Summary
Background
The hierarchical hemostasis response involves a self-inhibitory feature of von Willebrand Factor (VWF) that has not been fully characterized. The residues flanking the A1 domain of VWF are important in this self-inhibition by forming an autoinhibitory module (AIM) that masks A1.
Objectives
To delimit the AIM sequence and to evaluate the cooperative interplay between the discontinuous AIM regions.
Methods
ELISA, flow cytometry, thermal stability assay, and hydrogen-deuterium exchange (HDX) mass spectrometer were used to characterize recombinant VWF A1 fragments varying in length.
Results
The longest A1 fragment (rVWF1238-1493) showed a higher inactivity in binding platelet receptor GPIbα and greater thermostability than its shorter counterparts. The HDX results showed that most of the N-terminal residues and the residues 1459 to 1478 at C-terminus of rVWF1238-1493 have a slower deuterium uptake compared to its denatured counterpart, implying that these residues may interact with the A1 domain. In contrast, residues 1479 to 1493 exhibited less difference than the denatured form, indicating that these residues are unlikely involved in binding the A1 domain. The A1 fragment that lacks either the entire C-AIM, rVWF1238-1461, or the entire N-AIM, rVWF1271-1493, exhibited high GPIbα-binding affinity and low thermostability, suggesting that removal of either N- or C- terminal residues resulted in loss of AIM inhibition of the A1 domain.
Conclusion
AIM is likely composed of residues 1238–1271 (N-AIM) and 1459–1478 (C-AIM). Neither the N-AIM nor the C-AIM alone could fully inhibit the A1 domain binding to GPIbα.
Keywords: Blood Platelets, Tandem Mass Spectrometry, von Willebrand factor, Platelet Glycoprotein GPIb-IX Complex, Hemostasis
Von Willebrand Factor (VWF) is a large blood glycoprotein essential for normal hemostasis. Most plasma VWF molecules are secreted in a multimeric form by endothelial cells. These VWF molecules often adopt a loosely condensed “ball-of-yarn” shape under low shear conditions and remain hemostatically inactive[1]. Activation of VWF in vivo normally requires high shear conditions, where tension due to flow straightens VWF molecules in the direction of flow and subsequently exposes the platelet-binding domain in VWF–the A1 domain (residues 1272–1458). Binding of the VWF A1 domain to the platelet receptor glycoprotein (GP)Ibα triggers the downstream signaling pathway of platelets and leads to their activation. Deficiency or malfunction of VWF is a common cause of bleeding or thrombosis[2].
How VWF retains its hemostatic inactivity under low shear conditions remains ambiguous. Manifestly, the ball-of-yarn shape of VWF may shadow the A1 domain and therefore block the GPIbα binding. However, studies on isolated or recombinant A1 fragments showed that those A1 fragments containing residues flanking the A1 domain are as inactive as full-length VWF under low shear conditions[3–7]. But those A1 fragments containing less flanking residues are of high hemostatic activity in the absence of shear or agonist treatment[8, 9]. In addition, a recent study showed that elongation of VWF is not sufficient to promote the binding of GPIbα when tension force is below a certain threshold[10]. Obviously, these observations are against the VWF-shadowing model but instead raise the hypothesis that the self-inhibition of VWF is regulated by local interactions between the A1 domain and its flanking regions. The flanking regions at the N- and C-terminal of the A1 domain are held together by an intramolecular disulfide bond between Cys1272 and Cys1458, like two ribbons held in one hand (Fig. 1A). This organization renders a spatial advantage for these flanking residues to act in a cooperative manner and hypothetically bind the A1 domain and block the binding of GPIbα. However, the A1 crystal structures available to date have not addressed this hypothesis, partially because the flanking residues are not included or resolved in these studies.
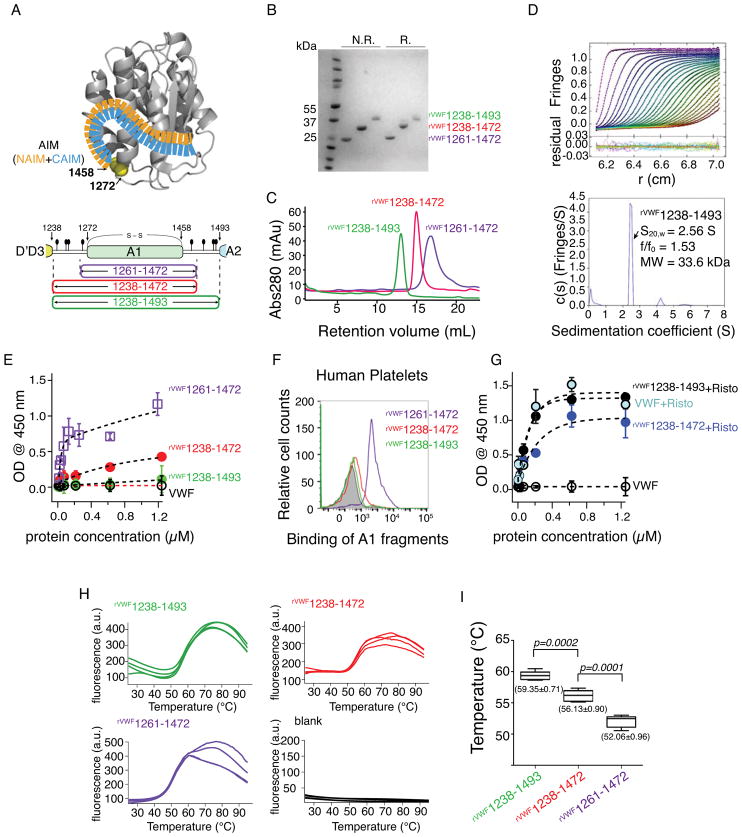
Figure 1.
