Abstract
There is an urgent need for developing more effective therapies for hepatocellular carcinoma (HCC) because of its aggressiveness. Guadecitabine (SGI-110) is a second-generation DNA methyltransferase inhibitor (DNMTi) currently in clinical trials for HCC and shows greater stability and performance over first generation DNMTis. In order to identify potential therapeutic targets of SGI-110 for clinical trials, HCC cell lines (SNU398, HepG2 and SNU475) were used to evaluate effects of transient SGI-110 treatment by an integrative analysis of DNA methylation, nucleosome accessibility, gene expression profiles and its clinical relevance by comparisons to TCGA HCC clinical data. Each HCC cell lines represents a different DNA methylation subtype of primary HCC tumors based on TCGA data. After SGI-110 treatment, all cell lines were sensitive to SGI-110 with prolonged anti-proliferation effects. Expression of up-regulated genes, including tumor suppressors, were positively correlated with nucleosome accessibility and negatively correlated with gene promoter DNA methylation. Alternatively, expression of down-regulated genes, such as oncogenes, were negatively correlated with nucleosome accessibility and positively correlated with gene body DNA methylation. SGI-110 can also act as dual inhibitor to down-regulate PRC2 complex genes by demethylating their gene bodies, resulting in re-activation of PRC2 repressed genes without involvement of DNA methylation. Furthermore, it can up-regulate endogenous retroviruses (ERVs) to reactivate immune pathways. Finally, about 48% of frequently altered genes in primary HCC tumors can be reversed by SGI-110 treatment. Conclusion: our integrative analysis has successfully linked the anti-tumor effects of SGI-110 to detailed epigenetic alterations in HCC cells, identified potential therapeutic targets, and provided rationale for combination treatments of SGI-110 with immune checkpoint therapies.
Keywords: DNA Methylation, Chromatin Accessibility, Gene Expression, Endogenous Retroviruses, Epigenetic Therapy
INTRODUCTION
Hepatocellular carcinoma (HCC) is an aggressive cancer with poor survival statistics and a growing rate of incidence related to hepatitis virus infections and other liver diseases in the population (1, 2). Patients with advanced HCC have very limited options for treatment (3). Sorafenib, a multi-tyrosine kinase inhibitor was the sole approved therapy for advanced HCC (4) until recently when regorefanib, another multitageted kinase inhibitor and niyolumab, an anti PD-1 antibody for treatment of patients who have been previously exposed to sorafenib. Despite these advances, the median overall survival of patients is still limited to about 10 months (5). Hence, there is an urgent need for developing new and more effective therapies for HCC.
A new therapeutic avenue has been emerged using DNA methyltransferase inhibitors (DNMTis) to reverse DNA hypermethylation and associated gene silencing. Indeed, DNA methylation alterations frequently occur in HCC and are integral to the initiation and progression of the disease (6–8). FDA-approved DNMTis, such as 5’-aza-2’deoxycytidine (5-aza-CdR or Decitabine) and 5-azacytidine (5-aza-CR or Vidaza), were developed and represented the first breakthroughs in the clinical application of epigenetic therapy. These DNMTis have shown significant efficacy in the treatment of myelodysplastic syndromes (MDS), chronic myelomonocytic leukemia (CMML) and acute myeloid leukemia (AML) (9–12). Recently DNMTis have been introduced in clinical trials for patients with solid tumors (13–15). However, those drugs exhibit very short in vivo half-lives following clinical administration due to deamination by cytidine deaminase (16–18). This inhibits their application in treating cancers, especially the solid tumors such as HCC.
SGI-110 (Guadecitabine), a second-generation DNMTi, is a dinucleotide consisting of 5-aza-CdR linked to deoxyguanosine. The dinucleotide configuration provides protection from deamination by cytidine deaminase, while maintaining the activity to inhibit DNMTs (19). SGI-110 exhibits a longer half-life and more extended exposure than 5-aza-CdR i.v. infusion (20, 21). In addition, in vitro and in vivo studies have shown that SGI-110 treatment in HCC cell lines resulted in inhibition of cell growth and delayed tumor growth in mouse xenograft models. Furthermore, combination treatments of SGI-110 with sorafenib or oxaliplatin showed improved anti-tumor effects (22, 23).
The traditional view on DNMTi treatment involves reactivation of tumor suppressor genes by DNA demethylation to suppress tumor growth and reverse the malignant phenotype (24, 25). However, the level of DNA demethylation does not always correlate with the level of gene reactivation and/or predict clinical outcome. Recent investigations including several from our group suggest that oncogene down-regulation via gene body DNA demethylation, activation of endogenous retroviral elements (ERVs) and an anti-viral defense response contribute to the clinical efficacy of DNMTis (26–28). Since DNA methylation has tissue- and cancer-specific profiles, which vary greatly between cancer types, the anti-tumor activities of DNMTis may be specific for individual tissues and/or cancer types.
Because the therapeutic targets of SGI-110 in HCC have not been well identified, we performed an integrative analysis of genome-wide DNA methylation, nucleosome accessibility, and gene expression in human HCC cell lines over time following transient SGI-110 treatment to identify the potential epigenetic driver events that contribute to drug efficacy. Nucleosome occupancy changes have been recently recognized as important epigenetic regulators of gene expression by facilitating or inhibiting the accessibility of chromatin to the transcription machinery (29–31). We used the AcceSssIble assay developed by our group to simultaneously analyze DNA methylation and nucleosome accessibility changes (32, 33) and then integrated these findings with gene expression data. Our study demonstrates a novel approach for identifying potential epigenetic therapeutic targets, and provide mechanistic rational for evaluating SGI-110 in treating HCC patients in the clinic.
EXPERIMENTAL PROCEDURES
Cell Culture and Drug Treatments
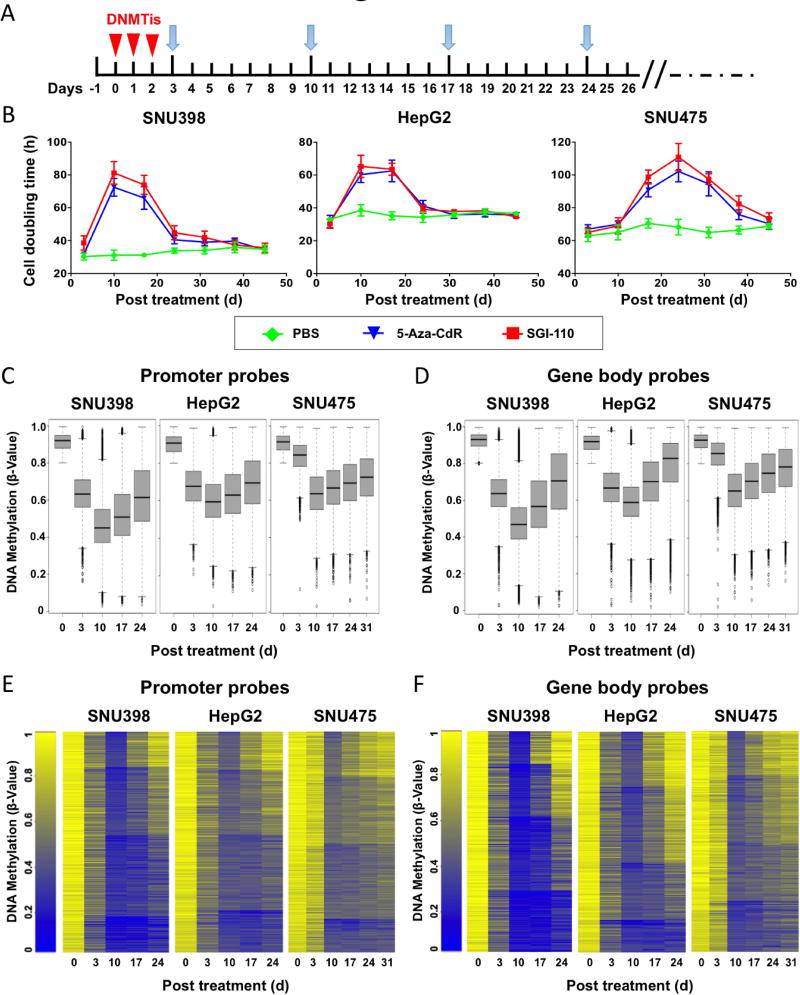
SNU398, SNU475 and HepG2 cells were purchased form the American Type Culture Collection (ATCC; www.atcc.org), and treated with three daily consecutive doses of SGI-110 (Astex Pharmaceuticals, Dublin, CA) at 100 nM (72 h total exposure) or GSK126 (EZH2 inhibitor, Active Biochem, LTD, Hong Kong) at 500 nM daily. Cells were harvested at the time points indicated in Figure 2A.
