Abstract
Objective(s):
To determine whether variation in cell associated (CA) unspliced (US) HIV RNA in HIV-infected individuals on antiretroviral therapy (ART) has a circadian basis.
Methods:
Prospective observational study of HIV-infected individuals on ART. Blood was collected on three occasions and CA-US HIV RNA and mRNA of the Circadian-locomotor-output-cycles-kaput (CLOCK)-associated genes quantified by real time PCR. CLOCK-associated proteins were over expressed in a cell line stably transfected with an HIV long terminal repeat (LTR) luciferase reporter.
Results:
Using a mixed effects model, there was a significant increase in log-CA-US RNA at the third visit compared to the first visit (effect size of 0.619 with standard error (SE) of 0.098, p<0.001) and an independent effect of time of blood draw (effect size 0.051, (SE 0.025), p=0.040). The CLOCK-associated gene, Brain-and-muscle-ARNT-like-1 (BMAL-1) had a significant relationship with log CA-US HIV RNA (effect size 8.508 (SE 3.777), p = 0.028) and also with time (p=0.045). Over expression of BMAL-1 and CLOCK in a cell line stably transfected with an HIV-LTR luciferase reporter resulted in an increase in luciferase expression and this was reduced following mutation of the second E-box in the HIV-LTR.
Conclusions:
The basal level of HIV transcription on ART can vary significantly and is modulated by the circadian regulator BMAL-1, amongst other factors.
Keywords: HIV, HIV latency, HIV transcription, Circadian rhythm, BMAL-1, Stress, unspliced RNA
Background
In HIV-infected individuals on suppressive antiretroviral therapy (ART), HIV persists indefinitely as long-lived latently-infected CD4+ T-cells [1]. Persistent HIV can be detected in blood or tissue as cell-associated unspliced (CA-US) HIV RNA, cell-associated HIV DNA or low level plasma HIV RNA [2]. While HIV DNA is a surrogate marker of the total number of infected cells, including both defective and intact viral genomes [3], CA-US HIV RNA is an important marker of HIV transcription [4]. CA-US HIV RNA has previously been used as a primary endpoint in clinical trials of interventions that activate transcription in latently-infected cells (referred to as latency reactivating agents (LRA)) as a strategy to eliminate HIV persistence on ART [5–11].
Multiple factors can drive changes in HIV transcription in latently-infected resting CD4+ T-cells, the most potent being T-cell activation leading to the availability of transcription factors but changes in T-cell differentiation, histone acetylation and methylation all play an important role in activating or suppressing HIV transcription in latently-infected cells (reviewed in [12]). In a recent prospective clinical trial in HIV-infected individuals on ART, we performed three baseline visits, with the third visit occurring just before the intervention [7]. CA-US HIV RNA, but not HIV DNA or plasma HIV RNA, was found to be consistently higher at the third baseline visit, which was earlier in the morning and on the day of, but prior to the administration of disulfram, an LRA. We hypothesised that in HIV-infected individuals on ART, there is time-dependent variation in CA-US HIV RNA from circadian changes in T-cell subsets or gene regulation.
Circadian regulation of gene expression is mediated by the Circadian-locomotor-output-cycles-kaput and Brain-and-muscle-ARNT-like-1 (CLOCK-BMAL1) heterodimer. Although circadian rhythm is centrally controlled in the suprachiasmatic nucleus and is entrained by light, peripheral cells have independent clock machinery (reviewed in [13]). This heterodimer binds to a six-nucleotide motif, the E-box [14, 15], to mediate transcription. HIV-1 is known to encode 4 potential E-Boxes in the promoter (5’ long terminal repeat, (LTR)) sequence [16] thus changes in HIV transcription may plausibly be mediated directly through CLOCK-BMAL1. In addition, CLOCK proteins have intrinsic histone acetyl transferase activity and can mediate chromatin remodelling [17] and therefore may also modulate HIV transcription through changes in histone acetylation.
Using blood collected from HIV-infected individuals on ART on three occasions, we quantified CA-US HIV RNA and the expression of multiple CLOCK associated genes and hormones that vary with time and stress. Using a mixed-effects model, we identified a significant increase in log-CA-US HIV RNA at the third visit compared to the first as well as an independent effect of time of blood draw measured in hours on log CA-US HIV RNA. We then included the relative expression of CLOCK-associated genes period (Per) 1 and 2 and cryptochrome 1, cortisol and thyroid stimulating hormone and T-cell subsets at each visit in a mixed-effects model and path analysis. We only identified the CLOCK-associated gene, BMAL-1 to have a significant relationship with log CA-US RNA and also with time. We propose a model to understand the independent effects of visit and time, potentially mediated through a direct effect of BMAL-1, on HIV transcription in HIV-infected individuals on suppressive on ART.
Methods
Study design
This observational clinical trial was a sub-study of a prospective clinical trial of disulfiram in HIV-infected individuals on ART and is registered on ClinicalTrials.gov (NCT01944371). Participants provided written informed consent, and the protocol was approved by the relevant Institutional Review Boards.
