This study investigates a group B Streptococcus LytR-CpsA-Psr enzyme which plays roles in multiple bacterial functions including biofilm formation, colonization, and innate immune resistance.
Keywords: Group B Streptococcus, Streptococcus agalactiae, biofilm, bacterial cell wall, virulence factor, innate immunity, neutrophil, RNA sequencing, gene regulation
Abstract
Background
Streptococcus agalactiae (group B Streptococcus [GBS]) asymptomatically colonizes approximately 20% of adults; however, GBS causes severe disease in susceptible populations, including newborns, pregnant women, and elderly individuals. In shifting between commensal and pathogenic states, GBS reveals multiple mechanisms of virulence factor control. Here we describe a GBS protein that we named “biofilm regulatory protein A” (BrpA) on the basis of its homology with BrpA from Streptococcus mutans.
Methods
We coupled phenotypic assays, RNA sequencing, human neutrophil and whole-blood killing assays, and murine infection models to investigate the contribution of BrpA to GBS physiology and virulence.
Results
Sequence analysis identified BrpA as a LytR-CpsA-Psr enzyme. Targeted mutagenesis yielded a GBS mutant (ΔbrpA) with normal ultrastructural morphology but a 6-fold increase in chain length, a biofilm defect, and decreased acid tolerance. GBS ΔbrpA stimulated increased neutrophil reactive oxygen species and proved more susceptible to human and murine blood and neutrophil killing. Notably, the wild-type parent outcompeted ΔbrpA GBS in murine sepsis and vaginal colonization models. RNA sequencing of ΔbrpA uncovered multiple differences from the wild-type parent, including pathways of cell wall synthesis and cellular metabolism.
Conclusions
We propose that BrpA is an important virulence regulator and potential target for design of novel antibacterial therapeutics against GBS.
Group B Streptococcus (GBS; Streptococcus agalactiae) is a member of mucosal microbiota of the intestinal and vaginal tracts, with a global prevalence of approximately 18% [1]. GBS is a preeminent agent of invasive neonatal infection, including early onset sepsis and meningitis, often in the first week of life [2]. Universal culture or risk factor–based screening of pregnant women to guide intrapartum antibiotic prophylaxis has reduced the incidence of early onset neonatal infections [2]. Currently, >90% of the GBS burden occurs outside of the perinatal period, and the incidence in adult and elderly populations is increasing [2, 3]. GBS bloodstream infections account for almost half of clinical manifestations [2, 3]. Acquisition of antibiotic resistance genes is increasing, along with mutations in penicillin-binding proteins [4].
GBS persistence within the host is a prerequisite for invasive disease. GBS capsular polysaccharide (CPS) promotes biofilm formation [5] and bloodstream survival across multiple serotypes [6, 7] and represents a primary antigen for host recognition. High maternal antibody titers are protective against infant transmission [8], providing rationale for the trivalent GBS glycoconjugate vaccine undergoing clinical trials [9].
GBS mechanisms promoting successful colonization, bloodstream dissemination, and antibiotic resistance, as well as the CPS-based vaccine strategy, all converge on the GBS cell wall. Gram-positive cell walls consist of a peptidoglycan-rich layer that serves as a molecular scaffold for anchoring CPS, wall teichoic acids (WTAs), and surface proteins. In GBS, peptidoglycan anchors CPS and the species-defining group B carbohydrate (GBC) [10]. In contrast to related pathogens, WTAs are absent in GBS; rather, GBC serves as a functional homologue [11]. Enzymatic pathways involved in cell wall anchoring of CPS and GBS are not fully known.
Recently, the LytR-CpsA-Psr (LCP) family of proteins have been identified as important mediators of cell wall integrity and maintenance, further contributing to biofilm formation, blood survival, autolysis, and antibiotic resistance [6, 12, 13]. LCP proteins help tether polysaccharides to the cell wall [14–16] and link WTAs to mature peptidoglycan [17]. GBS possesses 3 LCP family proteins, including the previously characterized cpsA, a member of the CPS synthesis operon [6].
Recent molecular insights have highlighted LCP proteins as targets for drug discovery to counteract antibiotic resistance [13, 17]. Here, we designate a previously uncharacterized GBS LCP protein as “biofilm regulatory protein A,” encoded by brpA, on the basis of its sequence and phenotypic similarities to Streptococcus mutans BrpA [18]. We describe a key role for BrpA in GBS cell chain length, biofilm formation, resistance to whole-blood and neutrophil-mediated killing, vaginal colonization, and systemic virulence, while providing a first look at the genome-wide transcriptional consequences of BrpA deficiency.
METHODS
Bacterial Strains and Growth Conditions
GBS CNCTC 10/84 serotype V (catalog no. 49447; American Type Culture Collection [ATCC]) and isogenic strains were grown to stationary phase at 37°C in Todd-Hewitt broth (THB; Hardy Diagnostics) supplemented with 50 µg/mL erythromycin when needed. Cultures were diluted in fresh THB and incubated at 37°C until mid-logarithmic phase (defined as an OD600 of 0.4).
Construction of the GBS brpA Insertional Mutant
Targeted insertional mutagenesis of brpA (GenBank accession no. AIX04136.1 and National Center for Biotechnology Information [NCBI] reference sequence WP_000708159.1) was performed using the vector pHY304 as described previously [19] and detailed in the Supplementary Methods.
GBS Chain Length, Growth, and Autolysis
Bright field images were obtained from random fields at 630 times the original magnification on a Zeiss Axio Scope.A1 with AxioCam mRm. Chain length was determined by counting 25 chains per strain per independent experiment, repeated twice. GBS cultures were grown in THB at pH 5.0, 6.0, or 7.0, and the OD600 was determined every 30 minutes for 5 hours. For autolysis, cultures were resuspended in phosphate-buffered saline (PBS) at an OD600 of 0.4, incubated at 37°C, with turbidity measured every 30 minutes, and normalized to T = 0.
Biofilm Formation and Microscopy
GBS biofilms were assessed as described elsewhere [20], with 1 adaption: assays were performed in a 200-µL volume, using 96-well plates. For fluorescence microscopy, biofilms were stained with 1 µM Syto13 (Molecular Probes) for 15 minutes, and fluorescent images were obtained at 100 times the original magnification on a Zeiss Axio Scope.A1 with AxioCam mRm.
Epithelial Adherence
Human vaginal epithelial cells (VK2; catalog no. CRL-2616; ATCC) were grown in keratinocyte serum-free medium (Life Technologies), and bladder epithelial cells (HTB-9; catalog no. 5637; ATCC) were grown in Roswell Park Memorial Institute 1640 medium with 10% fetal bovine serum. Cell lines were cultured at 37°C with 5% CO2. Adherence assays were conducted as described previously [21] at a multiplicity of infection (MOI; calculated as the GBS to cell ratio) of 10:1.