AcceSssIble and Gene Expression Analysis
For the AcceSssIble assay, nuclei preparation and M.SssI methyltransferase (New England BioLabs) treatment were performed as previously described (32, 33). The subsequent Infinium DNA methylation assay was performed at the University of Southern California Molecular Genomics Core Facility according to the manufacturer’s specifications (Illumina). Expression analysis was performed at Sanford-Burnham Medical Institute (La Jolla, CA) using the Illumina human genome-wide expression BeadChip (HumanHT-12_V4.0_R1) (Illumina). The detailed experimental procedures and data analysis are included as SI Materials and Methods in online supplementary files. All array-based DNA methylation and gene expression data utilized in this study have been deposited in the Gene Expression Omnibus (GEO) database under the accession number GSE105067.
Pathway Analysis
Gene Ontology (GO) and pathway enrichment analysis and functional annotation clustering were performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) database v6.8. Relevant GO lists were ranked according to their p-values as determined by DAVID.
Statistical analysis
Statistical tests were conducted using R software (R version 3.1.2, R Development Core Team, https://cran.r-project.org/). The R package “ploycor” was used to calculate a matrix consisting of p-values and Pearson correlations between gene expression and DNA methylation, as well as between gene expression and chromatin accessibility. Wilcoxon rank-sum test was used to assess statistical significance for RNA-seq gene expression differences of UHRF1, EZH2, EED and SUZ12 between TCGA normal and primary HCC tumor samples. P-values less than 0.05 were considered significant.
Accession codes
All data have been deposited at the Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) with accession code GSE105067.
A detailed description of all experimental methods is included in Supplementary Information.
RESULTS
The human HCC cell lines SNU398, HepG2 and SNU475 represent unique DNA methylation-based subgroups of primary hepatocellular carcinomas
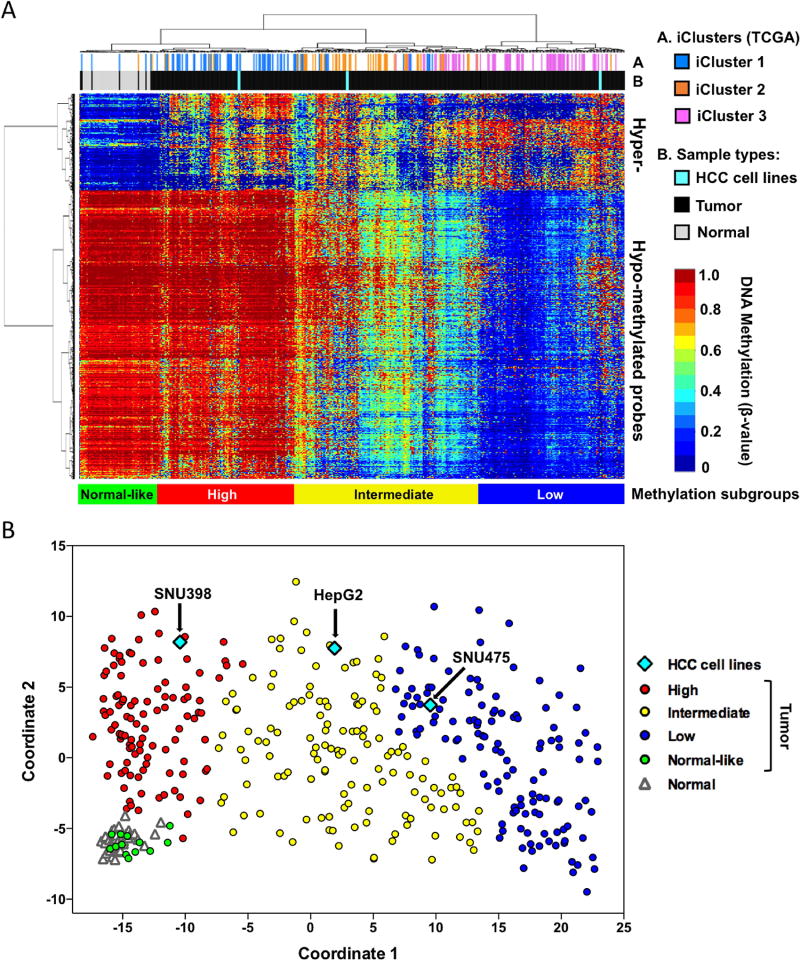
For preclinical studies, it is important to determine whether cell line DNA methylation profiles are comparable to primary tumors and can be used for determination of sensitivity of primary tumors to DNMTi-based therapies. In the aim of providing an experimental system in which the effects and potential therapeutic targets of the DNMTi SGI-110 could be evaluated for HCC patients, we first tested whether the DNA methylation status of cultured HCC cell lines mimic those of primary HCC tumor samples. We used the Illumina Infinium HumanMethylation450 BeadArray (HM450) platform to measure genome-scale DNA methylation of three HCC cell lines, HepG2, SNU398 and SNU475, and we then compared these findings to the publically available HM450 DNA methylation data of primary liver hepatocellular carcinoma patient samples from The Cancer Genome Atlas (TCGA) project (https://cancergenome.nih.gov/cancersselected/LiverHepatocellularCarcinoma) (8). The HM450 platform contains probes for 482,421 CpG sites across the human genome, covering 99% of RefSeq genes (more than 21,000 genes), and the probes are distributed in promoters, gene bodies, enhancers, and intergenic regions (34).
Unsupervised clustering of the TCGA HCC DNA methylation dataset identified three distinct DNA methylation-based tumor subgroups were identified by comparing 377 TCGA HCC tumors to 50 adjacent-normal tissues. One group contains mostly hypermethylated loci (“High”), one group harbors both hypermethylated and moderately hypomethylated loci (“Intermediate”) and the third group exhibits extensive DNA hypomethylation (“Low”) (Figure 1A). Several primary tumors were clustered with adjacent-normal tissues and labeled as “normal-like”. These subgroups show a high correspondence to the TCGA iClusters (8), with “high”, “intermediate” and “low” groups consist predominantly of iCluster 1, iCluster 2 and iCluster 3, respectively. Interestingly, SNU398, HepG2 and SNU475 cells were clustered within “High”, “Intermediate” and “Low” subgroups, respectively, and are closely clustered with HCC tumors from the three distinct iClusters. Multidimensional Scaling (MDS) analysis further confirmed that SNU398, HepG2 and SNU475 cells belong to three distinct DNA methylation subgroups (Figure 1B). Therefore, SNU398, HepG2 and SNU475 cells can be used as a preclinical model representing different HCC molecular subgroups to study how HCC cells respond to DNMTi treatment.