Participants were recruited at two sites – Alfred Hospital in Melbourne, Australia and San Francisco General Hospital, San Francisco, California. Blood was collected in acid citrate dextrose tubes and peripheral blood mononuclear cells (PBMC) prepared and stored as previously described [7]. Blood was collected prior to receiving the study treatment (screening [B1]), immediately before the first dose of an intervention, here being disulfiram [B3], and a time-point between [B2] [7]
Measurement of US RNA and CLOCK-associated genes by quantitative PCR
Following thawing of frozen PBMC, CD4+ T-cells were isolated by magnetic bead sorting, RNA was extracted and DNA and CA-US HIV RNA were amplified by qPCR using primers in the gag and LTR regions as previously described [4, 8, 18]. All HIV CA-US RNA samples were run in quadruplicate and the mean reported. The lower limit of detection for the assay was one copy per well. For any wells with no amplification of RNA, the value was counted as zero. If there was a signal detected but calculated copy number was <1, the value was included as 0.5 as we previously described [7].
CLOCK, BMAL1, period (Per)1–3, cryptochrome (Cry)1–2 and GADPH primer sets from Ackermann et al [19] and Yang et al [20] were used in a single round qPCR (Supplementary methods). The sequences of all primers are shown in Supplementary Table 1.
Analysis of T-cell subsets, activation and histone acetylation by flow cytometry
Endocrine parameters
Plasma cortisol and TSH were measured on the automated e601 Electro-ChemiLuminescence Immunoassay Analyser (ECLIA) (Roche Diagnostics GmbH (Mannheim, Germany)). Assay imprecision (expressed as coefficient of variance (CV)) was 2.8% and 3.5% for cortisol and TSH, respectively.
Plasmids and cell lines
cDNA for human Clock (AF011568) and Bmal1 (NM_001178) were synthesised (Bioneer) and cloned into the mammalian expression vector pcDNA3.1(+). The HIV TAT expression vector (pTargeT-HxB2-Tat) and HIV LTR-driven luciferase reporter vector (pGL3-HxB2 LTR) have been described previously [21, 22]. 293T cells were cultured in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% (vol/vol) fetal calf serum (FCS) and 100μg of penicillin and streptomycin per ml. Mutation of E-box elements in the HxB2 LTR were performed using the Quikchange Lightning Mutagenesis kit (Agilent). E-box elements (CANNTG) were mutated to GANNTC thereby ablating the site. All mutants were confirmed by sequencing.
LTR reporter activity assay
293T cells were seeded into 96-well plates at a concentration of 2×104 cells/well in 100μl and cultured for 24h at 37°C. Cells were then transfected by lipofectamine 2000 (Invitrogen) with 50ng of pGL3-LTR and either; 50ng of pTargeT-HxB2-Tat; 25ng of pCLOCK; 25ng pBMAL1 or; 25ng of pCLOCK and 25ng of pBMAL1. The level of LTR reporter activation was measured by luciferase activity in cell lysates (Promega). Luminescence was measured using a FLOUStar microplate reader (GmBH, Germany). Background activity was determined in mock transfected cells.
Statistical analyses
Comparisons between parameters at the three visits were made using the Wilcoxon matched pairs signed rank tests. Statistical analyses were performed using R v3.2.0.
We used a mixed effects model to determine factors that influence the logarithm of CA-US RNA that assumes a different (random) intercept for each individual and fixed effects for the other explanatory variables, such as time of day and visit, and other measures of the circadian genes. We also used a path model that replaced, in the mixed effects model, the time of blood draw with the expression levels of the CLOCK-associated genes, which included CLOCK, BMAL1, Per1–3 and Cry1–2. The aim was to demonstrate that time could be subsumed by the expression of the CLOCK-associated genes. Full description of the model is in the Supplementary methods.
Comparison of conditions in in vitro experiments were made using a linear regression model fit to the log-transformed data. p values < 0.05 were considered significant.
Results
Relationship of CA- US HIV RNA with time and visit
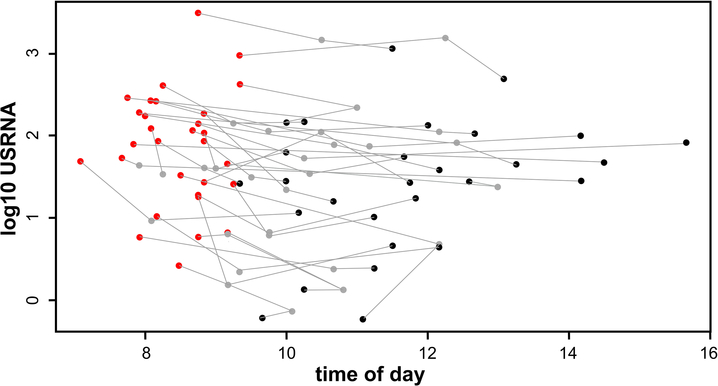
Demographic details have been previously provided but in brief, this was a male cohort, with a median age of 54 years and median baseline CD4 counts of 582 cells/ul [7]. All 30 participants were on ART including a non-nucleoside reverse transcriptase inhibitor (15/30), protease inhibitor (10/30), an integrase inhibitor (2/30) or a combination of these (3/30). We previously reported significantly higher levels of CA-US HIV RNA and earlier time of blood collection in visit B3 relative to visits B2 and B1 while there were no differences in HIV DNA or plasma HIV RNA across the three time-points [7]. There were no differences between visits in tubes used or time between blood collection and processing at either clinical site. All samples from each participant for the three visits were run on a single plate, making it unlikely that the differences could be explained by a technical issue. Figure 1 shows the relationship between visit (color of the dots), time and expression levels of US RNA for each individual (connected dots).
Figure 1.