Binding and thermostability analysis of A1 fragments. (A) An AIM model from the previous study [12] and an illustration of A1 fragments. The N- and C-terminal sequences flanking the von Willebrand factor (VWF) A1 domain were designated as NAIM (orange) and CAIM (celestial blue). The starting and ending residue numbers of each fragment are labeled. O-glycosylation sites (filled ovals) and the 1272-1458 disulfide bond are marked. (B) A Coomassie blue-stained polyacrylamide gel showing the purified proteins under non-reducing (N.R.) and reducing (R.) conditions. (C) Superimposed gel filtration chromatograms of A1 fragment from Superdex 200 10/300 GL SEC column. (D) Sedimentation velocity results for rVWF1238- 1493 showing the fitted interference scans and residuals (upper) and sedimentation coefficient distributions (lower). Every second scan and every third data point are shown for clarity. (E, G) ELISA measurements (n = 3) of the isotherms of A1 fragments and human plasma-derived VWF binding to immobilized GPIb-IX in the absence (E) and presence (G) of 1.5 mg/mL ristocetin. (F) Representative histogram of flow cytometry measurement to determine the binding of A1 fragments to platelets in platelet rich plasma. (H) SYPRO Orange fluorescence intensity changes over the temperature of each A1 fragment (n=4). Temperature increment is 1°C per minute. (a.u., arbitrary unit) (I) Whisker plot of the melting temperature measurements from (H). Mean value ± standard deviations are marked under each plot. A paired t-test was conducted using GraphPad Prism 5 (GraphPad Software, Inc. CA the USA) and the p-value was marked.
We recently tested the aforementioned hypothesis using hydrogen-deuterium exchange (HDX) mass spectroscopy–a powerful tool for probing a protein’s structure in solution in high resolution[11, 12]. We characterized two recombinant A1 fragments: rVWF1261-1472 and rVWF1238-1472, varying in the length of flanking residue sequence and exhibiting disparate binding activity to GPIbα[3, 8]. HDX results suggested that the residues flanking the A1 domain in rVWF1238-1472 (i.e., residues 1238–1271 and residues 1459–1472) bound the A1 domain cooperatively forming a discontinuous autoinhibitory module (AIM)[9]. The binding of AIM residues to the A1 domain provided a mechanism for the disparate GPIbα binding activity between rVWF1261-1472 and rVWF1238-1472. However, as rVWF1238-1472 contains only a portion of the C-terminal flanking region, the full spectrum of the AIM remained unclear. In addition, the cooperation of the discontinuous regions of AIM binding to the A1 domain was not directly verified. To delimit the AIM sequence and to evaluate the cooperative interaction between the discontinuous AIM regions, we characterized rVWF1238-1493, rVWF1238-1461 and rVWF1271-1493 in this study. rVWF1238-1493 contains the full-range flanking regions, whereas rVWF1238-1461 or rVWF1271-1493 contains either the entire N-terminal or the entire C-terminal but is missing the other half of flanking regions. Our results indicate that the range of the AIM likely stops at residue Gly1479 and that the cooperative interactions of the two discontinuous flanking regions of AIM are required for protecting A1.
Material and Methods
Plasma-derived VWF was purchased from Haematological Technologies (Essex Junction, VT). Ristocetin was purchased from MP Biomedicals (Santa Ana, CA). Recombinant GPIb-IX complex was purified in the lab as previously described[13]. SYPRO Orange dye was purchased from Thermo Fisher Scientific (Waltham, MA). Ammonium sulfate (protease free) was purchased from Fisher Scientific (Merelbeke, Belgium). 5-mL HisTrap Excel column, Superdex 200 10/300 GL SEC column and HiLoad Superdex 200pg 1660 size exclusion column were purchased from GE Healthcare Life Science (Pittsburgh, PA).
Protein purification
Expression and purification of rVWF1261-1472, rVWF1238-1472, rVWF1238-1461 and rVWF1271-1493 follow the same protocol as previously described[9]. rVWF1238-1493 was purified by using a two-step purification protocol. The cell culture media containing rVWF1238-1493 was centrifuged at 5,000 × g for 30 minutes at 4° C and sterile-filtered. Ammonium sulfate powder was added slowly with stirring and protein pellet between 50% ~70% ammonium sulfate concentration was collected and dialyzed overnight in 1X PBS, pH 7.4. The dialyzed sample was loaded to a pre-equilibrated 5-mL HisTrap Excel column (150 mM NaCl, 20 mM PO4, pH 7.4) at 1 mL/min. The column was washed (150 mM NaCl, 20 mM PO4, 20 mM imidazole, pH 7.4) until the UV280 was lower than 0.010 and protein was eluted (150 mM NaCl, 20 mM PO4, 500 mM imidazole, pH 7.4). Elution was concentrated using a Vivaspin20 (10,000 MWCO) and loaded to a HiLoad Superdex 200pg 1660 size exclusion column. Fractions containing the pure protein were pooled, dialyzed overnight in 1X PBS and concentrated in a Vivaspin20, 10,000 MWCO. Concentration was determined using the extinction coefficient of 0.585.
