Abstract
Background
Mutations in BRCA1 and BRCA2 are associated with better survival in ovarian cancer (OC) patients due to a better response to platinum-based chemotherapy. However, the impact of the BRCA1/2 mRNA-expression is not well characterized in OC.
Patients and methods
We investigated BRCA1/2 mRNA-expression in 12 non-neoplastic fallopian tubes and 201 epithelial OCs in relation to their clinical characteristics.
Results
We found higher BRCA1/2 mRNA-expression in OCs compared to controls (P = 0.011, P < 0.001, respectively). BRCA1 mutated OCs exhibited lower BRCA1 (P = 0.014) but higher BRCA2 mRNA-expression (P = 0.001). Low BRCA1-expression was associated with favorable overall survival (OS) (P = 0.012) and low BRCA2-expression with better progression-free survival (PFS) and OS (P = 0.004, P = 0.001, respectively). A subgroup-analysis showed that this effect was confined only to the BRCA1-wildtype cancers. Cox-regression confirmed the prognostic significance of BRCA1-expression for OS (P = 0.028). Independency of the prognostic value of BRCA2-expression for PFS (P = 0.045) and OS (P = 0.015) was restricted to high-grade serous OCs. Fully platinum-sensitivity was characterized by lower BRCA1/2 mRNA-expression in BRCA1-wildtype cancers in comparison to platinum-refractory OC.
Conclusion
Our findings may reflect higher platinum-sensitivity due to reduced capacity of DNA damage repair in tissues with low BRCA1/2-expression. In this context, especially in BRCA-wildtype cancers both parameters could also be potential predictors for PARP-sensitivity.
Subject terms: Translational research, Ovarian cancer, Molecular medicine
Introduction
BRCA1 and BRCA2 are tumor suppressor genes that are involved in cell growth inhibition, apoptosis, regulation of gene transcription and DNA damage repair through homologous recombination.1 Thus, germ line mutations in BRCA genes are considered to be associated with cancer susceptibility, especially with an increased risk of developing ovarian and breast cancer.2
Epithelial ovarian cancer (OC) is one of the leading causes of cancer death in women in the western world.3–5 Approximately 5–10% of all epithelial OC are hereditary and at least two-third of them are due to BRCA1/2 mutations.6,7
The current opinion is that OC patients who carry a pathogenic BRCA mutation show better survival rates possibly due to a better response to platinum-based chemotherapy and inhibitors of poly(ADP)-ribose polymerase (PARP).8,9 However, there are also conflicting data, showing worse survival in hereditary OC cases or no significant difference in survival rates between patients with BRCA associated and sporadic epithelial OC.10–12 Such discrepancies could be due to different duration of follow-up, different histological type or a death caused by secondary malignancies.
Besides BRCA mutation-status, the expression of these genes could contribute to the tumor pathogenesis and therapeutical response. Currently there is lack of data on BRCA1/2 expression and their clinical significance in epithelial OC patients. The implication of BRCA-expression in BRCA-wildtype epithelial OC is also poorly studied but could be clinically relevant. We wondered whether the expression of BRCA1/2 on the transcriptome level could be a reliable predictor for platinum response and thus for the clinical outcome in OC patients.
In our study, we evaluated BRCA1/2-mRNA-expression in frozen tissues of 201 epithelial OC patients. We analyzed progression-free survival (PFS), overall survival (OS), the association between the BRCA-expression and mutation-status and methylation-status as well as FIGO stage and achievement of a complete resection during debulking surgery.
Patients and methods
Patients and samples
Ovarian tissue samples from 201 patients with OC obtained at primary debulking (patients were 24–90 years old; median age at diagnosis was 62 years) and non-neoplastic tubal tissues from 12 patients obtained by elective salpingo-oophorectomy for benign conditions (patients were 30–73 years old, median age: 50 years) were collected and processed at the Department of Obstetrics and Gynecology of the Medical University of Innsbruck between 1989 and 2015 as described recently.13 Systemic treatment of OC patients consisted of six adjuvant cycles of platinum-based chemotherapy. We used a categorization which defines “platinum-refractory” as disease progressing during therapy or within one month after the last dose, “platinum-resistant” as disease progressing within 6 months, “partially platinum-sensitive” as disease progressing between 6 and 12 months, and “platinum-sensitive” as disease progressing with an interval of more than 12 months. Written informed consent was obtained from all patients before enrollment. The study was reviewed and approved by the Ethics committee of the Medical University of Innsbruck (reference number: AN2015-0038 346/4.17) and conducted in accordance with the Declaration of Helsinki. The median observation period of all patients was 1.6 years (0.03–26.4 years) regarding the progression-free survival and 3.6 years (0.1–26.4 years) concerning the median overall survival. Clinicopathological characteristics are shown in Table 1.
Table 1.