Relationship of CA-US HIV RNA in CD4+ T-cells, visit and time of blood collection (n=30). Results from three time points collected from each participant are connected by a line. The color of the symbol denotes the pre-intervention time-point. These were prior to receiving the study treatment (screening [B1; black]), immediately before the first dose of an intervention, here being disulfiram [B3; red], and a time-point between [B2; grey].
There was significant subject-to-subject variability in CA-US RNA, which suggested that additional factors beyond visit and time of blood collection may contribute the expression levels of CA-US RNA. Fitting a mixed effects model to this data confirmed this observation, and estimated the standard error for the subject random effect to be 0.71, which was more than twice the estimated standard error of 0.30 for the residual. The fitted model (Supp methods) further revealed a positive slope for the relationship between log CA-US HIV RNA and the time of blood collection with an effect size of 0.051 (SE 0.025) in the change in log CA-US HIV RNA per million cells per hour, p=0.040). The expression levels of log CA-US HIV RNA at the third visit was significantly higher than the expression levels at the first (effect size of 0.619 (SE 0.098), p<0.001; table 1). Taken together, these data demonstrated a very significant relationship between CA-US HIV RNA and visit and a more modest relationship with time.
Table 1.
Parameter estimates derived from the linear mixed effects model, Equation (1), for log10 CA-US HIV RNA/ million cells depending on time of blood collection and visit.
Here μ is a constant, γ is the coefficient of the linear dependence on time of blood draw, and β2 and β3 are the coefficients for the effect of visits B2 and B3 relative to the effect of B1 on log10 CA-US HIV RNA. The estimated standard deviation of the patient-specific random effect (αi) is 0.73 (see Supplemental Methods).
| Parameters | Effect estimate | Standard Error | P-value |
|---|---|---|---|
| μ, Intercept | 0.800 | 0.320 | 0.013 |
| γ, Time of blood collection (per hour of day) |
0.051 | 0.026 | 0.040 |
| β2, Visit 2 | 0.106 | 0.066 | 0.107 |
| β3, Visit 3 | 0.619 | 0.098 | 2.52 × 10−10 |
Relationship of CLOCK genes, CA-US HIV RNA and time
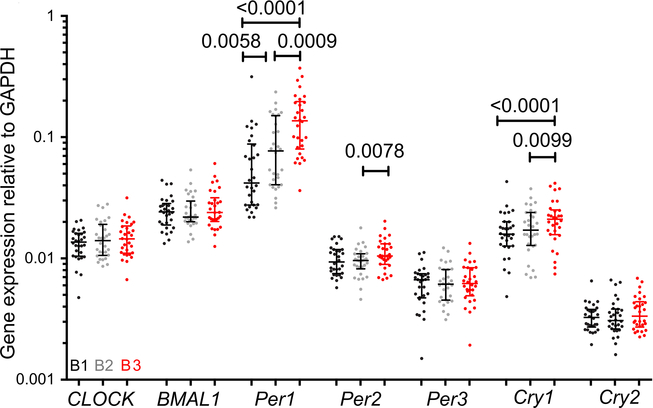
Given 9% of mammalian transcripts are circadian-controlled [23], we next measured regulators of circadian gene expression to explore whether HIV transcription may be mediated by similar mechanisms to human gene transcription. CLOCK-BMAL1 activity is harmonized by Per and Cry repressor feedback loops, endocrine and autonomic systems (reviewed in [24]). We quantified mRNA transcripts by RT-PCR for CLOCK, BMAL1 and the Per/Cry repressor-loop constituents (Per1–3, and Cry1–2) in CD4+ T-cells in blood collected at visits B1, B2 and B3. Per1, Per2 and Cry1 showed increased relative expression in samples collected at B3, the sample collected earlier in the day and where CA-US HIV RNA was shown to be higher (Figure 2A). Per1 and Cry1 expression were higher at B3 compared to B1 and B2. Per2 was higher at B3 compared to B2.
Figure 2.
Relationship of CLOCK-associated genes and visit. Expression of CLOCK-associated genes in HIV-infected individuals on suppressive ART at three time-points (n=30). Data were normalized to GAPDH expression level by the ΔΔCT method. The median and inter-quartile range are shown. Nominal p-values from Wilcoxon matched pairs signed rank tests are shown for significant associations. The color of the symbol denotes the pre-intervention time-point. These were prior to receiving the study treatment (screening [B1; black]), immediately before the first dose of an intervention, here being disulfiram [B3; red], and a time-point between [B2; grey].
To determine if expression of any of the CLOCK-associated genes was associated with the expression levels of CA-US HIV RNA, we performed a path analysis. Here, we replaced the time of blood draw in the mixed effects model, with the expression levels of the CLOCK-associated genes, including CLOCK, BMAL1, Per1, 2 and 3 and Cry1 and 2. Because the CLOCK-associated genes are themselves associated with time, the aim was to demonstrate that time could be subsumed by the expression of the CLOCK-associated genes. That argument had three components: first, that the expression of CLOCK-associated genes was a significant predictor of CA-US HIV RNA; second, that when time was added to that model, this relationship was no longer significant; and third, that time was a predictor for expression of the CLOCK-associated gene.
