Abstract
Myelination in the central nervous system takes place predominantly during the postnatal development of humans and rodents by myelinating oligodendrocytes (OLs), which are differentiated from oligodendrocyte progenitor cells (OPCs). We recently reported that Sox2 is essential for developmental myelination in the murine brain and spinal cord. It is still controversial regarding the role of Sox2 in oligodendroglial lineage progression in the postnatal murine spinal cord. Analyses of a series of cell- and stage-specific Sox2 mutants reveal that Sox2 plays a biphasic role in regulating oligodendroglial lineage progression in the postnatal murine spinal cord. Sox2 controls the number of OPCs for subsequent differentiation through regulating their proliferation. In addition, Sox2 regulates the timing of OL differentiation and modulates the rate of oligodendrogenesis. Our experimental data prove that Sox2 is an intrinsic positive timer of oligodendroglial lineage progression and suggest that interventions affecting oligodendroglial Sox2 expression may be therapeutic for overcoming OPC differentiation arrest in dysmyelinating and demyelinating disorders.
Keywords: Sox2, myelination, oligodendrocyte differentiation, oligodendrocyte progenitor cells (OPCs), proliferation, neural stem cells
Introduction
Oligodendrocytes are CNS myelin-forming cells. Oligodendroglial lineage progression encompasses multiple sequential steps including OPC generation, migration, proliferation, differentiation into premyelinating OLs (i.e. OL differentiation), and myelination by myelinating OLs [1]. Perturbation of any steps could result in myelination impairment.
In the CNS, the SRY (sex determining region Y)-box 2, Sox2 is highly expressed in NSCs [2, 3] and maintains the properties of neural stem cells (NSCs) and progenitor cells [4, 5]. In line with its role in NSC/progenitor cells, recent data have shown that overexpression of Sox2 converts differentiated astrocytes and OPCs back into neural progenitor cells in vivo under certain circumstances [6–8]. However, there are “conflicting” observations in the current literatures regarding the role of Sox2 in the spinal cord oligodendrocyte development. Hoffmann and colleagues reported that Sox2 is dispensable for OPC proliferation and migration, but essential for OL differentiation and myelination in the perinatal spinal cord [9]. A recent study by Zhao et al., reported that Sox2 seems dispensable for OL differentiation and myelination in the postnatal spinal cord [10]. However this study employed animal models of non-cell-specific Sox2 deletion driven by the actin promoter. More recently, we used animal models of cell specific Sox2 deletion driven by the Sox10 promoter (Sox10-Cre:Sox2fl/fl) and found that Sox2 is required for developmental myelination in the murine spinal cord and brain [11]. These discrepant observations prompted us to revisit the role of Sox2 in oligodendroglial lineage progression in the postnatal spinal cord by leveraging our cell- and stage-specific animal models of Sox2 deletion.
To this end, we used Sox10-Cre:Sox2fl/fl animal model to constitutively delete Sox2 in OPCs and tamoxifen-inducible Pdgfrα-CreERT2:Sox2fl/fl to delete Sox2 in OPCs at the neonatal ages. In addition, we used Cnp-Cre:Sox2fl/fl animal model to constitutively delete Sox2 in differentiation-committed OPCs and their progenies and tamoxifen-inducible Plp-CreERT2:Sox2fl/fl to time-conditionally delete Sox2 in differentiation-committed OPCs and their progenies. Through analysis and comparison of oligodendrocyte lineage phenotypes among these Sox2 mutants, we concluded that Sox2 is an intrinsic bi-phasic regulator of oligodendroglial proliferation and differentiation during developmental myelination of postnatal murine spinal cord, as we recently reported in the postnatal brain [11].
Results
Our recent data show that Sox2 is expressed in OPCs and upregulated in newly differentiated OLs during CNS developmental myelination [11]. Here, we employed the genetic approach of Cre-loxP-mediated labeling and demonstrated that Sox2 was expressed in all OPCs regardless of their CNS regional origins (Fig. 1).
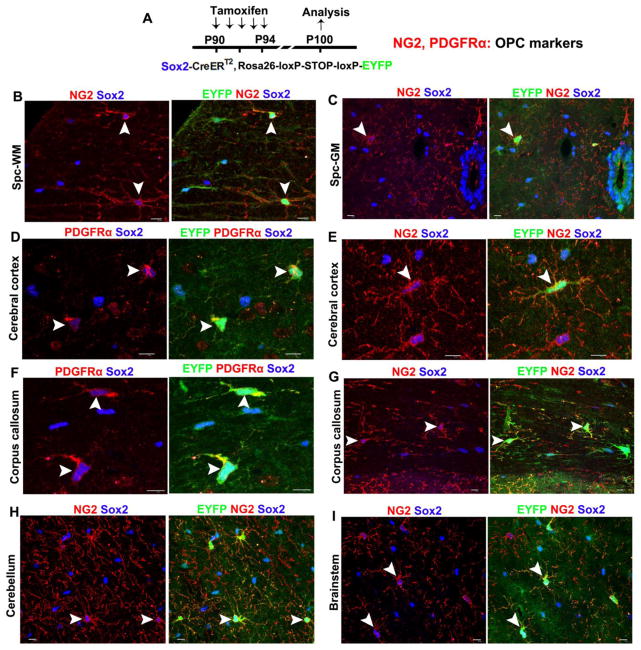
Figure 1. Sox2 is ubiquitously expressed in all adult OPCs throughout the CNS regions.
A, transgenic mice and experimental designs. Double transgenic mice carrying Sox2-CreERT2 and Rosa26-loxP-STOP-loxP-EYFP were injected intraperitoneally with tamoxifen daily from P90 through P94, and CNS tissues were processed at P100 for immunohistochemical analysis of EYFP and the OPC markers NG2 and PDGFRα. Note that the Sox2-CreERT2 transgene is homologously knocked in the locus of endogenous Sox2 gene. Therefore, tamoxifen inducible EYFP expression is a bona fide indicator of endogenous Sox2 promoter activity. B–I, triple immunohistochemistry of reporter EYFP, endogenous Sox2, and OPC markers PDGFRα (or NG2) unequivocally demonstrate that Sox2 is expressed in adult OPCs in the spinal cord white matter (WM) (B) and gray matter (GM) (C), cerebral cortex (D, E), corpus callosum (F, G), cerebellum (H), and brainstem (I). Arrowheads point to the examples of PDGFRα (or NG2)+ OPCs that have both endogenous Sox2 (blue) and transgenic EYFP expression. Note that 100% of EYFP+ cells (including astrocytes and OPCs) are Sox2 positive, validating that EYFP is a reliable indicator of endogenous Sox2 expression. Over 90% of NG2+ (or PDGFRα+) OPCs are EYFP+ using the tamoxifen injection paradigm (see Materials and Methods). Scale bars = 10 μm.
Sox2 is specifically and efficiently deleted in OPCs in the spinal cord of Sox10-Cre:Sox2fl/fl mutants
Sox10 expression is absent from embryonic NSCs (i.e. neuroepithelial cells) and starts in OPCs immediately after OPC specification from NSCs at ~ embryonic 12.5 days (E12.5) in the mouse spinal cord [12]. Accordingly, Sox10-Cre-mediated Sox2 deletion was observed in all PDGFRα+ OPCs (Fig. 2A1–A2) but not in the neuroepithelial cells at the ventricular zone (boxed areas in Fig. 2A1–A2).
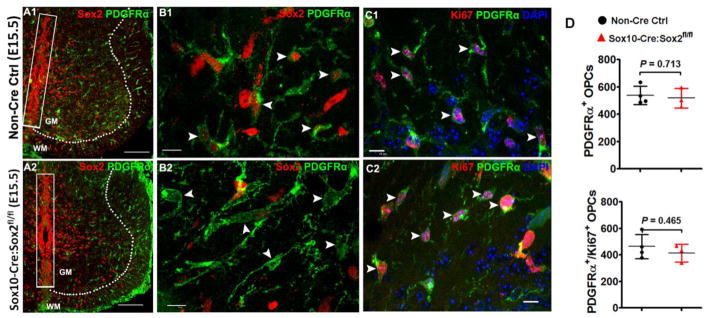
Figure 2. Sox2 ablation does not affect OPC migration and proliferation during embryonic spinal cord development.