Association of BRCA1 and BRCA2 mRNA-expression and mutations with clinicopathological characteristics in 201 ovarian cancer patients
| Variable | Number (%) | mRNA expression values (arbitrary units) | Somatic mutations | DNA methylation status | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BRCA1 | BRCA2 | BRCA1 | BRCA2 | BRCA1 | ||||||||||
| Median (IQR) | P value | Median (IQR) | P value | Non-mutated (%) | Mutated (%) | P value | Non-mutated (%) | Mutated (%) | P value | Unmethylated (%) | Methylated (%) | P value | ||
| Age | ||||||||||||||
| ≤62.3 years | 101 (50%) | 0.81 (0.53–1.30) | 0.491 | 3.72 (2.14–6.15) | 0.875 | 73 (73%) | 27 (27%) | 0.001 | 95 (94%) | 5 (5%) | 0.743 | 88 (88%) | 12 (12%) | 0.504 |
| >62.3 years | 100 (50%) | 0.86 (0.55–1.37) | 3.65 (2.47–6.01) | 90 (90%) | 9 (9%) | 93 (93%) | 6 (6%) | 90 (91%) | 9 (9%) | |||||
| FIGO stage | ||||||||||||||
| I/II | 50 (25%) | 0.91 (0.57–1.35) | 0.361 | 3.22 (2.08–4.87) | 0.089 | 42 (84%) | 7 (14%) | 0.426 | 48 (96%) | 1 (2%) | 0.219 | 48 (96%) | 2 (4%) | 0.081 |
| III/IV | 151 (75%) | 0.80 (0.53–1.30) | 4.02 (2.67–6.17) | 121 (80%) | 29 (19%) | 140 (93%) | 10 (7%) | 130 (87%) | 19 (13%) | |||||
| Tumor grade | ||||||||||||||
| 1 | 14 (7%) | 0.52 (0.43–1.01) | 0.099 | 2.07 (1.19–2.92) | <0.001 | 12 (86%) | 2 (14%) | 0.789 | 14 (100%) | 0 (0%) | 0.624 | 13 (93%) | 1 (7%) | 0.004 |
| 2 | 90 (45%) | 0.85 (0.56–1.26) | 3.55 (2.22–5.35) | 71 (80%) | 18 (20%) | 84 (94%) | 5 (6%) | 87 (97%) | 3 (3%) | |||||
| 3 | 95 (47%) | 0.82 (0.58–1.55) | 4.51 (3.00–7.30) | 78 (83%) | 16 (17%) | 88 (94%) | 6 (6%) | 76 (82%) | 17 (18%) | |||||
| n.a. | 2 (1%) | – | – | – | – | – | ||||||||
| Residual disease | ||||||||||||||
| Macroscopically tumor-free | 100 (50%) | 0.87 (0.58–1.34) | 0.261 | 3.36 (2.04–5.16) | 0.032 | 82 (82%) | 17 (17%) | 0.588 | 92 (92%) | 7 (7%) | 0.399 | 94 (95%) | 5 (5%) | 0.013 |
| Any tumor residual | 95 (47%) | 0.78 (0.51–1.33) | 3.99 (2.82–6.23) | 75 (79%) | 19 (20%) | 90 (95%) | 4 (4%) | 79 (84%) | 15 (16%) | |||||
| n.a. | 6 (3%) | |||||||||||||
| Histology | ||||||||||||||
| HGSOC | 129 (64%) | 0.79 (0.55–1.31) | 0.025 | 4.13 (2.59–6.16) | 0.005 | 102 (79%) | 25 (19%) | 0.848 | 117 (91%) | 10 (8%) | 0.314 | 114 (89%) | 14 (11%) | 0.993 |
| LGSOC | 12 (6%) | 0.48 (0.38–0.67) | 2.05 (1.16–2.87) | 10 (83%) | 2 (17%) | 12 (100%) | 0 (0%) | 11 (92%) | 1 (8%) | |||||
| Endometrioid | 45 (22%) | 1.05 (0.68–1.48) | 4.44 (2.91–6.40) | 37 (82%) | 8 (18%) | 44 (98%) | 1 (2%) | 40 (89%) | 5 (11%) | |||||
| Clear cell | 11 (5%) | 0.97 (0.74– 1.44) | 2.88 (2.00–4.56) | 10 (91%) | 1 (9%) | 11 (100%) | 0 (0%) | 9 (90%) | 1 (10%) | |||||
| Unknown | 4 (2%) | – | – | – | – | – | – | – | – | |||||
| Ovarian cancer type | ||||||||||||||
| Type I | 14 (7%) | 0.52 (0.43–1.00) | 0.034 | 2.07 (1.19–2.92) | 0.001 | 12 (86%) | 2 (14%) | 0.676 | 14 (100%) | 0 (0%) | 0.342 | 13 (93%) | 1 (7%) | 0.650 |
| Type II | 183 (91%) | 0.85 (0.57–1.36) | 4.09 (2.63–6.16) | 147 (80%) | 34 (19%) | 170 (93%) | 11 (6%) | 161 (89%) | 20 (11%) | |||||
| Unknown | 4 (2%) | – | – | – | – | – | – | |||||||
| BRCA1 mutation | ||||||||||||||
| Wild type | 163 (81%) | 0.90 (0.57–1.40) | 0.014 | 3.48 (2.23–5.48) | 0.001 | – | – | – | – | – | – | 141 (87%) | 21 (13%) | 0.024 |
| Mutate | 36 (18%) | 0.66 (0.42–0.89) | 5.92 (3.27–8.38) | – | – | – | – | 35 (100%) | 0 (0%) | |||||
| n.a. | 2 (1%) | – | – | |||||||||||
| BRCA2 mutation | ||||||||||||||
| Wild type | 188 (94%) | 0.82 (0.53–1.29) | 0.073 | 3.84 (2.46–6.16) | 0.346 | – | – | – | – | – | – | 166 (89%) | 20 (11%) | 0.862 |
| Mutate | 11 (6%) | 1.59 (0.59–2.39) | 3.40 (1.97–4.18) | – | – | – | – | 10 (91%) | 1 (9%) | |||||
| n.a. | 2 (1%) | – | – | |||||||||||
| BRCA1/2 mutation | ||||||||||||||
| Wild type | 152 (76%) | 0.88 (0.55–1.33) | 0.178 | 3.48 (2.26–5.51) | 0.015 | – | – | – | – | – | – | 131 (87%) | 20 (13%) | 0.033 |
| Mutate | 47 (24%) | 0.68 (0.44–1.35) | 5.14 (2.80–7.55) | 45 (98%) | 1 (2%) | |||||||||
| n.a. | 2 (1%) | – | – | – | – | – | – | |||||||
| BRCA1 DNA methylation | ||||||||||||||
| Unmethylated | 178 (89%) | 0.88 (0.58–1.35) | <0.001 | 3.41 (2.17–5.78) | 0.001 | – | – | – | – | – | – | – | – | – |
| Methylated | 21 (11%) | 0.20 (0.14–0.46) | 5.52 (3.99–8.21) | – | – | – | – | – | – | |||||
| BRCA2 DNA methylation | ||||||||||||||
| Unmethylated | 168 (100%) | 0.78 (0.53–1.28) | – | 3.68 (2.36–6.01) | – | – | – | – | – | – | – | |||
| Methylated | 0 (0%) | – | – | – | – | – | – | – | ||||||
Bold values indicates 0.05
RNA isolation and reverse transcription
Total cellular RNA extraction from and reverse transcription were performed as previously described.13
Quantitative real time PCR
Primers and probes for the TATA box-binding protein (TBP; endogenous RNA-control) were used as previously described.13 Primers and probes for BRCA2 [GenBank: NM_000059.3] were determined with the assistance of the computer program Primer Express (Life Technologies, Carlsbad, CA, USA). BRCA2 forward primer: 5′-GAA AAT CAA GAA AAA TCC TTA AAG GCT-3′; BRCA2 reverse-primer: 5′-GTA ATC GGC TCT AAA GAA ACA TGA TG-3′; BRCA2 TaqMan probe: 5′-FAM-AGC ACT CCA GAT GGC ACA ATA AAA GAT CGA AG-3′-TAMRA. Primers and probe for BRCA1 were purchased from Applied Biosystems (Foster City, CA, USA, Applied Biosystems Assay ID: Hs01556193_m1). PCR reactions were performed as previously described.13 Each experiment included a standard curve with five cDNA concentrations, a positive control sample (OVCAR-3 carcinoma cell-line), 40 patient samples and a no template control. The standard curves were generated using serially diluted solutions of standard cDNA derived from the HTB-77 carcinoma cell line. The target mRNA quantity in each sample was determined from the relative standard curve, data normalization was carried out against TBP, the endogenous RNA-control and expressed in arbitrary units corresponding to the dilution factors of the standard RNA preparation. Real-time PCR assays were conducted in duplicates for each sample, and the mean value was used for calculation.
