Abstract
Background
Several studies have shown that exposure to particulate matter (PM) may lead to increased systemic blood pressure, but the underlying biological mechanisms remain unknown. Emerging evidence shows that extracellular vesicle-enriched miRNAs (evmiRNAs) are associated with PM exposure and cardiovascular risk. In this study, we investigated the role of evmiRNAs in the association between PM and blood pressure, as well as their epigenetic regulation by DNA methylation.
Methods
Participants (n=22, men) were randomly selected from the Veterans Affairs Normative Aging Study (NAS). Long-term (1-year and 6-month average) PM2.5 exposure was estimated at 1×1-km resolution using spatio-temporal prediction models and BC was estimated using validated time varying land use regression models. We analyzed 31 evmiRNAs detected in ≥90% of all individuals and for statistical analysis, we used mixed effects models with random intercept adjusted for age, body mass index, smoking, C-reactive protein, platelets, and white blood cells.
Results
We found that per each 2-standard deviations increase in 6-month PM2.5 ambient levels, there was an increase in 0.19 mm Hg (95% Confidence Interval [95%CI]: 0.11, 0.28 mmHg; p<0.001) in systolic blood pressure (SBP). Per each 2-standard deviations increase in 1-year PM2.5 levels, there was an increase in 0.11 mm Hg (95% Confidence Interval [95% CI]: 0.03, 0.19 mmHg; p=0.012) in SBP in older male individuals. We also found that both miR-199a/b (β=6.13 mmHg; 95% CI: 0.87, 11.39; pinteraction=0.07) and miR-223–3p (β=30.17 mmHg; 95% CI: 11.96, 48.39 mmHg; pinteraction=0.01) modified the association between 1-year PM2.5 and SBP. When exploring DNA methylation as a potential mechanism that could epigenetically regulate expression of evmiRNAs, we found that PM2.5 ambient levels were negatively associated with DNA methylation levels at CpG (cg23972892) near the enhancer region of miR-199a/b (β=−13.11; 95% CI: −17.70, −8.52; pBonferroni<0.01), but not miR-223–3p.
Conclusions
Our findings suggest that expression of evmiRNAs may be regulated by DNA methylation in response to long-term PM2.5 ambient levels and modify the magnitude of association between PM2.5 and systolic blood pressure in older individuals.
Keywords: Exosomes, miRNAs, DNA methylation, air pollution, blood pressure
1. Introduction
High blood pressure (BP) affects more than 25% of adults and causes ~9.5 million premature deaths each year worldwide.(Kearney et al., 2005) Among all cardiovascular risk factors, high BP has the highest attributable mortality(Lim et al., 2012) and is linked to preventable lifestyle and environmental factors, including air pollution.(Yamamoto et al., 2014)
Ambient particulate matter (PM) with an aerodynamic diameter of <2.5 μm (PM2.5) are tiny air-suspended particles with well established toxic health effects.(Zanobetti et al., 2011) Black carbon (BC), a constituent of PM2.5 and a surrogate for traffic-related pollution, has also been linked to human diseases in epidemiologic studies.(Chen et al., 2015; Highwood and Kinnersley, 2006) Multiple studies have shown that exposure to PM leads to increases in systemic blood pressure (SBP).(Auchincloss et al., 2008; Wu et al., 2013) A recent meta-analysis has shown an increase of 1.39 mmHg (95% confidence interval (CI):0.87–1.91) in SBP and 0.90 mmHg (95% CI 0.49–1.30) in diastolic blood pressure (DBP) per 10 μg/m3 increase in PM2.5 ambient levels.(Liang et al., 2014) In addition, an interquartile range increase in long-term (1-year average) BC exposure (0.32 μg/m3) has been associated with an increase of 2.64 mmHg in SBP (95%CI 1.47 – 3.80) and 2.41 mmHg (95% CI 1.77 – 3.05) in DBP.(Mordukhovich et al., 2009)
Several physiological responses have been implicated in the association between PM and cardiovascular diseases (CVD), including the direct effect of PM on blood vessels,(Tamagawa et al., 2008) autonomic dysfunction,(Chuang et al., 2007) and inflammatory response after breaching endothelial integrity by PM.(Nemmar et al., 2001) Extracellular vesicles (EVs) are a recently characterized mechanism of cellular communication whereby active biological signals are being transferred between cells.(Sadik et al., 2018) Several studies have linked EVs and their cargo, including microRNAs (miRNAs), to several CVD-related processes such as cardiac fibrosis, hypertrophy, and coagulation.(Bei et al., 2017; Pergoli et al., 2017) We have recently reported changes in the levels of EV-enriched miRNAs (evmiRNAs) in response to long-term PM2.5 ambient levels (e.g., miR-126–3p, miR-199a/b, miR-223–3p, and miR-150–5p) and their implications in CVD-related biological pathways in older individuals.(Rodosthenous et al., 2016) In addition, Bollati et al. showed that the levels of evmiRNAs (miR-128 and miR-302c) are increased in the blood of metal workers in response to short-term exposure to higher PM levels.(Bollati et al., 2015) Other changes, including those in DNA methylation marks, have also been described associated to long-term PM exposure.(Baccarelli et al., 2009) Recently, Motta et al., showed that miRNAs in the blood may mediate the impact of short-term exposure to PM10 on blood pressure in obese individuals.(Motta et al., 2016) However, whether evmiRNAs are epigenetically modified by long-term PM2.5 exposures, and play a role in the association between long-term PM2.5 exposures and blood pressure has not been previously evaluated.
2. Material and methods
2.1. Study population
The profile of the Veterans Affairs Normative Aging Study (NAS) is described elsewhere.(Rodosthenous et al., 2016) We used existing DNA methylation data from NAS participants (n=563) who also had PM2.5 and BC data between years 2000–2008. Out of those, we randomly selected 22 participants and analyzed 42 serum samples for evmiRNAs. All 22 participants had a serum sample analyzed for evmiRNAs at first visit (n=22), and ten out of these participants had two additional serum samples analyzed for evmiRNAs that were collected during their second and third visit. Therefore, the total number of samples with evmiRNA data is n=42. Details of the study design, characteristics of the study participants, blood collection, EVs isolation and miRNA extraction, and miRNA profiling of this study have been published previously.(Rodosthenous et al., 2016) We screened for 800 known evmiRNAs in each serum sample and for statistical power reasons we only analyzed 31 evmiRNAs that were detected in ≥90% of all individuals. SBP and DBP of study participants were measured in seated position and we used the means of the measurements in both left and right arm. The NAS study was approved by the Institutional Review Boards of participating institutions (#14027–102) and all participants provided informed consent according to the Declaration of Helsinki.
2.2. PM2.5 and BC exposure assessment
We estimated daily PM2.5 ambient levels at 1×1-km resolution using our recently developed spatio-temporal prediction models that incorporate satellite aerosol optical depth data, local predictors and spatial smoothing, across the Great Boston Area.(Kloog et al., 2011; 2014) Ten-fold cross-validation showed very good model predictions with out-of-sample R2=0.88 for PM2.5 . BC was estimated using a time varying land use regression (R2=0.83 for daily measures).(Gryparis et al., 2007) We defined our PM2.5 and BC ambient levels as the average of daily estimates corresponding to 6-month and 1-year at the participant’s residential address prior to the date of visit (moving average) that each serum sample was analyzed (time-varying PM2.5 and BC measurement).