A1–A2, low magnification confocal images showing that Sox2-deficient PDGFRα+ embryonic OPCs are generated and migrate normally into the primitive gray and white matter (A2) in a similar manner to those in Sox2-intact OPCs (A1). Dashed lines demarcate gray matter (GM) and white matter (WM). Note that Sox2 is preserved in the neuroepithelial cells (boxed areas) aligning the ventricular zones in E15.5 Sox10-Cre, Sox2fl/fl (Sox2 cKO) spinal cord. B1–B2: high magnification confocal images showing similar distribution and equivalent density of PDGFRα+ embryonic OPCs in the whiter matter of E15.5 spinal cord of Sox2 cKO (B2, arrowheads) and non-Cre control (B1, arrowheads). C1–C2, the distribution and density of Ki67+PDGFRα+ proliferating OPCs are similar in Sox2 cKO (C2, arrowheads) spinal cord to those in non-Cre controls (C1, arrowheads) at E15.5. Blue in C1–C2 is DAPI nuclear staining. D, densities of OPCs and proliferating OPCs in the primitive WM of spinal cord at E15.5. Two tailed Student’s t test, t(5) = 0.3897 PDGFRα+, t(5) = 0.7903 PDGFRα+Ki67+. Scale bar: A1, A2, 100 μm; B1–C2, 10 μm.
OPC proliferation and migration in the embryonic spinal cord of Sox10-Cre:Sox2fl/fl mutants
Interestingly, Sox2-deficient embryonic OPCs migrated and proliferated normally in the spinal cord of Sox10-Cre:Sox2fl/fl embryos, indistinguishable from Sox2-intact OPCs in Sox2fl/fl controls (arrowheads in Fig. 2C1 vs Fig. 2C2, Fig. 2D) at E15.5, ~ 3 days after OPC specification. Our results suggest that Sox2 is dispensable for OPC proliferation and migration in the embryonic spinal cord, which is consistent with the previous data [9].
Hypomyelination in the adult spinal cord of Sox10-Cre:Sox2fl/fl mutants
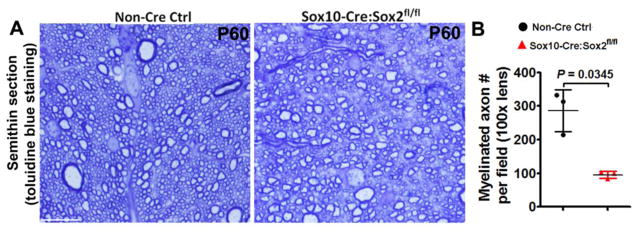
Our recent data show that Sox10-Cre:Sox2fl/fl mutants display tremors and ataxia (indicators of hypomyelination) and impaired performance on accelerating Rotarod [11] at P28, an approximate time point that oligodendrocyte differentiation and developmental myelination are close to completion in the murine spinal cord. In this study, we found that myelination impairment persisted into the adult ages, as assessed by diminished density of myelinated axons in CST of Sox10-Cre:Sox2fl/fl mutants at P60, compared with littermate control mice (Fig. 3). Our previous and current data suggest that Sox2 is essential for developmental myelination in the murine spinal cord.
Figure 3. Sox2 disruption impairs axonal myelination in the adult spinal cord of Sox10-Cre:Sox2fl/fl mutants.
A, representative images of toluidine blue staining of myelin in the corticospinal tract (CST) at P60. Scale bar = 10 μm. B, quantification of myelinated axon numbers per field. Two-tailed Student’s t test with Welch’s correction, t(2) = 5.231.
Time course analysis of OPC proliferation and OL differentiation in the postnatal spinal cord of Sox10-Cre:Sox2fl/fl mutants
Hypomyelination in Sox10-Cre:Sox2fl/fl mutant spinal cord indicates defects in the oligodendroglial lineage progression. To quantify cell number at the different developmental stages of oligodendroglial lineage cells, we used Sox10 or Olig2 to label all oligodendroglial lineage cells (OPCs and OLs), Sox10 (or Olig2) and PDGFRα to label OPCs, Sox10 (or Olig2) and CC1 to label differentiated OLs, and APC/TCF7l2 to label newly differentiated premyelinating OLs. We first confirmed that Sox2 was completely and specifically deleted from oligodendroglial lineage cells in the postnatal spinal white matter, as evedenced by the absence of Sox2 in Olig2+ oligodendroglial lineage cells (arrowheads in Fig. 4A left vs right panels) in the spinal cord of Sox10-Cre:Sox2fl/fl mutants.
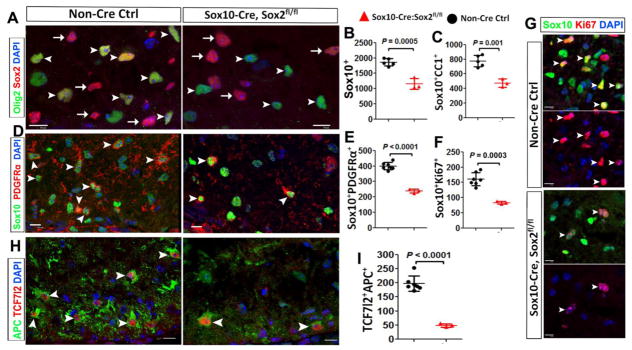
Figure 4. Sox2 deletion diminishes OPC number and proliferation and reduces the density of differentiated OLs in the spinal cord at postnatal day 3 (P3).
A, representative confocal images showing that Sox2 is expressed in Olig2+ oligodendroglial lineage cells (OPCs and OLs) in non-Cre controls and absent from Olig2+ cells in Sox10-Cre,Sox2fl/fl (Sox2 cKO) mice at P3 (A, arrowheads). Note that Sox2+/Olig2− astrocytic lineage cells (arrows) are equivalently present in both control and Sox2 cKO white matter. B–C, density (# per mm2) of Sox10+ oligodendroglial lineage cells and Sox10+CC1+ differentiated OLs in the spinal white matter at P3 (two-tailed Student t test, t(6) = 6.763 Sox10+, t(6) = 5.73 Sox10+CC1+). D–E, representative confocal images of the pan-oligodendroglial lineage marker Sox10 and the OPC marker PDGFRα and quantification. Two-tailed Student’s t test, t(8) = 10.93 Sox10+PDGFRα+. Arrowheads point to examples of Sox10+PDGFRα+ OPCs. F–G, density (per mm2) and representative images of Sox10 and Ki67. Two-tailed Student’s t test, t(8) = 6.02. Arrowheads point to examples of Sox10+Ki67+ cells. H–I, representative confocal images depicting TCF7l2+APC+ newly differentiated premyelinating OLs (H, arrowheads) and the density (per mm2) of TCF7l2+APC+ cells (I). Two-tailed Student’s t test, t(8) = 9.246. Blue, DAPI+ nuclei. Scale bars: 10 μm, applied to all.
At P3, the number of Sox10+ oligodendroglial lineage cells (Fig. 4B), Sox10+CC1+ OLs (Fig. 4C), and Sox10+PDGFRα+ OPCs (Fig. 4E) was significantly decreased in Sox10-Cre:Sox2fl/fl mutants compared with that in non-Cre controls. We used Sox10 and Ki67 to identify cycling OPCs (Fig. 4G, arrowheads) and found that the number of proliferating OPCs was also significantly reduced in the mutant white matter (Fig. 4F). We have shown that TCF7l2 [13] and APC [14] are upregulated and co-expressed in newly formed premyelinating OLs and dramatically downregulated in mature myelinating OLs. Therefore, the number of TCF7l2+APC+ cells at any given time provides a quantitative indication of the rate of OL generation from OPCs, or oligodendrogenesis. Our quantification data showed a 75% reduction of TCF7l2+APC+ newly formed premyelinating OLs in the white matter of Sox10-Cre:Sox2fl/fl mutants compared with non-Cre controls (Fig. 4H–I). These data indicate that Sox2 positively regulates oligodendroglial lineage progression and the rate of oligodendrogenesis in the early postnatal spinal cord.
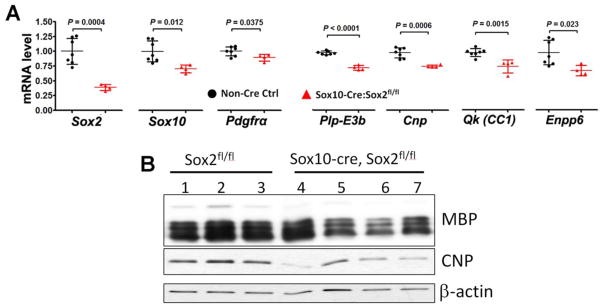
A recent study reported that oligodendroglial lineage progression was normal by the second postnatal week (P14) in the spinal cord of Sox2 ubiquitously ablated mice (Cag-CreER™:Sox2fl/fl) [10]. However, our recent data demonstrated that oligodendroglial lineage progression was persistently impaired in the spinal cord of Sox10-Cre:Sox2fl/fl mice even at later states of spinal cord development [11]. In line with our recent histological data of diminished number of OPCs and OLs, RT-qPCR quantification showed that the mRNA levels of OL lineage genes, Sox10, Pdgfrα, Plp-E3b, Cnp, CC1, and Enpp6, and OPC specific gene were significantly decreased in the P19 spinal cord of Sox10-Cre:Sox2fl/fl mice compared with non-Cre controls (Fig. 5A). Western blot verified the reduced protein level of myelin genes MBP and CNP (Fig. 5B). Therefore, our previous and current data suggest that oligodendroglial lineage progression is persistently impaired even during the later stages of spinal cord development.
