattern were identified from the loading plot; the greater the distance of a particular variable from the origin (0, 0), the greater is its contribution in distinguishing the groups. The relative metabolic changes were assessed using univariate (or box-plot) analysis, and statistical significance was determined by one-way analysis of variance (ANOVA) using Metaboanalyst.43 Unsupervised hierarchical clustering using Ward linkage was further employed to create the heat map consisting of 40 metabolite entities (with p < 0.001) that had the highest impact on separation of the different treatment groups. The resulted heat map was used to assess how similar or different the arsenic samples are compared with normal control samples on the basis of their metabolite profiles.
Acknowledgments
We acknowledge the support of biophysical instrumentation facilities at Institute instrumentation center (IIC), IIT-Roorkee. We are highly thankful for the access to 800 MHz NMR at CMBR, Lucknow.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.8b01692.
Detailed information regarding the IC50 values, the percentage removal of As(III, V) by Scenedesmus sp. IITRIND2 from the media and 2D NMR spectra of Scenedesmus sp. IITRIND2 annotated with different metabolites along with detailed experimental procedures (PDF)
Author Contributions
∥ N.A. and D.D. contributed equally to this work.
Authors are thankful for financial support DBT-SRF to N.A. (Grant No.: 7001-35-44) and UGC-SRF to M.S. K.M.P. acknowledges the receipt of DBT-IYBA fellowship, SERB-LS young scientist award, and grant no. GKC-01/2016-17/212/NMCG-Research from NMCG-MoWR, government of India (GoI).
The authors declare no competing financial interest.
Supplementary Material
References
- Dettmer K.; Hammock B. D. Metabolomics - A New Exciting Field within the “Omics” Sciences. Environ. Health Perspect. 2004, 112, 396–397. 10.1289/ehp.112-1241997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora N.; Pienkos P. T.; Pruthi V.; Poluri K. M.; Guarnieri M. T. Leveraging Algal Omics to Reveal Potential Targets for Augmenting TAG Accumulation. Biotechnol. Adv. 2018, 36, 1274–1292. 10.1016/j.biotechadv.2018.04.005. [DOI] [PubMed] [Google Scholar]
- Viant M. R.; Rosenblum E. S.; Tjeerdema R. S. NMR-Based Metabolomics: A Powerful Approach for Characterizing the Effects of Environmental Stressors on Organism Health. Environ. Sci. Technol. 2003, 37, 4982–4989. 10.1021/es034281x. [DOI] [PubMed] [Google Scholar]
- Verpoorte R.; Choi Y. H.; Kim H. K. NMR-Based Metabolomics at Work in Phytochemistry. Phytochem. Rev. 2007, 6, 3–14. 10.1007/s11101-006-9031-3. [DOI] [Google Scholar]
- Serkova N. J.; Niemann C. U. Pattern Recognition and Biomarker Validation Using Quantitative 1H-NMR-Based Metabolomics. Expert Rev. Mol. Diagn. 2006, 6, 717–731. 10.1586/14737159.6.5.717. [DOI] [PubMed] [Google Scholar]
- Markley J. L.; Brüschweiler R.; Edison A. S.; Eghbalnia H. R.; Powers R.; Raftery D.; Wishart D. S. The Future of NMR-Based Metabolomics. Curr. Opin. Biotechnol. 2017, 34–40. 10.1016/j.copbio.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D. S. Quantitative Metabolomics Using NMR. TrAC, Trends Anal. Chem. 2008, 27, 228–237. 10.1016/j.trac.2007.12.001. [DOI] [Google Scholar]