Human Whole-Blood and Neutrophil Killing Assays
Under approval from the UC–San Diego institutional review board, under protocol 070278X, venous blood specimens were obtained under informed consent. Hirudin and heparin were used as an anticoagulant for whole blood and neutrophil isolation, respectively. Neutrophils were isolated using PolymorphPrep (Axis-Shield). For the whole-blood killing assay, 90 µL of blood and 10 µL containing 1 × 106 colony-forming units (CFU) of GBS were incubated at 37°C with rotation for 30 minutes, and plated on THB agar, and GBS survival was calculated as a percentage of the inoculum. Neutrophil killing and oxidative burst assays were performed with GBS at a MOI of 1:1 as described previously [22].
Animal Experiments
Animal experiments were approved by the UC–San Diego Institutional Animal Care and Use Committee. Female CD-1 mice aged 3–4 months were purchased from Charles River Laboratories. Blood specimens were collected via cardiac puncture and killing assays conducted as described above. For neutrophil experiments, neutrophil migration was stimulated by injection of 1mL of 3% thioglycollate intraperitoneally, and 18 hours after injection, peritoneal lavage was performed using PBS with 5 mM ethylenediaminetetraacetic acid. Neutrophils composed 85%–90% of the total cell population, as determined by Giemsa staining; killing assays were conducted as described above. Vaginal colonization studies were conducted as described [23]. Wild-type (WT) and ΔbrpA bacteria were combined at a 1:1 ratio, and mice were inoculated with 10 μL (2 × 107 CFU total) into the vaginal tract. The vaginal lumen was swabbed daily, and recovered GBS were quantified on ChroMagar StrepB (DRG International) with or without 50µg/mL erythromycin, to distinguish strains. For sepsis studies, WT and ΔbrpA bacteria were combined at a 1:1 ratio, and mice were injected with 100 µL (2 × 107 CFU total) intraperitoneally. After 24 hours, blood specimens were collected via cardiac puncture, and kidneys and spleens were homogenized by adding PBS and silica beads (diameter, 1 mm; Biospec) and shaking at 6000 rpm for 60 seconds, using a MagNA Lyser (Roche). Blood and tissue homogenates were plated on agar with or without 50 µg/mL erythromycin, to distinguish strains.
RNA Sequencing and Bioinformatic Analysis
GBS strains were grown to an OD600 of 0.8 in THB at 37°C and subjected to RNA sequencing. Transcriptional analysis of WT and ΔbrpA strains is detailed in the Supplementary Methods.
Data Availability
The RNA sequencing data set is deposited at the NCBI Sequence Read Archive (accession no. SRP140532). Other data sets and materials generated and analyzed during this study are available from the corresponding author on request.
Statistical Analyses
All data were collected from at least 3 biological replicates performed in at least technical duplicate as detailed in Supplementary Methods. Statistical analyses were performed using GraphPad Prism, version 7.03. P values of <.05 were considered statistically significant.
RESULTS
GBS BrpA Is a Member of the Conserved LCP Family of Proteins
Mass spectrometry of GBS CNCTC 10/84 surface and secreted proteins identified multiple peptide sequences produced by the gene locus W903_0426. CNCTC 10/84 (Gene Expression Omnibus accession no. CP006910.1) locus W903_0426 (GenBank accession no. AIX04136.1; NCBI reference sequence WP_000708159.1), has a predicted 435–amino acid product, which we designate “biofilm regulatory protein A” (BrpA), encoded by brpA. Bioinformatic analysis revealed an LCP domain at residues 81–239 (PF03816) [24]. A transmembrane helix model predicted an N-terminal cytosolic domain (residues 1–6) and 1 transmembrane helix (residues 7–29) and that the remaining portion would be extracellular [25] (Supplementary Figure 1), in agreement with previous analyzes [26]. Within S. agalactiae (taxID: 1311), protein BLAST analysis of WP_000708159.1 found 35 additional unique sequences (representing 678 genomes) with ≥97% sequence identity. Protein-protein BLAST analysis showed 68% identity to Streptococcus pyogenes LytR (NCBI reference sequence WP_063811833.1). Subsequent alignment with S. pyogenes LytR, Streptococcus dysgalactiae LytR (NCBI reference sequence WP_042357980.1), and BrpA in S. mutans (WP_019316206.1) was performed (Figure 1). The LCP domain (amino acids 81–239) exhibited 75% identity across all 4 species. A previous cluster analysis categorized BrpA into LCP cluster F2 containing LytR/BrpA-like proteins of Firmicutes [26]. The other GBS LCP proteins, previously characterized CpsA (NCBI reference sequence WP_000064987.1), and a yet uncharacterized LCP protein (NCBI reference sequence WP_000089333.1) were grouped to cluster F3 and F1 (Supplementary Figure 1).
Figure 1.
Group B Streptococcus (GBS) biofilm regulatory protein A (BrpA) alignment with other Streptococcus species. Streptococcus agalactiae CNCTC 10/84 (Gene Expression Omnibus accession no. CP006910.1) biofilm regulatory protein A (GenBank accession no. AIX04136; National Center for Biotechnology Information [NCBI] reference sequence WP_000708159.1) aligned with Streptococcus pyogenes LytR family transcriptional regulator (NCBI reference sequence WP_063811833.1), Streptococcus dysgalactiae LytR family transcriptional regulator (NCBI reference sequence WP_042357980.1), and BrpA in Streptococcus mutans (NCBI reference sequence WP_019316206.1) was performed using the Alignment-Annotator web server. Amino acid residues with >74% identity are indicated in bold.