ELISA, analytical ultracentrifugation, and hydrogen-deuterium exchange (HDX) mass spectrometry were performed as described[9].
Flow cytometry measurement
The informed consent and related protocols were approved by Emory University institutional review boards. Citrated whole blood was obtained from healthy volunteers, from which platelet-rich plasma (PRP) was prepared. Platelet counts were measured using a Sysmex XP-300 analyzer (Lincolnshire, IL, USA). To measure platelet binding, PRP was incubated with defined concentration of proteins for 10 min at room temperature. After washing, the samples were mixed with 1 μg/mL APC-labeled anti-His-tag MAb, fixed with 4% paraformaldehyde solution and analyzed on a CytoFLEX S flow cytometer (Beckman, Boulevard Brea, CA). The signal from entire platelet population (10,000 platelets) was analyzed using FlowJo (FlowJo LLC, Ashland, OR).
Melting temperature (Tm) measurement
Tm was measured using a thermofluor assay operated on a real-time PCR. The proteins were incubated in a 96-well PCR plate with SYPRO Orange dye, which undergoes a significant increase in quantum yield upon binding hydrophobic elements exposed during protein denaturation. Four replicates from each protein were performed and the thermo denaturation curves (reflected as the change in fluorescence intensity) of these replicates were fit as described[14]. The final Tm value reflects the average of these measurements and the experimental error (standard deviation) was marked.
Results
Autoinhibitory activity and thermal stability of rVWF1238-1493
The migration speeds of rVWF1238-1493 on SDS-PAGE gel at non-reducing and reducing conditions are relatively close to each other, similar to that of rVWF1238-1472 and rVWF1261-1472 (Fig. 1B). In solution, rVWF1238-1493 stays primarily as a monomer as shown by the size-exclusion chromatography and analytical ultracentrifugation analyses (Fig. 1C and 1D). We compared the binding of rVWF1238-1493, rVWF1238-1472, rVWF1261-1472, and plasma-derived VWF to an immobilized GPIb-IX complex using ELISA. In the absence of agonist, rVWF1261-1472 tightly bound GPIb-IX, rVWF1238-1472 modestly bound, whereas rVWF1238-1493 and plasma-derived VWF did not bind up to 1.2 μM (Fig. 1E). The binding results were further examined by comparing the binding of these recombinant proteins with platelets in PRP (500 × 103/μL platelets) at 200 nM using flow cytometry. The results from flow cytometry measurement were consistent with the ELISA data (Fig. 1F). Similar results were obtained with washed platelets in mTyrodes’ buffer (134 mM NaCl, 0.34 mM Na2HPO4, 2.9 mM KCl, 1 mM MgCl2, 5 mM glucose, 12 mM NaHCO3, 20 mM HEPES, pH 7.35) as well (data not shown). In addition, adding ristocetin–an agonist that induces GPIbα-VWF interaction–stimulated the binding of plasma-derived VWF, rVWF1238-1493, and rVWF1238-1472 to GPIb-IX (Fig. 1G). All of these constructs contain the A1 domain with same conformation that is locked by the intramolecular disulfide bond[7, 8, 15–20]. The difference between these constructs is the number of AIM residues flanking the A1 domain. Therefore, we hypothesize that the decrease in GPIbα-binding activity of these constructs in the absence of agonist is due to the increase in the binding of AIM residues to the A1 domain. This binding likely affects the accessibility of the GPIbα-binding site on the A1 domain, but should entail minimal changes in the A1 domain structure. Since the binding of the AIM residues would increase the thermostability of the A1 domain, we then tested this hypothesis by measuring the thermostability of these samples. We measured the melting temperature (Tm) of these samples using a thermofluor assay. Because conformation of the A1 domain in these constructs is the same, the value of Tm can be used to compare the thermal stability of these constructs. During the thermal denaturation, the exposure of the hydrophobic elements in the A1 domain was probed by the change of the quantum yield of a fluorophore. The Tm was 59.35 ± 0.71°C for rVWF1238-1493, 56.13 ± 0.90°C for rVWF1238-1472, and 52.06 ± 0.96°C for rVWF1261-1472 (Fig. 1H and 1I). rVWF1261-1472 has the lowest Tm, suggesting it has the least coupling effect from the AIM residues. rVWF1238-1493 had a higher Tm than rVWF1238-1472, implying that it has a higher coupling effect than rVWF1238-1472. Since rVWF1238-1493 carries 22 more C-terminal flanking residues than rVWF1238-1472, the increased thermostability of rVWF1238-1493 may be attributed to the coupling of these extra flanking residues to the A1 domain. These results are consistent with our hypothesis that the coupling of AIM residues increases the thermostability and decreases the binding activity of A1.