Transient SGI-110 treatment of SNU398, HepG2 and SNU475 cells showed prolonged effects on cell growth inhibition and DNA demethylation
We then used these three cell lines with unique DNA methylation profiles to evaluate the potency of SGI-110 (Figure 1A and B). Because DNMTi incorporation into genomic DNA is dependent on cell doubling (28, 35), each cell line was treated with three consecutive 24 h doses (72 h total) of 100 nM SGI-110 to cover at least one cell doubling cycle before drug removal. Cells were then cultured in drug-free medium until there were no further differences in the population doubling times compared to untreated cells (Figure 2A). Depletion of DNMT1, DNMT3A and DNMT3B protein levels was observed at day 3 after the treatment with SGI-110, 5-Aza-CR or 5-Aza-CdR (Figure S1), suggesting that the current dose of SGI-110 is as potent as other DNA methylation inhibitors. Prolonged increases in cell doubling times occurred in all three cell lines by SGI-110, and peaked around 10 days for SNU398 and HepG2 cells, and 24 days for SNU475 cells (Figure 2B). We observed increases in apoptosis and cell cycle arrest at D10 after treatment of SNU398 cells (Figure S2), which may explain the anti-tumor effects observed as a consequence of SGI-110 treatment.
We then monitored DNA methylation changes after SGI-110 treatment using the HM450 DNA methylation array. Demethylation of methylated probes (β value >0.8 before treatment) was observed in all three cell lines, peaking at day 10 after SGI-110 treatment, and occurred for probes located in promoters (Figure 2C) or gene bodies (Figure 2D) or globally across the genome (Figure S3). Most of the demethylated promoter and gene body regions then were re-methylated at different kinetic rates with a population of regions that remained demethylated at day 24 after treatment (Figure 2E and 2F). However, the DNA remethylation rates of individual probes were not uniform. Interestingly, a population of probes remained demethylated, which may contribute to the prolonged growth inhibition effects after SGI-110 treatment (Figure 1B and Figure S3) (28).
SGI-110 treatment of SNU398, HepG2 and SNU475 cells increases chromatin accessibility of demethylated or unmethylated loci
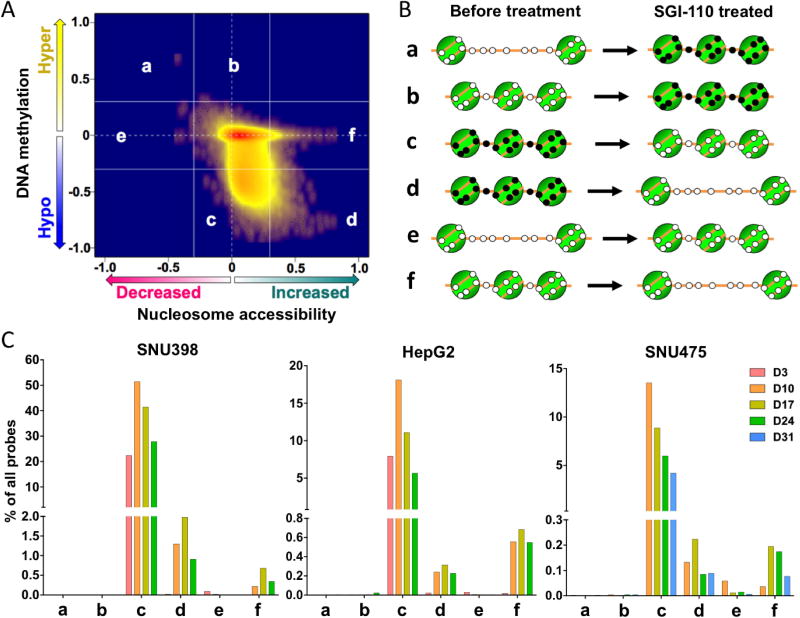
Our previous studies have demonstrated that most DNA demethylation events are “passenger” effects and do not result in functional gene expression changes due to retained chromatin inaccessibility after DNMTi treatment (32). Our goal is to identify “driver” epigenetic events as potential therapeutic targets induced by SGI-110 that could result in gene expression changes, thereby accounting for alterations of cellular phenotypes upon treatment. Since nucleosome positioning also plays an important role in regulating gene expression, we performed AcceSssIble analysis to simultaneously analyze DNA methylation and chromatin accessibility in HCC cells lines after SGI-110 treatment (32, 33). In this assay, chromatin accessibility levels at individual CpG loci were obtained by subtracting the endogenous DNA methylation beta value from the beta value after M.SssI treatment for each probe on the HM450 platform (Figure S4) (32).
We measured the DNA methylation and chromatin accessibility levels at each time point after SGI-110 treatment for all three HCC cell lines. We then plotted DNA methylation changes (Δβ-values) against the levels of chromatin accessibility changes (ΔΔβ-values) between untreated and SGI-110 treated cells in 2-D Kernel density scatter plots to examine the epigenetic changes at each time point after SGI-110 treatment at each time point (Figure 3A and S5). Six potential models (a, b, c, d, e, and f) can theoretically illustrate these changes (Figure 3A and 3B) (33). Groups a and b represent gains in DNA methylation with (group a) or without (group b) nucleosome accessibility changes after SGI110 treatment (Figure 3A and 3B). Groups c and d represent DNA demethylation without (group c) or with (group d) nucleosome accessibility changes after drug treatment. Finally, groups e and f represent nucleosome accessibility alterations at unmethylated loci. However, because SGI-110 mainly induces DNA demethylation, nearly all changes fell into groups c, d, and f (Figure 3C and S5).
SGI-110 treatment resulted in a majority of DNA demethylation events that were not associated with corresponding gain of nucleosome accessibility during the experimental time course (group c) (Figure 3C and S5). Interestingly, a small number of probes (< 2% of all probes) gained nucleosome accessibility with loss of DNA methylation (group d) and without involvement of DNA methylation (group f) (Figure 3C and S5). Therefore, the AcceSssIble assay allows for the discrimination of functional DNA demethylation events, namely those events that are accompanied by nucleosome accessibility changes, illustrated (group d in Figure 3B) from potential “passenger” effects of DNA demethylation with continued inaccessible chromatin (group c in Figure 3B). In addition, AcceSssIble also identified potential functional loci or genes after SGI-110 treatment without involvement of DNA methylation after SGI-110 treatment (group f) (Figure 3 and S5). In general, most DNA demethylation events occurred at day 10 after SGI-110 treatment (group c, Figure 3C and S5), while most nucleosome accessibility changes occurred at day 17 after treatment (group d and f, Figure 3C and S5) for all three cell lines, suggesting that DNA demethylation preceded the increase in nucleosome accessibility at functional loci.
Taken together, our results showed that the majority of demethylated DNA regions remained inaccessible as potential “passenger” changes, while a smaller subset of loci gained nucleosome accessibility with or without involvement of DNA methylation.
