SUMMARY
RNA-binding proteins Lin28a/b regulate cellular growth and tissue regeneration. Here, we investigated the role of Lin28 in the control of axon regeneration in postmitotic neurons. We find that Lin28a/b are both necessary and sufficient for supporting axon regeneration in mature sensory neurons through their regulatory partners, let-7 microRNAs (miRNAs). More importantly, overexpression of Lin28a in mature retinal ganglion cells (RGCs) produces robust and sustained optic nerve regeneration. Additionally, combined overexpression of Lin28a and downregulation of Pten in RGCs act additively to promote optic nerve regeneration, potentially by reducing the backward turning of regenerating RGC axons. Our findings not only reveal a vital role of Lin28 signaling in regulating mammalian axon regeneration but also identify a signaling pathway that can promote axon regeneration in the central nervous system (CNS).
In Brief
Axon regeneration in the mammalian CNS is a challenge. Wang et al. show that the Lin28/let-7 axis plays an important role in governing mammalian axon regeneration in the peripheral nervous system. More importantly, overexpression of Lin28a induces robust and sustained axon regeneration in the CNS.
Graphical Abstract
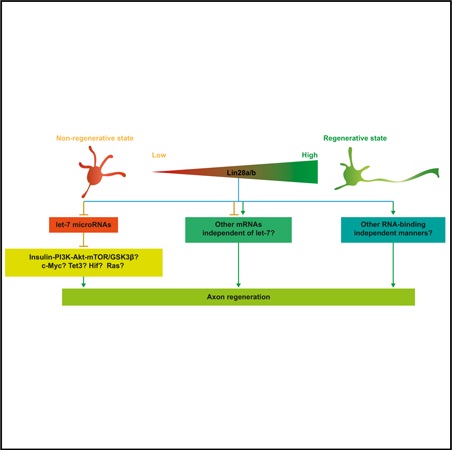
INTRODUCTION
Axon regeneration in the mammalian CNS has always been a challenge in biomedical studies. Patients suffering from spinal cord injury, traumatic brain injury, glaucoma, and various neurodegenerative diseases, including Parkinson’s disease, Alzheimer’s disease, amyotrophic lateral sclerosis, and multiple sclerosis, would greatly benefit from the discovery and development of approaches that can successfully help CNS axons regenerate. CNS neurons are terminally differentiated cells that have lost a variety of intrinsic abilities supporting axon growth during maturation. Recent studies have identified Klf4 and c-Myc, two reprogramming factors that can work together with other cohorts to reprogram differentiated cells back to induced pluripotent stem cells (iPSCs), as important regulators of axon regeneration (Belin et al., 2015; Moore et al., 2009). Sox11, a member of the Sox family to which another reprogramming factor, Sox2 belongs, has also been reported to promote axon regeneration in peripheral nerves (Jankowski et al., 2009; Jing et al., 2012) and non-a-retinal ganglion cells (RGCs) (Norsworthy et al., 2017). Thus, it is likely that, by manipulating reprogramming factors, mature CNS neurons can regain the intrinsic ability to support axon regeneration after nerve injury.
Lin28 RNA-binding protein, originally discovered in C. elegans as a heterochronic regulator of larval development (Ambros and Horvitz, 1984; Moss et al., 1997), has also been demonstrated to enhance the efficiency of the reprogramming from human somatic cells to iPSCs (Yu et al., 2007). A previous study in plants reported that PpCSP1, a Lin28 homolog in the moss Physcomitrella patens, can reprogram differentiated leaf cells to stem cells (Li et al., 2017), indicating that the cellular reprogramming function of Lin28 is evolutionarily conserved from plants to mammals. Recent studies have shown that Lin28a/b, two paralogs in vertebrates, play vital roles in many growth associated functions, such as body size and puberty control (Shinoda et al., 2013; Zhu et al., 2010), glucose metabolism (Shinoda et al., 2013; Zhu et al., 2011), and tissue regeneration (Shyh-Chang et al., 2013). In zebrafish, the reactivation of Lin28 was found to be essential for Muller glia dedifferentiation and€ cell cycle re-entry to become new neurons after retinal injury (Ramachandran et al., 2010). Abnormal expression of Lin28a/b has also been linked to different kinds of tumors (Nguyen et al., 2014; Tu et al., 2015; Urbach et al., 2014; Viswanathan et al., 2009; Wang et al., 2015; West et al., 2009). To our knowledge, the roles of Lin28 signaling in postmitotic neurons were only explored in three studies (Amen et al., 2017; Bhuiyan et al., 2013; Huang et al., 2012). Lin28a/b mainly functions through let-7 microRNA (miRNA) family-dependent manners by posttranscriptionally blocking the biogenesis of mature let-7 miRNAs (Heo et al., 2008; Nam et al., 2011; Newman et al., 2008; Rybak et al., 2008; Thornton et al., 2012; Viswanathan et al., 2008). Several let-7-independent mechanisms have also been discovered recently (Balzer et al., 2010; Shyh-Chang et al., 2013; Zeng et al., 2016). In C. elegans, let-7 has been reported to negatively regulate axon regeneration in anterior ventral microtubule (AVM) neurons (Zou et al., 2013). In mammals, although glial let-7 miRNAs have been shown to inhibit axon regeneration by targeting nerve growth factor in Schwann cells (Li et al., 2015), the function of neuronal let-7 miRNAs in axon regeneration remains unclear.
In our study, we found that Lin28a/b upregulation was rapidly induced in neurons of the peripheral nervous system (PNS) after axon injury to support axon regeneration through downregulation of let-7 miRNAs. As a result, downregulation of Lin28a/b or upregulation of let-7a resulted in reduced sensory axon regeneration in vitro and in vivo. Conversely, overexpression of Lin28a or Lin28b or inhibition of let-7 miRNAs enhanced PNS sensory axon regeneration in vivo. Most importantly, we showed that overexpression of Lin28a produced robust and persistent CNS axon regeneration after optic nerve injury. In addition, we found that upregulation of Lin28a/b led to Akt activation and GSK3β inactivation in mature sensory neurons as well as increased Ser235/ 236 phosphorylation of ribosomal protein S6 (pS6) in both sensory neurons and RGCs. We also showed that Lin28b overexpression increased the levels of c-Myc and Tet3 in dorsal root ganglia (DRGs). Last, combined Lin28a overexpression and Pten knockdown had an additive effect on promoting optic nerve regeneration, potentially through reducing the backward turning of regenerating axons. Our findings not only revealed Lin28a/b as positive regulators of mammalian PNS and CNS axon regeneration but also suggest that modulation of reprogramming factors might be an effective way to promote axon regeneration in mature CNS neurons.
RESULTS
Upregulation of Lin28a/b Is Both Necessary and Sufficient for Sensory Axon Regeneration In Vivo
We first tested the expression of Lin28a/b in mouse DRGs during development. CF-1 or CD-1 IGS mice were used in all DRG experiments unless otherwise stated. Lumbar 4 and 5 (L4/5) DRGs were dissected out from mice on embryonic day 15 (E15), E18, postnatal day 0 (P0), P3, P7, P14, P21, P28, and P56, and total RNA was isolated. We found that the mRNA levels of Lin28a/b dropped sharply from E15 to birth and remained low through adulthood (Figures S1A and S1B), indicating that Lin28a/b may be important in regulating axon growth of sensory neurons during early development. We next investigated whether the expression of Lin28a/b in sensory neurons changes in response to axon injury. Bilateral sciatic nerve axotomy or sham surgery was performed on mice. Seven days after the surgery, total RNA was isolated from L4/5 DRGs. The levels of Lin28a/b mRNAs were significantly upregulated in L4/5 DRGs following sciatic nerve injury (SNI) (Figure 1A). We also observed upregulated protein levels of Lin28a 1 or 3 days after SNI (Figures 1B and 1C). The anti-Lin28b antibody we used did not work well with mouse DRG tissue extracts. These results suggest that Lin28a/b might be positive regulators of sensory axon regeneration.