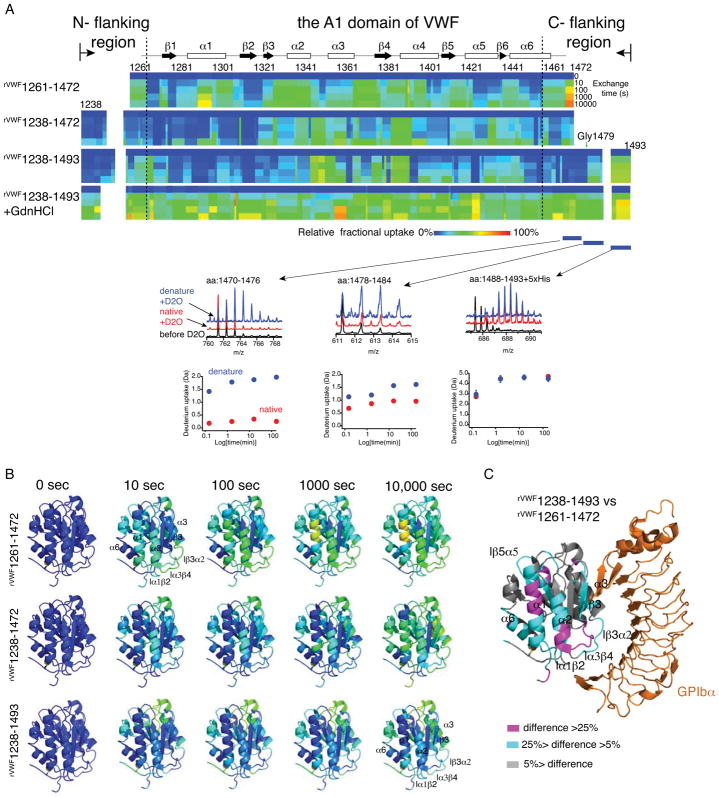
Many peripheral regions in rVWF1238-1493 are protected from HDX
To further verify the coupling of the flanking residues to the A1 domain and delimit the range of AIM in rVWF1238-1493, we conducted a similar HDX analysis of rVWF1238-1493 as done previously with rVWF1238-1472 and rVWF1261-1472[9]. In this approach, the deuterium exchange of the backbone proton on the amino acid chain of rVWF1238-1493 was evaluated as a function of time using a tandem mass spectrometer that can sequence the peptide fragments. The coupling of the flanking residues to the A1 domain would reduce the solvent accessibility of the affected backbone proton and thus decrease their deuterium exchange rate. 217 peptic fragments were identified and confirmed by MS/MS, covering ~97% of the protein sequence with a redundancy >10. No peptic fragment was identified covering the contiguous O-glycosylation sites T1255/T1266 and S1486/T1487, which was likely due to the known limitation for mass spectrometry in identifying glycopeptide. The deuterium uptake over time of each fragment in both native and denatured proteins was overlaid to the primary protein sequence to generate a heat map (Fig. 2A). Consistent with our expectation, the entire N- terminal flanking residues and residues 1459–1478 in the C- terminal flanking region in rVWF1238-1493 exhibited reduced HDX compared to their denatured counterparts (Fig. 2A), suggesting that these residues are coupled with the A1 domain. In comparison, residues after Gly1479 showed less difference in HDX compared to their denatured counterparts, implying that these residues may not bind the A1 domain. Three representative peptides around the Gly1479 were selected to show this transition. Peptide 1470–1476 showed low deuterium uptake in native rVWF1238-1493 but high uptake in the denatured form. This difference in deuterium uptake between native and denatured forms became smaller in peptide 1478–1484, and disappeared in peptide 1488–1493(+5His) (Fig. 2A). These results indicated that the AIM is likely composed of residues 1238–1271 (N-terminal flanking region, named N-AIM) and 1459–1478 (C-terminal flanking region, named C-AIM). Consistently, the flanking residues 1243–1271 and 1459–1481 were found to resist the protease-digestion in earlier studies[5, 21]. This resistance can be well explained by the binding of these residues to the A1 domain.
Figure 2.
Hydrogen-deuterium exchange (HDX) characterization of A1 fragments. (A) Residual HDX heat map of each A1 fragments from 10 s to 10 000 s. The line for 0 s denoted the results obtained without exchange. Relative fractional deuterium uptake was plotted using the rainbow color scale. Secondary structures of each region were labeled on top. Denaturation of rVWF1238-1493 (rVWF1238-1493 +GdnHCl) was performed as described [9]. Three representative peptides around Gly1479 were selected to show their raw mass spectra and deuterium uptake in the native (red) and denatured (blue) forms. (B) 3D projection of HDX data at the indicated exchange time. HDX data were mapped to the structure of the A1 domain (Protein Data Bank, 1SQ0, only residues 1269-1466 were shown in the structure) using PyMOL. (C) 3D projection of the difference in relative fractional uptake at 10 000 s of exchange between rVWF1238-1493 and rVWF1238-1472, as defined by the color code in the figure.
To find out how the binding of AIM residues to the A1 domain affects the GPIbα-binding activity of these constructs, we aligned the heat maps of rVWF1261-1472, rVWF1238-1472 and rVWF1238-1493 and projected them to a 3D structure of A1 (PDB: 1sq0) (Fig. 2B). As indicated, many peripheral regions in rVWF1261-1472 exhibited considerable deuterium exchange as early as in 10 seconds. In contrast, the same peripheral regions in rVWF1238-1472 and rVWF1238-1493 were largely not exchanged at 10 seconds. With the extension of exchange time, these peripheral regions in rVWF1238-1472 were slowly exchanged by deuterium, but many of these peripheral regions in rVWF1238-1493 were not exchanged even at the longest labeling time point (10,000 seconds). These results indicate a difference in solvent accessibility and dynamic flexibility of the peripheral regions in the A1 domain among these constructs. To further identify the coupling sites of AIM on the A1 domain in rVWF1238-1493, we compared the HDX between rVWF1238-1493 and rVWF1261-1472 and projected the difference to an A1/GPIbα complex structure (Fig. 2C). rVWF1261-1472 has the least AIM residues and exhibited no coupling effect. As indicated, the peripheral regions near the GPIbα-binding sites such as α1, α2, loopβ3α2, β3 and loopα3β4 were highlighted in this comparison, indicating that the AIM residues might bind at this area impairing the A1 domain binding to GPIbα. The complete time-dependent deuterium uptake data for every peptide in each presented recombinant A1 fragment is also included in Data S1.