Mutation analysis
Genomic DNA from pulverized, quick-frozen OC specimens was isolated using the DNeasy tissue-kit (Qiagen, Hilden, Germany). Targeted NGS was performed using the TruSight Cancer sequencing panel (Illumina, San Diego, USA). The analyses were performed on the Illumina MiSequ® and the NextSeq system (Illumina, CA, USA). Mutation analysis was performed using NextGene and Geneticist Assistant softwares.
DNA-methylation analysis
Bisulfite modification and MethyLight analysis were performed as described previously.14 For BRCA1 DNA-methylation two different MethyLight PCR primer sets were used, Primers and probes for BRCA1 were determined with the assistance of the computer program Primer Express version 2.0.0 (Applied Biosystems, Foster City, CA, USA) to produce a 86-base-pair PCR amplicon (located at +57 to +142 relative to transcription start site of BRCA1). Genomic DNA not treated with bisulfite (unmodified) was not amplified with the primers (data not shown). Primer sequences were: BRCA1 forward 5′-ATC CCC CGT CCA AAA AAT CT-3′, BRCA1 reverse 5′-TGG TAA CGG AAA AGC GCG-3′, BRCA1 Taq Man probe 5′FAM- CAC GCC GCG CAA TCG CAA -3′-BHQ1. For BRCA1 DNA-methylation analysis an additional MethyLight reaction was selected from literature, also primers and probes for BRCA2 were selected from literature.15 Cases were scored as positive if a percentage of methylated reference (PMR) value of ≥4.0% was obtained, according to studies published in the literature.16,17
Statistical analysis
To compare clinicopathological characteristics and BRCA1/2 mRNA-expression or BRCA1/2 mutation-status, the non-parametric Mann–Whitney U test or Kruskal–Wallis test or Chi-squared test were applied. The correlations between BRCA1/2 mRNA-expression were assessed by Spearman-rank correlation analyses. Progression-free survival (PFS) was defined as the time from diagnosis of the primary to tumor to the histopathological confirmation of recurrence or metastases and overall survival (OS) as the time from diagnosis of the primary to tumor to death from any cause or to the last clinical inspection. Univariate Kaplan-Meier analyses and multivariable Cox survival analyses were used to explore the association of BRCA1/2 expression or with PFS and OS. For survival analyses, patients were dichotomized into low and high mRNA-expression level groups by the optimal cut-off expression value calculated by the Youden’s index based on a receiver operating characteristic curve analysis for overall survival.18 P-values less than 0.05 were considered as statistically significant. Statistical analysis was performed using SPSS statistical software (version 20.0.0; SPSS Inc., Chicago, IL, USA).
Results
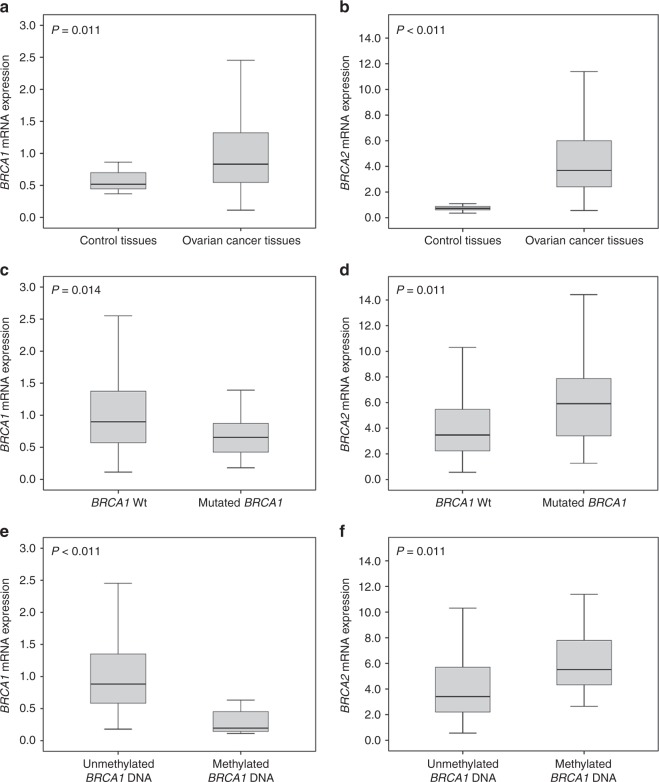
We analyzed BRCA1/2 mRNA-expression levels in 201 OC tissues and 12 non-neoplastic fallopian tubes. We found 1.6-fold higher BRCA1 and 5.0-fold higher BRCA2 mRNA-expression levels in OC samples in comparison to control tissues (P = 0.011, P < 0.001, Fig. 1a, b).
Fig. 1.
BRCA1 and BRCA2 mRNA-expression in ovarian tissues. a BRCA1 mRNA-expression in 12 non-neoplastic fallopian tubes and 192 OC tissues, b BRCA2 mRNA-expression in 11 non-neoplastic fallopian tubes and 168 OC tissues. c BRCA1 and d BRCA2 mRNA-expression according the BRCA1 mutation-status. e BRCA1 and f BRCA2 mRNA-expression according the BRCA1 DNA-methylation-status
Molecular and clinicopathological characteristics
Associations of BRCA1/2 mRNA-expression with clinicopathological characteristics are shown in Table 1.
We found that BRCA2 mRNA-expression was associated with poor tumor differentiation as it increases with tumor grade (P < 0.001; Table 1). Higher BRCA2 mRNA-expression was observed in patients with any residual disease (P = 0.032; Table 1) in comparison to patients with no residual disease. The highest BRCA1/2 mRNA-expression levels were identified in endometrioid OCs in comparison to the other histologic subtypes (P = 0.025, and P = 0.005, respectively; Table 1). Ninety-one percent of the patients included in this study had type II tumors (N = 183) which showed higher, intratumoral BRCA1/2 mRNA-expression compared to type I tumors (P = 0.034 and P = 0.001, respectively; Table 1).