- Brennan L. NMR-Based Metabolomics: From Sample Preparation to Applications in Nutrition Research. Prog. Nucl. Magn. Reson. Spectrosc. 2014, 83, 42–49. 10.1016/j.pnmrs.2014.09.001. [DOI] [PubMed] [Google Scholar]
- Jamers A.; Blust R.; Coen W. De; Griffin J. L.; Jones O. A. H. An Omics Based Assessment of Cadmium Toxicity in the Green Alga Chlamydomonas Reinhardtii. Aquat. Toxicol. 2013, 126, 355–364. 10.1016/j.aquatox.2012.09.007. [DOI] [PubMed] [Google Scholar]
- Sarı A.; Uluozlü Ö. D.; Tüzen M. Equilibrium, Thermodynamic and Kinetic Investigations on Biosorption of Arsenic from Aqueous Solution by Algae (Maugeotia genuflexa) Biomass. Chem. Eng. J. 2011, 167, 155–161. 10.1016/j.cej.2010.12.014. [DOI] [Google Scholar]
- Wang Y.; Wang S.; Xu P.; Liu C.; Liu M.; Wang Y.; Wang C.; Zhang C.; Ge Y. Review of Arsenic Speciation, Toxicity and Metabolism in Microalgae. Rev. Environ. Sci. Biotechnol. 2015, 14, 427–451. 10.1007/s11157-015-9371-9. [DOI] [Google Scholar]
- Podder M. S.; Majumder C. B. Phycoremediation of Arsenic from Wastewaters by Chlorella pyrenoidosa. Groundwater Sustainable Dev. 2015, 1, 78–91. 10.1016/j.gsd.2015.12.003. [DOI] [Google Scholar]
- Wang S.; Zhang D.; Pan X. Effects of Arsenic on Growth and Photosystem II (PSII) Activity of Microcystis Aeruginosa. Ecotoxicol. Environ. Saf. 2012, 84, 104–111. 10.1016/j.ecoenv.2012.06.028. [DOI] [PubMed] [Google Scholar]
- Rahman M. A.; Hogan B.; Duncan E.; Doyle C.; Krassoi R.; Rahman M. M.; Naidu R.; Lim R. P.; Maher W.; Hassler C. Toxicity of Arsenic Species to Three Freshwater Organisms and Biotransformation of Inorganic Arsenic by Freshwater Phytoplankton (Chlorella Sp. CE-35). Ecotoxicol. Environ. Saf. 2014, 106, 126–135. 10.1016/j.ecoenv.2014.03.004. [DOI] [PubMed] [Google Scholar]
- Veličković Z.; Vuković G. D.; Marinković A. D.; Moldovan M. S.; Perić-Grujić A. A.; Uskoković P. S.; Ristić M. Đ. Adsorption of Arsenate on Iron(III) Oxide Coated Ethylenediamine Functionalized Multiwall Carbon Nanotubes. Chem. Eng. J. 2012, 181–182, 174–181. 10.1016/j.cej.2011.11.052. [DOI] [Google Scholar]
- Mukherjee K. K.; Das D.; Samal A. C.; Santra S. C. Isolation and characterization of Arsenic tolerant fungal strains from contaminated sites around urban environment of Kolkata. IOSR J. Environ. Sci., Toxicol. Food Technol. 2014, 7, 33–37. 10.9790/2402-0753337. [DOI] [Google Scholar]
- Jasrotia S.; Kansal A.; Kishore V. V. N. Arsenic Phyco-Remediation by Cladophora Algae and Measurement of Arsenic Speciation and Location of Active Absorption Site Using Electron Microscopy. Microchem. J. 2014, 114, 197–202. 10.1016/j.microc.2014.01.005. [DOI] [Google Scholar]
- Foster S.; Thomson D.; Maher W. Uptake and Metabolism of Arsenate by Anexic Cultures of the Microalgae Dunaliella tertiolecta and Phaeodactylum tricornutum. Mar. Chem. 2008, 108, 172–183. 10.1016/j.marchem.2007.11.005. [DOI] [Google Scholar]
- Wang N. X.; Li Y.; Deng X. H.; Miao A. J.; Ji R.; Yang L. Y. Toxicity and Bioaccumulation Kinetics of Arsenate in Two Freshwater Green Algae under Different Phosphate Regimes. Water Res. 2013, 47, 2497–2506. 10.1016/j.watres.2013.02.034. [DOI] [PubMed] [Google Scholar]