Figure 1. Upregulation of Lin28a/b Is Both Necessary and Sufficient for Sensory Axon Regeneration In Vivo.
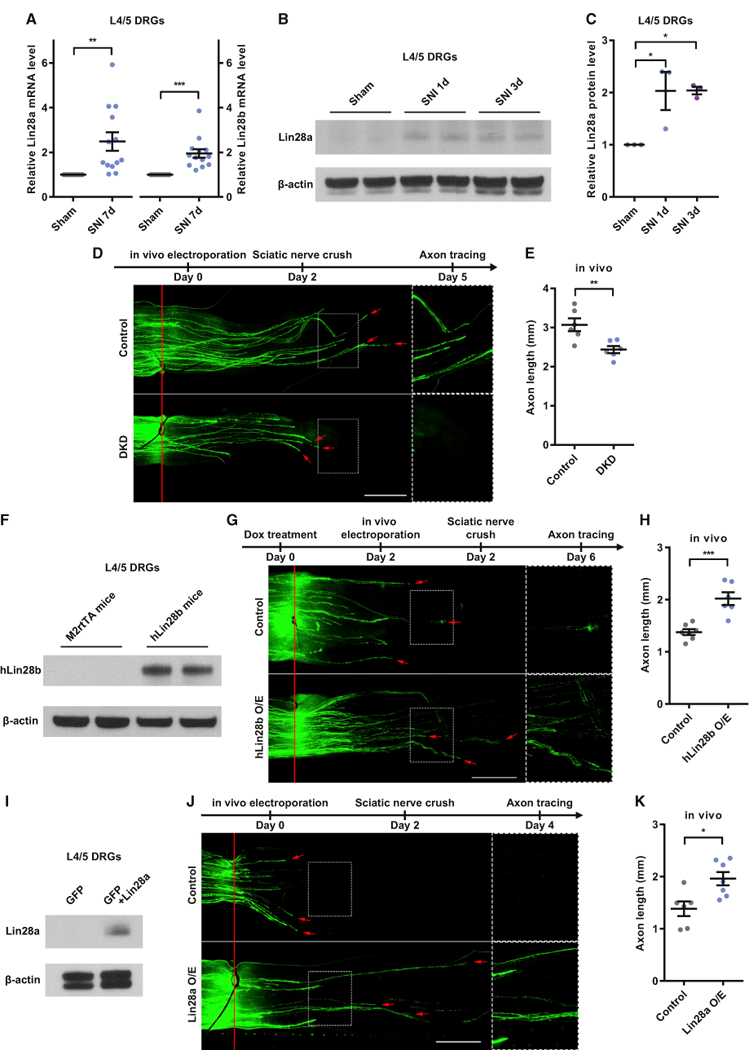
(A) Lin28a/b mRNA levels were significantly increased in L4/5 DRGs 7 days after SNI (one-sample t test, p = 0.0039 for Lin28a, p = 0.0003 for Lin28b, n = 13 independent experiments).
(B) Representative western blot result showing a markedly increased Lin28a protein level in L4/5 DRGs 1 or 3 days after SNI.
(C) Quantification of (B) (one-way ANOVA followed by Tukey’s multiple comparisons test, p = 0.0211, n = 3 independent experiments).
(D) Top: timeline of the experiment. Bottom: representative images showing that simultaneous knockdown of Lin28a/b impaired sensory axon regeneration in vivo. The right column shows enlarged images of the areas in the dashed white boxes on the left. The red line indicates the crush sites. Red arrows indicate axonal tips. Scale bar, 1 mm (left) and 0.5 mm (right).
(E) Quantification of (D) (unpaired Student’s t test, p = 0.0064, n = 6 mice in each group)
(F) Representative western blot result showing a markedly increased hLin28b protein level in L4/5 DRGs of hLin28b mice 2 days after induction of hLin28b by dox.
(G) Top: timeline of the experiment. Bottom: representative images showing that hLin28b overexpression promoted sensory axon regeneration in vivo. The right column shows enlarged images of the areas in the dashed white boxes on the left. The red line indicates the crush sites. Red arrows indicate axonal tips. Scale bar, 1 mm (left), 0.5 mm (right).
(H) Quantification of (G) (unpaired Student’s t test, p = 0.0004, n = 7 and 6 mice in the control group and hLin28b overexpression group, respectively).
(J) Top: timeline of the experiment. Bottom: representative images showing that Lin28a overexpression promoted sensory axon regeneration in vivo. The right column shows enlarged images of the areas in the dashed white boxes on the left. The red line indicates the crush sites. Red arrows indicate axonal tips. Scale bar, 1 mm (left), 0.5 mm (right).
(K) Quantification of (J) (unpaired Student’s t test, p = 0.0109, n = 6 and 7 mice in the control group and Lin28a overexpression group, respectively).
*p < 0.05, **p < 0.01, ***p < 0.001. Data are represented as mean ± SEM. KD, knockdown; DKD, double knockdown of Lin28a/b; O/E, overexpression. See also Figure S1
To further determine the functional roles of Lin28a/b in axon regeneration, we knocked down Lin28a and/or Lin28b in dissociated adult DRG neurons by electroporation of a GFP plasmid plus Lin28a small interfering RNA (siRNA) (siLin28a) and/or Lin28b siRNA (siLin28b) and cultured the neurons for 3 days. Electroporation of the GFP plasmid only was used as a control condition. The result showed that, compared with the control, single knockdown of either Lin28a or Lin28b had no effect on sensory axon regeneration, whereas double knockdown of Lin28a/b significantly inhibited sensory axon regeneration in vitro (Figures S1C and S1D), indicating that Lin28a/b play redundant roles in supporting sensory axon regeneration, resembling the discovery in the moss Physcomitrella patens that only quadruple deletion of all four closely related PpCSP genes results in attenuated function (Li et al., 2017). The efficacy of siLin28a or siLin28b was verified by detecting the mRNA levels of Lin28a and/or Lin28b in cultured DRG neurons and the protein levels in Cath.-a-differentiated (CAD) cells 3 days after the transfection of siRNAs. The results of both experiments demonstrated that siLin28a and siLin28b successfully brought down the level of the respective mRNA (Figure S1E) or protein (Figure S1F). According to our previous study, the transfection efficiency of siRNAs using in vitro electroporation is over 95% in adult DRG neurons (Jiang et al., 2015), so nearly all GFP+ neurons were also successfully transfected with siRNAs.
To extend the finding into in vivo model, we transfected siLin28a and/or siLin28b with the GFP plasmid into the left L4/5 DRGs of mice using in vivo electroporation, and a sciatic nerve crush (SNC) was performed on the left side of each mouse 2 days later. Control mice were transfected with the GFP plasmid only or the GFP plasmid plus non-targeting siRNA. A previous study found that DRG neurons switch into fast growth mode approximately 2 days after axon injury (Smith and Skene, 1997). Similarly, our unpublished data show that axon regeneration in the sciatic nerve reaches the fastest rate on the third day after the SNC (data not shown). Therefore, the left sciatic nerve of each mouse was collected 3 days after the crush. The lengths of all GFP+ regenerating axons in the whole-mount nerve were measured. Over 96% of neurons in a DRG were successfully transfected with siRNA by in vivo electroporation (Figures S1G and S1H), indicating that almost every GFP+ sensory neuron was also successfully transfected with siRNA. The result showed that, compared with GFP plasmid transfection only, neither nontargeting siRNA nor single knockdown of Lin28a or Lin28b had any effect on sensory axon regeneration (Figures S1I and S1J), whereas simultaneously knocking down Lin28a/b impaired sensory axon regeneration in vivo (Figures 1D and 1E), demonstrating that the SNI-triggered upregulation of Lin28a/b is necessary for in vivo axon regeneration.