GBS BrpA Deficiency Influences Cellular Chain Length, Pigmentation, and Acid Tolerance
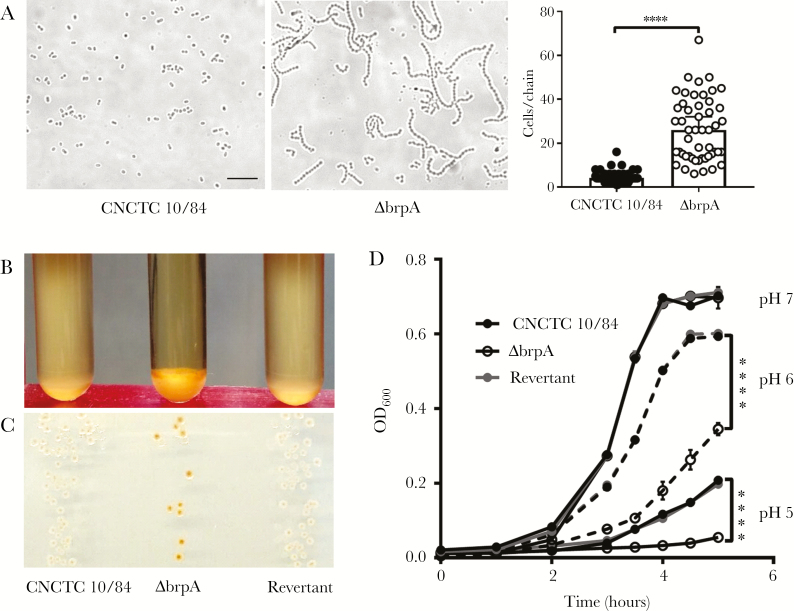
LCP proteins control cell envelope maintenance, chain length, and stress responses [13, 18, 27, 28]. To assess the impact of GBS BrpA, we generated an insertional mutant (ΔbrpA), using pHY304 to disrupt brpA [19]. Bright-field microscopy revealed a 6-fold increase (P < .0001) in the chain length of mid-logarithmic–phase GBS ΔbrpA (Figure 2A), with median chain length of 4 cells for the parental WT strain and 26 for the ΔbrpA strain. Transmission electron microscopy revealed no notable aberrant differences in cell shape, size, division planes, nucleoid structure, or cell-envelope electron density in the ΔbrpA strain (Supplementary Figure 2). Separation of GBS ΔbrpA dividing cells appeared similar to that of the WT strain; cells within chains were attached only by the outermost cell wall components (Supplementary Figure 2). Cellular morphology changes have been reported in LCP double or triple mutants [12, 13, 15], but deficiency of only 1 LCP protein may not impact cell division or morphology [12]. Complementation of ΔbrpA by using a full-length brpA construct under constitutive expression failed to yield viable colonies. Alternatively, the ΔbrpA mutant was serially passaged in the absence of Erm and screened for loss of the insertion (henceforth referred to as “revertant strains”).
Figure 2.
Group B Streptococcus (GBS) biofilm regulatory protein A (BrpA) contributes to cellular chain length, buoyancy, pigmentation, and acid tolerance. A, Bright-field images of wild-type (left) and ΔbrpA (center) strains in mid-log–phase growth, taken at 630× original magnification. Scale bar = 10 µm. Quantification of the chain length is shown at right, with data analyzed via the 2-tailed Mann-Whitney U test (n = 50). Median values with 95% confidence intervals are displayed. Experiments were performed twice independently, with representative images and combined data shown. B and C, Visualization of stationary-phase cultures (B) or colonies (C) grown for 18 hours at 37°C in Todd-Hewitt broth (THB) or on THB agar plates, respectively. D, Growth curves of GBS strains in THB at specified pH values. Independent replicates (n = 3) are shown, with lines indicating mean values ± standard errors of the mean. Data were analyzed using 2-way repeated measures analysis of variance with the Tukey multiple comparisons test. ****P < .0001.
LCP protein CpsA regulates capsule production in GBS [6], which influences buoyancy in liquid culture. Likewise, in planktonic culture, ΔbrpA organisms exhibited diminished buoyancy as compared to the WT and revertant strains (Figure 2B). Remarkably, ΔbrpA colonies possessed enhanced pigmentation with red/orange granadaene [29], compared with WT and revertant strains (Figure 2C). Since pigmentation and hemolytic activity have been positively correlated historically [19], we examined hemolytic activity of ΔbrpA, using live bacteria and hemolytic extracts (Supplementary Figure 3). Interestingly, there was no difference in hemolysis between the WT and ΔbrpA strains, with the nonhemolytic ΔcylE strain serving as a negative control. It has been proposed that granadaene and β-hemolysin are the same molecule [30]; however, we observed WT hemolytic extracts could be partially inactivated with a variety of proteases, suggesting a proteinaceous nature of β-hemolysin (Supplementary Figure 3). Although rare, hyperpigmented GBS strains that are not also hyperhemolytic have been reported [31].
LCP proteins influence tolerance to environmental and antimicrobial stressors [28, 32]. Although GBS ΔbrpA exhibited no growth defect at neutral pH level of 7.0, at acidic pH levels of 6.0 or 5.0 it demonstrated significantly impaired growth as compared to WT and revertant strains (Figure 2D). In contrast, WT and ΔbrpA strains had equivalent susceptibility to β-lactams antibiotics, chloramphenicol, LL-37, lysozyme, and hydrogen peroxide in minimum inhibitory concentration assays (Supplementary Table 1). In S. mutans, Psr deficiency likewise does not impact susceptibility to antimicrobials or oxidative stress [12], indicating that LCP proteins differentially influence susceptibility to stressors.
BrpA Supports Cell Survival, Biofilm Formation, and Epithelial Colonization
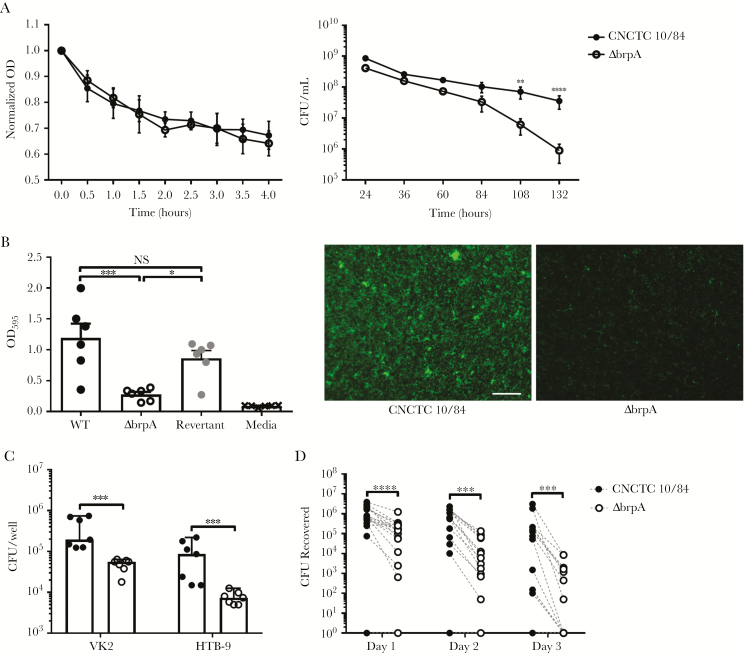
We observed similar rates of autolysis between WT (33%) and ΔbrpA (36%) strains in PBS (Figure 3A), but cell viability was significantly affected in planktonic cultures. BrpA deficiency led to a 10-fold reduction in viable CFU by day 4 of culture (P = .0055) and a 40-fold reduction (P < .0001) by day 5 (Figure 3A), although cultures achieved similar pH (approximately 4.5) at all time points (data not shown).
Figure 3.