- Zhang J.; Ding T.; Zhang C. Biosorption and Toxicity Responses to Arsenite (As[III]) in Scenedesmus quadricauda. Chemosphere 2013, 92, 1077–1084. 10.1016/j.chemosphere.2013.01.002. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Zhang C.; Zheng Y.; Ge Y. Phytochelatin Synthesis in Dunaliella salina Induced by Arsenite and Arsenate under Various Phosphate Regimes. Ecotoxicol. Environ. Saf. 2017, 136, 150–160. 10.1016/j.ecoenv.2016.11.002. [DOI] [PubMed] [Google Scholar]
- Arora N.; Gulati K.; Patel A.; Pruthi P. A.; Poluri K. M.; Pruthi V. A Hybrid Approach Integrating Arsenic Detoxification with Biodiesel Production Using Oleaginous Microalgae. Algal Res. 2017, 24, 29–39. 10.1016/j.algal.2017.03.012. [DOI] [Google Scholar]
- Zhang W.; Tan N. G. J.; Li S. F. Y. NMR-Based Metabolomics and LC-MS/MS Quantification Reveal Metal-Specific Tolerance and Redox Homeostasis in Chlorella vulgaris. Mol. Biosyst. 2014, 10, 149–160. 10.1039/C3MB70425D. [DOI] [PubMed] [Google Scholar]
- Zhang W.; Tan N. G. J.; Fu B.; Li S. F. Y. Metallomics and NMR-Based Metabolomics of Chlorella Sp. Reveal the Synergistic Role of Copper and Cadmium in Multi-Metal Toxicity and Oxidative Stress. Metallomics 2015, 7, 426–438. 10.1039/C4MT00253A. [DOI] [PubMed] [Google Scholar]
- Piotrowska-Niczyporuk A.; Bajguz A.; Talarek M.; Bralska M.; Zambrzycka E. The Effect of Lead on the Growth, Content of Primary Metabolites, and Antioxidant Response of Green Alga Acutodesmus obliquus (Chlorophyceae). Environ. Sci. Pollut. Res. 2015, 22, 19112–19123. 10.1007/s11356-015-5118-y. [DOI] [PubMed] [Google Scholar]
- Sytar O.; Kumar A.; Latowski D.; Kuczynska P.; Strzałka K.; Prasad M. N. V. Heavy Metal-Induced Oxidative Damage, Defense Reactions, and Detoxification Mechanisms in Plants. Acta Physiol. Plant. 2013, 35, 985–999. 10.1007/s11738-012-1169-6. [DOI] [Google Scholar]
- Tuzen M.; Sari A.; Mendil D.; Uluozlu O. D.; Soylak M.; Dogan M. Characterization of Biosorption Process of As(III) on Green Algae Ulothrix Cylindricum. J. Hazard. Mater. 2009, 165, 566–572. 10.1016/j.jhazmat.2008.10.020. [DOI] [PubMed] [Google Scholar]
- Chia M. A.; Lombardi A. T.; da Graça Gama Melão M.; Parrish C. C. Combined Nitrogen Limitation and Cadmium Stress Stimulate Total Carbohydrates, Lipids, Protein and Amino Acid Accumulation in Chlorella vulgaris (Trebouxiophyceae). Aquat. Toxicol. 2015, 160, 87–95. 10.1016/j.aquatox.2015.01.002. [DOI] [PubMed] [Google Scholar]
- Zoghlami L.; Djebali W.; Abbes Z.; Hediji H.; Maucourt M.; Moing A.; Brouquisse R.; Chaïbi W. Metabolite Modifications in Solanum lycopersicum Roots and Leaves under Cadmium Stress. African J. Biotechnol. 2013, 10, 567–579. [Google Scholar]
- Levy J. L.; Stauber J. L.; Adams M. S.; Maher W. A.; Kirby J. K.; Jolley D. F. Toxicity, Biotransformation, and Mode of Action of Arsenic in Two Freshwater Microalgae (Chlorella Sp. and Monoraphidium arcuatum). Environ. Toxicol. Chem. 2005, 24, 2630–2639. 10.1897/04-580R.1. [DOI] [PubMed] [Google Scholar]
- Afkar E.; Ababna H.; Fathi A. A. Toxicological Response of the Green Alga Chlorella vulgaris, to Some Heavy Metals. Am. J. Environ. Sci. 2010, 6, 230–237. 10.3844/ajessp.2010.230.237. [DOI] [Google Scholar]