OC tissues with BRCA1-mutations showed lower BRCA1 mRNA-expression (P = 0.014) but higher BRCA2 mRNA-expression (P = 0.001) (Table 1; Fig. 1c, d) in comparison to tissues without BRCA1 mutations. No association between BRCA2 mutation-status and BRCA1/2 mRNA-expression was identified (Table 1). Among 201 OC patients 36 patients (18%) presented BRCA1-mutations, 11 patients (6%) BRCA2-mutations. Interestingly, there was no correlation between BRCA mutation-status and any clinicopathological characteristics. In the herein investigated cohort, no differences in the expression of BRCA-1/2-mRNA could be revealed for the various subtypes of mutations detected in the BRCA1/2 genes (data not shown).
As expected, we found an inverse association between BRCA1 DNA-methylation-status and BRCA1 mRNA-expression (P < 0.001; Table 1; Fig. 1e). Interestingly we observed a direct association between BRCA1 DNA-methylation and BRCA2 mRNA-expression (P = 0.001; Table 1; Fig. 1f). Eighteen percent of undifferentiated tumors (tumor grade 3, N = 17) and 16% of tumors from patients with any residual disease (N = 15) were positive for BRCA1 DNA-methylation (P = 0.004, and P = 0.013; respectively; Table 1). Epigenetic silencing of BRCA1 was mutually exclusive with BRCA1 mutations (Table 1). No BRCA2 DNA-methylation was detected in the analyzed OC tissue samples.
Survival analysis of BRCA1 and BRCA2 mRNA-expression and DNA-methylation-status
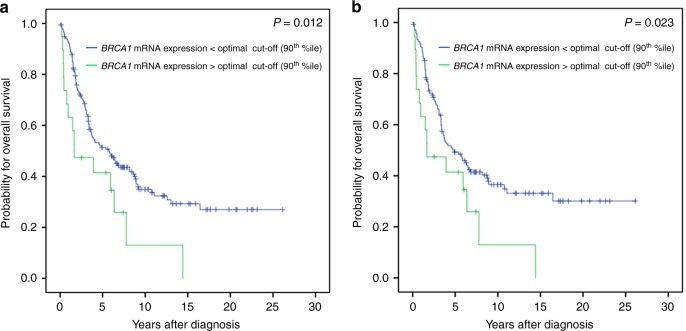
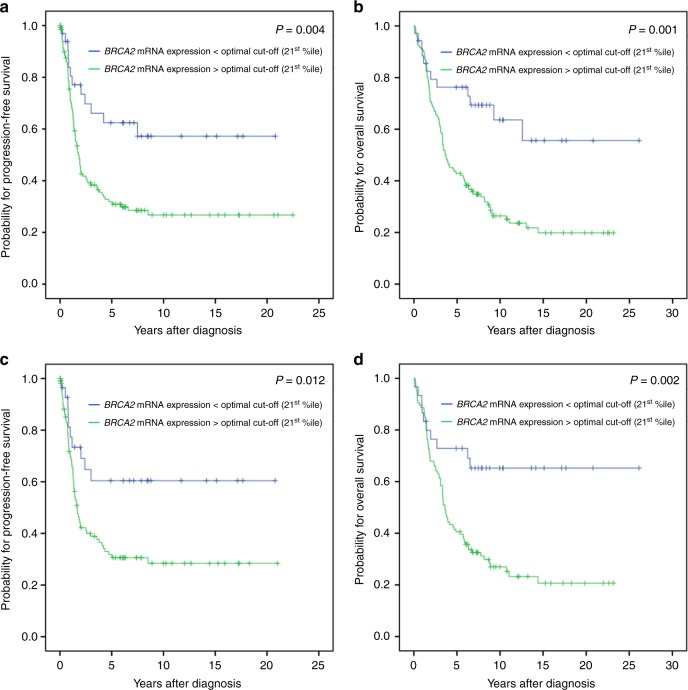
In order to investigate the prognostic value of BRCA1/2 mRNA-expression levels we identified the optimal threshold for “high” and “low” expression using Youden’s index.16 Univariate survival analysis in the entire cohort showed that a lower BRCA1 mRNA-expression (<90th percentile) was associated with a favorable OS (P = 0.012; Table 2A; Fig. 2a). This was also observed in the subgroups of high grade OC (P = 0.004; Table 2B) and high grade serous OC (P = 0.027; Table 2C). A detailed analysis revealed that these prognostic effects were only observed in patients with BRCA1 non-mutated (wildtype) tumors in all patients (P = 0.023; Table 2A; Fig. 2b) and in high grade OC patients (P = 0.011; Table 2B). Lower BRCA2-expression levels (<21st percentile) were associated with favorable PFS and OS in the entire cohort (P = 0.004; P = 0.001; Table 2A; Fig. 3a, b), in high grade OC (P = 0.006; P = 0.002; Table 2B) and in high grade serous OC (P = 0.006; P = 0.001; Table 2C). A detailed analysis showed again the prognostic relevance of low BRCA2 mRNA-expression only in patients with BRCA1 non-mutated tumors. This was true for the entire patient cohort (PFS: P = 0.012; OS: P = 0.002; Table 2A; Fig. 3c, d), high grade OC (PFS: P = 0.022; OS: P = 0.005; Table 2B) and high grade serous OC (PFS: P = 0.016; OS: P = 0.001; Table 2C). No impact of BRCA1 gene promoter methylation-status on progression-free survival and overall-survival rates was found (Table 2).
Table 2.