Figure 5. reduced expression of myelin-related genes in the spinal cord Sox10-Cre:Sox2fl/fl mutants at P19.
A, RT-qPCR quantification of spinal cord mRNA levels of Sox2, Sox10, Pdgfrα, Plp-E3b (exon3b-containing proteolipid protein), Cnp, Qk (also known as CC1), and premyelinating OL-specific Enpp6 at P19. Gene expression was normalized to the internal control Hsp90, and the level in non-Cre Ctrl was set to 1. N = 7 non-Cre control (black circles) and 4 Sox2 cKO (red triangles), two-tailed Student’s t test [t(9) = 5.397 Sox2, t(9) = 3.131 Sox10, t(9) = 2.437 Pdgfrα, t(9) = 11.71 Plp-E3b, t(9) = 5.145 Cnp, t(9) = 4.487 Qk, and t(9) = 2.734 Enpp6]. B, Western blotting of myelin proteins MBP and CNP, and the loading control β-actin in the P19 spinal cord.
Collectively, the time-course data obtained from Sox10-Cre:Sox2fl/fl mutants indicate that Sox2 is indispensable for oligodendroglial lineage progression during developmental myelination in the murine spinal cord.
Time course analysis of OPC proliferation and OL differentiation in postnatal spinal cord of tamoxifen inducible Pdgfrα-CreERT2:Sox2fl/fl mutants
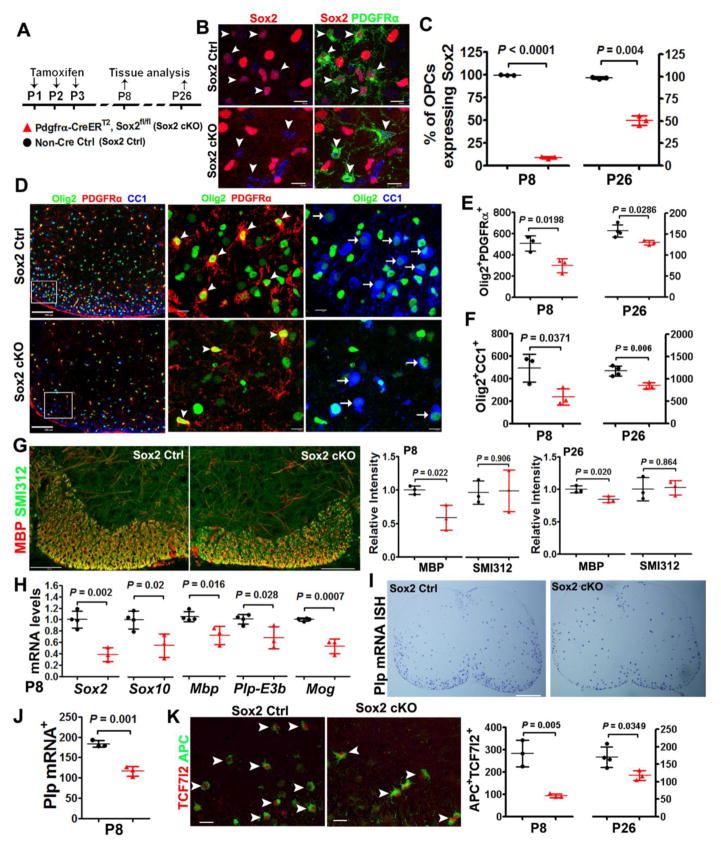
One can argue that the diminished OPC and OL population in the postnatal Sox10-Cre:Sox2fl/fl mutants is the secondary effects of Sox2 deletion in embryonic OPCs on postnatal development. To exclude this possibility, we employed a time-conditional paradigm to elicit Sox2 deletion. In this regard, we generated Pdgfrα-CreERT2:Sox2fl/fl (Sox2 cKO) transgenic hybrids. Tamoxifen was administered at P1, P2 and P3, and spinal cord was harvested at P8 and P26 (Fig. 6A). About 91.5% and 49.6% PDGFRα+ OPCs were Sox2-positive at P8 and P26, respectively in the Pdgfrα-CreERT2:Sox2 cKO mice (Fig. 6B, C) whereas nearly 100% of PDGFRα+ OPCs were positive for Sox2 at both time points in the non-Cre controls (Fig. 6C). Olig2+PDGFRα+ OPC population (Fig. 6D, arrowheads) was significantly reduced in the spinal cord of Pdgfrα-CreERT2:Sox2fl/fl mutants at both P8 and P26, compared with that of non-Cre littermate controls (41% reduction at P8 and 17% at P26) (Fig. 6E), suggesting that Sox2 is required for maintaining OPC population during postnatal spinal cord development.
Figure 6. Sox2 regulates OPC population supply and impacts OL differentiation in the murine postnatal spinal cord.
A, transgenic mice and experimental designs. Pdgfrα-CreERT2, Sox2fl/fl (Sox2 cKO) neonates and non-Cre controls were injected subcutaneously with tamoxifen at P1, P2 and P3 once a day and spinal cords were analyzed on P8 and P26. B, representative confocal images showing Sox2 is expressed in PDGFRα+ OPCs in non-Cre Ctrl (upper panels, arrowheads) and absent from PDGFRα+ OPCs in Sox2 cKO (lower panels, arrowheads) spinal cord at P8. Scale bars = 10 μm. C, percentage of PDGFRα+ OPCs that express Sox2 in the spinal white matter at P8 and P26. Two-tailed Student’s t test [t(4) = 116.2 P8, t(4) = 15.25 P26]. D, triple immunohistochemistry of pan-oligodendroglial lineage marker Olig2, OPC marker PDGFRα and OL marker CC1 in Sox2 Ctrl and cKO spinal cord at P8. Boxed areas in the first column are shown at higher magnification at the right. Arrowheads and arrows point to examples of Olig2+PDGFRα+ OPCs and Olig2+CC1+ OLs, respectively. Scale bars, 100 μm in the first column images, 10 μm in the rest images. E, quantification (per mm2) of Olig2+PDGFRα+ OPCs in spinal cord WM. Two-tailed Student’s t test [t(4) = 3.758 P8, t(5) = 3.044 P26]. F, density (per mm2) of Olig2+CC1+ OLs in the spinal cord WM. Two-tailed Student’s t test [t(4) = 3.077 P8, t(5) = 4.568 P26]. G, representative confocal images of MBP and axonal marker SMI312 (scale bars = 100 μm) in the ventral medial white matter of spinal cord (left) and quantification of MBP and SMI312 relative intensity (right) at P8 and P26. Two-tailed Student’s t test, t(4) = 3.644 MBP and t(4) = 0.126 SMI312 at P8; t(4) = 3.699 MBP and t(4) = 0.183 SMI312 at P26. H–I, representative bright-field images of Plp mRNA in situ hybridization (ISH) of spinal cord and quantification at P8. Two-tailed Student’s t test, t(4) = 7.941. Scale bars = 200 μm. J, RT-qPCR quantification of spinal cord mRNA levels of indicated genes at P8. The mRNA level was normalized to the internal control Hsp90, and the level in Sox2 Ctrl was set to 1. Two-tailed Student’s t test [t(5) = 5.880 Sox2, t(5) = 3.346 Sox10, t(5) = 3.552 Mbp, t(5) = 3.072 Plp-E3b, t(5) = 7.408 Mog]. K, representative confocal images of APC and TCF7l2 in the P8 spinal cord (left) and quantification (right). Scale bars = 20 μm. Two-tailed Student’s t test, t(4) = 5.526 P8, t(5) = 2.833 P26.
The decreased OPC population would lead to reduced number of differentiated oligodendrocytes. Accordingly, we found that OL differentiation and myelination were impaired in the spinal cord of Pdgfrα-CreERT2:Sox2 cKO mice, as evidenced by decreased number of Olig2+CC1+ OLs (Fig. 6D arrows and Fig. 6F) and hypomyelination (Fig. 6G). Consistently, the mRNA levels of myelin genes Mbp, Plp-E3b and Mog (Fig. 6H) and the density of Plp mRNA+ oligodendrocytes (Fig. 6I, J) were significantly reduced in Pdgfrα-CreERT2:Sox2 cKO mice at P8. Furthermore, the number of APC+TCF7l2+ newly formed premyelinating OLs was significantly reduced in the spinal cord of Pdgfrα-CreERT2:Sox2 cKO mice compared with non-Cre littermate controls at both P8 and P26 (Fig. 6K, right). Collectively, these data indicate that Sox2 promotes OPC population supply for subsequent OL differentiation.