The majority of gene expression alterations after SGI-110 treatment correlate with DNA methylation and/or nucleosome accessibility changes
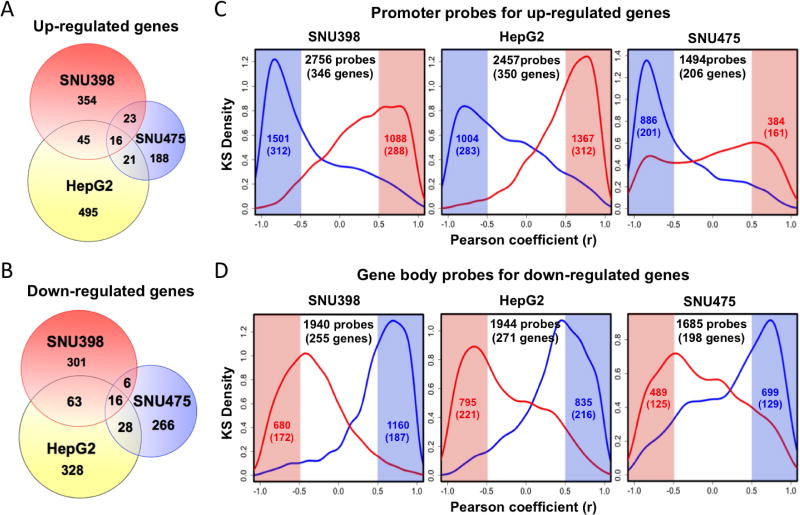
To further assess the functional consequences of SGI-110 induced DNA methylation and nucleosome accessibility changes, we integrated genome-wide expression, DNA methylation and chromatin accessibility data for all three cell lines. Using a two-fold expression change as a cutoff, we identified 438, 577 and 248 upregulated genes (Figure 4A), as well as 386, 435 and 316 down-regulated genes (Figure 4B), during the experimental time course after SGI-110 treatment in SNU398, HepG2 and SNU475 cells, respectively. The majority of these genes did not overlap (Figure 4A and 4B), which may be due to different DNA methylation profiles of each cell line before drug treatment (Figure 1).
We then examined the relationship between gene expression changes and alterations in nucleosome accessibility and DNA methylation at all the time points before and after SGI-110 treatment. We first examined the epigenetic changes at the promoters of genes that became upregulated after drug treatment (Figure 4A). As evident by the peak in the kernel density plot, there was a significant negative correlation; between gene expression and promoter DNA methylation, highlighted (blue, r<-0.5) and a positive correlation between gene expression and promoter accessibility (red, r > 0.5) for the majority of up-regulated genes after treatment in all cell lines (Figure 4C). This is consistent with the well-established model that promoter DNA demethylation and increased chromatin accessibility lead to gene activation (25, 32, 33).
More importantly, we also found that the majority of down-regulated genes after drug treatment exhibited a significant positive correlation (between expression and gene body DNA methylation (blue, r > 0.5) and a negative correlation between expression and gene body nucleosome accessibility (red, r < -0.5) (Figure 4D). Therefore, AcceSssIble analysis revealed that SGI-110 modulates gene expression in HCC cells through increasing nucleosome accessibility not only at gene promoters but also at gene body regions.
Integrative analysis identifies SGI-110 target genes that are frequently altered in primary HCC tumors
We further tested the number of genes targeted by SGI-110 that overlap with aberrantly expressed genes in primary HCCs, which may have important applications for preclinical studies, clinical trials and treatments in HCC patients. First, we identified the most frequently altered genes in primary HCCs based on the TCGA HCC gene expression dataset, and we compared these with SGI-110 targeted genes by intersecting the gene expression profiles of the three HCC cell lines before and after SGI-110 treatment.
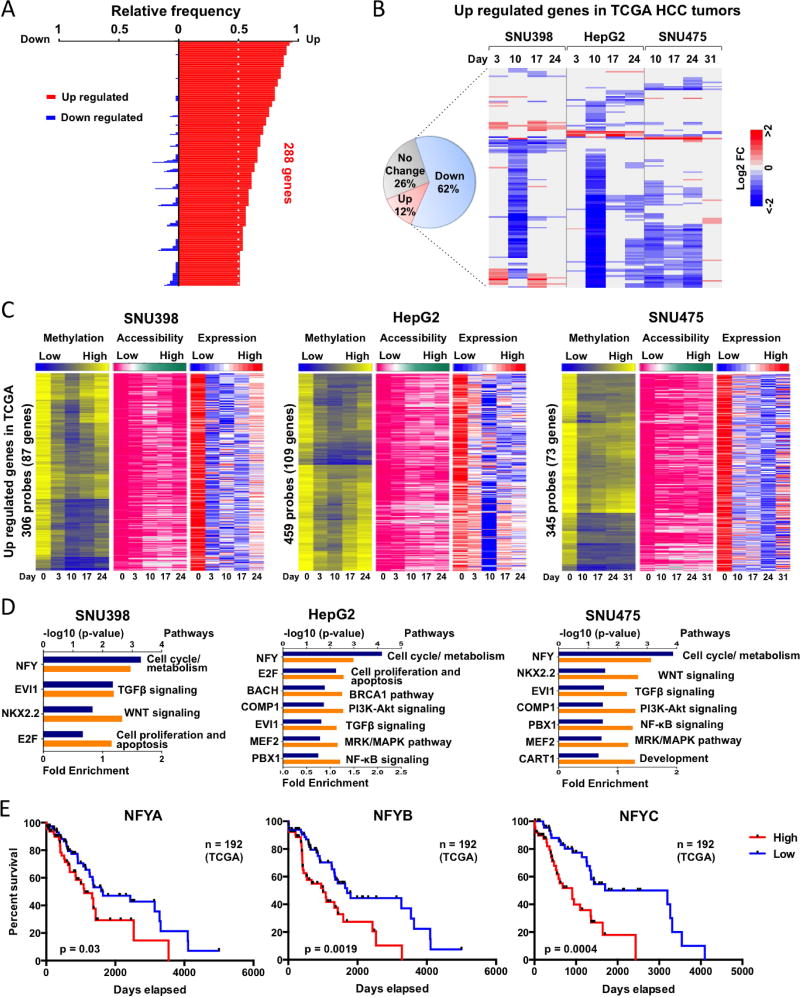
Using a 4-fold expression cut-off, we identified 288 over expressed genes in more than 50% of primary TCGA HCC tumors (Figure 5A). These genes were involved in various pathways including cell cycle regulation, DNA repair, chromatin organization, angiogenesis, etc. (Figure S6A and B). Importantly, several epigenetic regulators are over expressed in primary HCC tumors, including UHRF1 and the PRC2 complex members EZH2, EED and SUZ12 (Figure S7A). About 62% (148/238) became down-regulated greater than 2-fold after SGI-110 treatment in at least one HCC cell line (Figure 5B). In addition, the down-regulated expression of these genes in each HCC cell line was associated with gene body DNA demethylation and increased nucleosome accessibility at gene body regions (Figure 5C and S6C).
Remarkably, the top 10 most significant GO processes for these genes were mostly related to cell cycle regulation in all three HCC cell lines (Figure S6B), suggesting that SGI-110 treatment could revert the hyper-proliferative HCC phenotype through down-regulation of cell cycle related genes, such as the E2 factor (E2F) family of transcription factors. By searching the DAVID database (https://david.ncifcrf.gov) and using the “UCSC TFBS” program, these genes were regulated by transcription factors such as E2F, nuclear transcription factor Y (NFY), ecotropic viral integration site 1 (EVI1), and NK2 Homeobox 2 (NKX2.2) (Figure 5D), and are involved in key signaling pathways contributing to the hallmarks of HCC, including TGFβ, WNT, pRB-E2F and multiple cell cycle/metabolism (8, 36–38). Importantly, we found that the expression of the NFY family of transcription factors are linked with HCC survival, as patients with low NFY family expression showed improved survival outcome (Figure 5E), highlighting the important role of SGI-110 in down-regulating NFY target genes. In addition, the above-mentioned epigenetic regulators UHRF1, EZH2, EED and SUZ12, whose expression levels were reduced by gene body DNA demethylation after drug treatment (Figure S7B and C), showed restored expression levels upon remethylation as a function of gene body DNA re-methylation. Therefore, by altering gene body DNA methylation and nucleosome accessibility, SGI-110 reverted several well-known HCC related oncogenic pathways (Figure 5D and Figure S6A, B).