Univariate survival analysis in 201 ovarian cancer patients
| Variable | Progression-free survival | Overall survival | |||
|---|---|---|---|---|---|
| Median, years (95% CI) | P value | Median, years (95% CI) | P value | ||
| A | |||||
| Age | ≤62.3 years | 2.05 (1.47–2.63) | 0.805 | 8.20 (5.63–10.78) | 0.006 |
| >62.3 years | 1.81 (1.13–2.50) | 3.35 (2.68–4.02) | |||
| FIGO stage | I/II | n.r. | <0.001 | n.r. | <0.001 |
| III/IV | 1.48 (1.10–1.86) | 3.62 (3.06–4.18) | |||
| Tumor grade | 1/2 | 2.05 (1.23–2.87) | 0.221 | 6.24 (2.82–9.67) | 0.057 |
| 3 | 1.97 (1.16–2.78) | 3.62 (3.03–4.21) | |||
| Residual disease | Macroscopically tumor-free | n.r. | <0.001 | 13.03 (n.r.) | <0.001 |
| Any tumor residual | 1.25 (1.06–1.44) | 2.68 (1.83–3.53) | |||
| Histology | HGSOC | 1.77 (1.35–2.18) | 0.027 | 3.62 (3.13–4.12) | 0.006 |
| LGSOC | n.r. | n.r. | |||
| Endometrioid | 5.98 (n.r.) | 11.06 (n.r.) | |||
| Clear cell | 1.81 (1.10-2.53) | 2.72 (n.r.) | |||
| Ovarian cancer type | Type I | n.r. | 0.068 | n.r. | 0.022 |
| Type II | 1.91 (1.52–2.29) | 3.82 (2.11–5.52) | |||
| BRCA1 DNA methylation | No | 2.00 (1.41–2.59) | 0.850 | 4.54 (2.55–6.53) | 0.521 |
| Yes | 1.95 (0.46–3.45) | 4.89 (2.25–7.53) | |||
| BRCA1 mRNA expression | Low | 2.02 (1.38–2.65) | 0.183 | 5.74 (3.63–7.85) | 0.012 |
| High | 0.87 (0.00–1.82) | 1.66 (0.00–5.00) | |||
| Subgroup analysis | |||||
| BRCA1 non-mutated | Low | 2.06 (1.03–3.10) | 0.169 | 4.89 (2.88–6.90) | 0.023 |
| High | 0.87 (0.00–1.82) | 1.67 (0.00–5.00) | |||
| BRCA1 mutated | Low | 2.00 (1.60–2.39) | – | 8.20 (4.52–11.88) | – |
| High | – | – | |||
| BRCA2 mRNA expression | Low | n.r. | 0.004 | n.r. | 0.001 |
| High | 1.81 (1.41–2.22) | 3.70 (2.78–4.62) | |||
| Subgroup analysis | |||||
| BRCA1 non-mutated | Low | n.r. | 0.012 | n.r. | 0.002 |
| High | 1.65 (1.19–2.11) | 3.62 (3.09–4.14) | |||
| BRCA1 mutated | Low | 7.49 (n.r.) | 0.372 | 9.27 (n.r.) | 0.463 |
| High | 1.98 (1.62–2.34) | 6.03 (0.28–11.78) | |||
| B | |||||
| Age | ≤62.3 years | 1.98 (1.34–2.62) | 0.590 | 6.86 (4.23–9.50) | 0.012 |
| >62.3 years | 1.77 (1.27–2.26) | 3.32 (2.63–4.01) | |||
| FIGO stage | I/II | n.r. | <0.001 | n.r. | <0.001 |
| III/IV | 1.47 (1.10–1.84) | 3.43 (3.01–3.85) | |||
| Tumor grade | 2 | 1.90 (1.40–2.41) | 0.398 | 5.74 (2.79–8.68) | 0.177 |
| 3 | 1.95 (1.20–2.71) | 3.55 (3.01–4.08) | |||
| Residual disease | Macroscopically tumor-free | 5.98 (n.r.) | <0.001 | 13.03 (n.r.) | <0.001 |
| Any tumor residual | 1.25 (1.06–1.44) | 2.55 (1.58–3.51) | |||
| Histology | HGSOC | 1.77 (1.35–2.18) | 0.056 | 3.62 (3.13–4.12) | 0.029 |
| HGEOC | 5.11 (n.r.) | 8.94 (5.85–12.02) | |||
| HGCCOC | 1.81 (1.10–2.53) | 2.72 (n.r.) | |||
| BRCA1 DNA methylation | No | 1.91 (1.35–2.47) | 0.733 | 3.92 (2.09–5.76) | 0.531 |
| Yes | 1.95 (0.51–3.40) | 3.71 (1.65–5.77) | |||
| BRCA1 mRNA expression | Low | 1.98 (1.34–2.62) | 0.093 | 4.89 (2.74–7.04) | 0.004 |
| High | 0.87 (0.02–1.72) | 1.65 (0.68–2.63) | |||
| Subgroup analysis | |||||
| BRCA1 non-mutated | Low | 1.95 (1.07–2.84) | 0.094 | 3.94 (1.94–5.94) | 0.011 |
| High | 0.87 (0.02–1.72) | 1.65 (0.68–2.63) | |||
| BRCA1 mutated | Low | 2.00 (1.80–2.19) | – | 8.20 (3.28–13.12) | – |
| High | – | – | |||
| BRCA2 mRNA expression | Low | n.r. | 0.006 | n.r. | 0.002 |
| High | 1.81 (1.40–2.23) | 3.62 (2.97–4.27) | |||
| Subgroup analysis | |||||
| BRCA1 non-mutated | Low | n.r. | 0.022 | n.r. | 0.005 |
| High | 1.65 (1.19–2.11) | 3.43 (2.96–3.90) | |||
| BRCA1 mutated | Low | 7.49 (n.r.) | 0.326 | 12.58 (n.r.) | 0.461 |
| High | 1.98 (1.62–2.34) | 6.03 (0.28–11.78) | |||
| C | |||||
| Age | ≤62.3 years | 1.81 (1.20–2.42) | 0.650 | 5.74 (2.89–8.58) | 0.035 |
| >62.3 years | 1.68 (1.07–2.29) | 3.32 (2.74–3.89) | |||
| FIGO stage | I/II | n.r. | <0.001 | 7.78 (1.38–14.18) | 0.049 |
| III/IV | 1.47 (1.09–1.84) | 3.55 (3.15–3.94) | |||
| Tumor grade | 1/2 | 1.65 (1.10–2.20) | 0.519 | 3.82 (1.35–6.29) | 0.257 |
| 3 | 1.95 (1.49–2.42) | 3.55 (3.10–3.99) | |||
| Residual disease | Macroscopically tumor-free | 3.57 (0.00–7.23) | <0.001 | 8.17 (2.26–14.08) | <0.001 |
| Any tumor residual | 1.26 (1.09–1.42) | 2.94 (1.91–3.96) | |||
| BRCA1 DNA methylation | No | 1.77 (1.33–2.21) | 0.943 | 3.62 (3.07–4.17) | 0.750 |
| Yes | 1.95 (0.92–2.99) | 3.71 (2.02–5.40) | |||
| BRCA1 mRNA expression | Low | 1.84 (1.51–2.17) | 0.101 | 3.71 (2.73–4.69) | 0.027 |
| High | 0.87 (0.14–1.60) | 1.65 (0.42–2.88) | |||
| Subgroup analysis | |||||
| BRCA1 non-mutated | Low | 1.68 (1.20–2.17) | 0.117 | 3.62 (3.07–4.18) | 0.051 |
| High | 0.87 (0.14–1.60) | 1.65 (0.42–2.88) | |||
| BRCA1 mutated | Low | 1.98 (1.70–2.26)) | – | 6.03 (1.35–10.71) | – |
| High | – | – | |||
| BRCA2 mRNA expression | Low | n.r. | 0.006 | n.r. | 0.001 |
| High | 1.67 (1.31–2.03) | 3.39 (3.01–3.77) | |||
| Subgroup analysis | |||||
| BRCA1 non-mutated | Low | n.r. | 0.016 | n.r. | 0.001 |
| High | 1.46 (1.06–1.87) | 3.39 (2.87–3.91) | |||
| BRCA1 mutated | Low | 7.49 (n.r.) | 0.531 | 12.58 (n.r.) | 0.571 |
| High | 1.98 (1.69–2.27) | 4.08 (0.00–9.78) | |||
The significance level (P) was determined by log-rank test
HGCCOC high grade clear cell ovarian cancer, HGEOC high grade endometrioid ovarian cancer, HGSOC high grade serous ovarian cancer, LGSOC low grade serous ovarian cancer, n.r. not reached
A: Progression free and overall survival in 201 ovarian cancer patients
B: Subgroup analysis: progression-free and overall survival in 183 high grade OC patients
C: Subgroup analysis: progression-free and overall survival in 129 high grade serous OC patients. The optimal cutoff points for BRCA1/2 mRNA expression were calculated by the Youden’s index for overall survival (BRCA1 expression: low/ high:</>90th %ile; BRCA2 expression: low/ high:</>21st %ile). Bold values indicates 0.05
Fig. 2.