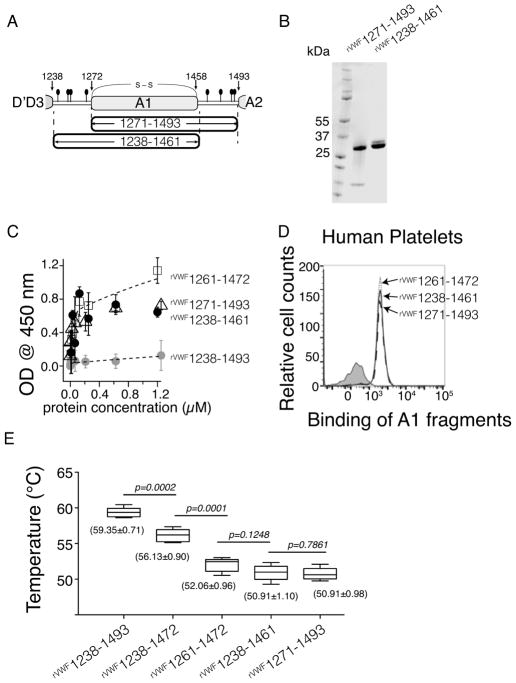
The N- and C- terminal AIM residues mask A1 in a cooperative manner
Although the cooperative interplay between the N- and C- terminal residues of AIM has been suggested in our previous study, generating the A1 fragments that contain only one side of AIM would be a direct way to verify this cooperation. We generated rVWF1238-1461 and rVWF1271-1493, two A1 fragments missing either the entire C- or entire N- terminal flanking region, respectively (Fig. 3A). Limited by the material we have, we did not run the AUC analysis on these proteins, but we think they are primarily in monomeric form on the basis of their elution time in the gel-filtration purification (Fig. S1). The migration speeds on SDS-PAGE gel of rVWF1238-1461 and rVWF1271-1493 are similar to each other (Fig. 3B), consistent with their similarity in molecular weight. Both rVWF1238-1461 and rVWF1271-1493 showed tight binding to the immobilized GPIb-IX complex similar to rVWF1261-1472 (Fig. 3C). The binding of these proteins with platelets was examined similarly to Figure 1, except that the protein concentration was 60 nM–the physiological concentration of VWF. The results indicated that at 60 nM rVWF1238-1461 and rVWF1271-1493 tightly associate with platelets in PRP, indistinguishable from that of rVWF1261-1472 (Fig. 3D). Similar results were obtained with washed platelets in mTyrodes’ buffer as well (data not shown). The Tm values of rVWF1238-1461 and rVWF1271-1493 were 50.91±0.98 °C and 50.91±1.10 °C, both akin to the Tm value of rVWF1261-1472 (Fig. 3E). These results strongly support the cooperation between the N- and C- terminal AIM in masking the A1 domain of VWF. Without the C-terminal AIM residues, the N-terminal AIM alone is not sufficient to protect A1.
Figure 3.
Cooperation of the N- and C- AIM is required for its inhibitory effect. (A) Illustration of rVWF1238- 1461 that misses the C-AIM. (B) Comparing the rVWF1271-1493 and rVWF1238-1461 by a coomassie blue- stained polyacrylamide gel under nonreducing conditions. (C) Comparing the binding to immobilized GPIb- IX among rVWF1261-1472, rVWF1271-1493, rVWF1238-1461, and rVWF1238-1493 by ELISA (n = 3). (D) Representative histogram of flow cytometry measurement to determine the binding of rVWF1261-1472, rVWF1271-1493, and rVWF1238-1461 to platelets in platelet rich plasma. (E) Whisker plot of the melting temperatures of A1 fragments. Mean value ± standard deviation (n=4) was marked under each plot. A paired t-test was conducted using GraphPad Prism 5 (GraphPad Software, Inc. CA the USA) and the pvalue was marked.
Discussion
It has long been thought that the tension-regulated VWF binding to GPIbα was through the lineation of the VWF from the “ball-of-yarn” shape. In the current study we demonstrate that the hemostatic activity of VWF is likely regulated by the residues flanking the A1 domain. The great consistency of the results from binding, thermostability and HDX assessments in our study indicates that the coupling of AIM residues to the A1 domain is critical for the GPIbα-binding activity of VWF. More importantly, our results suggest that the coupling between the flanking residues and the A1 domain might be a multivalent interaction whereas the flanking residues dynamically interact with multiple binding sites on the looped A1 domain. Shortening the AIM residues from rVWF1238-1493 to rVWF1238-1472 increased the GPIbα-binding activity but did not fully activate the A1 domain. Additionally, rVWF1238-1493 contained more HDX-protected regions than rVWF1238-1472. These observations suggest a multivalent binding between AIM and the A1 domain. As reported recently, the GPIbα-binding of VWF showed sigmoidal dependence on tensile force[10]. The exponential phase of the sigmoidal curve where the binding of GPIbα kept gradually increasing covered a wide force range from 10 pN to 40 pN. This observation can be well interpreted by the potentially stepwise uncoupling of the multivalent interaction between AIM and the A1 domain.