BMAL1 was the only gene that had a statistically significant effect on log CA-US HIV RNA (effect size of 8.508 (SE 3.777), p = 0.028). This reported p-value was not adjusted for multiple comparisons. Next, we performed a generalized likelihood ratio test to compare the model that adds time of blood draw to the mixed effect model with BMAL1, which produced a chi-square of 2.9201 (p-value of 0.09). This demonstrated that BMAL1 subsumed the observed relationship with time. Fitting the mixed effect model to the expression levels of BMAL1 showed that subject-to-subject variation was commensurate with the variability of the residual, a statistically significant increase in the expression level of BMAL-1 at the third visit compared to the two first visits (effect size of 7.43 × 10−3 (SE 3.04 × 10−3), p = 0.014), and a significant positive relationship between BMAL-1 and time of blood draw (effect size of 1.48 × 10−3 (SE= 0.74 × 10−3) p = 0.045). Taken together, these analyses reveal a relationship between the circadian-associated gene BMAL-1, time and log CA-US HIV RNA.
Changes in T-cell subsets, cortisol and TSH with time
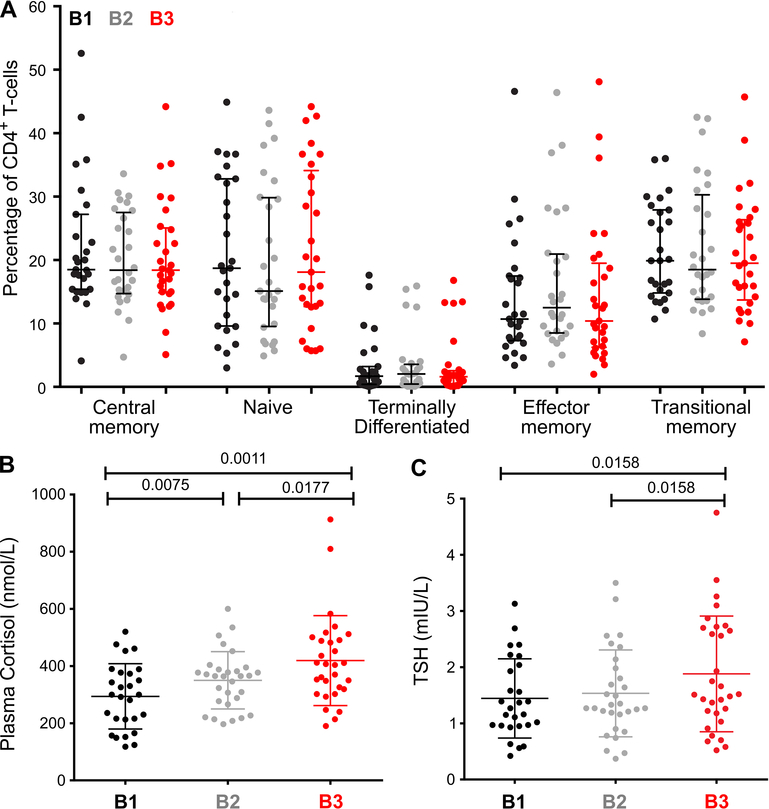
As cellular mobilization may contribute to increased frequency of T-cells with higher levels of CA-US HIV RNA, we next enumerated T-cell subsets by flow cytometry. The proportion of CD4+ or CD8+ T-cells did not vary between time-points, nor did the CD4:CD8 ratio (data not shown). In addition, the proportion of CD4+ T-cell subsets, including CD4+ TCM and TEM did not vary between the three time-points (Figure 3A) and there was no change in CD8+ T-cell subsets (data not shown). We also measured markers of CD4+ T-cell activation and observed increased expression of CD38+HLADR- on CD4+ and CD8+T-cells at B3 compared to B2 (p=0.016 and 0.040, respectively; Supplementary Figure 2). These data suggest that alteration in subsets was unlikely to explain the variation in HIV transcription.
Figure 3.
CD4+ T-cell composition and plasma cortisol and thyroid stimulating hormone (TSH) in HIV-infected individuals on suppressive ART. A: CD4+ T-cell subset proportions in participants are shown for n=28, 27 and 29 at B1, B2 and B3 respectively. The median and interquartile range are shown and comparisons are made with Wilcoxon matched pairs signed rank tests. B: Plasma cortisol and C: TSH in participants are shown for n=27 at B1 and 30 at B2 and B3. The mean and standard deviation are shown and comparisons are made with paired t-tests. The color of the symbol denotes the pre-intervention time-point. These were prior to receiving the study treatment (screening [B1; black]), immediately before the first dose of an intervention, here being disulfiram [B3; red], and a time-point between [B2; grey].
Given that B3 was collected immediately prior to administration of disulfiram and we saw a significant relationship between B3 and CA-US HIV RNA, we also considered whether acute stress may have contributed to changes in CA-US HIV RNA. Acute psychological stress induces a rise in cortisol and thyroid stimulating hormone (TSH) [25], whilst TSH may be low in severe stress [26]. Plasma cortisol measured at B3 was higher compared to B1 or B2 (Figure 3B) and TSH was higher at B3 compared to B1 or B2 (Figure 3C).
Mixed effects model of all measured variables and time
The mixed effects model for log CA-US HIV RNA showed an effect of visit and time-of-day (Table 1), as well as a relationship between log CA-US HIV RNA, time and the expression levels of BMAL-1. A similar mixed effects model was then fitted to all the other biomarkers measured in this study to determine if they had a relationship to time (Table 2). If time-of-day was a significant predictor for the observed expression level of CLOCK-related genes or hormones, then it is plausible that the relevant gene or hormone may drive the observed temporal variation in the expression levels of log CA-US HIV RNA. Only BMAL1 showed a statistically significant relationship with time (effect size of 8.51 (SE 3.78), p = 0.045). In addition, there was a weaker relationship between time and expression of Cry2, Per 3 and cortisol as measured by a generalized likelihood ratio test (p = 0.22, 0.38 and 0.14 respectively). In these models, the estimated parameters were nearly statistically different from zero. These biomarkers may become statistically significant if a larger dataset were available.