Kaplan Meier survival analysis and BRCA1 mRNA-expression in OC patients according the 90th percentile as cut-off value. Overall survival in a 192 OC patients, b 155 OC patients with BRCA1-wildtype tumors
Fig. 3.
Kaplan Meier survival analysis and BRCA2 mRNA-expression in OC patients according the 21st percentile as cut-off value. a Progression-free survival and b overall survival in 168 OC patients. c Progression-free survival and d overall survival in 136 patients with BRCA1-wildtype tumors
Cox-regression survival analysis confirmed the prognostic significance of BRCA1 mRNA-expression for OS in the whole cohort (HRdeath 2.0 (1.1–3.7), P = 0.028; Table 3A) but not in high grade serous OC (Table 3B). However, independency of the prognostic value of BRCA2 mRNA-expression was approved in patients with high grade serous OC, representing 64% of the entire cohort, as well for PFS (HRprogression 2.4 (1.0–5.7), P = 0.045) as for OS (HRdeath 2.9 (1.2–6.8), P = 0.015); (Table 3B).
Table 3.
Multivariable analysis in ovarian cancer patients
| Variable | Progression-free survival | Overall survival | |||
|---|---|---|---|---|---|
| HR of progression (95% CI) | P value | HR of death (95% CI) | P value | ||
| A | |||||
| Age | Low vs. high (<or>median age) | – | – | 1.9 (1.3–2.9) | 0.001 |
| FIGO stage | I/II vs. III/IV | 2.6 (1.2–5.5) | 0.013 | 1.1 (0.6–2.1) | 0.692 |
| Residual disease after surgery | No vs. yes | 2.7 (1.6–4.6) | <0.001 | 3.5 (2.1–6.0) | <0.001 |
| Histology | HGSOC vs. Others | 0.9 (0.6–1.5) | 0.675 | 0.6 (0.4–1.1) | 0.087 |
| Ovarian cancer type | Type I vs. Type II | – | – | 0.9 (0.3–2.7) | 0.829 |
| BRCA1 mRNA expression | Low vs. high (<or>optimal cut-off) | – | – | 2.0 (1.1–3.7) | 0.028 |
| BRCA2 mRNA expression | Low vs. high (<or>optimal cut-off) | 1.9 (1.0–3.7) | 0.061 | 1.9 (1.0–3.7) | 0.058 |
| B | |||||
| Age | Low vs. high (<or>median age) | – | – | 2.1 (1.3–3.4) | 0.002 |
| FIGO stage | I/II vs. III/IV | 2.3 (0.8–6.3) | 0.119 | 1.1 (0.5–2.4) | 0.812 |
| Residual disease after surgery | No vs. yes | 2.4 (1.3–4.7) | 0.008 | 3.2 (1.7–6.0) | <0.001 |
| BRCA1 mRNA expression | Low vs. high (<or>optimal cut-off) | – | – | 1.7 (0.8–3.5) | 0.151 |
| BRCA2 mRNA expression | Low vs. high (or>optimal cut-off) | 2.4 (1.0–5.7) | 0.045 | 2.9 (1.2–6.8) | 0.015 |
The significance level was determined by Cox regression analysis
HR hazard ratio. Bold values indicates 0.05
To answer the question whether the identified favorable survival in tumors with low BRCA1/2-mRNA expression may be interpreted by platinum-sensitivity we compared the expression levels in BRCA1-wildtype tumors from platinum-refractory and fully platinum-sensitive patients. We found statistically significant lower BRCA1- and BRCA2 mRNA-expression levels in platinum-sensitive tumors (P = 0.004 and P = 0.045; Supplemental Fig. 1).
Discussion
BRCA1/2 belong to genes that play key roles in the homologous recombination repair, which represents the main mechanism to repair DNA double-strand breaks.1 While BRCA1 is multifunctional, BRCA2 functions almost exclusively in homologous recombination by recruiting an essential homologous recombination protein RAD51C to double-strand break sites.19,20 Our investigations revealed higher BRCA1/2-expression on the transcriptome level in OC tissues in comparison with non-neoplastic fallopian tube tissue. The cause of this finding may be the higher proliferation rate in malignant tissues which together with genetic instability may increase the need for more DNA damage repair. In accordance higher BRCA1/2-expression was also found in high-grade (Type II) tumors. This notion is supported by Gudas et al. who suggest that the upregulation of BRCA1-expression by steroid hormones is caused indirectly by increasing proliferation of breast cancer cells.21
Multivariate Cox-regression analysis showed a favorable OS for low BRCA1-expression in the whole cohort of included patients. For BRCA2-expression in the subgroup of high grade serous OC an independent, prognostic value in terms of PFS and OS was confirmed. These findings could be explained by a reduced capacity of DNA damage repair via homologous recombination in cancers with low BRCA1/2 expression, enhancing the therapeutic effects of DNA-crosslinking agents such as platinum compounds. In fact, low BRCA1/2 mRNA-expression in BRCA1-wildtype cancers was associated with fully platinum-sensitive disease and high expression was evidenced in platinum-refractory disease.