Group B Streptococcus (GBS) biofilm regulatory protein A (BrpA) deficiency influences cell survival, biofilm formation, and epithelial cell adherence but not autolysis. (A) Autolysis (left) in cultures of wild-type (WT) and ΔbrpA organisms suspended in phosphate-buffered saline was measured spectrophotometrically (at OD600), using turbidity normalized to T = 0. Viability (right) of WT and ΔbrpA strains grown in Todd-Hewitt broth (THB) over time, as determined by plating on THB agar. Data from independent replicates (n = 3) are shown, with symbols representing mean values and lines indicating standard errors of the mean (SEMs). Data were analyzed using 2-way repeated measures analysis of variance with the Sidak multiple comparisons test. B, Biofilm formation of WT, ΔbrpA, and revertant strains quantified by crystal violet uptake (left). Independent replicates (n = 6) are shown, and lines represent mean values ± SEMs. Data were analyzed using 1-way ANOVA with the Holm-Sidak multiple comparisons test. Fluorescent images of in vitro biofilms (center and right) stained with Syto 13 (green), taken at 100× original magnification. Scale bar = 100 µm. Experiments were performed twice independently, with representative images shown. C, Adherence of WT (black symbols) and ΔbrpA (open circles) strains to vaginal (VK2) and bladder (HTB-9) epithelial cells. Independent replicates (n = 7) are shown, and lines represent median values ± 95% confidence intervals. Data were analyzed using the 2-tailed Mann-Whitney U test. D, In a competition model, CD1 mice were inoculated intravaginally with 1 × 107 CFU of WT and ΔbrpA organisms (2 × 107 CFU total/mouse), and bacterial burdens were quantified daily after infection. Symbols represent biological replicates (n = 18), and paired values (WT and ΔbrpA CFU) from each mouse are connected with dashed lines. Data were analyzed using the 2-tailed Wilcoxon matched-pairs signed rank test with Spearman rank-order correlation. ***P < .001, **P < .01, and *P < .05. NS, not significant.
To assess biofilms, WT and ΔbrpA strains were grown on tissue culture-treated polystyrene for 24h and adherent biomass determined by crystal violet staining [20]. GBS ΔbrpA was significantly attenuated for biofilm formation, compared with the WT (P = .0005) and revertant (P = .0174) strains (Figure 3B). Biomass did not differ between WT and revertant strains (P = .1840). Since modification of cell wall components, including the GBC and WTA, impact retention of crystal violet within the cytoplasm [11], we visualized biofilms, using the nucleic acid stain Syto13. Fluorescent images confirmed crystal violet quantification, with limited binding and sparse colonies formed by the ΔbrpA strain as compared to the WT strain (Figure 3B). GBS strains were incubated with human vaginal (VK2) or bladder (HTB-9) epithelial cells at a MOI of 10:1 for 30 minutes. In both cell types, the ΔbrpA strain showed significantly lower adherence, compared with the WT strain (P = .0006 for both VK2 and HTB-9 cells; Figure 3C). To characterize effects of BrpA-deficiency in vivo, we used a vaginal colonization model with WT and ΔbrpA strains administered in competition. Female CD-1 mice were vaginally inoculated with 1 × 107 CFU each of WT and ΔbrpA strains (2 × 107 CFU total/mouse). The vaginal lumen was swabbed daily to measure GBS persistence over 3 days. On all days tested, WT strain loads were significantly higher than ΔbrpA strain loads, with median differences of 5.5 × 105 CFU (P < .0001) on day 1, 3.6 × 105 CFU (P = .0002) on day 2, and 4.2 × 104 CFU (P = .0002) on day 3 (Figure 3D). Collectively, defects in biofilm formation, epithelial adherence, and vaginal persistence of the ΔbrpA mutant are evidence that BrpA is an important GBS determinant for successful mucosal colonization.
BrpA Deficiency Increases GBS Susceptibility to Human Blood and Neutrophil Killing
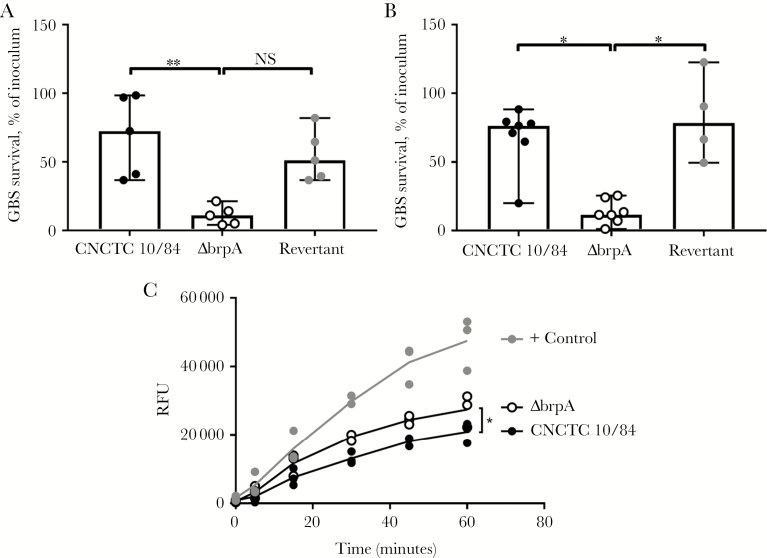
To assess BrpA in pathogenesis, ex vivo killing assays were performed in human blood specimens. Within 30 minutes, only a median of 10.9% of ΔbrpA organisms were recovered, compared with 72.4% of WT organisms (P = .0089) and 51.1% of revertant organisms (P = .0589; Figure 4A). Likewise, in neutrophil killing assays, only a median of 11.5% of ΔbrpA organisms were recovered, compared with 76.4% of WT organisms (P = .0152) and 78.4% of revertant organisms (P = .0140; Figure 4B). Neutrophil reactive oxygen species release was significantly greater upon challenge with the ΔbrpA strain as compared to the WT strain (P = .0421; Figure 4C), likely contributing to more-efficient killing of ΔbrpA bacteria. Thus, loss of BrpA compromises GBS resistance to blood immune components, particularly the microbicidal activity of neutrophils.
Figure 4.
Biofilm regulatory protein A (BrpA)–deficient group B Streptococcus (GBS) is more susceptible to killing by human whole blood and neutrophils. Human whole blood (A) and isolated neutrophil (B) killing of wild-type (WT), ΔbrpA, and revertant strains, expressed as a percentage of the inoculum. Biological replicates (n = 4–7) are shown, with lines indicating median values ± 95% confidence intervals. C, Reactive oxygen species production by neutrophils stimulated with WT, ΔbrpA, or phorbol 12-myristate 13-acetate as a positive control. Biological replicates (n = 3) are shown, with lines indicating mean values. Data were analyzed using the Kruskal-Wallis test with the Dunn multiple comparisons test (A and B) and repeated measures 2-way analysis of variance with the Tukey multiple comparisons test (C). **P < .01 and *P < .05. NS, not significant; RFU, relative fluorescent units.