We then explored whether overexpressing Lin28a or Lin28b sufficiently promotes sensory axon regeneration. Because single knockdown of Lin28a or Lin28b did not impair sensory axon regeneration, indicating that either Lin28a or Lin28b alone is sufficient to support axon regeneration, we first used a mouse strain carrying a copy of the human form Lin28b (hLin28b), which can be induced by a tetracycline transactivator, rtTA, located downstream of the Gt(Rosa26)Sor promoter (hLin28b mice). Littermates with the rtRA transgene only (M2rtTA mice) were used as a control (for details, see STAR Methods). After 2 days of treatment with the tetracycline analog doxycycline (dox), a high level of hLin28b was induced in L4/5 DRGs of hLin28b mice (Figure 1F). On the third day of dox treatment, we electroporated the GFP plasmid into L4/5 DRGs of M2rtTA mice and hLin28b mice to label the axons and performed the SNC 2 days later. As mentioned above, within the first 2 days after nerve injury, sensory axons regenerate at a lower rate (Smith and Skene, 1997). In addition, the injury-triggered spontaneous upregulation of Lin28a is already very strong 3 days after SNI (Figures 1B and 1C). Thus, the nerves were harvested 2 days after the SNC to evaluate the promoting effect. The result showed that hLin28b overexpression significantly enhanced sensory axon regeneration in vivo (Figures 1G and 1H). To rule out the possibility that such an effect was caused by hLin28b overexpression in other cells in the sciatic nerve, we specifically overexpressed Lin28a in L4/5 DRGs by in vivo electroporation of pCMV-Lin28a (Figure 1I). Control mice were electroporated with the GFP plasmid. Two days after the SNC, the regenerating axons overexpressing Lin28a were significantly longer than the control (Figures 1J and 1K). These results clearly demonstrate that overexpression of either Lin28a or Lin28b sufficiently promotes sensory axon regeneration in vivo. The potentiated axon growth ability is comparable with a previous study using the same model (Cho et al., 2013).
let-7 miRNAs Act Downstream of Lin28a/b to Regulate Sensory Axon Regeneration
As a renowned regulatory partner of Lin28a/b (Viswanathan et al., 2008), let-7 miRNA has been shown to be a negative regulator of axon regeneration in C. elegans (Zou et al., 2013). However, the role of the mammalian let-7 miRNA family in axon regeneration has never been elucidated. We found that, 7 days after SNI, the levels of let-7a/b in L4/5 DRGs were significantly downregulated (Figure 2A), indicating that low levels of let-7 miRNAs in sensory neurons might be essential for sensory axon regeneration. To explore the function of let-7 miRNAs in axon regeneration, we electroporated a let-7a or let-7b mimic plus the GFP plasmid into dissociated adult DRG neurons. Control neurons were only transfected with the GFP plasmid. The data in Figure S2A confirm the overexpression of mature let-7a or let-7b in the cultured DRG neurons 3 days after electroporation. After culturing the DRG neurons for 3 days, we discovered that overexpression of either let-7a or let-7b drastically impaired sensory axon regeneration in vitro (Figures S2B and S2C). Again, to confirm this finding in the in vivo model, we electroporated the let-7a mimic plus the GFP plasmid into L4/5 DRGs 2 days before the SNC and analyzed the axon lengths 3 days after the SNC. Control mice were electroporated with the GFP plasmid only or the GFP plasmid plus non-mammalian targeting miRNA. The result showed that, compared with GFP only, non-mammalian targeting miRNA had no effect on sensory axon regeneration (Figures S2D and S2E), whereas let-7a overexpression significantly reduced sensory axon regeneration in vivo (Figures 2B and 2C). The mature let-7a level was confirmed to be dramatically elevated in L4/5 DRGs 2 days after in vivo electroporation of the let-7a mimic (Figure 2D). These results demonstrate that mammalian let-7 miRNAs are negative regulators of axon regeneration.
Figure 2. let-7 miRNAs Act Downstream of Lin28a/b to Regulate Sensory Axon Regeneration.
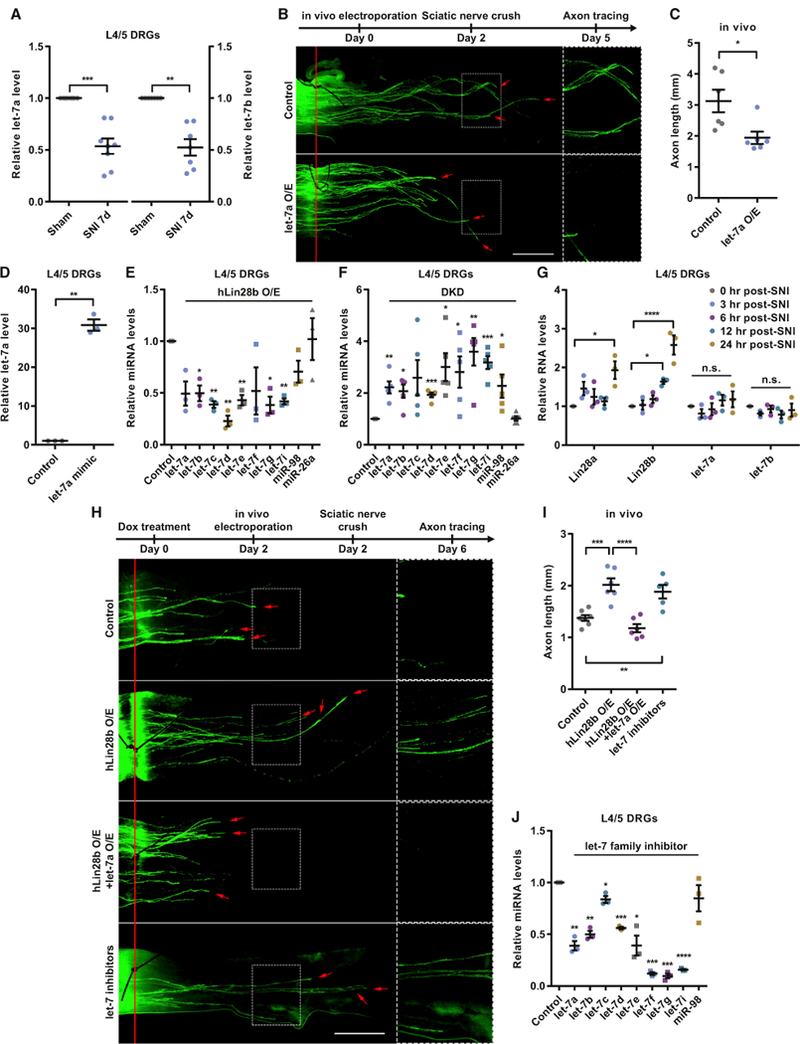
(A) let-7a/b levels were significantly decreased in L4/5 DRGs 7 days after SNI (one-sample t test, p = 0.0004 for let-7a, p = 0.0010 for let-7b, n = 8 and 7 independent experiments for let-7a and let-7b, respectively).
(B) Top: timeline of the experiment. Bottom: representative images showing that let-7a O/E impaired sensory axon regeneration in vivo. The right column shows enlarged images of the areas in the dashed white boxes on the left. The red line indicates the crush sites. Red arrows indicate axonal tips. Scale bar, 1 mm (left), 0.5 mm (right).
(C) Quantification of (B) (unpaired Student’s t test, p = 0.0175, n = 6 mice in each group).
(D) The let-7a level in L4/5 DRGs was significantly increased 2 days after in vivo electroporation of the let-7a mimic (one-sample t test, p = 0.0023, n = 3 independent experiments).