These BRCA mRNA-expression data are in line with the plethora of data showing that OC patients carrying a BRCA1 or 2 germline mutation exhibit high responsiveness to platinum-based chemotherapy consecutively associated with an improved clinical outcome. In recurrent OC, similar beneficial therapeutical effects in BRCA mutation carriers have been reported for PARP-inhibitors and for trabectedin a drug that is crosslinking the DNA in the minor grove.
Until to date there are only very few studies on BRCA-expression in OC and these are only dealing with the expression of BRCA1 but not with that of BRCA2. In a retrospective analysis of OC specimens obtained from patients included in the GOG-172 study comparing intraperitoneal (IP) with intravenous (i.v.) platinum/taxane chemotherapy, Lesnock et al. assessed BRCA1-expression on the protein level with regard to clinical outcome and responsiveness to chemotherapy considering especially the efficacy of the high loco-regional platinum doses reached by IP administration. The authors revealed that patients with cancers exhibiting BRCA1-immunostaining in less than 10% of the tumor cells was the only subgroup exhibiting a significant benefit in OS from a platinum-based IP chemotherapy.22 In addition, Carser et al. found also a strong response improvement to classical i.v. platinum-based chemotherapy in tumors with absent or low BRCA1-expression in immunohistochemistry. This effect was translated into a favorable PFS and OS in affected patients and the predictive value of BRCA1 immunostaining was confirmed in the multivariate analysis.23 These considerations were indirectly confirmed by Swisher et al., showing that in primary BRCA1-mutated OCs, recurrent platinum-resistant tumors exhibited secondary genetic changes within the BRCA1 gene, which interestingly were accompanied by restored expression of BRCA1-protein.24 Furthermore, Quinn et al. reported from an in vitro and in vivo approach that inhibition of BRCA1-expression via siRNA knock-down leads to increased sensitivity to platinum therapy but impaired responsiveness to anti-microtubule agents such as taxanes. In a small series of patients, they corroborated their in vitro data by showing a significant improved OS in patients with tumors exhibiting low levels of BRCA1-mRNA.25 Also in this study BRCA2-expression has not been accessed.
In breast cancers exhibiting low BRCA2 mRNA-levels, a significantly higher 5-year disease free survival rate was shown.26
Interestingly, our study emerged that in BRCA1-mutated OC the expression of BRCA1-transcripts was lower, but in contrary those of BRCA2 were significantly higher as compared with BRCA1-wildtype cancers. Furthermore, also down regulation of BRCA1-transcripts by methylation of the BRCA1-promotor was associated with increased BRCA2 mRNA levels. In contrast in cancers carrying a BRCA2-mutation, no up- or down-regulation of the BRCA1/2 mRNA was found. However, the latter findings should be interpreted with caution due to the low number of BRCA2-mutated cancers within our cohort. The reason of the “compensatory” upregulation of BRCA2-mRNA in low BRCA1-expressing cancers remains speculative because there is no exact knowledge on how the BRCA protein expression is regulated either in normal or in malignant tissues. High BRCA-expression could determine a distinct phenotype with a high constitutive expression or could reflect a transitory upregulation triggered by various situations (e.g., proliferative or genomic stress). Thus, it is theoretically possible that the functional loss of multifunctional BRCA1 is leading to genetically instable cancers requiring higher BRCA2 recruitment for repeated double-strand break repair.
In BRCA1 we found DNA-methylation in 11% of all tumors, which is in accordance with previously published data.27 We could not identify a prognostic relevance of BRCA1 DNA-methylation for PFS and OS consistent with recent studies.28,29
Our data show that low BRCA1/2 mRNA-expression confers platinum-hypersensitivity to OCs. As clinical studies in recurrent OC recently evidenced that the sensitivity of high grade serous OC to PARP-inhibitor maintenance therapy is particularly related to the response to the actual platinum-based chemotherapy,30 our data are tempting to speculate that BRCA1/2 mRNA levels may be reliable biomarkers to also predict responsiveness of cancers to PARP-inhibitors. The same may be true for other drugs whose effectivity is related to platinum-sensitivity such as trabectedin.
Electronic supplementary material
Acknowledgements
We thank Inge Gaugg, Martina Fleischer and Annemarie Wiedemair for excellent technical assistance. This work was supported by the Verein zur Krebsforschung in der Frauenheilkunde (no grant number is applicable), the Österreichische Krebshilfe - Krebsgesellschaft Tirol (15010/ 2015) and AstraZeneca (NCR-15-11443).
Author contributions
H.F., A.G.Z., and C.M. developed the study concept, H.F., A.G.Z., C.M., and I.T. designed the project and edited the manuscript, I.T., V.W., C.D., G.S., S.W., S.S., and S.F.L. were involved in data acquisition, I.T., V.W., C.D., G.S., S.W., S.S., S.F.L., and H.F. performed quality control of data and algorithms, I.T., V.W., C.D., G.S., S.W., S.S., S.F.L., C.M., H.F., and A.G.Z. analyzed and interpreted data and reviewed the final manuscript, I.T. and H.F. performed statistical analyses, I.T., H.F., and A.G.Z. prepared the manuscript.
Competing interests
The authors declare no competing interests.
Data availability
The datasets generated during and analyzed during the current study are available from the corresponding author on reasonable request.
Ethical approval
The study was reviewed and approved by the Ethics committee of the Medical University of Innsbruck (reference number: AN2015-0038 346/4.17) and conducted in accordance with the Declaration of Helsinki.
Contributor Information
Heidelinde Fiegl, Email: Heidelinde.Fiegl@i-med.ac.at.
Alain G. Zeimet, Email: Alain.Zeimet@i-med.ac.at
Electronic supplementary material
Supplementary information is available for this paper at 10.1038/s41416-018-0217-4.