Table 2:
Mixed effect regression analysis to predict the expression levels of candidate CLOCK genes and stress hormones as a function of time of day. If time-of-day is a significant predictor for the observed expression level of candidate CLOCK genes or stress hormones, then it is plausible that the gene or hormone may drive the observed temporal variations in the expression levels of log CA-US HIV RNA.
| Effect estimate | Standard Error | T-value | P-value | |
|---|---|---|---|---|
| cortisol | −2.05E+01 | 1.09E+01 | −1.877 | 0.061 |
| TSH | −2.60E-02 | 6.29E-02 | −0.413 | 0.680 |
| CLOCK | 9.00E-05 | 3.40E-04 | 0.276 | 0.783 |
| BMAL1 | 1.48E-03 | 7.40E-04 | 2.004 | 0.045 |
| Per3 | −2.60E-04 | 1.40E-04 | −1.856 | 0.063 |
| Per2 | −1.00E-04 | 2.30E-04 | −0.425 | 0.671 |
| Per1 | 8.62E-03 | 5.56E-03 | 1.551 | 0.121 |
| Cry2 | 1.80E-04 | 9.00E-05 | 1.929 | 0.054 |
| Cry1 | 7.00E-04 | 5.20E-04 | 1.338 | 0.181 |
These findings do not mean that the above selected biomarkers do not have a circadian rhythm. Their lack of statistical significance could be the result from (1) confounding between visit and time of day, so the effect of time of day is lessened, and (2) our fitted model assumes a linear effect over a limited observation window, which may only hold approximately. The combination of these two effects may explain why only BMAL-1 had a significant nonzero coefficient for CA-US HIV RNA.
CLOCK and BMAL1 upregulate HIV LTR activity
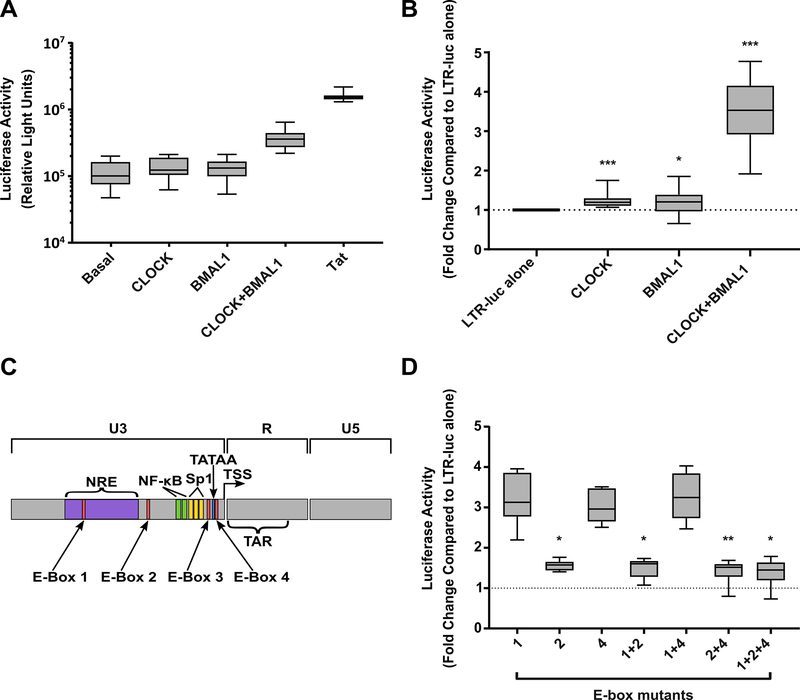
Given our finding of a relationship between CA-US HIV RNA and BMAL-1 and the presence of multiple E-boxes in the HIV LTR, we hypothesized that BMAL-1 may be acting directly on the HIV LTR to drive viral transcription. To address this, we transfected 293T cells with a plasmid encoding the luciferase gene under the control of the HIV LTR (HxB2) along with plasmids encoding for the human CLOCK and BMAL-1 proteins or the viral transactivator protein, Tat. Addition of Tat led to a 15.89-fold increase in luciferase activity, consistent with Tat enhancement of LTR transcription (Figure 4A). The presence of CLOCK (1.22-fold, p=0.004) or BMAL-1 (1.17-fold, p=0.018) alone lead to small but significant increases in luciferase activity (Figure 4B). However, the presence of CLOCK and BMAL-1 together led to a 3.32-fold increase in luciferase activity (p<0.0005), consistent with CLOCK and BMAL-1 acting as a heterodimer and upregulating HIV LTR activity.
Figure 4.