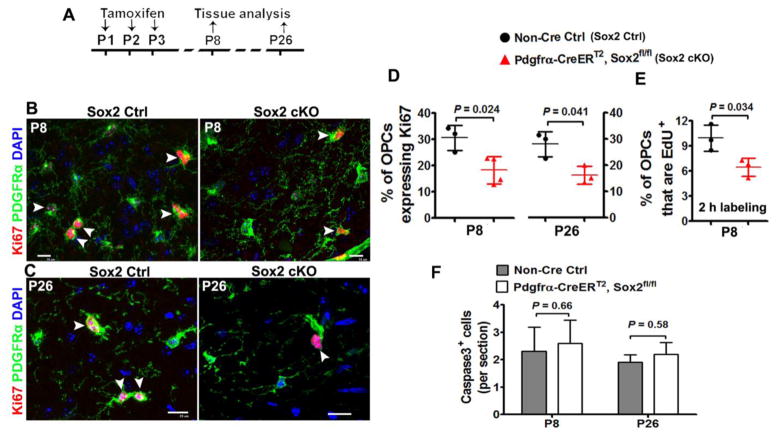
OPC population is maintained by proliferation, apoptosis, or both. We then analyzed OPC proliferation and apoptosis. Our data showed that the percentage of Ki67+ cycling OPCs was significantly reduced at both P8 (by 42%) and P26 (by 43%) in the spinal cord of Pdgfrα-CreERT2:Sox2 cKO mice (Fig. 7B, C, D), which is confirmed by two hours EdU pulse labeling (Fig. 7E). However, we found no alterations in the density of active Caspase 3+ apoptotic cells in the spinal cord of Pdgfrα-CreERT2:Sox2 cKO mice in comparison to non-Cre controls at P8 and P26 (Fig. 7F). Of note, our in vivo data are different from those collected from in vitro system in which Sox2 ablation significantly increases Caspase-dependent OPC apoptosis [10].
Figure 7. Sox2 regulates OPC proliferation in the postnatal mouse spinal cord.
A, transgenic mice and experimental designs for panels B–E. Thymidine analog EdU was injected intraperitoneally into Sox2 Ctrl and cKO mice 2 h before tissue harvesting. B–C, representative confocal images showing reduction of Ki67+PDGFRα+ cycling OPCs in the white matter of spinal cord at P8 (B) and P26(C). Arrowheads point to Ki67+PDGFRα+ proliferating cells. Scale bars = 10 μm. D, percentage of PDGFRα+ OPCs that are Ki67-positive in the spinal cord white matter at P8 and P26. Two-tailed Student’s t test, t(5) = 3.198 P8, t(4) = 3456 P26. E, percentage of PDGFRα+ OPCs that are EdU-positive (2 h pulse labeling) in the spinal cord white matter at P8. Two-tailed Student’s t test, t(4) = 3.175. F, active Caspase 3+ cells per section. Two-tailed Student’s t test, t(6) = 0.626 P8, t(4) = 0.702 P26.
Taken together, the data obtained from the time-conditional Pdgfrα-CreERT2:Sox2fl/fl mutants unequivocally demonstrate that Sox2 plays a crucial role in OPC proliferation and that Sox2 ablation in postnatal OPCs reduces their proliferative capacity, leading to a diminished density of OPCs and OLs.
Delayed OL differentiation in the spinal cord of Cnp-Cre:Sox2fl/fl mutants - a time-course analysis
To determine the role of Sox2 in OL differentiation without compromising OPC population, we analyzed the spinal cord of Cnp-Cre:Sox2fl/fl mice in which Cre-recombinase mediates gene deletion primarily in post-mitotic differentiation-committed OPCs and later stages of OLs [15–21].
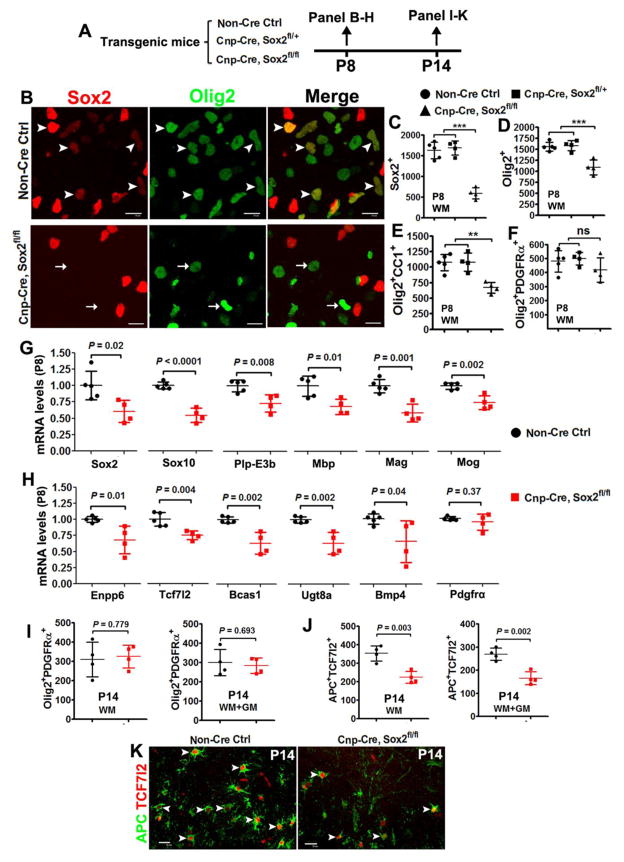
At P8, immunohistochemistry confirmed that Sox2 was specifically deleted in Olig2+ oligodendroglial lineage cells in the spinal cord of Cnp-Cre:Sox2fl/fl mutants (Fig. 8B, C). The density of Olig2+ pan-oligodendroglial lineage cells (Fig. 8D) and Olig2+CC1+ differentiated OLs (Fig. 8E) was significantly decreased in the spinal white matter (WM) of Cnp-Cre:Sox2fl/fl mutants compared with non-Cre control and one-allele deletion (Cnp-Cre:Sox2fl/+). In sharp contrast to the reduced OPC population in Sox10-Cre:Sox2fl/fl mutants, the number of Olig2+PDGFRα+ OPCs was indistinguishable between the three groups (Fig. 8F), indicating that Sox2 is primarily ablated in post-mitotic OPCs and later stages of OLs. Consistent with the histological data, we observed significant decreases in the mRNA levels of the pan-oligodendroglial lineage marker Sox10, mature OL-enriched genes Plp-E3b (exon3b-containing Plp), Mbp, Mag and Mog (Fig. 8G), and premyelinating OL-enriched genes Enpp6, Tcf7l2, Bcas1, Ugt8a and Bmp4 (Fig. 8H), indicating that OL differentiation is inhibited in the P8 spinal cord of Cnp-Cre:Sox2fl/fl mice.
Figure 8. Sox2 positively regulates the timing of OL differentiation in the postnatal mouse spinal cord.
A, transgenic mice and experimental designs. B, representative confocal images of Sox2 and pan-oligodendroglial marker Olig2 in the white matter of P8 spinal cord. Scale bars = 10 μm. Note that Sox2 is expressed in Olig2+ cells in non-Cre Ctrl (upper panels, arrowheads) and absent from most Olig2+ cells in Cnp-Cre, Sox2fl/fl mice (arrows in the lower panels). C–F, quantification of marker-positive cells in the white matter of P8 spinal cord of non-Cre Ctrl, Cnp-Cre, Sox2fl/+ (one-allele KO), and Cnp-Cre, Sox2fl/fl (Sox2 cKO) mice. One-way ANOVA, F(2,10) = 52.18 P < 0.0001 Sox2+, F(2,10) = 19.16 P = 0.0004 Olig2+, F(2,10) = 1.429 P = 0.285 Olig2+PDGFRα+, F(2,10) = 14.80 P = 0.001 Olig2+CC1+. Tukey post hoc test was used for pairwise comparisons, ** P < 0.001, *** P < 0.0001, ns, not significant. G–H, RT-qPCR quantification of mRNA expression of indicated genes in P8 spinal cord. mRNA level was normalized to the internal control Hsp90, and the level in Sox2 Ctrl was set to 1. Two-tailed Student’s t test [t(7) = 2.957 Sox2, t(7) = 8.307 Sox10, t(7) = 3.660 Plp-E3b, t(7) = 3.386 Mbp, t(7) = 5.252 Mag, t(7) = 4.782 Mog, t(7) = 3.331 Enpp6, t(7) = 4.129 Tcf7l2, t(7) = 4.688 Bcas1, t(7) = 3.950 Ugt8a, t(7) = 2.383 Bmp4, t(7) = 0.950 Pdgfrα]. I, densities (per mm2) of Olig2+PDGFRα+ OPCs in the WM and whole spinal cord (WM+GM) at P14. Two-tailed Student’s t test [t(6) = 0.293 WM, t(6) = 0.415 WM+GM]. J–K, quantification and representative confocal images showing substantial reduction of APC+TCF7l2+ premyelinating OLs in Cnp-Cre, Sox2fl/fl spinal cord at P14. Scale bars = 10 μm. Two-tailed Student’s t test [t(6) = 4.935 WM, t(6) = 5.448 WM+GM]. Arrowheads point to APC+TCF7l2+ premyelinating OLs.