2.3. DNA methylation in peripheral blood
Molecular mechanisms regulating the expression of evmiRNAs in the context of environmental exposures have been largely unexplored. To examine whether air pollution-associated evmiRNAs expression was epigenetically affected by DNA methylation in peripheral blood,(Soto-Reyes et al., 2012) we utilized 450K methylation (Infinium HumanMethylation450K BeadChip, Illumina, San Diego, CA) data from the NAS study. More specifically, we determined the association between long-term PM2.5 (moving average) ambient levels and DNA methylation levels in CpG sites located only within 2Mbp adjacent to promoter regions of the PM2.5/BC-associated evmiRNAs. A total of 1,022 blood samples collected from 563 participants between 2000 and 2008 were analyzed for DNA methylation using the Infinium HumanMethylation450K BeadChip. Each participant has between one to four DNA methylation measurements from follow up visits (mean=2.2 measurements per participant). DNA methylation profiles were calculated using the β-mixture quantile normalization (BMIQ) method. Control samples were included in each plate and were analyzed for batch effects. Cross-reactive probes, probes with detection p-value>0.01, or those associated to single nucleotide polymorphisms were excluded from the analysis.
2.4. Statistical analysis
To improve normality in the residuals, we log-transformed (log2) evmiRNAs data. Multivariate analysis was conducted between long-term (6-month and 1-year) PM2.5 and BC moving averages and evmiRNAs. For the association between PM2.5 and BC ambient levels and evmiRNAs, we included age (continuous), body mass index (continuous), tobacco smoking (current, never, former), number of platelets (continuous), C-reactive protein (≤10, >10), and white blood cell distribution (% of lymphocytes, monocytes, eosinophil, and basophil cells) as confounders. To determine associations between PM2.5 and BC ambient levels and evmiRNAs, we used mixed effects models with random intercept and we adjusted for multiple comparisons using Bonferroni correction.(Noble, 2009) Statistical significance after Bonferroni correction was defined p-value<0.05. All analyses were performed in SAS 9.4 software (SAS Institute Inc., Cary, NC). For methylation analysis, we used mixed models, in which the independent variable was 1-year PM2.5 ambient levels and as dependent variable the methylation values at 8,903 CpG sites for 6-month and 4,096 CpG sites from PM2.5-associated evmiRNAs (the number and the miRNAs were different for each model) as independent variables, using the same set of covariates and batch for adjustment. All p-values were adjusted for multiple comparisons using the Bonferroni method. DNA methylation analysis was done in R software (R Project for Statistical Computing, Wien, packages CpGassoc, minfi, and methylumi). In silico analysis to identify experimentally validated mRNA targets and their biological relevance was performed using Ingenuity Pathway Analysis (Ingenuity Systems®, Redwood City, CA).
3. Results
3.1. Characteristics of study participants
Study participants with evmiRNAs data available (n=22) were all males between 60 and 82 (mean=72.63, SD= 6.31) years old, 18/22 had a body mass index (BMI) ≥25 (mean BMI=26.72, SD=2.89), and 72.73% were former smokers, at baseline (Table 1). Study participants with DNA methylation data available, (n=563) were all males between 50 and 90 (mean=75.46, SD= 6.86) years old, 436/563 had a body mass index (BMI) ≥25 (mean BMI=27.43, SD=4.09), and 68.38% were former smokers, at baseline (Table 1).
Table 1.
Demographic, physical, clinical characteristics, PM2.5 and black carbon levels of participants at the baseline visit evaluated for extracellular vesicles-enriched microRNAs (n=22) and DNA methylation analysis (n=563).
| evmiRNAs analysis |
DNA methylation analysis | |||||||
|---|---|---|---|---|---|---|---|---|
| Mean (SD) | Mean (SD) | |||||||
| Characteristic | N (%) or Mean (SD) |
PM2.5, μg/m3 | BC†, μg/m3 | N (%) or Mean (SD) |
PM2.5 (μg/m3) | |||
| 6-month | 1-year | 6-month | 1-year | 6-month | 1-year | |||
| Age, years | 72.63 | 11.67 | 11.56 | 0.58 | 0.59 | 75.46 | 10.47 | 10.45 |
| (6.31) | (1.01) | (0.83) | (0.07) | (0.07) | (6.86) | (1.60) | (1.48) | |
| BMI*, kg/m2 | 26.72 | 27.43 | ||||||
| (2.89) | (4.09) | |||||||
| Platelets | 231.90 | 219.60 | ||||||
| (thousands/mm3) | (0.15) | (52.03) | ||||||
| C-reactive | 0.02 | 3.48 | ||||||
| protein (mg/L) | (0.15) | (8.52) | ||||||
| Tobacco | ||||||||
| smoking | ||||||||
| Never | 6 | 11.63 | 11.60 | 0.61 | 0.63 | 154 | 11.42 | 11.62 |
| (27.27%) | (0.71) | (0.89) | (0.06) | (0.07) | (27.35%) | (1.08) | (1.22) | |
| Current | 0 (0%) | - | - | - | - | 24 | 11.33 | 11.67 |
| (4.26%) | (0.96) | (1.15) | ||||||
| Former | 16 | 10.81 | 10.83 | 0.60 | 0.61 | 385 | 11.24 | 11.33 |
| (72.73%) | (1.47) | (1.23) | (0.08) | (0.07) | (68.38%) | (1.07) | (1.18) | |
BMI: Body mass index.
BC: Black carbon
3.2. Associations of long-term PM2.5 and BC ambient levels, evmiRNAs, and SBP
We found that evmiRNAs let-7g-5p, miR-1246, miR-126–3p, miR-142–3p, miR-150–5p, miR-15a-5p, miR-199a/b, and miR-223–3p were significantly associated with PM2.5 6-month ambient levels (Fig. 1, Panel a) and miR-130a-3p, miR-142–3p, miR-199a/b, miR-223–3p, and miR-23a-3p were significantly associated with PM2.5 1-year average (Fig. 1, Panel b). In these models, we used evmiRNA data from 36/42 samples because of missing PM2.5 exposure values. The full list of the association estimates between long-term PM2.5 and all evmiRNAs is shown in Supp. Table 1.
Fig 1.