The binding sites of these AIM residues are largely overlaid with the GPIbα-binding sites in the A1 domain. One should also note that due to the increased global stability of the A1 domain by the coupling of AIM residues, some regions that are not in direct contact with AIM could also exhibit slower HDX due to allosteric effect. It is not possible to distinguish the coupling effect from the allosteric effect because both of them are comprehensively contributed to slow HDX in this case[22]. In full-length VWF the potential interactions between the A1 domain and other domains such as the D domains or other A domains may also affect the GPIbα-binding activity of VWF[23–26]. These interactions may indeed exist, but they shall not contribute significantly to inhibit the A1 domain for two reasons. First, rVWF1238-1493 exhibited the same inactivity as full-length VWF, suggesting that it contains sufficient inhibitory elements compared to the full-length protein. Second, in low-tension force environment GPIbα does not bind VWF even when it is elongated, suggesting that the domain-domain interactions play a minor role in inhibiting the A1 domain[10]. But it is possible that these domain-domain interactions in VWF may affect the binding of AIM to the A1 domain and therefore may influence the GPIbα-binding activity of VWF.
The thermostability measurement has been applied in studying the folding/unfolding of different domains in VWF[27–30], but it is rarely used to study the interaction between VWF A1 domain and its ligands. According to our measurement, the melting temperature of the A1 domain is around 51 °C when it is de-protected. With the coupling of AIM, the melting temperature of the A1 domain could reach 60 °C. It is challenging technically to test the thermostability of the A1 domain in the full-length VWF monomer. However, it is reasonable to anticipate that in type 2b VWD mutants or in ultra-large multimers, where VWF is engaged with hyperactivity, the thermostability of the A1 domain is lower than that in wild-type VWF molecule due to the unmasking of A1.
We verified in the current study that the cooperative binding between the N- and C- AIM regions is required for inhibiting the A1 domain. Predictably, unzipping the AIM by mutations on either side of AIM could dampen the autoinhibition of VWF. Both the N- and C- AIM appear to simultaneously bind ristocetin as suggested by multiple studies[31–35]. Apparently, this could only be possible when the N- and C- AIM are cooperatively bound to the A1 domain. Interestingly, rVWF1238-1493 showed higher sensitivity to ristocetin-mediated GPIbα binding compared to rVWF1238-1472 (Fig. 1F), suggesting that the residues beyond 1472 in the C- AIM may participate in binding ristocetin. But a more rigorous characterization is required to determine how ristocetin interacts with VWF.
We did not have evidence for the mechanosensory behavior of AIM in the current study yet, but on the basis of its essential contribution to the activity of VWF A1 domain, we think AIM likely as in the center of the mechanosensing regulatory mechanism of VWF. For VWF activation, unzipping of AIM could be the determinant local transition to the state with high affinity for platelets. The elongation of VWF from compact to linear form, instead, occurs prior to its activation as shown in a recent study[10]. It is becoming increasingly clear that the activation of VWF is determined by local elements instead of global domain-domain interactions[9, 10]. Perhaps AIM is intrinsically disordered, and the local environment of VWF multimers dynamically regulates the multivalent binding of AIM to the A1 domain, which enables a unique shear-dependent regulatory mechanism. The exact force to unzipping AIM and the structure of AIM are the subjects for future studies.
In summary, the delimitation of the residues in the autoinhibitory module of VWF in current study solidifies the hypothesis that residues flanking the A1 domain of VWF regulate the hemostatic activity of VWF by directly masking A1. Uncoupling of the likely multivalent interactions of AIM to the A1 domain might be a general mechanism for VWF to act as a mechanosensor to have a different response to different shear environments in blood circulation.
Supplementary Material
Essentials.
The self-inhibitory mechanism of von Willebrand factor (VWF) remains unclear.
Residues flanking the A1 domain of VWF form a discontinuous autoinhibitory module (AIM).
rVWF1238-1493 exhibited greater thermostability and inactivity than its shorter counterparts.
The cooperative coupling between the N- and C- AIM regions are required for inhibiting A1.
Acknowledgments
We thank Emory HDX core facility for the help in collecting and analyzing data. This work is supported in part by the Emory WHSC Synergy award and National Institutes of Health grant HL082808. The department of pediatrics Grant and Manuscript Support Core contributed to the editing of this manuscript.
Footnotes
Addendum
W. Deng, P. Lollar, and R. Li designed research; W. Deng, K.M. Voos, J.K. Colucci, E.R. Legan, and P. Lollar performed research; W. Deng, E.A. Ortlund, P. Lollar, and R. Li analyzed the results; W. Deng wrote the paper.
Disclosure of Conflict of Interest
The authors state that they have no conflict of interest.