GBS ΔbrpA Contributes to GBS Survival In Vivo
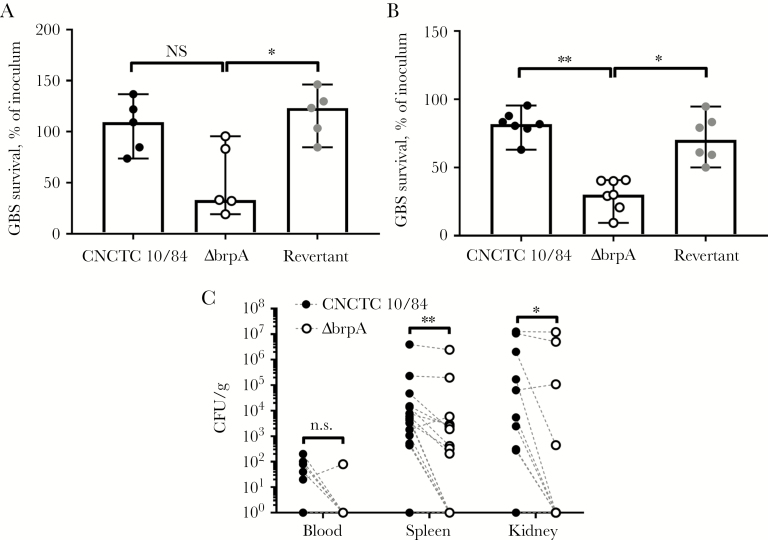
LCP proteins influence virulence in invertebrate and vertebrate models [6, 28, 33]. Mouse whole-blood specimens killed ΔbrpA organisms (median survival, 33.3% of the inoculum) more efficiently than WT organisms (109.3%; P = .1687) and revertant organisms (123.3%; P = .0327; Figure 5A). Similarly, murine peritoneal-derived leukocytes (85%–90% neutrophils) killed ΔbrpA organisms (median survival, 30.0% of the inoculum) more effectively than WT organisms (80.5%; P = .0012) and revertant organisms (70.1%; P = .0273; Figure 5B). To characterize BrpA-deficiency in vivo, we used a systemic infection model with WT and ΔbrpA bacteria administered in competition. Mice were infected intraperitoneally with 1 × 107 CFU each of the WT and ΔbrpA strains (2 × 107 CFU total/mouse), and 24 hours after infection, blood and tissue specimens were collected. Mice effectively cleared both WT and ΔbrpA organisms from the blood, with no bacteria detected in 10 of 15 and 14 of 15 mice, respectively (Figure 5C). However, WT bacterial loads were significantly higher than ΔbrpA bacterial loads in both spleens (median difference, 4941 CFU/g; P = .0012) and kidneys (297.4 CFU/g; P = .0273; Figure 5C).
Figure 5.
Biofilm regulatory protein A (BrpA)–deficient group B Streptococcus (GBS) is more susceptible to killing by murine whole blood and neutrophils and is attenuated in an in vivo sepsis model. Murine whole blood (A) or peritoneal-derived leukocytes (85% neutrophils; B) killing of wild-type (WT), ΔbrpA, and revertant strains, expressed as a percentage of the inoculum. Biological replicates (n = 5–7) are shown, with lines indicating median values ± 95% confidence intervals. C, In a competition model, CD1 mice were infected intraperitoneally with 1 × 107 colony-forming units (CFU) of WT and ΔbrpA strains (2 × 107 CFU total/mouse), and bacterial burdens were quantified 24 hours after infection. Symbols represent biological replicates (n = 15), and paired values (WT and ΔbrpA CFU) from each mouse are connected with dashed lines. Data were analyzed using the Kruskal-Wallis test with the Dunn multiple comparisons test (A and B) and the 2-tailed Wilcoxon matched-pairs signed rank test with Spearman rank-order correlation (C). **P < .01 and *P < .05. NS, not significant.
BrpA Deficiency Alters GBS Gene Expression
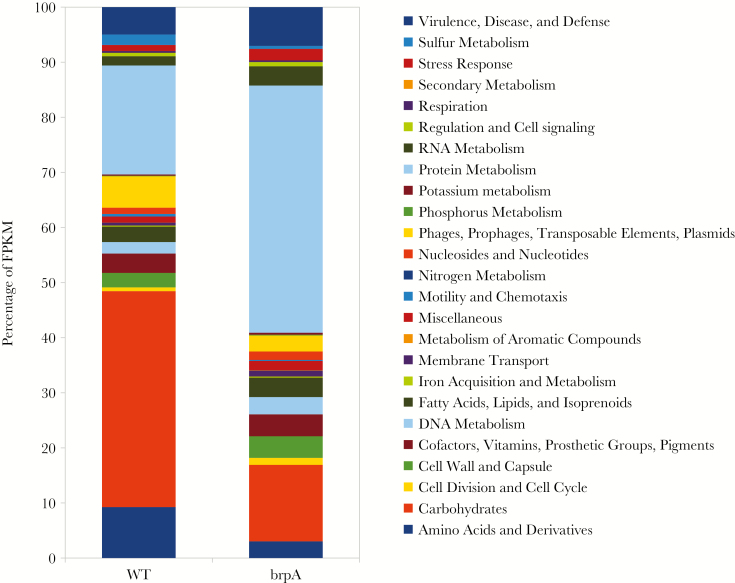
LCP proteins have been identified as transcriptional attenuators, with substantial changes of >200 genes reported [27, 28, 34]. RNA sequencing revealed 793 genes differentially expressed ≥2-fold in the ΔbrpA strain as compared to the WT strain (383 were upregulated and 410 were downregulated; P < .05; Supplementary Tables 2 and 3). Maximum changes in gene expression were approximately 50-fold. Genes were categorized into 25 functional groups on the basis of RAST annotations to determine molecular pathways differing between the WT and ΔbrpA strains (Figure 6).
Figure 6.
Transcriptome analysis of wild-type (WT) and ΔbrpA strains reveals multiple changes to bacterial physiology and metabolism. Late-logarithmic group B Streptococcus (GBS) cultures were subjected to RNA sequencing. Genes were categorized into RAST functional group and data expressed as the percentage of fragments per kilobase of transcript per million mapped reads (FPKM).
Metabolism
The most-pronounced differences between strains were genes involving protein metabolism (45% and 20% of fragments per kilobase of transcript per million mapped reads [FPKM] in ΔbrpA and WT strains, respectively) and carbohydrate uptake/fermentation (13% and 39% of FPKM, respectively). BrpA-deficient GBS showed a propensity for protein synthesis, with increased expression of ribosomal proteins, transfer RNA ligases, and amino acid transporters. Additionally, RNA metabolism was enhanced in the ΔbrpA strain (3.5% of FPKM), compared with the WT strain (1.7% of FPKM).