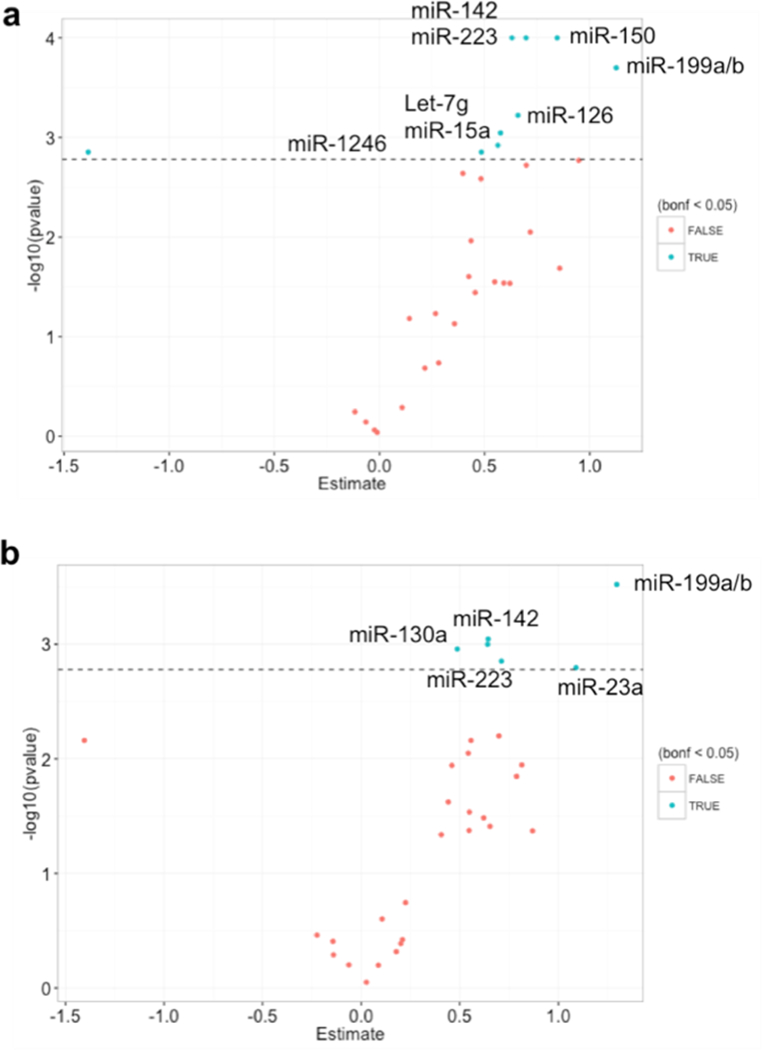
Associations between 6-month and 1-year exposure to PM2.5 ambient levels and extracellular vesicle-associated miRNAs (n=36). Dotted line represents the Bonferroni-adjusted threshold for statistical significance (a=0.05).
We also found that for each 2 standard deviations (2.76 μg/m3) increase in 6-month PM2.5, there was an increase of 0.19 mmHg (95% CI: 0.11, 0.28 mmHg; p<0.01) in SBP, whereas for each 2 standard deviations (2.39 μg/m3) increase in 1-year PM2.5 there was an increase of 0.12 mmHg (95%CI: 0.02, 0.19 mmHg; p=0.01) in SBP (Table 2). No associations were found between PM2.5-associated evmiRNAs and SBP in study participants (Supp. Table 2). When we focused on the PM2.5-associated evmiRNAs, we found significant interaction for miR-223–3p (β=30.17, SE=9.29, pinteraction=0.01 [no Bonferroni adjustment]) and borderline significant interaction (p-value lower than 0.1 and higher than 0.05) for miR-199a/b (β=6.13, SE=2.68, pinteraction=0.07 [no Bonferroni adjustment]) in the association of 1-year PM2.5 ambient levels and SBP (Table 3). No associations were found between long-term BC and any of the evmiRNAs after multiple comparisons adjustment (Bonferroni) (Supp. Table 3). For the BC models, we used evmiRNA data from 33/42 samples because of missing BC exposure values.
Table 2.
Linear mixed-effects models estimating the change in blood pressure of study participants associated with a 2-SD increase* in long-term exposure to PM2.5 ambient levels (n=36).
| PM2.5 6-month |
PM2.5 1-year |
||||||
|---|---|---|---|---|---|---|---|
| mmHg | 95% CI | p-value | mmHg | 95% CI | p-value | ||
| Systolic blood pressure | 0.19 | (0.11, 0.28) | <0.01 | 0.12 | (0.02, 0.19) | 0.01 | |
| Diastolic blood pressure | 0.11 | (−0.03, 0.28) | 0.13 | 0.05 | (−0.07, 0.17) | 0.43 |
Corresponding to a 2.76 μg/m3 increase in 6-month and to a 2.39 μg/m3 increase in PM2.5 levels. Models were adjusted for age, body mass index, smoking, C-reactive protein, white blood cells distribution (percentage of B-cells, monocytes, NK-cells, CD4, and CD8), and number of platelets. 95% CI: 95% Confidence interval.
Table 3.
Interaction of extracellular vesicle-associated miRNAs in the association between long-term PM2.5 ambient levels and systolic blood pressure in study participants (n=36).
| PM2.5 | microRNA | β* | 95% CI | pinteraction |
|---|---|---|---|---|
| 6-month | let-7g-5p | −1.40 | (−25.36, 22.55) | 0.93 |
| miR-1246 | −3.11 | (−9.17, 2.94) | 0.36 | |
| miR-126–3p | −8.92 | (−25.45, 7.62) | 0.46 | |
| miR-142–3p | −7.91 | (−32.28, 16.46) | 0.69 | |
| miR-150–5p | 6.89 | (−7.41, 21.20) | 0.42 | |
| miR-15a-5p | 11.03 | (−7.75, 29.82) | 0.21 | |
| miR-199a/b | 2.29 | (−1.01,5.59) | 0.14 | |
| miR-223–3p | 2.01 | (−14.52, 18.55) | 0.72 | |
| miR-23a-3p | −3.07 | (−14.23, 8.09) | 0.67 | |
| 1-year | miR-130a-3p | −2.01 | (−44.62, 40.60) | 0.92 |
| miR-142–3p | 3.77 | (−25.41, 32.94) | 0.76 | |
| miR-199a/b | 6.13 | (0.87, 11.39) | 0.07 | |
| miR-223–3p | 30.17 | (11.96, 48.39) | 0.01 | |
| miR-23a-3p | 5.46 | (−23.23, 34.14) | 0.74 |
β=Estimate for the interaction between long-term PM2.5 ambient levels and systolic blood pressure. Models were adjusted for age, body mass index, smoking, C-reactive protein, white blood cell distribution, and number of platelets.
3.3. MiR-223–3p and miR-199a/b target several proteins related to cardiovascular diseases
In silico analysis using experimentally validated findings showed that miR-223–3p targets several proteins involved in cardiovascular functions, including proliferation of cardiomyocytes (SLC8A1 and TGFBR3), dysfunction of the heart (MEF2C, SLC8A1, and CD36), blood vessels remodeling (LIF, FBXW7, and MEF2C), heart hypertrophy (IL6ST, LIF, HDAC4), vasculogenesis, abnormal morphology of blood vessels, hypertrophy of cardiac muscle and contractibility of heart ventricles (Fig. 2). In addition, in silico analysis showed that miR-199a/b may target proteins involved in heart fibrosis (PTGS2), coronary artery disease (PDE3B, MTOR), ventricular hypertrophy (PTGS2 and MTOR), among others (Fig.3).
Fig 2.