References
- 1.Slayter H, Loscalzo J, Bockenstedt P, Handin RI. Native conformation of human von Willebrand protein. Analysis by electron microscopy and quasi-elastic light scattering. The Journal of biological chemistry. 1985;260:8559–63. [PubMed] [Google Scholar]
- 2.Sadler JE. von Willebrand factor: two sides of a coin. Journal of thrombosis and haemostasis : JTH. 2005;3:1702–9. doi: 10.1111/j.1538-7836.2005.01369.x. [DOI] [PubMed] [Google Scholar]
- 3.Cruz MA, Handin RI, Wise RJ. The interaction of the von Willebrand factor-A1 domain with platelet glycoprotein Ib/IX. The role of glycosylation and disulfide bonding in a monomeric recombinant A1 domain protein. J Biol Chem. 1993;268:21238–45. [PubMed] [Google Scholar]
- 4.Miyata S, Goto S, Federici AB, Ware J, Ruggeri ZM. Conformational changes in the A1 domain of von Willebrand factor modulating the interaction with platelet glycoprotein Ibalpha. The Journal of biological chemistry. 1996;271:9046–53. doi: 10.1074/jbc.271.15.9046. [DOI] [PubMed] [Google Scholar]
- 5.Miura S, Fujimura Y, Sugimoto M, Kawasaki T, Ikeda Y, Titani K, Yoshioka A. Structural elements influencing von Willebrand factor (vWF) binding affinity for platelet glycoprotein Ib within a dispase-digested vWF fragment. Blood. 1994;84:1553–8. [PubMed] [Google Scholar]
- 6.Miura S, Li CQ, Cao Z, Wang H, Wardell MR, Sadler JE. Interaction of von Willebrand factor domain A1 with platelet glycoprotein Ibalpha-(1–289). Slow intrinsic binding kinetics mediate rapid platelet adhesion. The Journal of biological chemistry. 2000;275:7539–46. doi: 10.1074/jbc.275.11.7539. [DOI] [PubMed] [Google Scholar]
- 7.Blenner MA, Dong X, Springer TA. Structural basis of regulation of von Willebrand factor binding to glycoprotein Ib. The Journal of biological chemistry. 2014;289:5565–79. doi: 10.1074/jbc.M113.511220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huizinga EG, Tsuji S, Romijn RA, Schiphorst ME, de Groot PG, Sixma JJ, Gros P. Structures of glycoprotein Ibalpha and its complex with von Willebrand factor A1 domain. Science. 2002;297:1176–9. doi: 10.1126/science.107355. [DOI] [PubMed] [Google Scholar]
- 9.Deng W, Wang Y, Druzak SA, Healey JF, Syed AK, Lollar P, Li R. A discontinuous autoinhibitory module masks the A1 domain of von Willebrand factor. Journal of thrombosis and haemostasis : JTH. 2017;15:1867–77. doi: 10.1111/jth.13775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu H, Jiang Y, Yang D, Scheiflinger F, Wong WP, Springer TA. Flow-induced elongation of von Willebrand factor precedes tension-dependent activation. Nat Commun. 2017;8:324. doi: 10.1038/s41467-017-00230-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pirrone GF, Iacob RE, Engen JR. Applications of hydrogen/deuterium exchange MS from 2012 to 2014. Anal Chem. 2015;87:99–118. doi: 10.1021/ac5040242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marcsisin SR, Engen JR. Hydrogen exchange mass spectrometry: what is it and what can it tell us? Anal Bioanal Chem. 2010;397:967–72. doi: 10.1007/s00216-010-3556-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo SZ, Mo X, Afshar-Kharghan V, Srinivasan S, Lopez JA, Li R. Glycoprotein Ibalpha forms disulfide bonds with 2 glycoprotein Ibbeta subunits in the resting platelet. Blood. 2007;109:603–9. doi: 10.1182/blood-2006-05-024091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huynh K, Partch CL. Analysis of protein stability and ligand interactions by thermal shift assay. Curr Protoc Protein Sci. 2015;79:28 9 1–14. doi: 10.1002/0471140864.ps2809s79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang RH, Fremont DH, Diener JL, Schaub RG, Sadler JE. A structural explanation for the antithrombotic activity of ARC1172, a DNA aptamer that binds von Willebrand factor domain A1. Structure. 2009;17:1476–84. doi: 10.1016/j.str.2009.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tischer A, Campbell JC, Machha VR, Moon-Tasson L, Benson LM, Sankaran B, Kim C, Auton M. Mutational Constraints on Local Unfolding Inhibit the Rheological Adaptation of von Willebrand Factor. The Journal of biological chemistry. 2016;291:3848–59. doi: 10.1074/jbc.M115.703850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Celikel R, Ruggeri ZM, Varughese KI. von Willebrand factor conformation and adhesive function is modulated by an internalized water molecule. Nat Struct Biol. 2000;7:881–4. doi: 10.1038/79639. [DOI] [PubMed] [Google Scholar]
- 18.Fukuda K, Doggett T, Laurenzi IJ, Liddington RC, Diacovo TG. The snake venom protein botrocetin acts as a biological brace to promote dysfunctional platelet aggregation. Nat Struct Mol Biol. 2005;12:152–9. doi: 10.1038/nsmb892. [DOI] [PubMed] [Google Scholar]
- 19.Maita N, Nishio K, Nishimoto E, Matsui T, Shikamoto Y, Morita T, Sadler JE, Mizuno H. Crystal structure of von Willebrand factor A1 domain complexed with snake venom, bitiscetin: insight into glycoprotein Ibalpha binding mechanism induced by snake venom proteins. The Journal of biological chemistry. 2003;278:37777–81. doi: 10.1074/jbc.M305566200. [DOI] [PubMed] [Google Scholar]