Transcriptional Regulation
We observed significant changes to 37% of the genome in GBS ΔbrpA, which was more than reported for other LCP mutants [27, 33]. These marked changes may largely be attributed to changes in global transcriptional regulators. GBS has 21 two-component systems (TCS) critical for controlling virulence, adherence, resistance to host defenses, and bacterial metabolism [35]. CovRS, which regulates approximately 27% of the entire genome [35], was not impacted by BrpA deficiency. However, 7 TCS were downregulated in GBS ΔbrpA, including LytST, FspSR, NsrRK, and 4 TCS, which remain uncharacterized. One TCS, dtlRS, was upregulated in ΔbrpA organisms. Global regulator codY was upregulated in GBS ΔbrpA. CodY has roles in biofilms, acid tolerance, and the stringent response and production of (p)ppGpp [36]. Increased codY expression may in part explain increased pigmentation of the ΔbrpA strain, as a recent study reported decreased pigmentation in ΔcodY [7].
Stress Response
Transcripts for stress responses were twice as abundant in the ΔbrpA strain (2% of FPKM), compared with the WT strain (1% of FPKM), matching phenotypic data of decreased tolerance for acid stress. GBS homologues (arcABCD) of the arginine deiminase system, a well-known streptococcal mechanism of countering acid stress, were >10-fold downregulated in GBS ΔbrpA.
Cell Wall Synthesis
Transcriptional expression of cell wall synthesis pathways was moderately expanded in ΔbrpA organisms (3.9% of FPKM), compares with WT organisms (2.6% of FPKM), which may be compensatory if these components are not appropriately anchored. The GBC, CPS, and peptidoglycan biosynthesis operons were increased in GBS ΔbrpA by >2-fold. Related cell surface proteins penicillin binding protein 2 (Pbp2) and Pbp2X were upregulated 4-fold and 2-fold, respectively, in the ΔbrpA strain. Enzymes predicted to synthesize LTA were increased 2–4-fold in ΔbrpA organisms, as was the dlt operon responsible for d-alanine incorporation into LTA, and iagA, which forms the diglucosyldiacylglycerol membrane anchor for LTA [37].
Pathogenesis and Host Interactions
Virulence gene expression was expanded in GBS ΔbrpA (7.0% of FPKM) as compared to WT GBS (4.9% of FPKM). Several adhesins were upregulated in the ΔbrpA strain, including PilB, Srr1, fibrinogen binding proteins FsbA and FbsB, and the fibronectin-binding protein sfbA. Additionally, factors important to complement evasion, C5a peptidase and C3-degrading proteinase, were upregulated. Members of the cyl operon, with roles in β-hemolysin and pigment production, were downregulated approximately 4-fold, as was Camp factor, by approximately 3-fold.
DISCUSSION
GBS remains an important pathogen in vulnerable populations, including neonates and elderly individuals [2]. Current antibiotic prophylaxis and treatment and the success of future vaccine and therapeutic strategies depend on targeting GBS surface physiology. We characterize BrpA, an LCP family protein, and demonstrate roles in cell separation, biofilm formation, colonization, and virulence. LCP expression is growth-phase dependent and typically highest in the early stationary or mid-log phases of growth [18, 28, 32]; similarly, GBS brpA expression is 7-fold higher at the mid-log phase, compared with expression during the stationary phase [38]. GBS brpA expression is downregulated in human blood [39] but is not effected by pH [40] or induction of the stringent response [7]. Furthermore, brpA is not regulated by TCS LiaRS [41], RgfAC [42], or CovRS [40]. Prior screens had suggested that brpA is not an essential gene [43], and we have confirmed this here by insertional disruption.
We noted several changes in GBS physiology with loss of brpA organisms, including sedimentation during growth. Loss of buoyancy can result from either GBC [11] or capsule [6] deficiency or can arise due to self-aggregation from loss of d-alanine residues in LTA [44]. LCP proteins affect capsule expression in S. pneumonia [45], S. aureus [46], and GBS [6], resulting in self-agglutination and sedimentation [13, 33]. Similar to other LCP mutants, including GBS CpsA [6, 18], ΔbrpA strain elongated chains led to a 6-fold CFU reduction at an OD identical to that of the WT strain; thus, assays yielding small CFU differences (<10-fold) between WT and ΔbrpA strains are difficult to interpret. When feasible, we made adjustments for the impact of chain length by expressing CFU data as a percentage of CFU input (Figures 4 and 5).
In gram-positive bacteria, longer chains correspond to reduced activity of autolytic enzymes [47], and, accordingly, we found that autolysin/murein hydrolase LrgA was downregulated in the ΔbrpA strain. Many LCP mutants display accelerated autolysis [12–14, 18, 34], but under our conditions, BrpA deficiency did not alter autolysis, as reported for MsrR deficiency in S. aureus [32, 33]. Over days, GBS BrpA deficiency accelerated death in static culture, and upregulation of RecA and ClpXP protease homologues support involvement of apoptosis-like programmed cell death pathways [48]. Aberrant stress responses may contribute, since ΔbrpA organisms also demonstrated reduced acid tolerance, a common trait of LCP mutants [12, 27].
Loss of BrpA influenced GBS-host interactions in both colonization and disease, including a severe biofilm defect. Several surface constituents, including pilus type 2a [49], antigens I/II [20], and CPS [5], promote biofilms, yet all of these constituents are aberrantly expressed in GBS ΔbrpA, so identification of precise components responsible for the biofilm deficit is a limitation of this study. BrpA plays a prominent role in GBS survival in human whole blood and GBS resistance to neutrophils, likely through modification of CPS. GBS CPS is vital to survival in blood [7], and its abundant terminal sialic acids suppress neutrophil function through engagement of inhibitory sialic acid–binding immunoglobulin-like lectins (ie, Siglecs) [35]. In vivo, GBS ΔbrpA was recovered in significantly lower abundance in host tissues in the presence of WT organisms, implying that bacterial fitness, rather than host immune response, is the main contributor of decreased virulence of BrpA-deficient GBS.
BrpA deficiency resulted in significant transcriptional changes to approximately 48% of the entire genome, with a ≥2-fold change in 37%. These changes appear specific to the loss of brpA and not to polar effects, as expression of immediately downstream loci were not different from findings for WT organisms (Supplementary Figure 4). Of note, transcript expression of several cellular processes contrasted with phenotypic results, a discordance all too familiar in biology. Specifically, while levels of several adhesin transcripts were increased, GBS ΔbrpA consistently displayed decreased epithelial adherence (Figure 3). Possible explanations include increased capsular expression in GBS ΔbrpA, which itself inhibits adherence [50], or improper anchoring of adhesins. Additionally, expression of the cyl operon responsible for granadaene [29] was downregulated in ΔbrpA organisms, yet colony pigmentation was increased (Figure 2). None of the known cyl regulators, including CovRS and RovS, were altered in ΔbrpA organisms, suggesting there remain undescribed transcriptional or posttranslational regulators of the cyl operon. Additionally, the disconnect between phenotypic pigmentation and hemolytic activity of the ΔbrpA strain (Supp. Figure 3) adds to the complexity of the unique nature of GBS pigmentation and β-hemolysis. Genetic manipulation of brpA will serve as a tool for future discoveries.