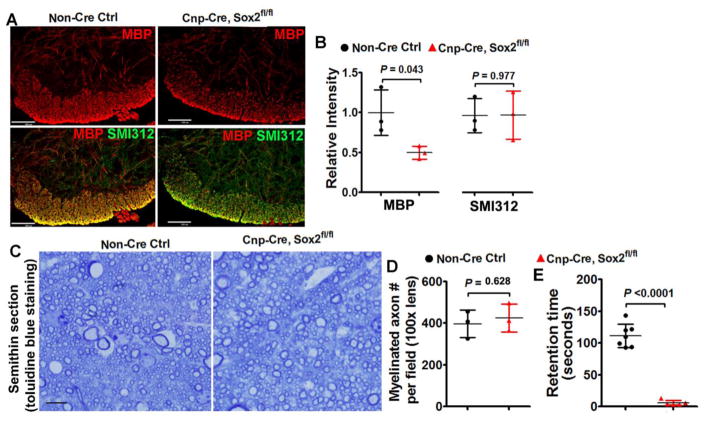
Our recent data report that oligodendrocyte number and myelin gene expression are normal in the P14 spinal cord of Cnp-Cre:Sox2fl/fl mice [11]. In the current study, we showed that OPC density was unchanged in the P14 spinal cord of Cnp-Cre:Sox2fl/fl mice compared with that in the non-Cre controls (Fig. 8I). Interestingly, the number of APC+/TCF7l2+ newly formed premyelinating OLs (Fig. 8K, arrowheads) was significantly decreased in the spinal cord WM and GM of Cnp-Cre:Sox2fl/fl mice at P14 (Fig. 8J), indicating that the rate of oligodendrogenesis is impaired. Consistently, myelination was inhibited in the spinal cord at P8 (Fig. 9A, B) and seemed normal in the adult of Cnp-Cre:Sox2fl/fl mice (Fig. 9C, D). Accelerating Rotarod test showed that Cnp-Cre:Sox2fl/fl mutants still exhibited impaired motor coordination at the adult age compared with non-Cre control mice (Fig. 9E), which is presumably due to the myelin abnormalities in the peripheral nervous system.
Figure 9. myelination assessment in the spinal cord of Cnp-Cre:Sox2fl/fl mutants at P8 and P60.
A, immunohistochemistry of MBP and axonal marker SMI312 in the ventral medial white matter at P8, scale bar = 100μm. Note the SMI312+ axons are highly myelinated in the ventral medial white matter of spinal cord at P8, thus rendering the overlapped images yellow. B, quantification of the relative intensity of MBP and SMI312 in the ventral medial white matter at P8. Two-tailed Student’s t test, t(4) = 2.922 MBP, t(4) = 0.030 SMI312. C–D, representative images of toluidine blue staining of myelin on semithin (500 nm) sections in the corticospinal tract at P60 (C, scale bar=10μm) and quantification of myelination axon numbers (D). Two-tailed Student’s t test, t(4) = 0.524. E, retention time on the accelerating Rotarod of Cnp-Cre:Sox2fl/fl mutant and control mice at P60. Two-tailed Student’s t test with Welch’s correction, t(6) = 14.85.
Taken together, time-course analyses of Cnp-Cre:Sox2fl/fl transgenic hybrids suggest that Sox2 additionally regulates the timing of oligodendrocyte differentiation and enhances the rate of oligodendrocyte generation.
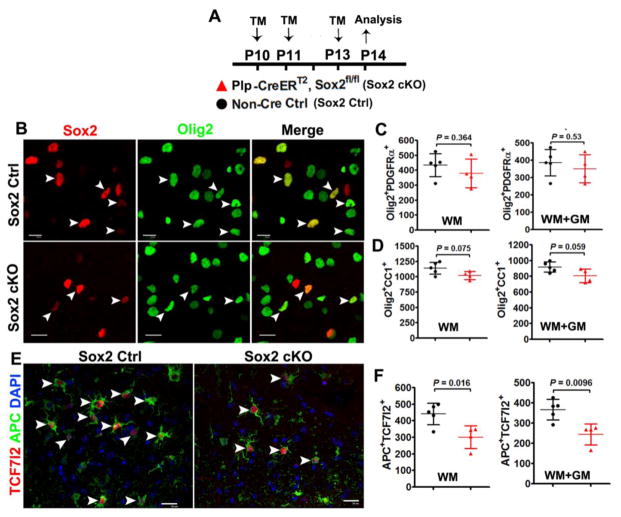
Reduced rate of oligodendrocyte generation in the P14 spinal cord of Plp-CreERT2:Sox2fl/fl mutants
To corroborate the conclusion that Sox2 enhances the rate of oligodendrocyte generation, we created the tamoxifen-inducible, time-conditional Plp-CreERT2:Sox2fl/fl transgenic mice. Tamoxifen was injected to Plp-CreERT2:Sox2fl/fl mice and non-Cre littermate controls at P10, P11 and P13, and spinal tissues were assessed at P14 (Fig. 10A). Sox2 and Olig2 immunostaining confirmed that Sox2 was deleted in most Olig2+ oligodendroglial lineage cells (Fig. 10B). Our quantification data showed that Olig2+/PDGFRα+ OPCs were similar (Fig. 10C) whereas Olig2+/CC1+ OLs were reduced in the spinal cord of Plp-CreERT2:Sox2fl/fl mice although this reduction did not reach statistical significance (Fig. 10D). In line with the data derived from P14 Cnp-Cre:Sox2fl/fl hybrids, the number of APC+TCF7l2+ newly formed premyelinating OLs (Fig. 10E, arrowheads) was significantly reduced in the spinal cord (WM and GM) of Plp-CreERT2:Sox2fl/fl mice (Fig. 10F), indicating that Sox2 ablation perturbs the rate of oligodendrocyte generation.
Figure 10. Sox2 regulates the rate of oligodendrocyte production – evidences from Plp-CreERT2:Sox2fl/fl transgenic mice.
A, transgenic mice and experimental designs for Panels B–F. Plp-CreERT2:Sox2fl/fl (Sox2 cKO) and non-Cre Ctrl mice were injected intraperitoneally with tamoxifen at P10, P11 and P13 once a day, and sacrificed 24 h after the last injection. B, representative confocal images showing Sox2 is expressed in Olig2+ oligodendroglial lineage cells in non-Cre Ctrl (upper panels, arrowheads) and absent from most Olig2+ cells in P14 spinal cord of Sox2 cKO mice (lower panels, arrowheads). Scale bars = 10 μm. C, densities (per mm2) of Olig2+PDGFRα+ OPCs. Two-tailed Student’s t test [t(7) = 0.972 WM, t(7) = 0.661 WM+GM]. D, densities (mm2) of Olig2+CC1+ OPCs. Two-tailed Student’s t test [t(7) = 2.088 WM, t(7) = 2.258 WM+GM]. E, representative confocal images showing reduction of APC+TCF7l2+ premyelinating OLs in P14 spinal cord. Scar bars = 10 μm. F, density (per mm2) of APC+TCF7l2+ cells. Two-tailed Student’s t test [t(7) = 3.163 WM, t(7) = 3.528 WM+GM].
Discussion
The current study resolved a key question regarding the role of Sox2 in the oligodendroglial lineage progression in the postnatal murine spinal cord. Two previous studies reported discrepant observations. Using the Sox10-Cre:Sox2fl/fl model to constitutively delete Sox2 in OPCs, Hoffmann and colleagues demonstrated that Sox2 is required for spinal cord myelination [9]. Axonal myelination occurs predominantly during postnatal development in the CNS of both human and rodents. Unfortunately, this study did not analyze spinal myelination beyond the neonatal ages. A subsequent study reported that Sox2 seems dispensable for myelination in the postnatal spinal cord [10]. However, this study used a non-cell-specific Sox2 deletion paradigm. In the current study, we analyzed myelination in the postnatal spinal cord of the cell-specific Sox2 mutants (Sox10-Cre:Sox2fl/fl) and conclude that Sox2 is indispensable for spinal cord myelination. The experimental data from the current and our previous studies [11] suggest that Sox2 is an intrinsic positive regulator for CNS myelination, weakening a CNS region-specific (brain vs spinal cord) role of Sox2 in developmental myelination.