The circadian transcription factors CLOCK and BMAL1 upregulate HIV LTR activity. A: Raw luciferase activity in 293T cells stably transfected with an HIV-LTR (HXB2)-luciferase reporter and then co-transfected with CLOCK, BMAL1, CLOCK and BMAL1 or Tat. B: Fold change in luciferase activity. Comparisons are made to the HIV-LTR alone. C: Schematic of the HIV LTR highlighting the position of E-box elements. D: The effect of E-box mutations on upregulation of HIV LTR luciferase activity following co-transfection of CLOCK/BMAL-1. Fold change in luciferase activity of the mutated LTRs in the presence of CLOCK and BMAL1 compared to baseline expression. Box plots are shown which demonstrate the median, 25th and 75th percentiles (box) and minimum and maximum values (whiskers), n=13 (A,B) and n=5 (D) experiments are shown. Comparisons were made using a linear regression analysis. * p<0.05, **p<0.01, ***p<0.001. WT, wild type
To determine if CLOCK and BMAL-1 were directly binding to LTR and driving transcription, we mutated each E-box in the HxB2 LTR alone or in combination and assessed the ability of CLOCK and BMAL-1 to upregulate transcription from these LTRs. The HxB2 LTR has three intact E-boxes; E-box 1 (HxB2 nucleotides 151–156); E-box 2 (288–293); and E-box 4 (433–438) (Figure 4C). An additional E-box (E-box 3, 416–421), is not present in the HxB2 strain. Mutation of E-box 2 led to a 63% reduction in CLOCK/BMAL1 mediated LTR activity compared to the wild type LTR (p<0.001). Additional mutations of E-box 1, E-box 4 or both E1 and E4, led to a 69%, 77% and 72% reduction respectively (all p<0.001), did not increase this effect. Mutation of E-box 1 alone (p=0.361) and E-box 4 alone (p=0.627) had no significant impact on LTR activity. Restoration of E-box 3 in the HxB2 LTR did not further increase CLOCK/BMAL1 mediated LTR activity (p=0.183; (Figure 4D showing fold change luciferase expression with mutant LTR in the presence of CLOCK/BMAL-1). Taken together, this data strongly suggests that the circadian transcription factors CLOCK and BMAL-1 can drive HIV transcription via binding to the E-box 2 motif in the HIV LTR.
Discussion
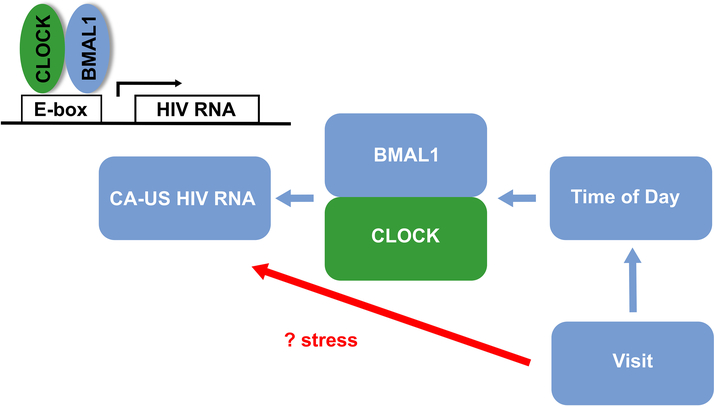
Cell-associated US HIV RNA is frequently detected in CD4+ T-cells from HIV-infected individuals on ART and is an important marker of HIV transcription. Here we show significant differences in CA-US HIV RNA and the relative expression of CLOCK-associated genes, specifically Per 1 and 2 and Cry 1 in blood collected at three time points from HIV-infected individuals on ART. Using a linear mixed effects model we found there was a highly significant association between CA-US HIV RNA and study visit (p<0.001), which was independent of time; and a modest relationship between CA-US HIV RNA with time and the circadian gene BMAL-1. Furthermore, given BMAL-1 forms a heterodimer with CLOCK which binds to a six-nucleotide motif, the E-box [14, 15] and there are multiple E-Boxes in the HIV-LTR sequence [16], using an in vitro reporter system, we demonstrated that CLOCK and BMAL-1 can activate HIV LTR driven gene expression. This activation was partially dependent on an E-box upstream of the NF-KB binding site. We propose a model where time impacts the level of circadian genes such as BMAL-1, which in combination with CLOCK can directly activate the HIV LTR. In addition, other factors can impact on CA-US HIV RNA that remain unexplained, but could potentially be related to stress (Figure 5).
Figure 5.
Proposed model for interaction of visit, time and circadian genes in HIV transcription on ART. Effects of visit alone on CA-US RNA remains unexplained but could potentially be secondary to stress. The CLOCK-BMAL-1 heterodimer binds to a six-nucleotide motif, the E-box [14, 15], to mediate transcription. HIV-1 is known to encode multiple E-Boxes in the LTR [16] and therefore changes in HIV transcription on ART, may plausibly be mediated directly through CLOCK-BMAL-1.
Other investigators have previously investigated the relationship between HIV transcription and time of day. In HIV-infected individuals not on ART, plasma HIV RNA has been previously shown to fluctuate significantly over a 24-hour period in one study [27] but not in another [28]. A previous study in HIV-infected individuals on suppressive ART suggested that CA-US HIV RNA was relatively stable over six months, but the time of day of blood collection was not accounted for in this study [29].
Our data demonstrates that CLOCK and BMAL1 can upregulate LTR activity likely by binding the E-box 2 motif. Each protein led to an increase in transcription but this was significantly enhanced when co-administered. Interestingly, ablation of E-box 2 markedly reduced but did not completely abrogate CLOCK/BMAL-1 upregulation. This may point to an additional indirect role where CLOCK/BMAL-1 complexes upregulate other genes which can affect HIV LTR activity. This could be a consequence of the histone acetyl transferase activity of CLOCK, which is enhanced by dimerization with BMAL-1 [17]. Given our results, the CLOCK/BMAL-1 axis provides a novel pathway for exploration of HIV latency reversal.