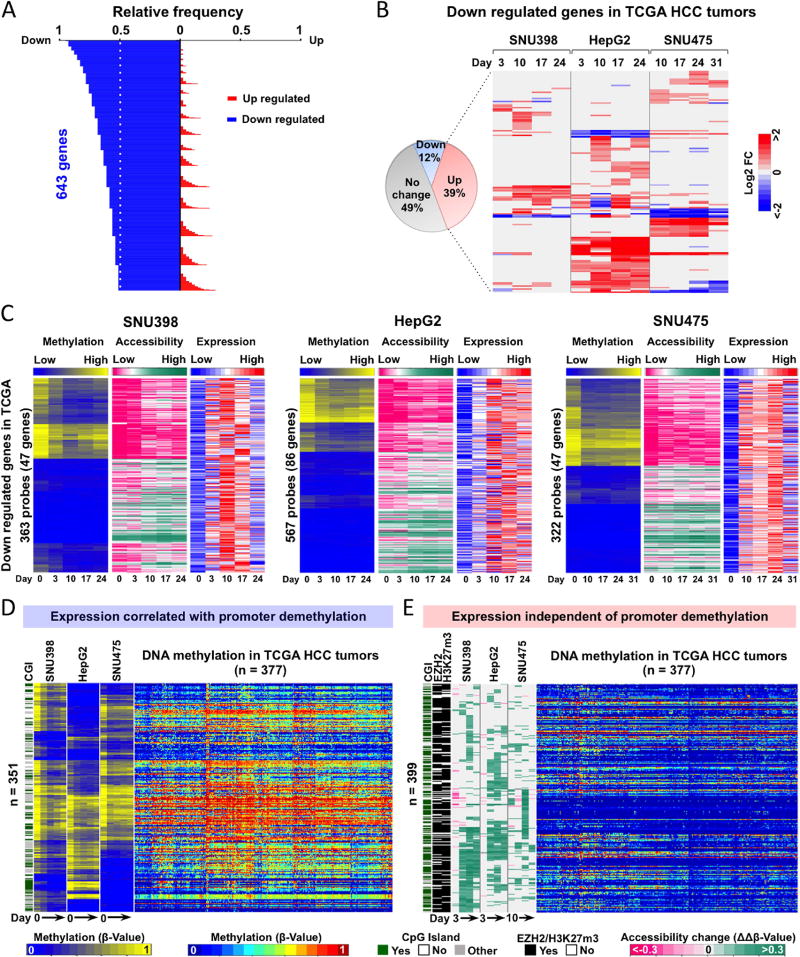
Using the same approach, we identified 643 down-regulated genes in TCGA HCC tumors (Figure 6A) that are also associated with hallmarks of HCC by dysregulation of cellular differentiation, resistance to apoptosis, metabolic reprogramming, and immune destruction avoidance, as previously described (Figure S8A) (8, 36–38). About 39% (147/377) were activated >2-fold after SGI-110 treatment in at least one HCC cell line (Figure 6B). Although the majority of these genes were differentially up-regulated between the individual HCC cell lines (Figure 6B), their functions largely overlapped (Figure S8B), suggesting that SGI-110 treatment may reverse HCC hallmarks by activating different genes within the same signaling pathways.
Next, we found that the genes that were up-regulated after SGI-110 treatment (Figure 6C and S8C) fell into two groups: 1) genes that correlated with promoter DNA demethylation and increased chromosome accessibility, and 2) reactivated genes that were independent of promoter DNA demethylation but demonstrated increased chromosome accessibility (Figure 6C and S8C). We found the first group of genes displayed differential promoter DNA methylation and were also largely methylated in TCGA HCC tumors (Figure 6D), suggesting these genes may be direct targets of SGI-110 through promoter DNA demethylation. The second group of genes exhibited only increased chromatin accessibility after SGI-110 treatment, and were mostly unmethylated in TCGA HCC tumors (Figure 6E). We found that the majority of these genes were EZH2 targets (PRC2 complex) after cross-referencing with the ENCyclopedia of DNA Elements (ENCODE) database for histone modifications, as 87% of the probes were occupied by EZH2 or marked by H3K27me3 in HepG2 cells (Figure 6E). Importantly, many of these EZH2 targets are well-known tumor suppressors, such as SFRP1, MT2A, ALPL, RND3, MT1F and MT1M. These findings indicated that this group of genes could be activated via down regulation of EZH2, EED and SUZ12 upon SGI-110 treatment (Figure S7), and suggest that SGI-110 functions as a dual inhibitor, not only for DNA demethylation but also reduced H3K27me3 occupancy via down-regulated expression of PRC2 complex genes. To further test this hypothesis, we treated all three HCC cell lines with an EZH2 inhibitor (GSK126) and tested whether this group of SGI-110 target genes could be up-regulated. We found that EZH2 inhibitor treatment did indeed up-regulate a panel of SGI-110 targeted genes independent of DNA methylation (Figure S9). Therefore, SGI-110 as a dual inhibitor reactivates down-regulated or silenced genes in two unique ways: 1) direct DNA demethylation and increased chromatin accessibility of methylated gene promoters; and 2) indirectly increased accessibility of PRC2-targeted gene promoters in a manner independent of DNA methylation but through down-regulation of EZH2, EED and SUZ12. In addition, this finding also demonstrated that AcceSssIble analysis identified gene regulation changes through altering nucleosome accessibility with or without involvement of DNA methylation, which would otherwise be unidentifiable by DNA methylation analysis alone. In conclusion, these findings suggest that a large portion of aberrantly regulated genes (about 48%, 295/615) in primary HCC can be reversed by SGI-110 treatment and are the therapeutic targets of SGI-110.