- 20.Dumas JJ, Kumar R, McDonagh T, Sullivan F, Stahl ML, Somers WS, Mosyak L. Crystal structure of the wild-type von Willebrand factor A1-glycoprotein Ibalpha complex reveals conformation differences with a complex bearing von Willebrand disease mutations. The Journal of biological chemistry. 2004;279:23327–34. doi: 10.1074/jbc.M401659200. [DOI] [PubMed] [Google Scholar]
- 21.Andrews RK, Gorman JJ, Booth WJ, Corino GL, Castaldi PA, Berndt MC. Cross-linking of a monomeric 39/34-kDa dispase fragment of von Willebrand factor (Leu-480/Val-481-Gly-718) to the N-terminal region of the alpha-chain of membrane glycoprotein Ib on intact platelets with bis(sulfosuccinimidyl) suberate. Biochemistry. 1989;28:8326–36. doi: 10.1021/bi00447a010. [DOI] [PubMed] [Google Scholar]
- 22.Chalmers MJ, Busby SA, Pascal BD, West GM, Griffin PR. Differential hydrogen/deuterium exchange mass spectrometry analysis of protein-ligand interactions. Expert Rev Proteomics. 2011;8:43–59. doi: 10.1586/epr.10.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ulrichts H, Udvardy M, Lenting PJ, Pareyn I, Vandeputte N, Vanhoorelbeke K, Deckmyn H. Shielding of the A1 domain by the D’D3 domains of von Willebrand factor modulates its interaction with platelet glycoprotein Ib-IX-V. The Journal of biological chemistry. 2006;281:4699–707. doi: 10.1074/jbc.M513314200. [DOI] [PubMed] [Google Scholar]
- 24.Martin C, Morales LD, Cruz MA. Purified A2 domain of von Willebrand factor binds to the active conformation of von Willebrand factor and blocks the interaction with platelet glycoprotein Ibalpha. Journal of thrombosis and haemostasis : JTH. 2007;5:1363–70. doi: 10.1111/j.1538-7836.2007.02536.x. [DOI] [PubMed] [Google Scholar]
- 25.Obert B, Houllier A, Meyer D, Girma JP. Conformational changes in the A3 domain of von Willebrand factor modulate the interaction of the A1 domain with platelet glycoprotein Ib. Blood. 1999;93:1959–68. [PubMed] [Google Scholar]
- 26.Zhang C, Kelkar A, Nasirikenari M, Lau JTY, Sveinsson M, Sharma UC, Pokharel S, Neelamegham S. The physical spacing between the von Willebrand factor D’D3 and A1 domains regulates platelet adhesion in vitro and in vivo. Journal of thrombosis and haemostasis : JTH. 2017 doi: 10.1111/jth.13927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Auton M, Sowa KE, Smith SM, Sedlak E, Vijayan KV, Cruz MA. Destabilization of the A1 domain in von Willebrand factor dissociates the A1A2A3 tri-domain and provokes spontaneous binding to glycoprotein Ibalpha and platelet activation under shear stress. The Journal of biological chemistry. 2010;285:22831–9. doi: 10.1074/jbc.M110.103358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Auton M, Sedlak E, Marek J, Wu T, Zhu C, Cruz MA. Changes in thermodynamic stability of von Willebrand factor differentially affect the force-dependent binding to platelet GPIbalpha. Biophys J. 2009;97:618–27. doi: 10.1016/j.bpj.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lynch CJ, Lane DA. N-linked glycan stabilization of the VWF A2 domain. Blood. 2016;127:1711–8. doi: 10.1182/blood-2015-09-672014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jakobi AJ, Mashaghi A, Tans SJ, Huizinga EG. Calcium modulates force sensing by the von Willebrand factor A2 domain. Nat Commun. 2011;2:385. doi: 10.1038/ncomms1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Flood VH, Friedman KD, Gill JC, Morateck PA, Wren JS, Scott JP, Montgomery RR. Limitations of the ristocetin cofactor assay in measurement of von Willebrand factor function. Journal of thrombosis and haemostasis : JTH. 2009;7:1832–9. doi: 10.1111/j.1538-7836.2009.03594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mohri H, Fujimura Y, Shima M, Yoshioka A, Houghten RA, Ruggeri ZM, Zimmerman TS. Structure of the von Willebrand factor domain interacting with glycoprotein Ib. The Journal of biological chemistry. 1988;263:17901–4. [PubMed] [Google Scholar]
- 33.Berndt MC, Ward CM, Booth WJ, Castaldi PA, Mazurov AV, Andrews RK. Identification of aspartic acid 514 through glutamic acid 542 as a glycoprotein Ib-IX complex receptor recognition sequence in von Willebrand factor. Mechanism of modulation of von Willebrand factor by ristocetin and botrocetin. Biochemistry. 1992;31:11144–51. doi: 10.1021/bi00160a027. [DOI] [PubMed] [Google Scholar]
- 34.Dong JF, Berndt MC, Schade A, McIntire LV, Andrews RK, Lopez JA. Ristocetin-dependent, but not botrocetin-dependent, binding of von Willebrand factor to the platelet glycoprotein Ib-IX-V complex correlates with shear-dependent interactions. Blood. 2001;97:162–8. doi: 10.1182/blood.v97.1.162. [DOI] [PubMed] [Google Scholar]
- 35.De Luca M, Facey DA, Favaloro EJ, Hertzberg MS, Whisstock JC, McNally T, Andrews RK, Berndt MC. Structure and function of the von Willebrand factor A1 domain: analysis with monoclonal antibodies reveals distinct binding sites involved in recognition of the platelet membrane glycoprotein Ib-IX-V complex and ristocetin-dependent activation. Blood. 2000;95:164–72. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.