(E) The levels of let-7 miRNAs in L4/5 DRGs of hLin28b mice were decreased 2 days after induction of hLin28b by dox (one-sample t test; p = 0.0501, 0.0221,0.0029, 0.0045, 0.0074, 0.1702, 0.0167, 0.0124, 0.1090, and 0.9294 for let-7a, let-7b, let-7c, let-7d, let-7e, let-7f, let-7g, let-7i, miR-98, and miR-26a, respectively; n = 3 independent experiments).
(F) The levels of let-7 miRNAs in L4/5 DRGs were increased 2 days after DKD of Lin28a/b (one-sample t test; p = 0.0064, 0.0135, 0.0819, 0.0005, 0.0202, 0.0402,0.0082, 0.0009, 0.0474, and 0.8325 for let-7a, let-7b, let-7c, let-7d, let-7e, let-7f, let-7g, let-7i, miR-98, and miR-26a, respectively; n = 5 independent experiments).
(G) The increase of Lin28a/b mRNA levels occurred within 12–24 hr after SNI, whereas no significant change in let-7a or let-7b level was seen within 24 hr after SNI (one-way ANOVA followed by Tukey’s multiple comparisons test, p = 0.0164 for Lin28a, p < 0.0001 for Lin28b, p = 0.3634 for let-7a, p = 0.5388 for let-7b, n = 3 independent experiments).
(H) Top: timeline of the experiment. Bottom: representative images showing that let-7a O/E abolished the enhanced sensory axon regeneration induced byhLin28b O/E in vivo and that knocking down let-7 miRNAs sufficiently promoted sensory axon regeneration in vivo. The right column shows enlarged images of the areas in the dashed white boxes on the left. The red line indicates the crush sites. Red arrows indicate axonal tips. Scale bar, 1 mm (left), 0.5 mm (right).
(I) Quantification of (H) (one-way ANOVA followed by Tukey’s multiple comparisons test, p < 0.0001, n = 7 mice in the control group, n = 5 mice in the let-7 inhibitors group, n = 6 mice in the other two groups). Note that the control group and the hLin28b O/E group are identical to those in Figure 1G.
(J) Levels of let-7 miRNAs in L4/5 DRGs were decreased 2 days after in vivo electroporation of let-7 miRNA family inhibitors (one-sample t test; p = 0.0055, 0.0046, 0.0386, 0.0007, 0.0238, 0.0002, 0.0006, and 0.3509 for let-7a, let-7b, let-7c, let-7d, let-7e, let-7f, let-7g, and miR-98, respectively; p < 0.0001 for let-7i, n = 3 independent experiments).
n.s., not significant. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 compared with the control if not designated. Data are represented as mean ± SEM. See also Figure S2.
We next investigated the relationship between Lin28a/b and the let-7 miRNA family in the regulation of axon regeneration. We found that the levels of almost all let-7 miRNAs were significantly decreased in L4/5 DRGs of hLin28b mice compared with M2rtTA mice after 2 days of dox treatment (Figure 2E), consistent with the roles of Lin28a/b in suppressing mature let-7 biogenesis. Conversely, increased levels of let-7 miRNAs were detected in L4/5 DRGs 2 days after electroporation of siLin28a and siLin28b (Figure 2F). No change in the level of miR-26a, a non-let-7 family miRNA, was seen under either condition (Figures 2E and 2F). Because Lin28a/b and let-7 miRNAs usually respond to each other and form a double-negative feedback loop (Viswanathan and Daley, 2010), we examined which one of them responds first to SNI and causes the subsequent change of the other one. We collected L4/5 DRGs 0, 3, 6, 12, and 24 hr after SNI and tested the levels of Lin28a/b mRNAs and let7a/b. The result showed that the Lin28b mRNA level started to escalate as early as 12 hr post-SNI and that both Lin28a/b mRNAs were significantly increased 24 hr post-SNI (Figure 2G). In contrast, let-7a and let-7b remained at the baseline level throughout the 24 hr (Figure 2G), indicating that SNI first induced the upregulation of Lin28a/b, which then caused the subsequent downregulation of let-7 miRNAs. Thus, we think that Lin28a/b upregulation acts upstream of let-7 downregulation in adult sensory neurons to regulate axon regeneration in response to SNI.
To test this hypothesis, we overexpressed let-7a in L4/5 DRGs of dox-treated hLin28b mice via in vivo electroporation and made the SNC 2 days later. Sensory axon regeneration in these mice was analyzed 2 days after the SNC. The result showed that let7a overexpression significantly blocked sensory axon regeneration promoted by hLin28b overexpression (Figures 2H and 2I). Furthermore, we tested whether direct inhibition of let-7 miRNAs sufficiently promotes sensory axon regeneration. Two days after in vivo electroporation of the let-7 miRNA family inhibitor in L4/5 DRGs, the levels of most let-7 miRNAs were significantly knocked down (Figure 2J). We found that, 2 days after the SNC, sensory axon regeneration was boosted to a degree comparable with hLin28b overexpression, indicating that knockdown of let-7 miRNAs in L4/5 DRGs can phenocopy hLin28b overexpression (Figures 2H and 2I). These results provide strong evidence that Lin28a/b support sensory axon regeneration through their inhibition of the let-7 miRNA family.
Upregulation of Lin28a in RGCs Induces Robust and Sustained Optic Nerve Regeneration
In light of the promising promoting effect of Lin28a/b on PNS sensory axon regeneration, we hypothesized that their overexpression might enhance CNS axon regeneration as well. Therefore, we introduced the optic nerve regeneration model into our study. We crushed the right optic nerves of M2rtTA and hLin28b mice on the day dox treatment started. Because over half of the hLin28b mice would die on the tenth day of hLin28b induction, probably because of a glucose metabolism defect (Zhu et al., 2011), we assessed optic nerve regeneration 9 days after the optic nerve crush (ONC). In fact, one hLin28b mouse survived for 2 weeks (Figure S3A). The RGC axons were anterogradely labeled with Alexa 594-conjugated cholera toxin subunit B (CTB), which was injected into the vitreous humor 2 days prior to tissue harvest. The fixed whole-mount nerves underwent a tissue clearing procedure and were imaged with confocal microscopy. Only a very limited number of axons crossed the crush site in the control mice 9 days after the ONC. On the contrary, hLin28b overexpression induced evident optic nerve regeneration (Figures S3A and S3B). Induction of hLin28b in the retinae of hLin28b mice was confirmed after 2 days of dox treatment (Figure S3C). To exclude the likelihood that such an effect was the result of global overexpression of hLin28b and to investigate whether stronger regeneration can be stimulated by extending the length of time and keeping the mice in a healthy state without metabolic disorder, we injected AAV2-GFP or AAV2-Lin28a-FLAG into the vitreous humors of C57BL/6 mice and performed the ONC 2 weeks after the injection. Two weeks after the ONC, the nerves were collected, and axon regeneration was evaluated. The virus transduction rate was about 88% (Figures S3D and S3F), indicating that most RGCs overexpressed Lin28a. We found that, compared with the GFP group, robust optic nerve regeneration was produced by Lin28a overexpression (Figures 3A and3B). To further explore whether Lin28a overexpression can induce sustainable optic nerve regeneration on a longer timescale, we extended the regeneration time to 4 weeks. The result showed that Lin28a overexpression significantly increased not only the length but also the number of regenerating axons compared with the 2-week group (Figures 3A and 3B). Assessment of RGC survival showed no difference between the GFP and Lin28a overexpression groups either 2 weeks or 4 weeks after the ONC but, instead, revealed a significantly lower RGC survival rate 4 weeks after the ONC compared with 2 weeks after the ONC (Figures S3E and S3G), indicating that the death of RGCs continued throughout the 4 weeks, and Lin28a promoted optic nerve regeneration by boosting the regeneration potential in RGCs that survived the ONC rather than by protecting RGCs from dying of the ONC. We also estimated the shrinkage rate of the optic nerves caused by tissue clearing. The nerves shrank to 82.0% ± 2.81% in length (Figures S3H and S3I) and 56.3% ± 6.58% in diameter (Figures S3H and S3J), resulting in a total shrinkage of 73.7% ± 5.72% in volume (estimated using the formula for the volume of a cylinder) (Figures S3H and S3K). Thus, the axon length observed in our experiments was actually underestimated by about 18%. Overall, these results demonstrate that overexpression of Lin28a can induce robust and sustainable optic nerve regeneration without affecting the survival rate of RGCs.