- Kieffer P.; Planchon S.; Oufir M.; Ziebel J.; Dommes J.; Hoffmann L.; Hausman J. F.; Renaut J. Combining Proteomics and Metabolite Analyses to Unravel Cadmium Stress-Response in Poplar Leaves. J. Proteome Res. 2009, 8, 400–417. 10.1021/pr800561r. [DOI] [PubMed] [Google Scholar]
- Yancey P. H.; Clark M. E.; Hand S. C.; Bowlus R. D.; Somero G. N. Classes of Intracellular Osmolyte Systems and Their Distributions Living with Water Stress: Evolution of Osmolyte Systems. Science 1982, 217, 1214–1222. 10.1126/science.7112124. [DOI] [PubMed] [Google Scholar]
- Chatzimitakos T. G.; Stalikas C. D. Qualitative Alterations of Bacterial Metabolome after Exposure to Metal Nanoparticles with Bactericidal Properties: A Comprehensive Workflow Based On 1H NMR, UHPLC-HRMS, and Metabolic Databases. J. Proteome Res. 2016, 15, 3322–3330. 10.1021/acs.jproteome.6b00489. [DOI] [PubMed] [Google Scholar]
- Arora N.; Patel A.; Pruthi P. A.; Pruthi V. Boosting TAG Accumulation with Improved Biodiesel Production from Novel Oleaginous Microalgae Scenedesmus Sp. IITRIND2 Utilizing Waste Sugarcane Bagasse Aqueous Extract (SBAE). Appl. Biochem. Biotechnol. 2016, 180, 109–121. 10.1007/s12010-016-2086-8. [DOI] [PubMed] [Google Scholar]
- Smith E. J.; Davison W.; Hamilton-taylor J. Methods for Preparing Synthetic Freshwaters. Water Res. 2002, 36, 1286–1296. 10.1016/S0043-1354(01)00341-4. [DOI] [PubMed] [Google Scholar]
- Bahar M. M.; Megharaj M.; Naidu R. Toxicity, Transformation and Accumulation of Inorganic Arsenic Species in a Microalga Scenedesmus Sp. Isolated from Soil. J. Appl. Phycol. 2013, 25, 913–917. 10.1007/s10811-012-9923-0. [DOI] [Google Scholar]
- Arora N.; Patel A.; Pruthi P. A.; Poluri K. M.; Pruthi V. Utilization of Stagnant Non-Potable Pond Water for Cultivating Oleaginous Microalga Chlorella minutissima for Biodiesel Production. Renewable Energy 2018, 126, 30–37. 10.1016/j.renene.2018.03.033. [DOI] [Google Scholar]
- Fitzpatrick M. A.; McGrath C. M.; Young S. P. Pathomx: An Interactive Workflow-Based Tool for the Analysis of Metabolomic Data. BMC Bioinf. 2014, 15, 396. 10.1186/s12859-014-0396-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J.; Bjorndahl T. C.; Tang P.; Wishart D. S. MetaboMiner - Semi-Automated Identification of Metabolites from 2D NMR Spectra of Complex Biofluids. BMC Bioinf. 2008, 9, 507. 10.1186/1471-2105-9-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J.; Psychogios N.; Young N.; Wishart D. S. MetaboAnalyst: A Web Server for Metabolomic Data Analysis and Interpretation. Nucleic Acids Res. 2009, 37, W652–W660. 10.1093/nar/gkp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckonert O.; Keun H. C.; Ebbels T. M. D.; Bundy J.; Holmes E.; Lindon J. C.; Nicholson J. K. Metabolic Profiling, Metabolomic and Metabonomic Procedures for NMR Spectroscopy of Urine, Plasma, Serum and Tissue Extracts. Nat. Protoc. 2007, 2, 2692–2703. 10.1038/nprot.2007.376. [DOI] [PubMed] [Google Scholar]
- Xia J.; Wishart D. S. Using Metaboanalyst 3.0 for Comprehensive Metabolomics Data Analysis. Curr. Protoc. Bioinf. 2016, 14.10.1–14.10.91. 10.1002/cpbi.11. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.