References
- 1.Roy R, Chun J, Powell SN. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat. Rev. Cancer. 2011;12:68–78. doi: 10.1038/nrc3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saha S, et al. Decreased expression of BRCA2 accelerates sporadic breast cancer progression. Indian J. Surg. Oncol. 2015;6:378–383. doi: 10.1007/s13193-015-0449-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. Cancer J. Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 4.Holschneider CH, Berek JS. Ovarian cancer: epidemiology, biology, and prognostic factors. Semin. Surg. Oncol. 2000;19:3–10. doi: 10.1002/1098-2388(200007/08)19:1<3::AID-SSU2>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 5.Johannsson OT, Ranstam J, Borg A, Olsson H. Survival of BRCA1 breast and ovarian cancer patients: a population-based study from southern Sweden. J. Clin. Oncol. 1998;16:397–404. doi: 10.1200/JCO.1998.16.2.397. [DOI] [PubMed] [Google Scholar]
- 6.Risch HA, et al. Prevalence and penetrance of germline BRCA1 and BRCA2 mutations in a population series of 649 women with ovarian cancer. Am. J. Hum. Genet. 2001;68:700–710. doi: 10.1086/318787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Permuth-Wey J, Sellers TA. Epidemiology of ovarian cancer. Methods Mol. Biol. 2009;472:413–437. doi: 10.1007/978-1-60327-492-0_20. [DOI] [PubMed] [Google Scholar]
- 8.Harter P, et al. BRCA1/2 mutations associated with progression-free survival in ovarian cancer patients in the AGO-OVAR 16 study. Gyn. Oncol. 2016;140:443–449. doi: 10.1016/j.ygyno.2015.12.027. [DOI] [PubMed] [Google Scholar]
- 9.Ledermann JA. PARP inhibitors in ovarian cancer. Ann. Oncol. 2016;27:i40–i44. doi: 10.1093/annonc/mdw094. [DOI] [PubMed] [Google Scholar]
- 10.Pharoah PD, Easton DF, Stockton DL, Gayther S, Ponder BA. Survival in familial, BRCA1-associated, and BRCA2-associated epithelial ovarian cancer. United Kingdom Coordinating Committee for Cancer Research (UKCCCR) Familial Ovarian Cancer Study Group. Cancer Res. 1999;59:868–871. [PubMed] [Google Scholar]
- 11.Sabatier R, et al. Ovarian cancer patients at high risk of BRCA mutation: the constitutional genetic characterization does not change prognosis. Fam. Cancer. 2016;15:497–506. doi: 10.1007/s10689-016-9873-9. [DOI] [PubMed] [Google Scholar]
- 12.Kotsopoulos J, et al. Ten-year survival after epithelial ovarian cancer is not associated with BRCA mutation status. Gyn. Oncol. 2016;140:42–47. doi: 10.1016/j.ygyno.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 13.Goebel G, et al. Elevated mRNA expression of CHAC1 splicing variants is associated with poor outcome for breast and ovarian cancer patients. Br. J. Cancer. 2012;106:189–198. doi: 10.1038/bjc.2011.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Notaro S, et al. Evaluation of folate receptor 1 (FOLR1) mRNA expression, its specific promoter methylation and global DNA hypomethylation in type I and type II ovarian cancers. BMC Cancer. 2016;16:589. doi: 10.1186/s12885-016-2637-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weisenberger DJ, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat. Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 16.Press JZ, et al. Ovarian carcinomas with genetic and epigenetic BRCA1 loss have distinct molecular abnormalities. BMC Cancer. 2008;8:17. doi: 10.1186/1471-2407-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eads CA, et al. Epigenetic patterns in the progression of esophageal adenocarcinoma. Cancer Res. 2001;61:3410–3418. [PubMed] [Google Scholar]
- 18.Youden WJ. Index for rating diagnostic tests. Cancer. 1950;3:32–35. doi: 10.1002/1097-0142(1950)3:1<32::AID-CNCR2820030106>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 19.Moynahan ME, Pierce AJ, Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol. Cell. 2001;7:263–272. doi: 10.1016/S1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- 20.Takaoka M, Miki Y. BRCA1 gene: function and deficiency. Int. J. Clin. Oncol. 2017;23:36–44. doi: 10.1007/s10147-017-1182-2. [DOI] [PubMed] [Google Scholar]
- 21.Gudas JM, Nguyen H, Li T, Cowan KH. Hormone-dependent regulation of BRCA1 in human breast cancer cells. Cancer Res. 1995;55:4561–4565. [PubMed] [Google Scholar]
- 22.Lesnock JL, et al. BRCA1 expression and improved survival in ovarian cancer patients treated with intraperitoneal cisplatin and paclitaxel: a Gynecologic Oncology Group Study. Br. J. Cancer. 2013;108:1231–1237. doi: 10.1038/bjc.2013.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carser JE, et al. BRCA1 is both a prognostic and predictive biomarker of response to chemotherapy in sporadic epithelial ovarian cancer. Gyn. Oncol. 2011;123:492–498. doi: 10.1016/j.ygyno.2011.08.017. [DOI] [PubMed] [Google Scholar]
- 24.Swisher EM, et al. Secondary BRCA1 mutations in BRCA1-mutated ovarian carcinomas with platinum resistance. Cancer Res. 2008;68:2581–2586. doi: 10.1158/0008-5472.CAN-08-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quinn JE, et al. BRCA1 mRNA expression levels predict for overall survival in ovarian cancer after chemotherapy. Clin. Cancer Res. 2007;13:7413–7420. doi: 10.1158/1078-0432.CCR-07-1083. [DOI] [PubMed] [Google Scholar]
- 26.Egawa C, Miyoshi Y, Taguchi T, Tamaki Y, Noguchi S. High BRCA2 mRNA expression predicts poor prognosis in breast cancer patients. Int. J. Cancer. 2002;98:879–882. doi: 10.1002/ijc.10231. [DOI] [PubMed] [Google Scholar]
- 27.Cunningham JM, et al. Clinical characteristics of ovarian cancer classified by BRCA1, BRCA2, and RAD51C status. Sci. Rep. 2014;4:4026. doi: 10.1038/srep04026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang D, et al. Association of BRCA1 and BRCA2 mutations with survival, chemotherapy sensitivity, and gene mutator phenotype in patients with ovarian cancer. JAMA. 2011;306:1557–1565. doi: 10.1001/jama.2011.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruscito I, et al. BRCA1 gene promoter methylation status in high-grade serous ovarian cancer patients—a study of the tumour Bank ovarian cancer (TOC) and ovarian cancer diagnosis consortium (OVCAD) Eur. J. Cancer. 2014;50:2090–2098. doi: 10.1016/j.ejca.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 30.Mirza MR, et al. Niraparib maintenance therapy in platinum-sensitive, recurrent ovarian cancer. N. Engl. J. Med. 2016;375:2154–2164. doi: 10.1056/NEJMoa1611310. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and analyzed during the current study are available from the corresponding author on reasonable request.