Figure 3. Upregulation of Lin28a in RGCs Induces Robust and Sustained Optic Nerve Regeneration.
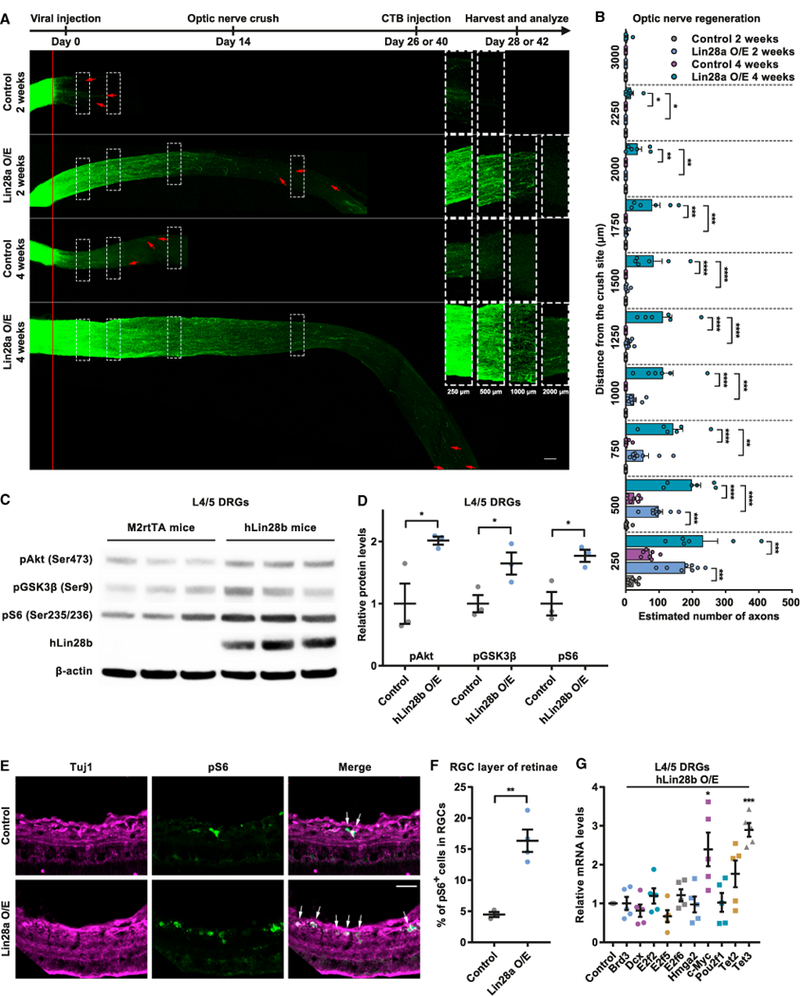
(A) Top: timeline of the experiment. Bottom: representative images showing that Lin28a O/E in RGCs induced drastic and persistent axon regeneration in optic nerves 2 and 4 weeks after the ONC. The right column shows enlarged images of nerves 250, 500, 1,000, and 2,000 μ distal to the crush sites, which are marked by dashed white boxes on the left. The red line indicates the crush sites. Red arrows show the longest axons of each nerve. Scale bar, 100 μ (left), 50 μ (right).
(B) Quantification of (A) (one-way ANOVA followed by Tukey’s multiple comparisons test; p < 0.0001 at 250, 500, 750, 1,000, 1,250, 1,500, and 1,750 μ; p = 0.0005, 0.0193, and 0.3113 at 2,000, 2,250, and 3,000 μ, respectively; n = 7 mice in 2-week and 4-week control groups; n = 8 and 6 mice in 2-week and 4-week Lin28a O/E groups, respectively).
(C) Representative western blot result showing elevated hLin28b O/E elevated levels of pAkt, pGSK3β, and pS6 in L4/5 DRGs.
(D) Quantification of (C) (unpaired Student’s t test; p = 0.0372, 0.0217, and 0.0453 for pAkt, pGSK3β, and pS6, respectively; n = 3 independent experiments).
(E) Representative images showing that Lin28a O/E markedly increased the percentage of pS6+ RGCs in the RGC layer. Retinal cryosections were stained with anti-pS6 (green) and anti-tubulin β3 (far-red) antibodies. White arrows indicate pS6+ RGCs. Scale bar, 50 μ.
(F) Quantification of (E) (unpaired Student’s t test, p = 0.0027, n = 3 mice in the control group, n = 4 mice in the Lin28a O/E group, 3–5 non-adjacent retinal sectionsanalyzed for each mouse).
(G) Levels of c-Myc and Tet3 miRNAs in L4/5 DRGs of hLin28b mice were significantly increased 2 days after induction of hLin28b by dox (one-sample t test;p = 0.9999, 0.3074, 0.3740, 0.1001, 0.2228, 0.9051, 0.0326, 0.9180, 0.0914, and 0.0004 for Brd3, Dcx, E2f2, E2f5, E2f6, Hmga2, c-Myc, Pou2f1, Tet2, and Tet3, respectively; n = 5 independent experiments).
*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 compared with control if not designated. Data are represented as mean ± SEM. See also Figure S3.
The Lin28/let-7 axis has previously been proven to regulate glucose metabolism through the insulin-phosphatidylinositol 3-kinase (PI3K)-Akt pathway (Zhu et al., 2011). Additionally, the increased level of pS6, downstream of the PI3K-Akt-mTOR (mammalian target of rapamycin) pathway, was linked to Pten knockout-induced optic nerve regeneration (Park et al., 2008). GSK3β, the phosphorylation at Ser9 of which by Akt inhibits its activity, has been shown to negatively control axon regeneration (Guo et al., 2016; Leibinger et al., 2017). Thus, it is reasonable to speculate that Lin28-induced PNS and CNS axon regeneration is mediated by Akt, GSK3β, and/or mTOR signaling. We thus collected L4/5 DRGs from hLin28b mice and M2rtTA mice after 2 days of dox treatment and tested the level of Ser473 phosphorylation of Akt (pAkt), Ser9 phosphorylation of GSK3β (pGSK3β), and pS6. We found that hLin28b overexpression significantly increased the levels of pAkt, pGSK3β, and pS6 (Figures 3C and 3D). Importantly, we also observed that, 2 weeks after the ONC, the percentage of pS6+ RGCs increased by about 3-fold in the Lin28a overexpression group compared with the control group (Figures 3E and 3F). These results suggest that the Lin28/let-7 axis might regulate axon regeneration, at least partially, through Akt activation, GSK3β inactivation, and/or activation of the mTOR pathway.
As a reprogramming factor, Lin28 has been shown to promote postnatal tissue regeneration by reprogramming cellular metabolism (Shyh-Chang et al., 2013). Thus, we tested the mRNA levels of some well-characterized or potential let-7 target genes that are also involved in cell reprogramming or epigenetic remodeling in L4/5 DRGs of M2rtTA mice and hLin28b mice. We found that, after 2 days of dox treatment, the mRNA levels of c-Myc and Tet3, two genes previously reported to be positive regulators of axon regeneration (Belin et al., 2015; Weng et al., 2017), were increased by hLin28b overexpression (Figure 3G), indicating that Lin28a/b may enhance axon regeneration by reprogramming the cellular state of mature neurons to reclaim axon growth ability.