In the CNS, Sox2 is highly expressed in NSCs and downregulated or absent upon differentiation into neural progenitors [2]. Recent studies have reached the consensus that OPCs express Sox2 in the embryonic and early postnatal CNS in vivo [9, 10, 22]. However, whether OPCs in the adult CNS Sox2 expression is still controversial [10, 23]. By leveraging Cre-LoxP-based genetic labeling (Sox2-CreERT2:Rosa-loxP-STOP-loxP-EYFP), we demonstrate that adult OPCs from different CNS regions exhibit endogenous Sox2 promoter activity and express Sox2 albeit at a lower level, thus arguing against a CNS region-dependent Sox2 expression in adult OPCs. This notion is consistent with recent single cell RNA-seq data demonstrating that adult OPCs in different CNS regions are homogenous at the transcription level [21]. At the functional level, our previous and current data have unequivocally demonstrated that Sox2 is essential for OPC proliferation in the postnatal brain and spinal cord. It is well-established that OPCs are uniformly distributed throughout CNS regions and represent the largest population of proliferating cells in the adult CNS [24], and adult OPCs are responsible for oligodendrogenesis albeit at a slower rate, myelin turnover and/or adaptive myelination [25–27]. It is plausible to speculate that Sox2 regulates the homeostasis of adult OPCs, adult oligodendrogenesis, and/or adaptive myelination. Future studies are needed to prove or falsify this prediction.
Our recent data [11] report that oligodendrocyte number and myelin gene expression are unchanged at P14 in the spinal cord of Cnp-Cre:Sox2fl/fl mutants compared to non-Cre controls. Based on these observations, one can argue that Sox2 is dispensable for OL differentiation in the postnatal spinal cord, as concluded by previous study [10]. Alternatively, it is possible that Sox2 may regulate the timing of OL differentiation in the postnatal spinal cord. If this possibility happens, oligodendrocyte population should be reduced at some time points before P14. To this end, we analyzed oligodendrocyte differentiation and myelin gene expression at P8, the approximate peak of OL differentiation in the murine spinal cord, and found that OL differentiation and myelination are significantly decreased in the P8 spinal cord of Cnp-Cre:Sox2fl/fl mutants. The time course analysis of Cnp-Cre:Sox2fl/fl hybrids favors the latter possibility that Sox2 is an intrinsic positive timer regulating the timing of oligodendrocyte differentiation in the postnatal spinal cord. Another novel finding in the current study is that Sox2 regulates OPC proliferation in a developmental stage-dependent (embryonic vs spinal cord) manner. Using Sox10-Cre:Sox2fl/fl embryos, we confirmed that Sox2 does not regulate OPC proliferation in the embryonic spinal cord, a conclusion that is in line with the one by Hoffmann et al [9]. Interestingly, our data show that Sox2 plays a crucial role in postnatal OPC proliferation in the spinal cord.
How Sox2 promotes oligodendrocyte differentiation and myelination is an open question. Sox2 is a transcription factor that either activates or silences its target genes depending on recruiting its partners of co-activators or co-repressors, respectively in different contexts [28–31]. Previous study suggests that Sox2 represses miR145, thus preventing miR145 from inhibiting several pro-differentiation factors [9]. Our preliminary study of co-immunoprecipitation in combination with mass spectrometry (CoIP-MS) showed that, in the Oli-neu oligodendrocyte cell line [32], Sox2 interacted with transcriptional repressors HDAC1/2 and YY1 and transcriptional activators TCF7l2, and Olig1/2 (data not shown), all of which have been shown to promote oligodendrocyte differentiation and myelination [13, 15, 22, 33–35]. Recent ChIP-Seq data show that Sox2 may target the signaling pathways of Wnt, Akt, and Notch in the adult cerebral cortex [36], all of which have been shown to regulates OL differentiation and myelination [37]. Since Sox2-expressing cells in the adult cerebral cortex are astrocytes [36] and OPCs (Fig. 1D), it is not quite clear whether Sox2 targets these pathways in astrocytes or in OPCs. Further experimental data are needed to link Sox2 with those pathways, if any, in the context of oligodendroglial lineage cells, and importantly, how Sox2 modulates those pathways also warrants more investigations.
In summary, our data from genetic approaches unequivocally prove that the pluripotency factor Sox2 plays a crucial role in coordinating OPC proliferation and OL differentiation and is required for developmental myelination in the postnatal murine spinal cord. Our study provides justification that Sox2 may be a therapeutic target for overcoming OPC differentiation arrest in CNS dysmyelinating and demyelinating disorders.
Materials and Methods
Animals
Animals and procedures used in the study were covered and approved by the Institutional Animal Care and Use Committee (IACUC) of UC Davis. Animals were housed at 12h light/dark cycle with access to food and drink and both male and female mice were used in this study. All transgenic mice were crossed and maintained on a C57BL/6J background. The transgenic lines and their identification are: Sox2-CreERT2 (RRID: IMSR_JAX:017593), Pdgfrα-CreERT2 (RRID: IMSR_JAX:018280), Sox10-Cre (RRID: IMSR_JAX:025807), Cnp-Cre (RRID: MGI_3051754), Plp-CreERT2 (RRID: IMSR_JAX:005617), Sox2fl/fl (RRID: IMSR_JAX:013093), and Rosa26-LoxP-STOP-LoxP-EYFP (referred to as Rosa-EYFP) (RRID: IMSR_JAX:006148). All Cre lines including breeders and study mice were maintained as heterozygosity (i.e. Cre+/−). In this study, Sox2 conditional knockout (cKO) mice were referred as to Cre+/−:Sox2fl/fl whereas non-Cre controls were referred as to Sox2fl/+ or Sox2fl/fl that do not carry Cre transgene. For studies using timed pregnant mice (Fig. 2), we placed male and female mice into one cage around 5:00 pm in the late afternoon, and the female mice were designated as pregnancy if the sperm plugs were seen by 10:00 am of the next day, and the embryos were assigned as embryonic day 0.5 (E0.5). For postnatal studies, the day when pups were born was designated as postnatal day 0 (P0).
Tamoxifen, BrdU and Edu treatment
Tamoxifen (TM) (T5648; Sigma-Aldrich) was dissolved in mixture of ethanol and sunflower oil (1:9 by volume) at a concentration of 30 mg/ml [13, 14]. Tamoxifen was administrated intraperitoneally to adult Sox2-CreERT2:Rosa-EYFP mice (Fig. 1) at a dose of 1 mg (~35 μl) daily for 5 consecutive days, subcutaneously to neonatal pups (Pdgfrα-CreERT2:Sox2fl/fl and non-Cre controls) on P1, P2 and P3 daily at a dose of 10 μl (300 μg) once a day (Fig. 6 and Fig. 7), and intraperitoneally to Plp-CreERT2:Sox2fl/fl and non-Cre controls daily on P10, P11, and P13 at a dose of 20 μl (600 μg) (Fig. 9). Animals were sacrificed at time points indicated in each figure. BrdU or EdU (10mg/ml, dissolved in saline) was intraperitoneally injected to Sox2 cKO and control littermates at a dose of 100 ug/g body weight 2 hours prior to sacrifice.
Tissue harvesting and processing for histology
After anesthetized by ketamine/xylazine mixture, mice were transcardially perfused with ice-cold PBS. Spinal cord was dissected out and immediately placed on dry ice (for protein or RNA extraction) or post-fixed in 4% paraformaldehyde (PFA) (for histological study). After post-fix in 4% PFA for 2 hours at room temperature, the spinal cord was washed with PBS, three times, 15 minutes each time. Tissue samples were then cryoprotected in 30% sucrose (dissolved in PBS) for 20 hours and cut into 16-μm-thick sections on Leica Cryostats. Sections were stored under −80°C until used for immunohistochemistry or mRNA in situ hybridization.
Immunohistochemistry and mRNA in situ hybridization
Immunohistochemistry (IHC) and mRNA in situ hybridization (ISH) were performed as previously described [13, 14]. The following antibodies were used in immunohistochemistry: Sox2 (sc-17320, RRID: AB_2286684, 1:500; Santa Cruz Biotechnology), Olig2 (AF2418, RRID: AB_2157554, 1:100; R&D Systems; AB9610, AB_10141047,1:500; Millipore), CC1 (OP80, RRID:AB_213434, 1:200;Calbiochem), TCF7l2 (2569S, RRID: AB_2199816,1:200; Cell Signaling Technology; sc-8632, RRID: AB_2199825,1;100; Santa Cruz Biotechnology), PDGFRα (AF1062, AB_2236897,1:200; R&D System), EYFP (06-896, RRID: AB_310288, 1;500; Millipore), NG2 (AB5320, RRID:AB_91789, 1:200; Millipore), Ki67(9129, RRID: AB_10989986,1:200; Cell Signaling Technology), Sox10 (sc-17342, RRID: AB_2195374, 1:100; Santa Cruz Biotechnology), BrdU (sc-70441, RRID: AB_1119696, 1:100; Santa Cruz Biotechnology), APC (sc-896, RRID: AB_2057493,1:100; Santa Cruz Biotechnology). DyLight 488- or DyLight 549-conjugated secondary antibodies were from Jackson ImmunoResearch. Brdu, Edu (Click-iT EdU imaging kits, Invitrogen C10339) immunostaining were performed as previous study [13, 14].