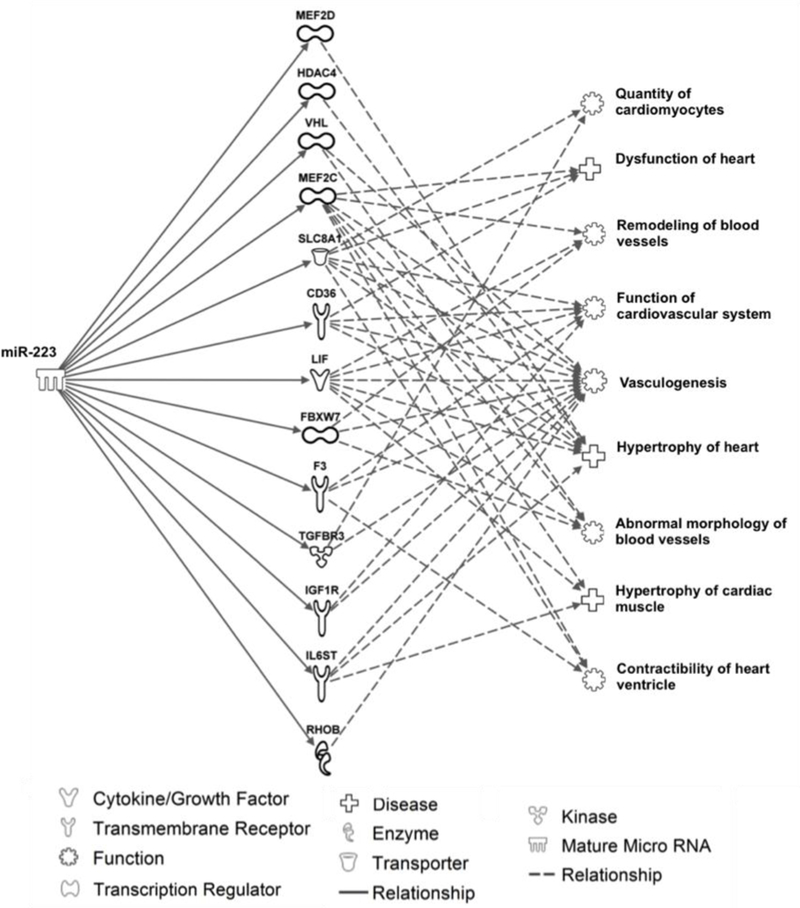
In silico analysis of experimentally validated targets and cardiovascular pathways linked to miR-223 using Ingenuity Pathway Analysis®.
Fig 3.
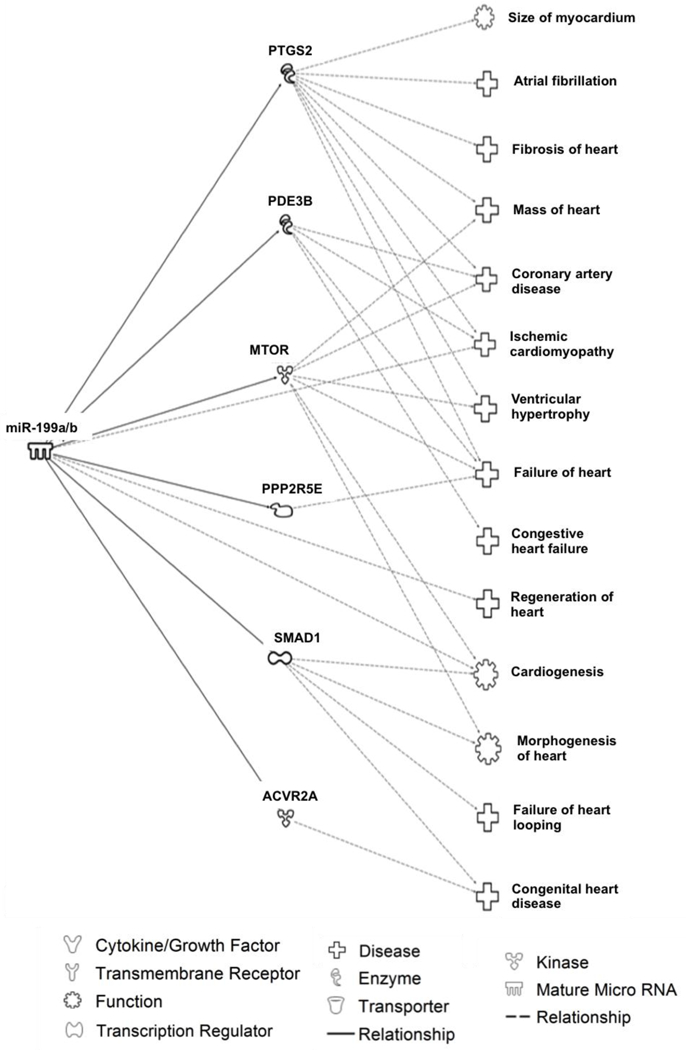
In silico analysis of experimentally validated targets and cardiovascularpathways linked to miR-4199a/b using Ingenuity Pathway Analysis®.
3.4. Long-term PM2.5 ambient levels are associated with DNA methylation in the enhancer region near miR-199a/b promoter
We found that DNA methylation levels of CpG cg23972892 were negatively associated with both 6-month (β=−4.61, SE=1.69, pBonferroni<0.05) and 1-year (β=−13.11, SE=2.34, pBonferroni <0.01) PM2.5 ambient levels (Fig. 4). This CpG site is located in a region enriched with histone-3 acetylation in lysine 27 (H3K27Ac) on chromosome 19 within the nearest miR-199a/b promoter region (chr19:10,928,102–10,928,172) (Fig. 5), which we found to be positively associated with both 6-month and 1-year PM2.5 levels.
Fig 4.
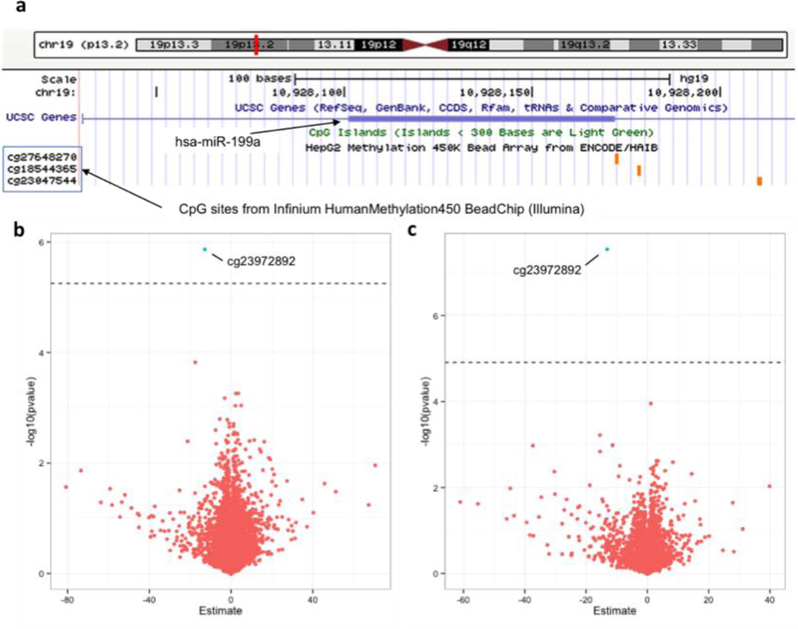
Association between 6-month and 1-year PM2.5 ambient levels and CpG methylation within 2Mbp adjacent to extracellular vesicle-associated miRNAs (evmiRNAs) promoter in peripheral blood of study participants (n=563). (a) Diagram (adapted from USCS-Genome Browser) showing miR-199a/b near-promoter CpG sites included in Infinium HumanMethylation450 BeadChip (Illumina). (b) Volcano plot showing the association between 6-month PM2.5 ambient levels and CpG sites within 2Mbp adjacent to evmiRNAs promoter (8,903 CpG sites); (c) Volcano plot showing the association between 1-year PM2.5 ambient levels and CpG sites within 2Mbp adjacent to evmiRNAs promoter (4,096 CpG sites). Dotted line represents the Bonferroni-adjusted threshold for statistical significance (a=0.05).
Fig 5.

Representation of CpG (cg23972892) near miR-199a/b promoter. Upper panel: Location at chromosome level (chr19:10,928,102–10,928,172). Lower panel: Zoom-in to show Histone 3 Lysine 24 acetylated (H3K27Ac) mark and other transcription factors (e.g. CTCF, TBP, ZNF143, RUNX3) in cells H1-hESC cells, suggesting a regulatory region. Adapted from UCSC-Genome Browser.