However, despite the findings of a relationship between HIV transcription and time and a direct effect of circadian proteins, we also found that the relationship between the B3 visit and CA-US HIV RNA was highly significant and independent of time. Stress could be one explanation given the participants in our study were all about to receive an interventional drug disulfiram at the B3 visit. The elevated TSH and cortisol at B3 could potentially support this interpretation, however both hormones are also known to undergo circadian variation [30], although only cortisol showed a trend towards being associated with time in this study.
Several lines of evidence suggest that physiological stress responses may influence HIV replication and latent infection. In vitro, hydrocortisone increases HIV-replication in activated PBMC [31]. Alternatively, the effects of hydrocortisone may be mediated by altering production of cytokines that can regulate HIV transcription. The neuroendocrine system may also influence HIV replication and latency via the autonomic nervous system (ANS). Heightened ANS reactivity is associated with greater plasma HIV RNA among HIV-infected men [32] and norepinephrine increases HIV replication in vitro 5- to 10-fold [33].
In summary, we demonstrate natural variation in CA-US HIV RNA in CD4+ T-cells in HIV-infected individuals on suppressive ART as well as a clear effect of the circadian proteins CLOCK and BMAL-1 on driving transcription from the LTR by binding to E-box2. This pathway could potentially be exploited to enhance the effects of or finding novel latency reversing agents. The relationship between CA-US HIV RNA and study visit was highly significant and remains unexplained suggesting that other unmeasured factors other than time, are likely contributing to the level of CA-US HIV RNA. Further work is needed to understand the biological basis of the natural variation in CA-US HIV RNA in individuals on ART.
Supplementary Material
Acknowledgments
We acknowledge the patients and staff at Alfred Hospital, UCSF, and The Peter Doherty Institute for Infection and Immunity. This study was supported by the National Institutes of Health (NIH) Delaney AIDS Research Enterprise (DARE U19 AI096109 and UM1 AI126611–01), NIH grants R01-AI028433 and R01-OD011095 (ASP), The American Foundation for AIDS Research (amFAR) Research consortium on HIV eradication (ARCHE) and the National Health and Medical Research Council (NHMRC) of Australia (CCC, MR and JHE – Early Career Fellowships; and SRL - Practitioner Fellowship). These data were presented in part at the International AIDS Society Conference, Vancouver, Canada on 20 July 2015 - MOAA0106LB) and the Australasian 2017 HIV&AIDS Conference, Canberra, Australia on 6 November 2017.
Footnotes
The authors have declared that no conflict of interest exists.
References
- 1.Chun TW, Carruth L, Finzi D, Shen X, DiGiuseppe JA, Taylor H, et al. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 1997; 387(6629):183–188. [DOI] [PubMed] [Google Scholar]
- 2.Lewin SR, Rouzioux C. HIV cure and eradication: how will we get from the laboratory to effective clinical trials? Aids 2011; 25(7):885–897. [DOI] [PubMed] [Google Scholar]
- 3.Eriksson S, Graf EH, Dahl V, Strain MC, Yukl SA, Lysenko ES, et al. Comparative analysis of measures of viral reservoirs in HIV-1 eradication studies. PLoS pathogens 2013; 9(2):e1003174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewin SR, Vesanen M, Kostrikis L, Hurley A, Duran M, Zhang L, et al. Use of real-time PCR and molecular beacons to detect virus replication in human immunodeficiency virus type 1-infected individuals on prolonged effective antiretroviral therapy. J Virol 1999; 73(7):6099–6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Archin NM, Bateson R, Tripathy MK, Crooks AM, Yang KH, Dahl NP, et al. HIV-1 expression within resting CD4+ T cells after multiple doses of vorinostat. J Infect Dis 2014; 210(5):728–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Archin NM, Liberty AL, Kashuba AD, Choudhary SK, Kuruc JD, Crooks AM, et al. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature 2012; 487(7408):482–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elliott JH, McMahon JH, Chang CC, Lee SA, Hartogensis W, Bumpus N, et al. Short-term administration of disulfiram for reversal of latent HIV infection: a phase 2 dose-escalation study. Lancet HIV 2015; 2(12):e520–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elliott JH, Wightman F, Solomon A, Ghneim K, Ahlers J, Cameron MJ, et al. Activation of HIV transcription with short-course vorinostat in HIV-infected patients on suppressive antiretroviral therapy. PLoS pathogens 2014; 10(10):e1004473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rasmussen TA, Tolstrup M, Brinkmann CR, Olesen R, Erikstrup C, Solomon A, et al. Panobinostat, a histone deacetylase inhibitor, for latent-virus reactivation in HIV-infected patients on suppressive antiretroviral therapy: a phase 1/2, single group, clinical trial. Lancet HIV 2014; 1(1):e13–21. [DOI] [PubMed] [Google Scholar]