SGI-110 up-regulates interferon-responsive genes by activating endogenous retrovirus (ERV) expression
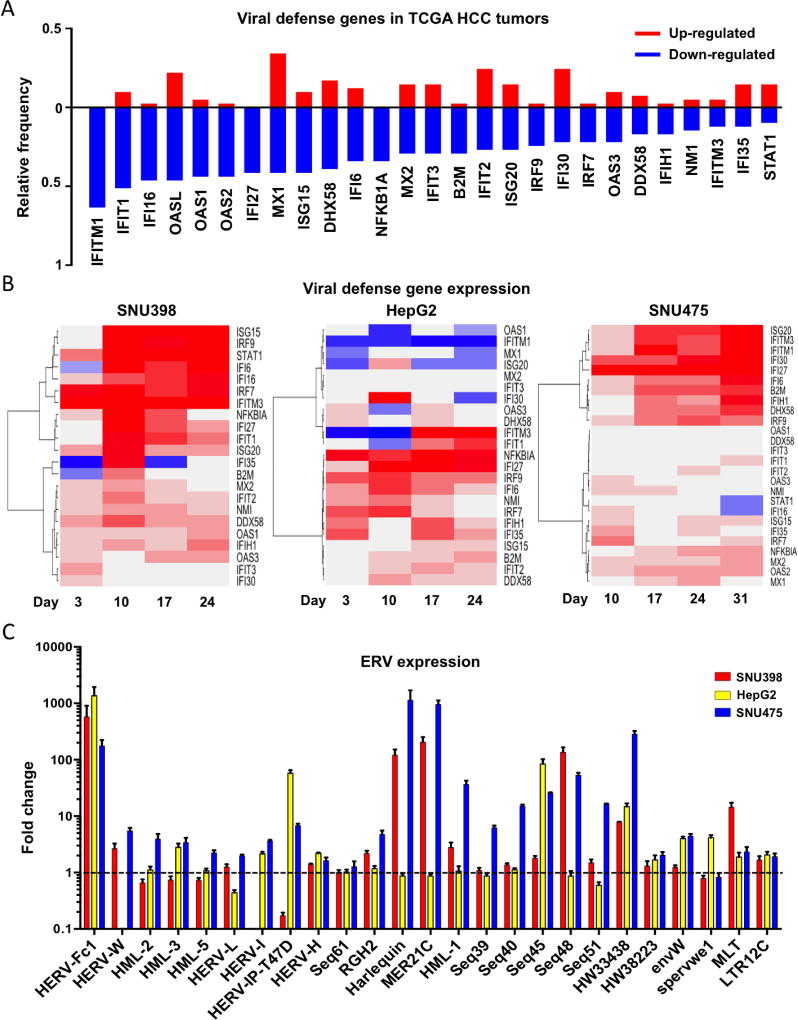
We and others have demonstrated that ERV reactivation to stimulate an innate immune response in cancer cells is a key mechanism underlying the anti-tumor activities of DNMTis (26, 27, 39). Gene ontology (GO) analysis of the down-regulated genes in TCGA HCC tumors also revealed an impaired immune response (Figure S7A). We analyzed the expression of 27-curated viral defense genes from previous publications (26, 27, 39) in 41 pairs of TCGA HCC tumors. All of these genes were down-regulated in 10–60% of TCGA HCC tumors (Figure 7A), indicating that this innate viral defense pathway is frequently down-regulated in HCC. In contrast, more than 75% of the viral defense genes were up regulated in HCC cell lines after SGI-110 treatment (Figure 7B).
We then examined the expression status of a panel of 25 ERVs at the day 10 (D10) time point after SGI-110 treatment, the time in which an elicit immune response may occur. Indeed, ERVs were differentially up-regulated in all three HCC cell lines (Figure 7C). Among these ERVs, HERV-Fc1, HW33438, MLT and LTC12C were up-regulated more than 2-fold in all three HCC cell lines after drug treatment (Figure 7C).
DISCUSSION
The majority of targeted therapeutic approaches for HCC have focused on individual pathways that are known to be disrupted in HCC or harboring genetic alterations but with limited success (4, 40). Although epigenetic alterations in general are pharmacologically reversible and DNA methylation-dependent epigenetic driver genes have been identified in HCC (41), epigenetic alterations are overlooked as potential therapeutic targets in cancer therapy. The FDA-approved DNMTi Decitabine and Vidaza represented breakthroughs in the clinical application of epigenetic therapy (42). In the area of solid tumors, SGI-110 was evaluated as a safe single agent in clinic trails in patients with advanced HCC, but target genes and pathway alterations to maximize the potential benefit of this approach.
Our integrative analysis of DNA methylation, nucleosome accessibility and gene expression has revealed the great potential of SGI-110 in treating HCC. We identified that three common HCC cell lines (SNU398, HepG2 and SNU475) were appropriate pre-clinical models for HCC since they represent the three DNA methylation-based subgroups of primary HCC tumors based on TCGA data. Furthermore, all three cell lines responded to transient, low-dose SGI-110 treatment and exhibited prolonged cell growth inhibition together with prolonged DNA demethylation, suggesting that SGI-110 targets HCC cells irrespective of their DNA methylation status. As previously demonstrated, prolonged cell inhibition may be dependent on extended DNA demethylation-based gene regulation, which may be caused by changes in histone modifications at gene body regions such as those with H3K36me3 occupancy and recruitment of DNMT3B recruitment (28). However, our results also suggest the prolonged DNA demethylation may also be explained by UHRF1 down-regulation due to DNA demethylation of its gene body after SGI-110 treatment. Importantly, UHRF1 acts as an accessory protein in recruiting DNMT1 and binding hemimethylated DNA for maintenance DNA methylation through DNA replication (43), and UHRF1 overexpression has been identified as a driver of HCC tumorigenesis (44). Thus, SGI-110 can not only down-regulate DNMTs at protein level but also directly down-regulate UHRF1 at the transcription level, leading to prolonged DNA dememthylation, and thereby exerting an antitumor effect.
Our previous work has demonstrated the importance of nucleosome accessibility in regulating gene expression (32, 33). Here, we showed similar findings in all three HCC cell lines. Specifically, we showed that functional DNA demethylation or potential therapeutic targets represents only a very small portion of demethylated loci, that together with increasing chromatin accessibility, are highly correlated with up-regulation of silenced genes or down-regulation of over-expressed genes by promoter and gene body DNA demethylation, respectively. Interestingly, we found that SGI-110, similar to an EZH2 inhibitor, directly increases nucleosome accessibility of unmethylated promoters initially silenced by PRC2 occupancy, thereby leading to altered gene expression. This may be due to the direct removal of chromatin-based PRC2 proteins to turn on gene expression. Indeed, we have demonstrated that SGI-110, by comparison to the effects of an EZH2 inhibitor, can up-regulate or re-activate PRC2-silenced tumor suppressor genes in a manner independent of DNA methylation by direct down-regulation of EZH2, and EED and SUZ12 via gene body DNA demethylation. Thus, for the first time, we demonstrate that SGI-110 acts as dual inhibitor not only for DNA methyltransferases but also for PRC2 complex in HCC.
We successfully tested this in vitro system to determine whether the expression of well-known tumor suppressors, oncogenes, and other tumorigenesis-related genes can be corrected by SGI-110 treatment. Indeed, most of the aberrantly expressed genes in HCC were corrected by SGI-110 treatment based on the HCC cell line and TCGA datasets. Another notable finding is that SGI-110 preferentially down-regulates over-expressed genes, especially cell cycle-related genes, by gene body DNA demethylation rather than reactivation of down-regulated/silenced genes, as we previously demonstrated, but is still largely overlooked (28). These genes are regulated by transcription factors such as NFY, E2F and EVI1, and belong to pathways such as WNT, TGFβ and PI3K signaling, which were shown to be associated with HCC or other malignancies (8, 36–38, 45). In addition, we also find that high expression levels of the NFY family of genes correlate with worse clinical outcome in HCC patients.
Our results indicate that SGI-110 reverses a large portion of abnormally transcribed genes in HCC tumors and can up-regulate ERVs to stimulate an innate immune response (26, 27, 39). Thus, we expect that SGI-110 may be even more efficacious in combination with other cancer treatment drugs in up-regulating ERVs or transposable elements and subsequently increase sensitivity to immune checkpoint therapies (46) and re-sensitize drug-resistant tumor cells (47).
Taken together, our findings suggest SGI-110, acting as dual inhibitor, has a significant anti-tumor effect by: 1) reshaping the cancer cell expression profile into a “normal like” status based on epigenetic reprogramming with a dependence on chromatin accessibility alterations; and 2) re-activating ERVs to stimulate cancer cell immune response pathways, which provide the rationale for combination treatments with immune checkpoint therapies or chemotherapies to re-sensitize drug resistant cancer cells. These data also provide mechanistic evidence to support clinic trials of SGI-110 in treating patients with HCC or other types of malignancies.