The magnitude of transcriptional changes during brpA deficiency may explain our failure to obtain viable complementation under constitutive expression. Conversely, constitutive expression of cpsA is not lethal [6]. LCP proteins play distinct roles in cell wall maintenance but may also compensate for each other [16, 17], and increased expression of cpsA (by >2-fold) in ΔbrpA organisms corroborates this. Deletion of all 3 LCP enzymes in Staphylococcus aureus does not appear to influence surface protein anchoring [15], but this cannot yet be ruled out in GBS, since expression of several sortases were increased in GBS ΔbrpA. Contribution of each GBS LCP protein to proper surface anchoring of GBC and CPS is a topic of future study.
LCP proteins are exclusive to bacteria and universally found in gram-positive organisms [26]. As such, they are appealing targets for antimicrobial therapies. Our observations support that the LCP protein BrpA is one such target, owing to its critical involvement in GBS cell wall modification to promote both successful colonization and blood survival within its human host.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank Michael Florio, for breeding and maintaining the mice, and the UC–San Diego vivarium staff, for their assistance.
K. A. P. and V. N. conceived and designed experiments. K.A.P., J.D., N.A., M.M.A., and A.V. performed experiments. K. A. P., M. M. A., and V. N. analyzed and interpreted results. J. D. L, K. Z., and D. J. G. contributed reagents, materials, analyses, discussions, and manuscript edits. K. A. P. and V. N. drafted the manuscript.
Financial support. This work was supported by the National Institutes of Health (grants HL107150 and HD090259 to V. N. and grant AR071731 to KZ) and the University of California Chancellor’s Postdoctoral Fellowship Program (fellowship to K. A. P.)
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Presented in part: 20th Lancefield International Symposium on Streptococci and Streptococcal Diseases, 16–20 October 2018, Denarau Island, Fiji; 6th International Conference on Gram-Positive Pathogens, 10 October 2016, Omaha, Nebraska.
References
- 1. Russell NJ, Seale AC, O’Driscoll M, et al. ; GBS Maternal Colonization Investigator Group Maternal colonization with Group B streptococcus and serotype distribution worldwide: systematic review and meta-analyses. Clin Infect Dis 2017; 65:100–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Phares CR, Lynfield R, Farley MM, et al. Epidemiology of invasive Group B streptococcal disease in the United States, 1999–2005. JAMA 2008; 299:2056–65. [DOI] [PubMed] [Google Scholar]
- 3. Skoff TH, Farley MM, Petit S, et al. Increasing burden of invasive Group B streptococcal disease in nonpregnant adults, 1990–2007. Clin Infect Dis 2009; 49:85–92. [DOI] [PubMed] [Google Scholar]
- 4. Metcalf BJ, Chochua S, Gertz RE Jr., et al. Short-read whole genome sequencing for determination of antimicrobial resistance mechanisms and capsular serotypes of current invasive Streptococcus agalactiae recovered in the USA. Clin Microbiol Infect 2017; 23:574 e7–e14. [DOI] [PubMed] [Google Scholar]
- 5. Rosini R, Margarit I. Biofilm formation by Streptococcus agalactiae: influence of environmental conditions and implicated virulence factors. Front Cell Infect Microbiol 2015; 5:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hanson BR, Runft DL, Streeter C, Kumar A, Carion TW, Neely MN. Functional analysis of the CpsA protein of Streptococcus agalactiae. J Bacteriol 2012; 194:1668–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hooven TA, Catomeris AJ, Bonakdar M, et al. Streptococcus agalactiae stringent response enhances virulence and persistence in human blood. Infect Immun 2018; 86:e00612–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Le Doare K, Faal A, Jaiteh M, et al. Association between functional antibody against Group B streptococcus and maternal and infant colonization in a Gambian cohort. Vaccine 2017; 35:2970–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Madhi SA, Koen A, Cutland CL, et al. Antibody kinetics and response to routine vaccinations in infants born to women who received an investigational trivalent Group b streptococcus polysaccharide CRM197-conjugate vaccine during pregnancy. Clin Infect Dis 2017; 65:1897–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Deng L, Kasper DL, Krick TP, Wessels MR. Characterization of the linkage between the type III capsular polysaccharide and the bacterial cell wall of Group B streptococcus. J Biol Chem 2000; 275:7497–504. [DOI] [PubMed] [Google Scholar]
- 11. Caliot É, Dramsi S, Chapot-Chartier MP, et al. Role of the Group B antigen of Streptococcus agalactiae: a peptidoglycan-anchored polysaccharide involved in cell wall biogenesis. PLoS Pathog 2012; 8:e1002756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bitoun JP, Liao S, McKey BA, et al. Psr is involved in regulation of glucan production, and double deficiency of BrpA and Psr is lethal in Streptococcus mutans. Microbiology 2013; 159:493–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Over B, Heusser R, McCallum N, et al. LytR-CpsA-Psr proteins in Staphylococcus aureus display partial functional redundancy and the deletion of all three severely impairs septum placement and cell separation. FEMS Microbiol Lett 2011; 320:142–51. [DOI] [PubMed] [Google Scholar]
- 14. Massidda O, Kariyama R, Daneo-Moore L, Shockman GD. Evidence that the PBP 5 synthesis repressor (psr) of Enterococcus hirae is also involved in the regulation of cell wall composition and other cell wall-related properties. J Bacteriol 1996; 178:5272–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chan YG, Frankel MB, Dengler V, Schneewind O, Missiakas D. Staphylococcus aureus mutants lacking the LytR-CpsA-Psr family of enzymes release cell wall teichoic acids into the extracellular medium. J Bacteriol 2013; 195:4650–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chan YG, Kim HK, Schneewind O, Missiakas D. The capsular polysaccharide of Staphylococcus aureus is attached to peptidoglycan by the LytR-CpsA-Psr (LCP) family of enzymes. J Biol Chem 2014; 289:15680–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schaefer K, Matano LM, Qiao Y, Kahne D, Walker S. In vitro reconstitution demonstrates the cell wall ligase activity of LCP proteins. Nat Chem Biol 2017; 13:396–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chatfield CH, Koo H, Quivey RG Jr. The putative autolysin regulator LytR in Streptococcus mutans plays a role in cell division and is growth-phase regulated. Microbiology 2005; 151:625–31. [DOI] [PubMed] [Google Scholar]
- 19. Pritzlaff CA, Chang JC, Kuo SP, Tamura GS, Rubens CE, Nizet V. Genetic basis for the beta-haemolytic/cytolytic activity of Group B streptococcus. Mol Microbiol 2001; 39:236–47. [DOI] [PubMed] [Google Scholar]