4. Discussion
In this study, we found that EV-associated miR-223–3p and miR-199a/b modify positively the association between 1-year PM2.5 moving average ambient levels and blood pressure in older male individuals, suggesting that the magnitude of the effect of PM2.5 levels on blood pressure differs depending on the levels of these evmiRNAs. In silico analysis showed that EV-associated miRNA-223–3p and miR-199a/b can potentially target several proteins implicated in important cardiovascular functions. We also found that higher 1-year PM2.5 levels were associated with lower DNA methylation in an enhancer region near miR-199a/b promoter and higher expression levels of EV- associated miR-199a/b circulating in the blood of the study participants. Together, these findings suggest that the effect of PM2.5 on blood pressure may be modified by PM2.5-asssociated epigenetic and expression changes of evmiRNAs in the blood circulation.
We have previously showed that long-term (6-month and 1-year) ambient PM2.5 levels were associated with increased levels of multiple evmiRNAs in serum, including miR-223–3p, miR-199a/b, miR-23a, miR-150, miR-15a, miR-191, and let-7a.(Rodosthenous et al., 2016) In addition, Bollatti et al. found that short-term exposure to occupational PM was associated with increased levels of evmiRNAs miR-128 and miR-302, showing also an in vitro dose-response release of miR-128.(Bollati et al., 2015) Short-term exposure to PM (PM2.5 and PM10) has also been associated with the number of platelet-derived and annexin V-binding EV release in peripheral blood.(Emmerechts et al., 2012) Studies from Liu et al. found that PM increases release of macrophages EVs and induces selective neurotoxicity.(Liu et al., 2015) Although evmiRNAs have a high potential to mediate cell-to-cell communication and potentially might link PM2.5 induced-damage and cardiovascular diseases, no previous study had reported the interaction of evmiRNAs between long-term PM2.5 exposure and blood pressure, especially in older individuals.
In agreement with our findings, several studies have previously reported changes in miR-223 expression levels in response to PM2.5, as well as other environmental exposures such as including smoking and ozone.(Herberth et al., 2014) In the blood, miR-223 is one of the most abundant miRNAs released primarily from monocytes, macrophages and platelets after cell activation.(Gatsiou et al., 2012; Johnnidis et al., 2008) In atherosclerotic mice models, endothelial cells express high levels of miR-223, which attenuates the toll-like receptor 4 (TLR4)-dependent inflammatory response, lipid accumulation, and show anti-angiogenic activity through the RPS6KB1/hif-1a and β1 integrin pathway.(Shi et al., 2013) Dai et al., found that miR-223 inhibits angiogenesis in ischemic cardiac microvascular endothelial cells, suggesting involvement in the RPS6KB1/hif-1a signaling pathway, which affects endothelial migration and proliferation.(Dai et al., 2014) In addition, Wang et al. have also reported that miR-223 protects cardiomyocytes against hypertrophy by targeting cardiac troponin I-interacting kinase.(Y.-S. Wang et al., 2015) Last, other studies have shown that miR-223 regulates macrophage polarization,(Zhuang et al., 2012) suppress tissue factor expression, regulates thrombosis on disrupted atherosclerotic plaques, and inhibits its pro-coagulant activity.(Li et al., 2014) Altogether, these findings suggest that changes in miR-223 expression in the blood in response to toxic environmental exposures may modulate their effect on human health, including the effect of PM2.5 on blood pressure.
We also found that EV-associated miR-199a/b expression levels are positively associated with long-term PM2.5 levels. To the best of our knowledge no previous studies have associated miR-199a/b to PM2.5 ambient levels, except from our previous study in the same population where increased levels of miR199a/b were associated with higher long-term PM2.5 ambient levels.(Rodosthenous et al., 2016) In the present study, we used a higher resolution approach (1×1 km model) to assess PM2.5 levels, confirming that miR-199a/b is associated with long-term PM2.5 levels. In addition, we found that miR-199a/b is a potential effect modifier in the association between PM2.5 and SBP. However, other exposures have been associated with this miRNA, including ozone(Fry et al., 2014) and in occupational workers exposed to perfluorooctanoic acid, suggesting a potential role as stress-induced biomarker.(J. Wang et al., 2012) This stress-induced expression may have a beneficial effect in the reduction of cardiovascular damage. In studies from Jansen et al, EV-associated miR-199a has been associated with a lower risk of major adverse cardiovascular events and revascularization in patients with stable coronary disease. They also demonstrated that the major source of EV-associated miR-199a was platelets.(Jansen et al., 2014) But stress-induced miR-199a overexpression may contribute also to angiogenesis and cardiac regeneration. Shatseva et al. have reported that miR-199a-3p promotes proliferation and survival of endothelial cells.(Shatseva et al., 2011) Remarkably, miR-199a promoted cell cycle re-entry of adult cardiomyocytes ex vivo in animal models, and promoted cardiomyocyte proliferation in both neonatal and adult animals. In fact, after myocardial infarction, miR-199a stimulated marked cardiac regeneration and almost completed recovery of cardiac functional parameters.(Eulalio et al., 2012) In cardiac hypoxia models, blocking miR-199a improves cardiac function and restores mitochondrial fatty acid oxidation.(Azzouzi et al., 2013) These findings suggest a potential beneficial effect of miR-199 after PM2.5 exposure and its effect on blood pressure.
To further explore whether the increase we observed in expression levels of PM2.5-associated evmiRNAs was epigenetically regulated, we examined the association between PM2.5 levels and DNA methylation levels in the promoter regions and up to 2Mbp adjacent to the promoter of each PM2.5-associated evmiRNAs. We found a negative association between both 6-month and 1-year PM2.5 ambient levels and DNA methylation in a CpG site (cg23972892) near miR-199a/b promoter. To our knowledge, this is the first study showing an epigenetic change linked to microRNAs in the context of PM levels. In silico analysis co-localized this CpG site with histone 3 acetylation in lysine 27 (H3K27Ac), a known mark for active enhancers.(Guo et al., 2014) These findings suggest a link between PM2.5 ambient levels and DNA methylation changes with potential downstream effects for evmiRNAs expression levels. Previous studies have reported similar associations between PM exposure and DNA methylation changes in both repetitive sequences and other genes, which are in agreement with our findings that increased PM2.5 levels are associated with decreased DNA methylation levels.(Hou et al., 2010)
Our study has certain limitations. To maximize our ability to detect as many evmiRNAs as possible we decided to use all the available sample volume, such that we could not for further validate our data using another method (e.g., real-time qPCR). Nevertheless, Knutsen et al., reported a relatively high correlation range (r = 0.7–0.8) between miRNA levels when measured with NanoString nCounter® and and other platforms, including real-time qPCR.(Knutsen et al., 2013) Also, although there is a difference in sample size when analyzing PM2.5 effect on evmiRNAs than PM2.5 effect on DNA methylation, demographic and clinical characteristics were similar among participants (Supp. Tables 1 and 2), except for CRP values which were higher in the methylation dataset (mean: 3.48 [SD: 8.52] vs. 2.77 [SD: 4.11] in the microRNAs dataset) but did not reach statistical significance (p=0.056). We also acknowledge the fact that evmiRNAs and DNA methylation cellular origin was not determined in our study. However, we evaluated whether the number of red blood cells, white blood cells, and platelets could confound our data, and whenever appropriate we adjusted for it in the analysis.