Supplementary Material
Acknowledgments
We thank Dr. Peter A. Jones for helpful discussions and support for this project. We also thank Dr. William G. Corey for his passion and contributions towards liver cancer research.
Financial Support:
This work was supported by the Vicky Joseph Cancer Research Foundation (G.L.) and NIH/NCI P30 CA014089 (to D.J.W.)
Abbreviations
- HCC
Hepatocellular Carcinoma
- DNMTi
DNA Methyltransferase Inhibitor
- SGI-110
Guadecitabine
- TCGA
The Cancer Genome Atlas project
- ERVs
Endogenous Retroviruses
- PRC2
Polycomb Repressive Complex 2
- UHRF1
Ubiquitin Like With PHD and Ring Finger Domains 1
- EED
Embryonic Ectoderm Development
- EZH2
Enhancer Of Zeste 2 Polycomb Repressive Complex 2 Subunit
- SUZ12
SUZ12 Polycomb Repressive Complex 2 Subunit
- E2F
E2 factor
- NFY
Nuclear Transcription Factor Y
- EVI1
Ecotropic Viral Integration Site 1
- NKX2.2
NK2 Homeobox 2
- TGFβ
Transforming growth factor beta
- WNT
Wingless-type MMTV Integration Site Family
- ENCODE
the ENCyclopedia of DNA Elements
- GO
Gene ontology
Footnotes
Contributors
ML designed and performed experiments, analyzed and interpreted data, wrote paper. GL designed experiments, interpreted data, wrote paper. ZL and HL performed experiments, analyzed and interpreted data. DJW interpreted the data and wrote paper. TH, WZ, HD, AE, CO, JD, TD, QL interpreted and discussed the data.
Competing interests
DJW is a consultant for Zymo Research Corporation (Irvine, CA)
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Shibata T, Aburatani H. Exploration of liver cancer genomes. Nat Rev Gastroenterol Hepatol. 2014;11:340–349. doi: 10.1038/nrgastro.2014.6. [DOI] [PubMed] [Google Scholar]
- 3.Llovet JM, Burroughs A, Bruix J. Hepatocellular carcinoma. Lancet. 2003;362:1907–1917. doi: 10.1016/S0140-6736(03)14964-1. [DOI] [PubMed] [Google Scholar]
- 4.Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, de Oliveira AC, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 5.He AR, Goldenberg AS. Treating hepatocellular carcinoma progression following first-line sorafenib: therapeutic options and clinical observations. Therap Adv Gastroenterol. 2013;6:447–458. doi: 10.1177/1756283X13498540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calvisi DF, Ladu S, Gorden A, Farina M, Lee JS, Conner EA, Schroeder I, et al. Mechanistic and prognostic significance of aberrant methylation in the molecular pathogenesis of human hepatocellular carcinoma. J Clin Invest. 2007;117:2713–2722. doi: 10.1172/JCI31457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen J, Wang S, Zhang YJ, Kappil M, Wu HC, Kibriya MG, Wang Q, et al. Genome-wide DNA methylation profiles in hepatocellular carcinoma. Hepatology. 2012;55:1799–1808. doi: 10.1002/hep.25569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cancer Genome Atlas Research Network. Electronic address wbe, Cancer Genome Atlas Research N. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell. 2017;169:1327–1341. doi: 10.1016/j.cell.2017.05.046. e1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kantarjian HM, Thomas XG, Dmoszynska A, Wierzbowska A, Mazur G, Mayer J, Gau JP, et al. Multicenter, randomized, open-label, phase III trial of decitabine versus patient choice, with physician advice, of either supportive care or low-dose cytarabine for the treatment of older patients with newly diagnosed acute myeloid leukemia. J. Clin. Oncol. 2012;30:2670–2677. doi: 10.1200/JCO.2011.38.9429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kantarjian H, Issa JP, Rosenfeld CS, Bennett JM, Albitar M, DiPersio J, Klimek V, et al. Decitabine improves patient outcomes in myelodysplastic syndromes: results of a phase III randomized study. Cancer. 2006;106:1794–1803. doi: 10.1002/cncr.21792. [DOI] [PubMed] [Google Scholar]
- 11.Issa JP, Roboz G, Rizzieri D, Jabbour E, Stock W, O'Connell C, Yee K, et al. Safety and tolerability of guadecitabine (SGI-110) in patients with myelodysplastic syndrome and acute myeloid leukaemia: a multicentre, randomised, dose-escalation phase 1 study. Lancet Oncol. 2015;16:1099–1110. doi: 10.1016/S1470-2045(15)00038-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fenaux P, Mufti GJ, Hellstrom-Lindberg E, Santini V, Gattermann N, Germing U, Sanz G, et al. Azacitidine prolongs overall survival compared with conventional care regimens in elderly patients with low bone marrow blast count acute myeloid leukemia. J. Clin. Oncol. 2010;28:562–569. doi: 10.1200/JCO.2009.23.8329. [DOI] [PubMed] [Google Scholar]
- 13.Li H, Chiappinelli KB, Guzzetta AA, Easwaran H, Yen RW, Vatapalli R, Topper MJ, et al. Immune regulation by low doses of the DNA methyltransferase inhibitor 5-azacitidine in common human epithelial cancers. Oncotarget. 2014;5:587–598. doi: 10.18632/oncotarget.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Juo YY, Gong XJ, Mishra A, Cui X, Baylin SB, Azad NS, Ahuja N. Epigenetic therapy for solid tumors: from bench science to clinical trials. Epigenomics. 2015;7:215–235. doi: 10.2217/epi.14.73. [DOI] [PubMed] [Google Scholar]
- 15.Zahnow CA, Topper M, Stone M, Murray-Stewart T, Li H, Baylin SB, Casero RA., Jr Inhibitors of DNA Methylation, Histone Deacetylation, and Histone Demethylation: A Perfect Combination for Cancer Therapy. Adv Cancer Res. 2016;130:55–111. doi: 10.1016/bs.acr.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 16.Chabot GG, Bouchard J, Momparler RL. Kinetics of deamination of 5-aza-2'-deoxycytidine and cytosine arabinoside by human liver cytidine deaminase and its inhibition by 3-deazauridine, thymidine or uracil arabinoside. Biochem Pharmacol. 1983;32:1327–1328. doi: 10.1016/0006-2952(83)90293-9. [DOI] [PubMed] [Google Scholar]
- 17.Chabner BA, Drake JC, Johns DG. Deamination of 5-azacytidine by a human leukemia cell cytidine deaminase. Biochem Pharmacol. 1973;22:2763–2765. doi: 10.1016/0006-2952(73)90137-8. [DOI] [PubMed] [Google Scholar]
- 18.van Groeningen CJ, Leyva A, O'Brien AM, Gall HE, Pinedo HM. Phase I and pharmacokinetic study of 5-aza-2'-deoxycytidine (NSC 127716) in cancer patients. Cancer Res. 1986;46:4831–4836. [PubMed] [Google Scholar]
- 19.Yoo CB, Jeong S, Egger G, Liang G, Phiasivongsa P, Tang C, Redkar S, et al. Delivery of 5-aza-2'-deoxycytidine to cells using oligodeoxynucleotides. Cancer Res. 2007;67:6400–6408. doi: 10.1158/0008-5472.CAN-07-0251. [DOI] [PubMed] [Google Scholar]
- 20.Tellez CS, Grimes MJ, Picchi MA, Liu Y, March TH, Reed MD, Oganesian A, et al. SGI-110 and entinostat therapy reduces lung tumor burden and reprograms the epigenome. Int J Cancer. 2014;135:2223–2231. doi: 10.1002/ijc.28865. [DOI] [PubMed] [Google Scholar]