Lin28a Overexpression and Pten Knockdown Have an Additive Effect on Optic Nerve Regeneration
We next tested whether Lin28a overexpression and Pten knockdown have an additive effect on optic nerve regeneration. Viruses expressing small hairpin RNA (shRNA) against Pten (AAV2-shPten) and/or AAV2-Lin28a-FLAG were injected into the vitreous humors of C57BL/6 mice on day 0 and day 2 of the experiment, respectively. Control mice were injected with AAV2-GFP. The ONC was made 2 weeks after the viral injections. Two weeks after the ONC, we found that combined Lin28a overexpression and Pten knockdown induced faster optic nerve regeneration (4A and4B). Specifically, compared with Lin28a overexpression only and Pten knockdown only, the combination of Lin28a overexpression and Pten knockdown induced significantly stronger axon regeneration at 750, 1,000, 1,250, 1,500, 1,750, and even 2,000 μ from the crush site (Figure 4B). These results indicate that Lin28a overexpression and Pten knockdown act additively in promoting optic nerve regeneration.
Figure 4. Lin28a O/E and Pten KD Have an Additive Effect on Optic Nerve Regeneration.
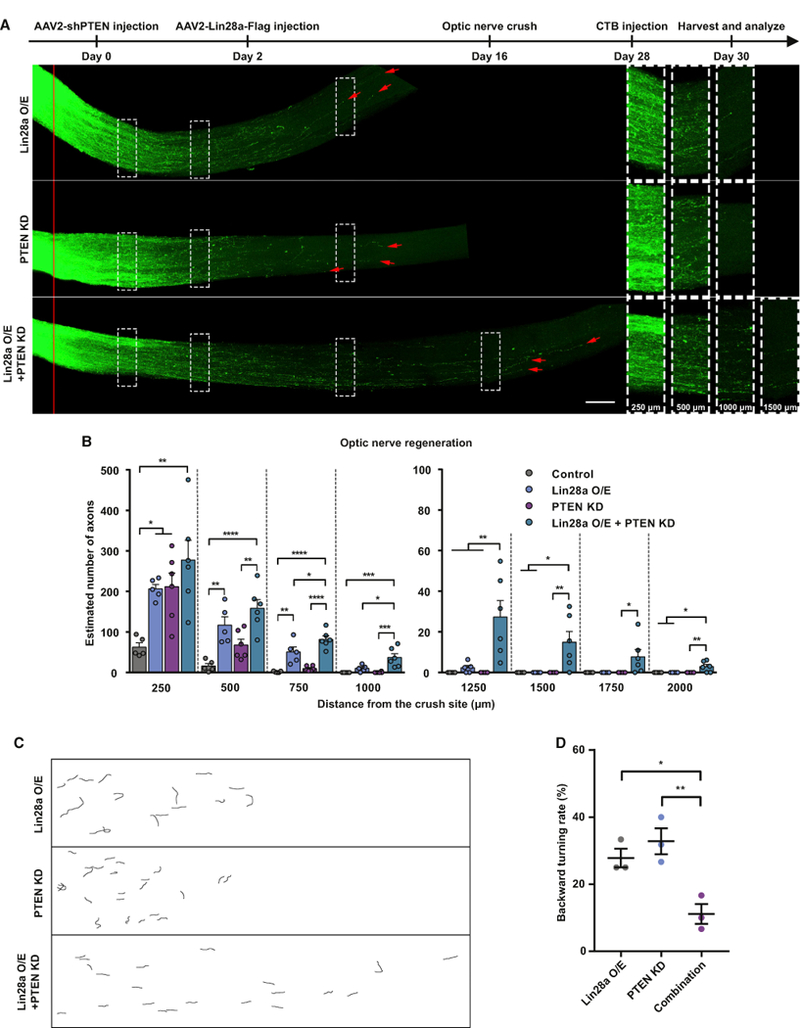
(A). Top: timeline of the experiment. Bottom: representative images showing that combined Lin28a O/E and Pten KD in RGCs induced faster axon regeneration in optic nerves 2 weeks after the ONC. The right column shows enlarged images of nerves 250, 500, 1,000, and 1,500 μ distal to the crush sites, which are marked by dashed white boxes on the left. The red line indicates the crush sites. Red arrows show the longest axons of each nerve. Scale bar, 100 μ (left), 50 μ (right).
(B) Quantification of (A) (one-way ANOVA followed by Tukey’s multiple comparisons test; p = 0.0025, 0.0001, 0.0003, 0.0006, 0.0028, 0.0242, and 0.0039 at 250, 500, 1,000, 1,250, 1,500, 1,750 and 2,000 μ, respectively; p < 0.0001 at 750 μ, n = 5 mice in the control group and the Lin28a O/E group; n = 6 mice in other two groups).
(C) Representative reconstructed graphs of distal axon trajectories showing that the combination of Lin28a O/E and Pten KD significantly reduced the backward turning rate of axons.
(D) Quantification of (C) (one-way ANOVA followed by Tukey’s multiple comparisons test, p = 0.0075, n = 3 mice in each group).
*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Data are represented as mean ± SEM.
We quantified the backward turning rate of regenerating axons under each condition. The result showed that about 30% of regenerating axons induced by either Lin28a overexpression or Pten knockdown alone made backward turns (Figures 4C and 4D). Combination of Lin28a overexpression and Pten knockdown significantly reduced the percentage (Figures 4C and 4D), suggesting that the reduced backward turning rate at least partially explains the additive effect of Lin28a overexpression and Pten knockdown.
DISCUSSION
During development, stem cells undergo many steps to turn into differentiated cells. In such processes, the whole gene expression profile changes drastically, with stemness-related genes shutting down and only genes relevant for the differentiated cell type expressed. The emerging idea is that the regulation of gene expression during differentiation is largely achieved through epigenetic modifications, including DNA methylation, histone modifications, and non-coding RNAs. Importantly, differentiated cells (e.g., fibroblasts) can be reprogrammed back to iPSCs by overexpressing several reprogramming factors, such as Sox2, c-Myc, Klf4, Oct4, Nanog, and Lin28 (Takahashi and Yamanaka, 2006; Yu et al., 2007), leading to global epigenetic remodeling. Do mammalian CNS neurons undergo similar epigenetic changes during maturation to lose their ability to support axon growth? If so, can we manipulate reprogramming factors to help CNS neurons reclaim such an ability?
Indeed, Klf4 (Moore et al., 2009), c-Myc (Belin et al., 2015), and Sox11 (Norsworthy et al., 2017) are all important regulators of axon regeneration. A previous study reported that the reactivation of Lin28a can promote mammalian tissue regeneration in postnatal tissues by reprogramming cellular metabolism (Shyh-Chang et al., 2013). This led us to hypothesize that CNS neurons can also regain the potential to support axon regeneration by reintroduction of Lin28a and/or Lin28b. Our study clearly showed that Lin28a/b are not only required for intrinsic axon regeneration ability in PNS sensory neurons but can also promote both PNS and, more importantly, CNS axon regeneration in vivo when overexpressed. Our findings support the notion that mature CNS neurons can restore axon regeneration ability by modulating reprogramming factors. Moreover, because cell reprogramming can now be achieved by using small-molecule compounds (Hou et al., 2013; Zhao et al., 2015), an easy-to-manage method without genome editing, new translational opportunities of enhancing CNS axon regeneration may be available. However, further studies are required to reveal how Lin28 signaling reprograms epigenetic state and gene expression in mature mammalian neurons. A recent study showed that, during iPSC reprogramming, Lin28 acted to regulate mitochondrial function, nucleotide metabolism, and histone methylation (Zhang et al., 2016). In our study, we found that hLin28b overexpression induced upregulation of c-Myc and Tet3 in L4/5 DRGs. c-Myc is one of the four Yamanaka factors (Oct4, Sox2, Klf4, and c-Myc) that can reprogram mature cells into iPSCs (Takahashi and Yamanaka, 2006) and can be functionally replaced by Lin28 (Yu et al., 2007). It has also been verified as a target gene of let-7 (Kumar et al., 2007). Tet3 is a methylcytosine dioxygenase that plays an important role in chromatin modification and epigenetic reprogramming by catalyzing the demethylation of DNA. Although it was never proven to be regulated by the Lin28/let-7 axis, evidence showed that Thompson factor (Oct4, Sox2, Nanog, and Lin28)-induced iPSCs express higher Tet3 levels than Yamanaka factor-induced iPSCs (Planello et al., 2014). Interestingly, another study showed that Lin28a functions to recruit Tet1 to regulate DNA methylation and gene expression (Zeng et al., 2016). These findings imply that Lin28a/b play key roles in regulating epigenetic remodeling through Tet enzymes.