Strengths of this study include the use of higher resolution (1×1-km) PM2.5 and BC models, which enabled us to assess ambient levels in our study population more precisely. In addition, the combination of DNA methylation data from 1,022 blood samples (n=563 participants) with evmiRNA expression data from 42 serum samples in a smaller subset (n=22 participants) enabled us to investigate the impact of PM2.5 at both the epigenetic and expression levels of PM2.5-associated evmiRNAs.
Conclusions
In conclusion, our findings suggest that evmiRNAs (i.e., miR-223–3p and miR-199a/b) are positively associated with long-term PM2.5 ambient levels and can modify the association between long-term PM2.5 levels and blood pressure in older individuals. Our findings also suggest that long-term PM2.5 levels is associated with lower DNA methylation levels in regulatory regions of PM-associated evmiRNAs that may explain their increase in expression levels in the peripheral blood. We encourage further research to determine the interplay between PM2.5, epigenetic regulation of PM2.5-associated evmiRNAs, and their impact on blood pressure in older individuals.
Supplementary Material
Highlights.
PM leads to increases in systemic blood pressure (SBP) by unknown mechanisms.
Long-term PM2.5 ambient levels were associated with evmiRNAs levels.
evmiRNAs levels modified the magnitude of the association between PM2.5 and SBP.
evmiRNAs involved between PM5 and SBP could be regulated byepigenetic mechanisms.
ACKNOWLEDGMENTS
We thank the VA-NAS and the Massachusetts Veterans Epidemiology Research and Information Center (MAVERIC). The views expressed in this paper are those of the authors and do not necessarily represent the views of the U.S. Department of Veterans Affairs. The contents of this work are solely the responsibility of the grantee and do not necessarily represent the official views of the funding institutions. Further, the funding institutions do not endorse the purchase of any commercial products or services mentioned in the publication. We thank Clementina Castro-Hernández for the critical review of this manuscript.
FUNDING
This work was supported by National Institutes of Health [grants R01ES015172, R21ES021895, R01ES021733, R21ES021895, R01ES020836, R01ES021357, R21ES027087, P30ES009089, and P30ES000002]; and by the U.S. Environmental Protection Agency [RD-83479801]. The VA Normative Aging Study is supported by the Cooperative Studies Program/Epidemiology Research and Information Center of the US. Department of Veterans Affairs and is a component of the Massachusetts Epidemiology Research and Information Center, Boston, MA. Additional funding to the VA Normative Aging Study was provided by the U.S. Department of Agriculture, Agricultural Research Service [contract 53-K06-510]. DP was financially supported by Consejo Nacional de Ciencia y Tecnología (CONACYT) and the Fundación México en Harvard from the program: “Postdoctoral Fellowships in the Sciences at Harvard University: Mexico” and by the grant CONACYT-FOSISS [grant 289503].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
COMPETING INTERESTS
Authors declare no financial conflicts of interest.
REFERENCES
- Auchincloss AH, Diez-Roux AV, Dvonch JT, Brown PL, Barr RG, Daviglus ML, Goff DC, Kaufman JD, O’Neill MS, 2008. Associations between recent exposure to ambient fine particulate matter and blood pressure in the Multi-ethnic Study of Atherosclerosis (MESA). Environ Health Perspect 116, 486–491. doi: 10.1289/ehp.10899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzouzi el, H., Leptidis S, Dirkx E, Hoeks J, van Bree B, Brand K, McClellan EA, Poels E, Sluimer JC, van den Hoogenhof MMG, Armand A-S, Yin X, Langley S, Bourajjaj M, Olieslagers S, Krishnan J, Vooijs M, Kurihara H, Stubbs A, Pinto YM, Krek W, Mayr M, da Costa Martins PA, Schrauwen P, De Windt LJ, 2013. The hypoxia-inducible microRNA cluster miR-199a∼214 targets myocardial PPARδ and impairs mitochondrial fatty acid oxidation. Cell Metab 18, 341–354. doi: 10.1016/j.cmet.2013.08.009 [DOI] [PubMed] [Google Scholar]
- Baccarelli A, Wright RO, Bollati V, Tarantini L, Litonjua AA, Suh HH, Zanobetti A, Sparrow D, Vokonas PS, Schwartz J, 2009. Rapid DNA methylation changes after exposure to traffic particles. Am J Respir Crit Care Med 179, 572–578. doi: 10.1164/rccm.200807-1097OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bei Y, Das S, Rodosthenous RS, Holvoet P, Vanhaverbeke M, Monteiro MC, Monteiro VVS, Radosinska J, Bartekova M, Jansen F, Li Q, Rajasingh J, Xiao J, 2017. Extracellular Vesicles in Cardiovascular Theranostics. Theranostics 7, 4168–4182. doi: 10.7150/thno.21274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollati V, Angelici L, Rizzo G, Pergoli L, Rota F, Hoxha M, Nordio F, Bonzini M, Tarantini L, Cantone L, Pesatori AC, Apostoli P, Baccarelli AA, Bertazzi PA, 2015. Microvesicle-associated microRNA expression is altered upon particulate matter exposure in healthy workers and in A549 cells. J Appl Toxicol 35, 59–67. doi: 10.1002/jat.2987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Qiao L, Li H, Zhao Y, Zhang Y, Xu W, Wang C, Wang H, Zhao Z, Xu X, Hu H, Kan H, 2015. Fine Particulate Matter Constituents, Nitric Oxide Synthase DNA Methylation and Exhaled Nitric Oxide. Environ. Sci. Technol 49, 11859–11865. doi: 10.1021/acs.est.5b02527 [DOI] [PubMed] [Google Scholar]
- Chuang K-J, Chan C-C, Su T-C, Lee C-T, Tang C-S, 2007. The effect of urban air pollution on inflammation, oxidative stress, coagulation, and autonomic dysfunction in young adults. Am J Respir Crit Care Med 176, 370– 376. doi: 10.1164/rccm.200611-1627OC [DOI] [PubMed] [Google Scholar]
- Dai G-H, Ma P-Z, Song X-B, Liu N, Zhang T, Wu B, 2014. MicroRNA-223–3p inhibits the angiogenesis of ischemic cardiac microvascular endothelial cells via affecting RPS6KB1/hif-1a signal pathway. PLoS ONE 9, e108468. doi: 10.1371/journal.pone.0108468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmerechts J, Jacobs L, Van Kerckhoven S, Loyen S, Mathieu C, Fierens F, Nemery B, Nawrot TS, Hoylaerts MF, 2012. Air pollution-associated procoagulant changes: the role of circulating microvesicles. J. Thromb. Haemost 10, 96–106. doi: 10.1111/j.1538-7836.2011.04557.x [DOI] [PubMed] [Google Scholar]