Protein extraction and Western Blot
The frozen mouse spinal cord was homogenized in RIPA buffer (Cell Signaling Technology, #9806) supplemented with the protease/phosphatase inhibitor cocktail buffer (Cell Signaling Technology, #5872) and PMSF (Cell Signaling Technology, #8553) at a ratio of 100 mg of tissue to 1 ml of buffer. Lysates were sonicated, incubated on ice for 30 min, and centrifuged at 14,000 g for 10 min. The supernatants were assessed for protein concentration using the BCA protein assay kit (Thermo Scientific, #23235). Western blot was performed according to the protocols in our previous studies [13, 14]. A total of 30 μg protein extracts was separated by anykD precast polyacrylamide gel (Bio Rad, #4569033) and transferred to nitrocellulose immobilization membranes (Bio Rad, #1704158) using Trans-Blot Turbo transfer system (Bio Rad, #1704150). Membranes were blocked for 1 hour in 5% BSA (Cell Signaling Technology, #9998) at room temperature, and incubated with the primary antibody overnight at 4°C. Subsequently, membranes were washed 3 times 10 min each in TBST buffer (Tris 25 mM, KCI 3 mM, NaCl 140 mM, PH 7.4, and Tween 0.1%) and incubated with secondary antibodies for 1 hour at room temperature. Membranes were washed 3 times in the same wash buffer, and detection was performed by exposing the membranes to Pewmium X-Ray Film (PHENIX, #F-BX57).
The following antibodies were used for Western Blot: MBP (NB600-717, RRID: AB_2139899, 1:1000; Novus), Cnp (C5922, RRID: AB_823640, 1:1000; Sigma), and beta-actin (3700, RRID: AB_2242334, 1:1000; Cell Signaling Technology).
RNA extraction, cDNA preparation and real time quantitative PCR (RT-qPCR)
Total spinal cord RNA was extracted by using the commercial isolation kit (Qiagen catalog#:74804) with additional on-column DNase I digestion to eliminate genomic DNA contamination. RNA quality was assessed by Agilent Bioanalyzer 2100. Only RNAs with RNA Integrity Number (RIN) greater than 6.8 were used for subsequent complementary DNA (cDNA) preparation. cDNA was prepared by Qiagen Omniscript RT Kit (catalog#: 205111) with 2 μg RNA in 20 μl reaction buffer. The resulting 20 μl cDNA was further diluted with TE buffer (pH8.0) to a total volume of 400 μl (380 μl TE buffer to 20 μl cDNA stock solution), and 1 microliter of the diluted cDNA solution (equivalent to 5ng RNA) was used as templates for RT-qPCR. We used Sybr green-based method (QIAGEN catalog#: 204143) for qPCR. In our study, the Ct (cycle threshold) of the internal control gene Hsp90 was approximately 20 – 21. The mRNA levels of indicated genes were normalized to that of Hsp90 and calculated by the equation 2^(Ct value for Hsp90 – Ct value for indicated genes). The mRNA level in non-Cre control spinal cord was designated to 1. Most of the primer sets used in this study were from PrimerBank (https://pga.mgh.harvard.edu/primerbank/) and listed here: Hsp90 (F: AAACAAGGAGATTTTCCTCCGC, R: CCGTCAGGCTCTCATATCGAAT), Sox2 (F: CGGCACAGATGCAACCGAT, R: CCGTTCATGTAGGTCTGCG), Sox10 (F: ACACCTTGGGACACGGTTTTC, R: TAGGTCTTGTTCCTCGGCCAT), Mag (F: CTGCCGCTGTTTTGGATAATGA, R: CATCGGGGAAGTCGAAACGG), Enpp6 (F: CAGAGAGATTGTGAACAGAGGC, R: CCGATCATCTGGTGGACCT), Bcas1 (F: AGAAGCGAAAGGCTCGGAAG, R: AGGGACAGAATAACTCAGAGTGT), Mbp (F: GGCGGTGACAGACTCCAAG, R: GAAGCTCGTCGGACTCTGAG), Cnp (F: TTTACCCGCAAAAGCCACACA, R: CACCGTGTCCTCATCTTGAAG), Qk or CC1 (F: CTGGACGAAGAAATTAGCAGAGT, R: ACTGCCATTTAACGTGTCATTGT), Ugt8a (F: GAACATGGCTTTGTCCTGGT, R: CATGGCTTAGGAAGGCTCTG), Plp-E3b (targeting the exon 3b of Plp1 gene, which is only expressed in myelinating OLs) (F: GTTCCAGAGGCCAACATCAAG, R: CTTGTCGGGATGTCCTAGCC), Mog (F: AGCTGCTTCCTCTCCCTTCTC, R: ACTAAAGCCCGGATGGGATAC), Bmp4: (F: TTCCTGGTAACCGAATGCTGA, R: CCTGAATCTCGGCGACTTTTT), Tcf7l2, (F: GGAGGAGAAGAACTCGGAAAA, R: ATCGGAGGAGCTGTTTTGATT).
Confocal Image acquisition and quantification
Nikon laser-scanning confocal microscope was used to acquire confocal images. Laser settings remained consistent across different mouse genotypes of histological sections. Nikon EZ-C1 FreeViewer v3.90 was used for cell quantification and area calculation. To quantify cell density, four projected confocal images at 20× (oil lens) magnification (10 μm optical thickness from 11 consecutive stacked 1 μm confocal optical slices, ~330,000 μm2 per 20× image) were obtained from same locations in the spinal cord of Sox2 cKO and non-Cre control mice. To increase the accuracy of OPC and OL quantification, we included the nuclear Sox10 (or Olig2) immunostaining with the OPC marker PDGFRα (or NG2) and the differentiated OL marker CC1. Only PDGFRα+ or CC1+ cells with nuclear Sox10 (or Olig2) signals were counted as individual OPCs or OLs. Similarly, to increase the accuracy of premyelinating OL quantification, we used double immunohistochemistry of the cell surface antigen APC and the nuclear antigen TCF7l2. Only APC+ cells with nuclear TCF7l2 signals were counted as individual premyelinating OLs. The NIH Image J software was used to quantify the relative intensity of MBP and SMI312 immunoreactivity.
Accelerating Rotarod test
Motor performance and coordination were measured by accelerating rotarod test machine (Columbus instruments, Rotamex-5 Rota rod). The following parameter settings were used: starting speed 4 rotations per minute (rpm), speed increased by 1.2 rpm every 10 seconds, maximal speed 40 rpm. The retention time was recorded for each mouse which is defined as the time duration that a mouse is retained on the rorating rods prior to falling off.
Semi-thin section preparation and toluidine blue staining of myelin
Mice were anesthetized by ketamine/xylazine mixture, then perfused with 60 ml 4% paraformaldehyde (dilute with phosphate buffer, EMS, 11601-40, pH 7.4) and 60 ml 3% glutaraldehyde (Electron Microscopy Science, 16130, dilute in PBS, pH 7.4) at a speed of 5 ml per minute. Spinal cord was carefully dissected out and fixed with 3% glutaraldehyde overnight followed by wash with sodium cacodylate (Electron Microscopy Science, EMS, 11653) twice, 10 minutes each time, and then post-fixed with 2% (w/v) osmium tetroxide (Electron Microscopy Science, 19152) for 1.5 hours and wash with sodium cacodylate twice, 10 minutes each time. The resulting spinal cord was dehydrated with gradient ethanol of 50%, 70%, 90%, and 100% followed by wash with propylene oxide for 3 times, 30 min each time, and incubation with 1:1 mixture of propylene oxide: Eponate resin overnight, and with 3:1 mixture of propylene oxide: Eponate resin for 10 hr, and incubation with 100% Eponate resin overnight. Transverse sections were embedded in EMBED812-Resin for 2 days at 65 degrees. Semi-thin (500 nm) were cut by using a Leica EM UC7 microtome. Semi-thin sections were dry and incubated with 2% toluidine blue (Cat#19451, Ted Pella Inc) at 100°C degrees for 2 min, followed by imaging on Olympus BX61 microscope.