The major downstream signaling mediator of Lin28 is the let-7 miRNA family, which has been broadly studied because of its phylogenetically conserved expression patterns and functions. It has been reported previously in C. elegans that let-7 inhibits axon regeneration in older AVM neurons, whereas its loss of function reactivates AVM axon regeneration (Zou et al., 2013). In our study, we found that let-7 gain of function inhibited axon regeneration and abrogated the hLin28b overexpressioninduced acceleration of axon regeneration in injured mouse sciatic nerves, whereas let-7 loss of function significantly improved axon regeneration, indicating that the role of let-7 as a negative regulator in axon regeneration is also evolutionarily conserved between nematodes and mammals. However, distinct roles played by different let-7 miRNAs in the regulation of mammalian axon regeneration are yet to be elucidated in future studies.
Here, we found that Lin28a upregulation led to Akt activation, GSK3β inactivation, and mTOR activation in mature sensory neurons and RGCs. These results suggest that Lin28-promoted optic nerve regeneration might be partially mediated by the AktmTOR and/or GSK3β pathways. Future studies using rapamycin or active mutants of GSK3β are required to determine whether these pathways functionally act downstream of Lin28 to promote axon regeneration. We also showed that Lin28a overexpression and Pten knockdown acted additively to promote optic nerve regeneration, indicating that these two modifications have some different underlying mechanisms, although they both regulate the Akt-mTOR and GSK3β pathways. Previous studies have shown that, although robust optic nerve regeneration can be achieved by stimulating intrinsic growth ability, long-distance regeneration and precise functional targeting are still largely hindered by axonal misguidance (Pernet et al., 2013; Pernet and Schwab, 2014; Yungher et al., 2015). Here, we found that the combination of Lin28a overexpression and Pten knockdown effectively prevented regenerating axons from turning back. However, the molecular mechanisms of how such a combination can significantly reduce backward turning are still unknown. Moreover, the trajectories of regenerating RGC axons were only captured at a single time point. It is hard to determine for sure whether the axons have turned sufficiently unless timelapse in vivo imaging can be done.
Tissue-clearing strategies have been widely adopted nowadays because of their irreplaceable advantages in imaging. Different approaches, however, may yield different tissue shrinkage rates, resulting in inconsistencies in studies involving measurement of tissue size. Our observations suggest that the tissue shrinkage rate should be taken into consideration when comparing results from different tissue-clearing techniques.
STAR⋆METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, FengQuan Zhou (fzhou4@jhmi.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Primary cell cultures
Culture of adult mouse DRG neurons were performed based on procedures described previously (Hur et al., 2011; Saijilafu et al., 2013) with minor changes. DRGs were first dissected out from 6–8-week-old female CF-1 mice and digested with 1 mg/ml type I collagenase (Thermo Fisher Scientific) and 5 mg/ml dispase II (Thermo Fisher Scientific) at 37C for 70 min, followed by 3 times of wash with HBSS. The digested DRGs were then dissociated into single cells in MEM containing 10% fetal bovine serum and 1x penicillin/streptomycin (Thermo Fisher Scientific). The dissociated cells were filtered with a 100 μm cell strainer and centrifuged at 1000 rpm for 8.5 minutes. The pelleted cells were then used for in vitro electroporation (for details, see below). After electroporation, the cells were immediately mixed with desired volume of pre-warmed medium mentioned above and plated onto glass coverslips pre-coated with a mixture of 100 μg/ml poly-D-lysine (Sigma-Aldrich) and 10 μg/ml laminin (Thermo Fisher Scientific). After the cells fully attached to the coverslips (about 6 hr after plating), the medium was totally removed to get rid of the electroporation buffer and dead cells and replaced with MEM containing 5% fetal bovine serum, 1x GlutaMAX-I (Thermo Fisher Scientific), 1x penicillin/streptomycin and antimitotic reagents (20 μM 5-Fluoro-2′-deoxyuridine and 20μM uridine, both from Sigma-Aldrich). Cells were cultured for 3 days with no additional growth factors added into the culture medium.
Mice
All animal experiments were performed according to the animal protocol approved by the Institutional Animal Care and Use Committee of the Johns Hopkins University. Unless otherwise noted, female 6–8-week-old CF-1 or CD-1 IGS mice purchased from Charles River Laboratories were used in experiments involving DRG and sciatic nerve, and 6-week old C57BL/6 mice of both sexes from The Jackson Laboratory were used in experiments involving retina and optic nerve.
The hLin28b mutant mouse strain (JAX stock#023911) (Zhu et al., 2011) was a kind gift from Dr. George Daley’s laboratory at Harvard Medical School. The M2rtTA transgenic line (JAX stock#006965) (Hochedlinger et al., 2005) was obtained from Dr. Angelika Doetzlhofer’s laboratory at Johns Hopkins University School of Medicine. Male M2rtTAtg/tg mice were crossed with female hLin28btg/+ mice to generate M2rtTAtg/+; hLin28btg/+ mice and M2rtTAtg/+ mice, which were used as hLin28b mice and M2rtTA control mice in experiments, respectively. Six-week old mice of both sexes were used. Genotypes of the mice were determined by PCR using primers provided by The Jackson Laboratory. Doxycycline-containing food (Bio-Serv) was used to induce the expression of hLin28b.
All animal surgeries were performed under anesthesia induced by intraperitoneal injection of ketamine (100 mg/kg) and xylazine (10 mg/kg) diluted in sterile saline. Details of the surgeries are described below.
Cell line
CAD (Cath.-a-differentiated) cells were maintained in DMEM/F12 (1:1, Thermo Fisher Scientific) supplemented with 10% fetal bovine serum and 1x penicillin/streptomycin in a standard humidified 5% CO2, 37C tissue culture incubator. Cells were transfected with siRNAs targeting Lin28a and/or Lin28b using Lipofectamine 2000 (Thermo Fisher Scientific) and Opti-MEM I reduced serum medium (Thermo Fisher Scientific) at 80% confluency and cultured for 3 days in serum-free DMEM/F12 (1:1) before total protein was isolated.