- Eulalio A, Mano M, Dal Ferro M, Zentilin L, Sinagra G, Zacchigna S, Giacca M, 2012. Functional screening identifies miRNAs inducing cardiac regeneration. Nature 492, 376–381. doi: 10.1038/nature11739 [DOI] [PubMed] [Google Scholar]
- Fry RC, Rager JE, Bauer R, Sebastian E, Peden DB, Jaspers I, Alexis NE, 2014. Air toxics and epigenetic effects: ozone altered microRNAs in the sputum of human subjects. Am. J. Physiol. Lung Cell Mol. Physiol 306, L1129–37. doi: 10.1152/ajplung.00348.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatsiou A, Boeckel J-N, Randriamboavonjy V, Stellos K, 2012. MicroRNAs in platelet biogenesis and function: implications in vascular homeostasis and inflammation. Curr Vasc Pharmacol 10, 524–531. [DOI] [PubMed] [Google Scholar]
- Gryparis A, Coull BA, Schwartz J, Suh HH, 2007. Semiparametric latent variable regression models for spatiotemporal modelling of mobile source particles in the greater Boston area. J Royal Statistical Soc C 56, 183–209. doi: 10.1111/j.1467-9876.2007.00573.x [DOI] [Google Scholar]
- Guo L, Byun H-M, Zhong J, Motta V, Barupal J, Zheng Y, Dou C, Zhang F, McCracken JP, Díaz A, Marco S-G, Colicino S, Schwartz J, Wang S, Hou L, Baccarelli AA, 2014. Effects of short-term exposure to inhalable particulate matter on DNA methylation of tandem repeats. Environ. Mol. Mutagen 55, 322–335. doi: 10.1002/em.21838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herberth G, Bauer M, Gasch M, Hinz D, Röder S, Olek S, Kohajda T, Rolle-Kampczyk U, Bergen, von M, Sack U, Borte M, Lehmann I, Lifestyle and Environmental Factors and Their Influence on Newborns Allergy Risk study group, 2014. Maternal and cord blood miR-223 expression associates with prenatal tobacco smoke exposure and low regulatory T-cell numbers. J. Allergy Clin. Immunol 133, 543–550. doi: 10.1016/j.jaci.2013.06.036 [DOI] [PubMed] [Google Scholar]
- Highwood EJ, Kinnersley RP, 2006. When smoke gets in our eyes: the multiple impacts of atmospheric black carbon on climate, air quality and health. Environ Int 32, 560–566. doi: 10.1016/j.envint.2005.12.003 [DOI] [PubMed] [Google Scholar]
- Hou L, Zhu Z-Z, Zhang X, Nordio F, Bonzini M, Schwartz J, Hoxha M, Dioni L, Marinelli B, Pegoraro V, Apostoli P, Bertazzi PA, Baccarelli A, 2010. Airborne particulate matter and mitochondrial damage: a cross-sectional study. Environ Health 9, 48. doi: 10.1186/1476-069X-9-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen F, Yang X, Proebsting S, Hoelscher M, Przybilla D, Baumann K, Schmitz T, Dolf A, Endl E, Franklin BS, Sinning J-M, Vasa-Nicotera M, Nickenig G, Werner N, 2014. MicroRNA expression in circulating microvesicles predicts cardiovascular events in patients with coronary artery disease. J Am Heart Assoc 3, e001249. doi: 10.1161/JAHA.114.001249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, Lam MH, Kirak O, Brummelkamp TR, Fleming MD, Camargo FD, 2008. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature 451, 1125–1129. doi: 10.1038/nature06607 [DOI] [PubMed] [Google Scholar]
- Kearney PM, Whelton M, Reynolds K, Muntner P, Whelton PK, He J, 2005. Global burden of hypertension: analysis of worldwide data. Lancet 365, 217–223. doi: 10.1016/S0140-6736(05)17741-1 [DOI] [PubMed] [Google Scholar]
- Kloog I, Koutrakis P, Coull BA, Lee HJ, Schwartz J, 2011. Assessing temporally and spatially resolved PM2.5 exposures for epidemiological studies using satellite aerosol optical depth measurements. Atmospheric Environment 45, 6267–6275. doi: 10.1016/j.atmosenv.2011.08.066 [DOI] [Google Scholar]
- Kloog I, Nordio F, Zanobetti A, Coull BA, Koutrakis P, Schwartz JD, 2014. Short term effects of particle exposure on hospital admissions in the Mid-Atlantic states: a population estimate. PLoS ONE 9, e88578. doi: 10.1371/journal.pone.0088578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutsen E, Fiskaa T, Ursvik A, Jørgensen TE, Perander M, Lund E, Seternes OM, Johansen SD, Andreassen M, 2013. Performance comparison of digital microRNA profiling technologies applied on human breast cancer cell lines. PLoS ONE 8, e75813. doi: 10.1371/journal.pone.0075813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Chen H, Ren J, Geng Q, Song J, Lee C, Cao C, Zhang J, Xu N, 2014. MicroRNA-223 inhibits tissue factor expression in vascular endothelial cells. Atherosclerosis 237, 514–520. doi: 10.1016/j.atherosclerosis.2014.09.033 [DOI] [PubMed] [Google Scholar]
- Liang R, Zhang B, Zhao X, Ruan Y, Lian H, Fan Z, 2014. Effect of exposure to PM2.5 on blood pressure: a systematic review and meta-analysis. J. Hypertens 32, 2130–2141. doi: 10.1097/HJH.0000000000000342 [DOI] [PubMed] [Google Scholar]
- Lim SS, Vos T, Flaxman AD, Danaei G, Shibuya K, Adair-Rohani H, Amann M, Anderson HR, Andrews KG, Aryee M, Atkinson C, Bacchus LJ, Bahalim AN, Balakrishnan K, Balmes J, Barker-Collo S, Baxter A, Bell ML, Blore JD, Blyth F, Bonner C, Borges G, Bourne R, Boussinesq M, Brauer M, Brooks P, Bruce NG, Brunekreef B, Bryan-Hancock C, Bucello C, Buchbinder R, Bull F, Burnett RT, Byers TE, Calabria B, Carapetis J, Carnahan E, Chafe Z, Charlson F, Chen H, Chen JS, Cheng AT-A, Child JC, Cohen A, Colson KE, Cowie BC, Darby S, Darling S, Davis A, Degenhardt L, Dentener F, Jarlais, Des DC, Devries K, Dherani M, Ding EL, Dorsey ER, Driscoll T, Edmond K, Ali SE, Engell RE, Erwin PJ, Fahimi S, Falder G, Farzadfar F, Ferrari A, Finucane MM, Flaxman S, Fowkes FGR, Freedman G, Freeman MK., Gakidou E, Ghosh S, Giovannucci E, Gmel G, Graham K, Grainger R, Grant B, Gunnell D, Gutierrez HR, Hall W, Hoek HW, Hogan A, Hosgood HD, Hoy D, Hu H, Hubbell BJ, Hutchings SJ, Ibeanusi SE, Jacklyn GL, Jasrasaria R, Jonas JB, Kan H, Kanis JA, Kassebaum N, Kawakami N, Khang Y-H, Khatibzadeh S, Khoo J-P, Kok C, Laden F, Lalloo R, Lan Q, Lathlean T, Leasher JL, Leigh J, Li Y, Lin JK, Lipshultz SE, London S, Lozano R, Lu Y, Mak J, Malekzadeh R, Mallinger L, Marcenes W, March L, Marks R, Martin R, McGale P, McGrath J, Mehta S, Mensah GA, Merriman TR, Micha R, Michaud C, Mishra V, Mohd Hanafiah K, Mokdad AA, Morawska L, Mozaffarian D, Murphy T, Naghavi M, Neal B, Nelson PK, Nolla JM, Norman R, Olives C, Omer SB, Orchard J, Osborne R, Ostro B, Page A, Pandey KD, Parry CDH, Passmore E, Patra J, Pearce N, Pelizzari PM, Petzold M, Phillips MR, Pope D, Pope CA, Powles J, Rao M, Razavi H, Rehfuess EA, Rehm JT, Ritz B, Rivara