- 10.Sogaard OS, Graversen ME, Leth S, Olesen R, Brinkmann CR, Nissen SK, et al. The Depsipeptide Romidepsin Reverses HIV-1 Latency In Vivo. PLoS pathogens 2015; 11(9):e1005142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ke R, Lewin SR, Elliott JH, Perelson AS. Modeling the Effects of Vorinostat In Vivo Reveals both Transient and Delayed HIV Transcriptional Activation and Minimal Killing of Latently Infected Cells. PLoS pathogens 2015; 11(10):e1005237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dahabieh MS, Battivelli E, Verdin E. Understanding HIV latency: the road to an HIV cure. Annual review of medicine 2015; 66:407–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cermakian N, Sassone-Corsi P. Multilevel regulation of the circadian clock. Nat Rev Mol Cell Biol 2000; 1(1):59–67. [DOI] [PubMed] [Google Scholar]
- 14.Darlington TK, Wager-Smith K, Ceriani MF, Staknis D, Gekakis N, Steeves TD, et al. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science 1998; 280(5369):1599–1603. [DOI] [PubMed] [Google Scholar]
- 15.Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, et al. Role of the CLOCK protein in the mammalian circadian mechanism. Science 1998; 280(5369):1564–1569. [DOI] [PubMed] [Google Scholar]
- 16.Ou SH, Garcia-Martinez LF, Paulssen EJ, Gaynor RB. Role of flanking E box motifs in human immunodeficiency virus type 1 TATA element function. J Virol 1994; 68(11):7188–7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doi M, Hirayama J, Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell 2006; 125(3):497–508. [DOI] [PubMed] [Google Scholar]
- 18.Pasternak AO, Adema KW, Bakker M, Jurriaans S, Berkhout B, Cornelissen M, et al. Highly sensitive methods based on seminested real-time reverse transcription-PCR for quantitation of human immunodeficiency virus type 1 unspliced and multiply spliced RNA and proviral DNA. Journal of clinical microbiology 2008; 46(7):2206–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ackermann K, Dehghani F, Bux R, Kauert G, Stehle JH. Day-night expression patterns of clock genes in the human pineal gland. J Pineal Res 2007; 43(2):185–194. [DOI] [PubMed] [Google Scholar]
- 20.Yang SL, Yu C, Jiang JX, Liu LP, Fang X, Wu C. Hepatitis B virus X protein disrupts the balance of the expression of circadian rhythm genes in hepatocellular carcinoma. Oncol Lett 2014; 8(6):2715–2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gray LR, Cowley D, Crespan E, Welsh C, Mackenzie C, Wesselingh SL, et al. Reduced basal transcriptional activity of central nervous system-derived HIV type 1 long terminal repeats. AIDS Res Hum Retroviruses 2013; 29(2):365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gray LR, Cowley D, Welsh C, Lu HK, Brew BJ, Lewin SR, et al. CNS-specific regulatory elements in brain-derived HIV-1 strains affect responses to latency-reversing agents with implications for cure strategies. Mol Psychiatry 2016; 21(4):574–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akhtar RA, Reddy AB, Maywood ES, Clayton JD, King VM, Smith AG, et al. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr Biol 2002; 12(7):540–550. [DOI] [PubMed] [Google Scholar]
- 24.Curtis AM, Bellet MM, Sassone-Corsi P, O’Neill LA. Circadian clock proteins and immunity. Immunity 2014; 40(2):178–186. [DOI] [PubMed] [Google Scholar]
- 25.Richter SD, Schurmeyer TH, Schedlowski M, Hadicke A, Tewes U, Schmidt RE, et al. Time kinetics of the endocrine response to acute psychological stress. J Clin Endocrinol Metab 1996; 81(5):1956–1960. [DOI] [PubMed] [Google Scholar]
- 26.Sumita S, Ujike Y, Namiki A, Watanabe H, Kawamata M, Watanabe A, et al. Suppression of the thyrotropin response to thyrotropin-releasing hormone and its association with severity of critical illness. Crit Care Med 1994; 22(10):1603–1609. [PubMed] [Google Scholar]
- 27.Zeichner SL, Mueller BU, Pizzo PA, Dimitrov DS. Kinetics of HIV-1 RNA concentration changes in pediatric patients. Pathobiology : journal of immunopathology, molecular and cellular biology 1996; 64(6):289–294. [DOI] [PubMed] [Google Scholar]
- 28.Deeks SG, Coleman RL, White R, Pachl C, Schambelan M, Chernoff DN, et al. Variance of plasma human immunodeficiency virus type 1 RNA levels measured by branched DNA within and between days. J Infect Dis 1997; 176(2):514–517. [DOI] [PubMed] [Google Scholar]
- 29.Leth S, Nymann R, S JO, Olesen R, Rasmussen TA, Ostergaard L, et al. HIV-1 transcriptional activity during frequent longitudinal sampling in aviremic patients on ART. Aids 2015. [DOI] [PubMed] [Google Scholar]
- 30.Collomp K, Baillot A, Forget H, Coquerel A, Rieth N, Vibarel-Rebot N. Altered diurnal pattern of steroid hormones in relation to various behaviors, external factors and pathologies: A review. Physiology & behavior 2016; 164(Pt A):68–85. [DOI] [PubMed] [Google Scholar]
- 31.Markham P, Salahuddin S, Veren K, Orndorff S, Gallo R. Hydrocortisone and some other hormones enhance the expression of HTLV-III. International journal of cancer Journal international du cancer 1986; 37(1):67–72. [DOI] [PubMed] [Google Scholar]
- 32.Cole S, Naliboff B, Kemeny M, Griswold M, Fahey J, Zack J. Impaired response to HAART in HIV-infected individuals with high autonomic nervous system activity. Proc Natl Acad Sci U S A 2001; 98(22):12695–12700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cole SW, Korin YD, Fahey JL, Zack JA. Norepinephrine accelerates HIV replication via protein kinase A-dependent effects on cytokine production. J Immunol 1998; 161(2):610–616. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.