Statistical analyses
Quantification was performed by blinded observers. At least 3 sections from each mouse were used, and 3–7 mice were quantified. Data were presented as mean ± s.d. except stated otherwise. We used graphs of scatter plots to present the quantification data throughout our manuscript, and each circle, square or triangle represents one mouse. Shapiro-Wilk approach was used for testing data normality. The statistical methods were described in the figure legends. For two-tailed Student’s t test, P value was presented in each graph, and t value and degree of freedom (df) were presented as t(df) in figure legends. Welch’s correction was used in Student’s t test if the variance of two groups of datasets was not equal. For comparisons among three or more groups, one-way ANOVA followed by Tukey post hoc test was used. We used GraphPad Prism version 5.0 to perform all statistical analyses and data plotting. P value less than 0.05 was considered as significant. ns stands for not significant with P value greater than 0.05.
Acknowledgments
Grant support: Shriners Hospitals for Children (F.G., S.Z.) 2016YFC0503200 (X. Z.), and NIH (R01NS094559, R21NS093559 to F.G.).
References
- 1.Franklin RJ, Gallo V. The translational biology of remyelination: Past, present, and future. Glia. 2014 doi: 10.1002/glia.22622. [DOI] [PubMed] [Google Scholar]
- 2.Graham V, et al. SOX2 functions to maintain neural progenitor identity. Neuron. 2003;39(5):749–65. doi: 10.1016/s0896-6273(03)00497-5. [DOI] [PubMed] [Google Scholar]
- 3.Lee KE, et al. Positive feedback loop between Sox2 and Sox6 inhibits neuronal differentiation in the developing central nervous system. Proc Natl Acad Sci U S A. 2014;111(7):2794–9. doi: 10.1073/pnas.1308758111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kondo T, Raff M. Chromatin remodeling and histone modification in the conversion of oligodendrocyte precursors to neural stem cells. Genes Dev. 2004;18(23):2963–72. doi: 10.1101/gad.309404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lyssiotis CA, et al. Inhibition of histone deacetylase activity induces developmental plasticity in oligodendrocyte precursor cells. Proc Natl Acad Sci U S A. 2007;104(38):14982–7. doi: 10.1073/pnas.0707044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niu W, et al. In vivo reprogramming of astrocytes to neuroblasts in the adult brain. Nature cell biology. 2013;15(10):1164–75. doi: 10.1038/ncb2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Su Z, et al. In vivo conversion of astrocytes to neurons in the injured adult spinal cord. Nat Commun. 2014;5:3338. doi: 10.1038/ncomms4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heinrich C, et al. Sox2-Mediated Conversion of NG2 Glia into Induced Neurons in the Injured Adult Cerebral Cortex. Stem Cell Reports. 2014;3(6):1000–14. doi: 10.1016/j.stemcr.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffmann SA, et al. Stem cell factor Sox2 and its close relative Sox3 have differentiation functions in oligodendrocytes. Development. 2014;141(1):39–50. doi: 10.1242/dev.098418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao C, et al. Sox2 Sustains Recruitment of Oligodendrocyte Progenitor Cells following CNS Demyelination and Primes Them for Differentiation during Remyelination. J Neurosci. 2015;35(33):11482–99. doi: 10.1523/JNEUROSCI.3655-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang S, et al. Sox2 is essential for oligodendroglial proliferation and differentiation during postnatal brain myelination and CNS remyelination. J Neurosci. 2018 doi: 10.1523/JNEUROSCI.1291-17.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stolt CC, et al. Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes & development. 2002;16(2):165–70. doi: 10.1101/gad.215802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammond E, et al. The Wnt effector transcription factor 7-like 2 positively regulates oligodendrocyte differentiation in a manner independent of Wnt/beta-catenin signaling. J Neurosci. 2015;35(12):5007–22. doi: 10.1523/JNEUROSCI.4787-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lang J, et al. Adenomatous polyposis coli regulates oligodendroglial development. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2013;33(7):3113–30. doi: 10.1523/JNEUROSCI.3467-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao C, et al. Dual regulatory switch through interactions of Tcf7l2/Tcf4 with stage-specific partners propels oligodendroglial maturation. Nat Commun. 2016;7:10883. doi: 10.1038/ncomms10883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dugas JC, et al. Dicer1 and miR-219 Are required for normal oligodendrocyte differentiation and myelination. Neuron. 2010;65(5):597–611. doi: 10.1016/j.neuron.2010.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moyon S, et al. Functional Characterization of DNA Methylation in the Oligodendrocyte Lineage. Cell Rep. 2016 doi: 10.1016/j.celrep.2016.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moyon S, Casaccia P. DNA methylation in oligodendroglial cells during developmental myelination and in disease. Neurogenesis (Austin) 2017;4(1):e1270381. doi: 10.1080/23262133.2016.1270381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishii A, et al. ERK1/ERK2 MAPK signaling is required to increase myelin thickness independent of oligodendrocyte differentiation and initiation of myelination. J Neurosci. 2012;32(26):8855–64. doi: 10.1523/JNEUROSCI.0137-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishii A, Furusho M, Bansal R. Sustained activation of ERK1/2 MAPK in oligodendrocytes and schwann cells enhances myelin growth and stimulates oligodendrocyte progenitor expansion. J Neurosci. 2013;33(1):175–86. doi: 10.1523/JNEUROSCI.4403-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marques S, et al. Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science. 2016;352(6291):1326–1329. doi: 10.1126/science.aaf6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dai J, et al. Olig1 function is required for oligodendrocyte differentiation in the mouse brain. J Neurosci. 2015;35(10):4386–402. doi: 10.1523/JNEUROSCI.4962-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen S, et al. Age-dependent epigenetic control of differentiation inhibitors is critical for remyelination efficiency. Nat Neurosci. 2008;11(9):1024–34. doi: 10.1038/nn.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernandez-Castaneda A, Gaultier A. Adult oligodendrocyte progenitor cells - Multifaceted regulators of the CNS in health and disease. Brain Behav Immun. 2016;57:1–7. doi: 10.1016/j.bbi.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Young KM, et al. Oligodendrocyte dynamics in the healthy adult CNS: evidence for myelin remodeling. Neuron. 2013;77(5):873–85. doi: 10.1016/j.neuron.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiao L, et al. Rapid production of new oligodendrocytes is required in the earliest stages of motor-skill learning. Nat Neurosci. 2016;19(9):1210–1217. doi: 10.1038/nn.4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tripathi RB, et al. Remarkable Stability of Myelinating Oligodendrocytes in Mice. Cell Rep. 2017;21(2):316–323. doi: 10.1016/j.celrep.2017.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reiprich S, Wegner M. From CNS stem cells to neurons and glia: Sox for everyone. Cell and tissue research. 2014 doi: 10.1007/s00441-014-1909-6. [DOI] [PubMed] [Google Scholar]
- 29.Reiprich S, Wegner M. Sox2: A multitasking networker. Neurogenesis (Austin) 2014;1(1):e962391. doi: 10.4161/23262125.2014.962391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu YR, et al. Sox2 acts as a transcriptional repressor in neural stem cells. BMC Neurosci. 2014;15:95. doi: 10.1186/1471-2202-15-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Engelen E, et al. Sox2 cooperates with Chd7 to regulate genes that are mutated in human syndromes. Nat Genet. 2011;43(6):607–11. doi: 10.1038/ng.825. [DOI] [PubMed] [Google Scholar]
- 32.Jung M, et al. Lines of murine oligodendroglial precursor cells immortalized by an activated neu tyrosine kinase show distinct degrees of interaction with axons in vitro and in vivo. Eur J Neurosci. 1995;7(6):1245–65. doi: 10.1111/j.1460-9568.1995.tb01115.x. [DOI] [PubMed] [Google Scholar]
- 33.He Y, et al. The transcription factor Yin Yang 1 is essential for oligodendrocyte progenitor differentiation. Neuron. 2007;55(2):217–30. doi: 10.1016/j.neuron.2007.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ye F, et al. HDAC1 and HDAC2 regulate oligodendrocyte differentiation by disrupting the beta-catenin-TCF interaction. Nat Neurosci. 2009;12(7):829–38. doi: 10.1038/nn.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu QR, et al. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell. 2002;109(1):75–86. doi: 10.1016/s0092-8674(02)00678-5. [DOI] [PubMed] [Google Scholar]
- 36.Chen C, et al. Astrocyte-Specific Deletion of Sox2 Promotes Functional Recovery After Traumatic Brain Injury. Cereb Cortex. 2017:1–16. doi: 10.1093/cercor/bhx303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wheeler NA, Fuss B. Extracellular cues influencing oligodendrocyte differentiation and (re)myelination. Exp Neurol. 2016;283(Pt B):512–30. doi: 10.1016/j.expneurol.2016.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]