- 21.Chuang JC, Warner SL, Vollmer D, Vankayalapati H, Redkar S, Bearss DJ, Qiu X, et al. S110, a 5-Aza-2'-deoxycytidine-containing dinucleotide, is an effective DNA methylation inhibitor in vivo and can reduce tumor growth. Mol Cancer Ther. 2010;9:1443–1450. doi: 10.1158/1535-7163.MCT-09-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuang Y, El-Khoueiry A, Taverna P, Ljungman M, Neamati N. Guadecitabine (SGI-110) priming sensitizes hepatocellular carcinoma cells to oxaliplatin. Mol Oncol. 2015;9:1799–1814. doi: 10.1016/j.molonc.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jueliger S, Lyons J, Cannito S, Pata I, Pata P, Shkolnaya M, Lo Re O, et al. Efficacy and epigenetic interactions of novel DNA hypomethylating agent guadecitabine (SGI-110) in preclinical models of hepatocellular carcinoma. Epigenetics. 2016:1–12. doi: 10.1080/15592294.2016.1214781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 26.Chiappinelli KB, Strissel PL, Desrichard A, Li H, Henke C, Akman B, Hein A, et al. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell. 2015;162:974–986. doi: 10.1016/j.cell.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roulois D, Loo Yau H, Singhania R, Wang Y, Danesh A, Shen SY, Han H, et al. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell. 2015;162:961–973. doi: 10.1016/j.cell.2015.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang X, Han H, De Carvalho DD, Lay FD, Jones PA, Liang G. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell. 2014;26:577–590. doi: 10.1016/j.ccr.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.You JS, Jones PA. Cancer genetics and epigenetics: two sides of the same coin? Cancer Cell. 2012;22:9–20. doi: 10.1016/j.ccr.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andreu-Vieyra CV, Liang G. Nucleosome occupancy and gene regulation during tumorigenesis. Adv Exp Med Biol. 2012;754:109–134. doi: 10.1007/978-1-4419-9967-2_5. [DOI] [PubMed] [Google Scholar]
- 31.Shen H, Laird PW. Interplay between the cancer genome and epigenome. Cell. 2013;153:38–55. doi: 10.1016/j.cell.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pandiyan K, You JS, Yang X, Dai C, Zhou XJ, Baylin SB, Jones PA, et al. Functional DNA demethylation is accompanied by chromatin accessibility. Nucleic Acids Res. 2013;41:3973–3985. doi: 10.1093/nar/gkt077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Becket E, Chopra S, Duymich CE, Lin JJ, You JS, Pandiyan K, Nichols PW, et al. Identification of DNA Methylation-Independent Epigenetic Events Underlying Clear Cell Renal Cell Carcinoma. Cancer Res. 2016;76:1954–1964. doi: 10.1158/0008-5472.CAN-15-2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sandoval J, Heyn H, Moran S, Serra-Musach J, Pujana MA, Bibikova M, Esteller M. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics. 2011;6:692–702. doi: 10.4161/epi.6.6.16196. [DOI] [PubMed] [Google Scholar]
- 35.Tsai HC, Li H, Van Neste L, Cai Y, Robert C, Rassool FV, Shin JJ, et al. Transient low doses of DNA-demethylating agents exert durable antitumor effects on hematological and epithelial tumor cells. Cancer Cell. 2012;21:430–446. doi: 10.1016/j.ccr.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coulouarn C, Gomez-Quiroz LE, Lee JS, Kaposi-Novak P, Conner EA, Goldina TA, Onishchenko GE, et al. Oncogene-specific gene expression signatures at preneoplastic stage in mice define distinct mechanisms of hepatocarcinogenesis. Hepatology. 2006;44:1003–1011. doi: 10.1002/hep.21293. [DOI] [PubMed] [Google Scholar]
- 37.Hoshida Y, Nijman SM, Kobayashi M, Chan JA, Brunet JP, Chiang DY, Villanueva A, et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 2009;69:7385–7392. doi: 10.1158/0008-5472.CAN-09-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allain C, Angenard G, Clement B, Coulouarn C. Integrative Genomic Analysis Identifies the Core Transcriptional Hallmarks of Human Hepatocellular Carcinoma. Cancer Res. 2016;76:6374–6381. doi: 10.1158/0008-5472.CAN-16-1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu M, Ohtani H, Zhou W, Orskov AD, Charlet J, Zhang YW, Shen H, et al. Vitamin C increases viral mimicry induced by 5-aza-2'-deoxycytidine. Proc Natl Acad Sci U S A. 2016;113:10238–10244. doi: 10.1073/pnas.1612262113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Forner A, Llovet JM, Bruix J. Chemoembolization for intermediate HCC: is there proof of survival benefit? J Hepatol. 2012;56:984–986. doi: 10.1016/j.jhep.2011.08.017. [DOI] [PubMed] [Google Scholar]
- 41.Villanueva A, Portela A, Sayols S, Battiston C, Hoshida Y, Mendez-Gonzalez J, Imbeaud S, et al. DNA methylation-based prognosis and epidrivers in hepatocellular carcinoma. Hepatology. 2015;61:1945–1956. doi: 10.1002/hep.27732. [DOI] [PubMed] [Google Scholar]
- 42.Liang G, Weisenberger DJ. DNA methylation aberrancies as a guide for surveillance and treatment of human cancers. Epigenetics. 2017;12:416–432. doi: 10.1080/15592294.2017.1311434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duymich CE, Charlet J, Yang X, Jones PA, Liang G. DNMT3B isoforms without catalytic activity stimulate gene body methylation as accessory proteins in somatic cells. Nat Commun. 2016;7:11453. doi: 10.1038/ncomms11453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mudbhary R, Hoshida Y, Chernyavskaya Y, Jacob V, Villanueva A, Fiel MI, Chen X, et al. UHRF1 overexpression drives DNA hypomethylation and hepatocellular carcinoma. Cancer Cell. 2014;25:196–209. doi: 10.1016/j.ccr.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bard-Chapeau EA, Jeyakani J, Kok CH, Muller J, Chua BQ, Gunaratne J, Batagov A, et al. Ecotopic viral integration site 1 (EVI1) regulates multiple cellular processes important for cancer and is a synergistic partner for FOS protein in invasive tumors. Proc Natl Acad Sci U S A. 2012;109:2168–2173. doi: 10.1073/pnas.1119229109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Orskov AD, Treppendahl MB, Skovbo A, Holm MS, Friis LS, Hokland M, Gronbaek K. Hypomethylation and up-regulation of PD-1 in T cells by azacytidine in MDS/AML patients: A rationale for combined targeting of PD-1 and DNA methylation. Oncotarget. 2015;6:9612–9626. doi: 10.18632/oncotarget.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guler GD, Tindell CA, Pitti R, Wilson C, Nichols K, KaiWai Cheung T, Kim HJ, et al. Repression of Stress-Induced LINE-1 Expression Protects Cancer Cell Subpopulations from Lethal Drug Exposure. Cancer Cell. 2017 doi: 10.1016/j.ccell.2017.07.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.