- 20. Chuzeville S, Dramsi S, Madec JY, Haenni M, Payot S. Antigen I/II encoded by integrative and conjugative elements of Streptococcus agalactiae and role in biofilm formation. Microb Pathog 2015; 88:1–9. [DOI] [PubMed] [Google Scholar]
- 21. Patras KA, Wescombe PA, Rösler B, Hale JD, Tagg JR, Doran KS. Streptococcus salivarius K12 limits Group B streptococcus vaginal colonization. Infect Immun 2015; 83:3438–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Patras KA, Coady A, Olson J, et al. Tamm-Horsfall glycoprotein engages human Siglec-9 to modulate neutrophil activation in the urinary tract. Immunol Cell Biol 2017; 95:960–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Patras KA, Doran KS. A murine model of Group B streptococcus vaginal colonization. J Vis Exp 2016. Nov 16; (117). doi: 10.3791/54708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Finn RD, Coggill P, Eberhardt RY, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 2016; 44:D279–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 2001; 305:567–80. [DOI] [PubMed] [Google Scholar]
- 26. Hübscher J, Lüthy L, Berger-Bächi B, Stutzmann Meier P. Phylogenetic distribution and membrane topology of the LytR-CpsA-Psr protein family. BMC Genomics 2008; 9:617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wen ZT, Baker HV, Burne RA. Influence of BrpA on critical virulence attributes of Streptococcus mutans. J Bacteriol 2006; 188:2983–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bitoun JP, Liao S, Yao X, et al. BrpA is involved in regulation of cell envelope stress responses in Streptococcus mutans. Appl Environ Microbiol 2012; 78:2914–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rosa-Fraile M, Rodríguez-Granger J, Haidour-Benamin A, Cuerva JM, Sampedro A. Granadaene: proposed structure of the Group B streptococcus polyenic pigment. Appl Environ Microbiol 2006; 72:6367–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Whidbey C, Harrell MI, Burnside K, et al. A hemolytic pigment of Group B streptococcus allows bacterial penetration of human placenta. J Exp Med 2013; 210:1265–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lupo A, Ruppen C, Hemphill A, Spellerberg B, Sendi P. Phenotypic and molecular characterization of hyperpigmented Group B Streptococci. Int J Med Microbiol 2014; 304:717–24. [DOI] [PubMed] [Google Scholar]
- 32. Rossi J, Bischoff M, Wada A, Berger-Bächi B. MsrR, a putative cell envelope-associated element involved in Staphylococcus aureus sarA attenuation. Antimicrob Agents Chemother 2003; 47:2558–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hübscher J, McCallum N, Sifri CD, et al. MsrR contributes to cell surface characteristics and virulence in Staphylococcus aureus. FEMS Microbiol Lett 2009; 295:251–60. [DOI] [PubMed] [Google Scholar]
- 34. Lazarevic V, Margot P, Soldo B, Karamata D. Sequencing and analysis of the Bacillus subtilis lytRABC divergon: a regulatory unit encompassing the structural genes of the N-acetylmuramoyl-L-alanine amidase and its modifier. J Gen Microbiol 1992; 138:1949–61. [DOI] [PubMed] [Google Scholar]
- 35. Patras KA, Nizet V. Group B streptococcal maternal colonization and neonatal disease: molecular mechanisms and preventative approaches. Front Pediatr 2018; 6:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lemos JA, Nascimento MM, Lin VK, Abranches J, Burne RA. Global regulation by (p)ppGpp and CodY in Streptococcus mutans. J Bacteriol 2008; 190:5291–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Doran KS, Engelson EJ, Khosravi A, et al. Blood-brain barrier invasion by Group B Streptococcus depends upon proper cell-surface anchoring of lipoteichoic acid. J Clin Invest 2005; 115:2499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sitkiewicz I, Musser JM. Analysis of growth-phase regulated genes in Streptococcus agalactiae by global transcript profiling. BMC Microbiol 2009; 9:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mereghetti L, Sitkiewicz I, Green NM, Musser JM. Extensive adaptive changes occur in the transcriptome of Streptococcus agalactiae (Group B streptococcus) in response to incubation with human blood. PLoS One 2008; 3:e3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Santi I, Grifantini R, Jiang SM, et al. CsrRS regulates Group B Streptococcus virulence gene expression in response to environmental pH: a new perspective on vaccine development. J Bacteriol 2009; 191:5387–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Klinzing DC, Ishmael N, Dunning Hotopp JC, et al. The two-component response regulator LiaR regulates cell wall stress responses, pili expression and virulence in Group B Streptococcus. Microbiology 2013; 159:1521–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gendrin C, Lembo A, Whidbey C, et al. The sensor histidine kinase RgfC affects Group B streptococcal virulence factor expression independent of its response regulator RgfA. Infect Immun 2015; 83:1078–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hooven TA, Catomeris AJ, Akabas LH, et al. The essential genome of Streptococcus agalactiae. BMC Genomics 2016; 17:406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Poyart C, Lamy MC, Boumaila C, Fiedler F, Trieu-Cuot P. Regulation of D-alanyl-lipoteichoic acid biosynthesis in Streptococcus agalactiae involves a novel two-component regulatory system. J Bacteriol 2001; 183:6324–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Eberhardt A, Hoyland CN, Vollmer D, et al. Attachment of capsular polysaccharide to the cell wall in Streptococcus pneumoniae. Microb Drug Resist 2012; 18:240–55. [DOI] [PubMed] [Google Scholar]
- 46. Wang Y, Sun D, Song F, Hu Y, Smith DE, Jiang H. Expression and regulation of the proton-coupled oligopeptide transporter PhT2 by LPS in macrophages and mouse spleen. Mol Pharm 2014; 11:1880–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Buist G, Kok J, Leenhouts KJ, Dabrowska M, Venema G, Haandrikman AJ. Molecular cloning and nucleotide sequence of the gene encoding the major peptidoglycan hydrolase of Lactococcus lactis, a muramidase needed for cell separation. J Bacteriol 1995; 177:1554–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bayles KW. Bacterial programmed cell death: making sense of a paradox. Nat Rev Microbiol 2014; 12:63–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rinaudo CD, Rosini R, Galeotti CL, et al. Specific involvement of pilus type 2a in biofilm formation in Group B Streptococcus. PLoS One 2010; 5:e9216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tamura GS, Nittayajarn A. Group B streptococci and other gram-positive cocci bind to cytokeratin 8. Infect Immun 2000; 68:2129–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA sequencing data set is deposited at the NCBI Sequence Read Archive (accession no. SRP140532). Other data sets and materials generated and analyzed during this study are available from the corresponding author on request.