FP, Roberts T, Robinson C, Rodriguez-Portales JA, Romieu I, Room R, Rosenfeld LC, Roy A, Rushton L, Salomon JA, Sampson U, Sanchez-Riera L, Sanman E, Sapkota A, Seedat S, Shi P, Shield K, Shivakoti R, Singh GM, Sleet DA, Smith E, Smith KR, Stapelberg NJC, Steenland K, Stöckl H, Stovner LJ, Straif K, Straney L, Thurston GD, Tran JH, Van Dingenen R, van Donkelaar A, Veerman JL, Vijayakumar L, Weintraub R, Weissman MM, White RA, Whiteford H, Wiersma ST, Wilkinson JD, Williams HC, Williams W, Wilson N, Woolf AD, Yip P, Zielinski JM, Lopez AD, Murray CJL, Ezzati M, AlMazroa MA, Memish ZA 2012. A comparative risk assessment of burden of disease and injury attributable to 67 risk factors and risk factor clusters in 21 regions, 1990–2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 380, 2224–2260. doi: 10.1016/S0140-6736(12)61766-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Huang Y, Zhang F, Chen Q, Wu B, Rui W, Zheng JC, Ding W, 2015. Macrophages treated with particulate matter PM2.5 induce selective neurotoxicity through glutaminase-mediated glutamate generation. J. Neurochem 134, 315–326. doi: 10.1111/jnc.13135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordukhovich I, Wilker E, Suh H, Wright R, Sparrow D, Vokonas PS, Schwartz J, 2009. Black carbon exposure, oxidative stress genes, and blood pressure in a repeated-measures study. Environ Health Perspect 117, 1767–1772. doi: 10.1289/ehp.0900591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motta V, Favero C, Dioni L, Iodice S, Battaglia C, Angelici L, Vigna L, Pesatori AC, Bollati V, 2016. MicroRNAs are associated with blood-pressure effects of exposure to particulate matter: Results from a mediated moderation analysis. Environ Res 146, 274–281. doi: 10.1016/j.envres.2016.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemmar A, Vanbilloen H, Hoylaerts MF, Hoet PH, Verbruggen A, Nemery B, 2001. Passage of intratracheally instilled ultrafine particles from the lung into the systemic circulation in hamster. Am J Respir Crit Care Med 164, 1665–1668. doi: 10.1164/ajrccm.164.9.2101036 [DOI] [PubMed] [Google Scholar]
- Noble WS, 2009. How does multiple testing correction work? Nat. Biotechnol 27, 1135–1137. doi: 10.1038/nbt1209-1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pergoli L, Cantone L, Favero C, Angelici L, Iodice S, Pinatel E, Hoxha M, Dioni L, Letizia M, Albetti B, Tarantini L, Rota F, Bertazzi PA, Tirelli AS, Dolo V, Cattaneo A, Vigna L, Battaglia C, Carugno M, Bonzini M, Pesatori AC, Bollati V, 2017. Extracellular vesicle-packaged miRNA release after short-term exposure to particulate matter is associated with increased coagulation. Part Fibre Toxicol 14, 32. doi: 10.1186/s12989-017-0214-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodosthenous RS, Coull BA, Lu Q, Vokonas PS, Schwartz JD, Baccarelli AA, 2016. Ambient particulate matter and microRNAs in extracellular vesicles: a pilot study of older individuals. Part Fibre Toxicol 13, 13. doi: 10.1186/s12989-016-0121-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadik N, Cruz L, Gurtner A, Rodosthenous RS, Dusoswa SA, Ziegler O, Van Solinge TS, Wei Z, Salvador-Garicano AM, Gyorgy B, Broekman M, Balaj L, 2018. Extracellular RNAs: A New Awareness of Old Perspectives. Methods Mol. Biol 1740, 1–15. doi: 10.1007/978-1-4939-7652-2_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatseva T, Lee DY, Deng Z, Yang BB, 2011. MicroRNA miR-199a-3p regulates cell proliferation and survival by targeting caveolin-2. J. Cell. Sci 124, 2826–2836. doi: 10.1242/jcs.077529 [DOI] [PubMed] [Google Scholar]
- Shi L, Fisslthaler B, Zippel N, Frömel T, Hu J, Elgheznawy A, Heide H, Popp R, Fleming I, 2013. MicroRNA-223 antagonizes angiogenesis by targeting β1 integrin and preventing growth factor signaling in endothelial cells. Circ. Res 113, 1320–1330. doi: 10.1161/CIRCRESAHA.113.301824 [DOI] [PubMed] [Google Scholar]
- Soto-Reyes E, González-Barrios R, Cisneros-Soberanis F, Herrera-Goepfert R, Pérez V, Cantú D, Prada D, Castro C, Recillas-Targa F, Herrera LA, 2012. Disruption of CTCF at the miR-125b1 locus in gynecological cancers. BMC Cancer 12, 40. doi: 10.1038/ng.869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamagawa E, Bai N, Morimoto K, Gray C, Mui T, Yatera K, Zhang X, Xing L, Li Y, Laher I, Sin DD, Man SFP, van Eeden SF, 2008. Particulate matter exposure induces persistent lung inflammation and endothelial dysfunction. Am. J. Physiol. Lung Cell Mol. Physiol 295, L79–85. doi: 10.1152/ajplung.00048.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhang Y, Zhang W, Jin Y, Dai J, 2012. Association of perfluorooctanoic acid with HDL cholesterol and circulating miR-26b and miR-199–3p in workers of a fluorochemical plant and nearby residents. Environ. Sci. Technol 46, 9274–9281. doi: 10.1021/es300906q [DOI] [PubMed] [Google Scholar]
- Wang Y-S, Zhou J, Hong K, Cheng X-S, Li Y-G, 2015. MicroRNA-223 displays a protective role against cardiomyocyte hypertrophy by targeting cardiac troponin I-interacting kinase. Cell. Physiol. Biochem 35, 1546–1556. doi: 10.1159/000373970 [DOI] [PubMed] [Google Scholar]
- Wu S, Deng F, Huang J, Wang H, Shima M, Wang X, Qin Y, Zheng C, Wei H, Hao Y, Lv H, Lu X, Guo X, 2013. Blood pressure changes and chemical constituents of particulate air pollution: results from the healthy volunteer natural relocation (HVNR) study. Environ Health Perspect 121, 66–72. doi: 10.1289/ehp.1104812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto SS, Phalkey R, Malik AA, 2014. A systematic review of air pollution as a risk factor for cardiovascular disease in South Asia: limited evidence from India and Pakistan. Int J Hyg Environ Health 217, 133–144. doi: 10.1016/j.ijheh.2013.08.003 [DOI] [PubMed] [Google Scholar]
- Zanobetti A, Baccarelli A, Schwartz J, 2011. Gene-air pollution interaction and cardiovascular disease: a review. Prog Cardiovasc Dis 53, 344–352. doi: 10.1016/j.pcad.2011.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang G, Meng C, Guo X, Cheruku PS, Shi L, Xu H, Li H, Wang G, Evans AR, Safe S, Wu C, Zhou B, 2012. A novel regulator of macrophage activation: miR-223 in obesity-associated adipose tissue inflammation. Circulation 125, 2892–2903. doi: 10.1161/CIRCULATIONAHA.111.087817 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