METHOD DETAILS
Constructs
The Lin28a open reading frame was PCR-amplified from pMSCV-mLin28A using a forward primer that incorporated a 5′ EcoRI restriction site (5′-CGGAATTCATGGGCTCGGTGTCCAACC-3′) and a reverse primer that incorporated a 3′ NotI restriction site (5′-ATAAGAATGCGGCCGCTCAATTCTGGGCTTCTGGG-3′). The amplified sequence was then used to replace the EGFP open reading frame in pCMV-GFP with standard digestion and ligation to generate pCMV-Lin28a. pCMV-GFP was a gift from Dr. Connie Cepko (Addgene plasmid # 11153). pMSCV-mLin28A was a gift from Dr. George Daley (Addgene plasmid # 26357). The Lin28a-Flag open reading frame with a 5′ BamHI restriction site and a 3′ EcoRV restriction site was synthesized (codon optimized, gBlocks of Integrated DNA Technologies) and used to replace the EYFP open reading frame in pAAV-Ef1a-EYFP. pAAV-Ef1a-EYFP was a gift from Dr. Hongjun Song. pAAV-shPten was a gift from Dr. David Turner and Dr. Kevin Park. All restriction endonucleases and T4 DNA ligase were purchased from New England Biolabs. Plasmids were amplified using DH5α competent cells (Thermo Fisher Scientific) and purified with Endofree plasmid maxi kit (QIAGEN).
in vivo electroporation of adult DRG neurons
The in vivo electroporation of adult mouse DRGs was performed as previously described (Saijilafu et al., 2011). Briefly, after a mouse was anesthetized, a small dorsolateral laminectomy was performed on the left side to expose the left L4/5 DRGs. Plasmids (2 mg per kind) and/or RNA oligos (0.1 nmol per kind) were injected into each DRG with a glass micropipette pulled from a glass capillary (World Precision Instruments) connected to a Picospritzer III (20-psi pressure, 6-ms duration, Parker Hannifin). Right after the injection, in vivo electroporation was performed by applying five electric pulses (35 V, 15-ms duration, 950-ms interval) using a platinum tweezertrode (BTX) powered by the ECM 830 Electro Square Porator (BTX). The wound was then closed and the mouse was allowed 2–3 days to recover and then the left sciatic nerve was exposed right below pelvis and crushed with Dumont #5 forceps. The crush site was marked with 10–0 nylon epineural sutures. After 2 or 3 days, the mouse was transcardially perfused with PBS followed by icecold 4% PFA. The sciatic nerve segment was then dissected out and post-fixed in 4% PFA overnight at 4C. On the next day, the nerve segment was mounted onto a glass slide and covered with a coverslip and flattened before imaging. Mice without a clearly identifiable crush site or an epineural suture were excluded from data analysis. The siRNAs targeting mouse Lin28a/b, mimics of let-7a and let-7b, non-targeting siRNA control and non-mammalian targeting miRNA control were purchased from Dharmacon. The let-7 miRNA family inhibitor was purchased from QIAGEN. The fluorescent siRNA control was purchased from Sigma-Aldrich. All target sequences of siRNAs used are listed in Table S2.
Analysis of in vivo axon regeneration
For quantification of in vivo axon regeneration, fluorescent tiled images of a whole-mount sciatic nerve segment were first captured with a CCD camera connected to an inverted fluorescence microscope controlled by the AxioVision software (Zeiss) using the MosaiX module. The tiles were stitched to generate an image of the whole-mount nerve segment. Each identifiable GFP+ axon in the nerve segment was then manually traced from the crush site to the distal axonal tip to determine the length. The mean length of all axons traced in one nerve was used as the average axon length of this nerve. Only nerves with at least 10 identifiable axons were included in data analysis. Measurement was done by experimenters blinded to the conditions.
Analysis of in vivo siRNA transfection rate
Two days after in vivo electroporation of fluorescent siRNA control, mice were perfused and the electroporated L4/5 DRGs were collected. Frozen DRG sections of 10 μ were obtained with a cryostat and warmed on a slide warmer at 37C for 1 hr. After being blocked with PBS containing 10% goat serum and 0.3% Triton X-100 at room temperature for 1 hr, the sections were immunostained with anti-tubulin β3 primary antibody (TUJ1, 1:500, Biolegend) overnight at 4C, followed by Alexa Fluor 488 conjugated secondary antibody (1:500, Thermo Fisher Scientific) at room temperature for 1 hr. All antibodies were diluted with the blocking buffer. Four times of 15-min wash with PBS containing 0.3% Triton X-100 was performed following each antibody incubation. The DRG sections were mounted with Fluoroshield histology mounting medium (Sigma-Aldrich) and imaged with the inverted fluorescent microscope mentioned above. Four or five non-adjacent sections from each DRG were used for analysis. For each DRG, the transfection rate was calculated by dividing the of number of red fluorescent siRNA control+/TUJ1+ cells by the number of TUJ1+ cells.
in vitro electroporation of adult DRG neurons
The pelleted cells obtained from digested DRGs (for details, see Primary Cell Cultures) were resuspended with 100 mL electroporation buffer (mouse neuron nucleofector kit, Lonza) containing RNA oligos (0.2 nmol per kind) and/or GFP plasmid (10 mg). The mixture of cells, GFP plasmid and/or RNA oligos was transferred to a 2.0-mm electroporation cuvette and electroporated with the Nucleofector II (Lonza).
Analysis of in vitro axon growth
Neurons cultured for 3 days were first fixed with 4% PFA and washed with PBS for 3 times, and then blocked in PBS containing 2% bovine serum albumin and 0.1% Triton X-100 for 1 hr. The neurons were then sequentially immunostained with anti-tubulin β3 (TUJ1, 1:1000, BioLegend) and anti-GFP (1:1000, Thermo Fisher Scientific) primary antibodies, and corresponding Alexa Fluor conjugated secondary antibodies (1:500, Thermo Fisher Scientific) for 1 hr each at room temperature. All antibodies were diluted with the blocking buffer. Three times of 10-min wash with PBS was performed after each antibody incubation. The coverslips were then mounted onto glass slides with Fluoroshield histology mounting medium (Sigma-Aldrich) and fluorescent images of the neurons were captured with the inverted fluorescent microscope mentioned above. The longest axon of each neuron was manually traced and measured with the built-in ‘‘measure/curve spline’’ function of the AxioVision. Only neurons with axons longer than twice the diameter of their cell bodies were included. In each independent experiment, at least 50 neurons were measured in each condition.
Optic nerve regeneration model
Intravitreal viral injection, optic nerve crush and RGC axon labeling were performed as previously described (Park et al., 2008). Briefly, under anesthesia, 1.5 mL of AAV2 virus was injected into the right vitreous humor of a mouse with glass micropipette connected to a Picospritzer III (15-psi pressure, 4-ms duration). The position and direction of the injection were well-controlled to avoid lens damage. Two weeks after viral injection, the right optic nerve of the mouse was exposed intraorbitally and crushed with Dumont #5 fine forceps (Fine Science Tools) for 5 s at approximately 1 mm behind the optic disc. To label RGC axons in the optic nerve, 1.5 mL of CTB conjugated with Alexa Fluor 594 (2 mg/ml, Thermo Fisher Scientific) was injected into the right vitreous humor with glass micropipette and Picospritzer III 2 days before the mouse was sacrificed by transcardial perfusion. Both retinae and the right optic nerve were dissected out and post-fixed in 4% PFA overnight at 4C. AAV2-GFP was purchased from SignaGen Laboratories. AAV2-Lin28a-Flag was packaged by SignaGen Laboratories. AAV2-shPten was packaged by Vigene Biosciences. All virus used had titers > 1 × 1013.
Optic nerve dehydration and clearing
Dehydration and clearing of optic nerves were done based on previous studies (Erturk et al., 2012; Luo et al., 2013€ ). Briefly, fixed optic nerves were first dehydrated in incremental concentrations of tetrahydrofuran (TFH, 50%, 70%, 80%, 100% and 100%, v/v % in distilled water, 20 min each, Sigma-Aldrich) in amber glass bottles. Incubations were performed on an orbital shaker at room temperature. Then the nerves were incubated with benzyl alcohol/benzyl benzoate (BABB, 1:2 in volume, Sigma-Aldrich) clearing solution for 20 min. The nerves were protected from light during the whole process to reduce photo bleaching of the fluorescence.
Analysis of RGC axon regeneration
The cleared whole-mount nerves were imaged with a 20x objective on an LSM 510 confocal microscope (Zeiss) equipped with a motorized stage and an LSM 510 software (Zeiss). For each optic nerve, Z stack function was used to acquire a stack of 1.6-mm-thick slices, and tiling function with 10% overlap between adjacent tiles was used to scan the whole nerve. The tiles from each 1.6-mmthick plane were then stitched to obtain an image of the whole-mount slice. Quantification of regenerating axons in the optic nerve was done as previous described (Park et al., 2008). Specifically, every 10 continuous slices were Z-projected with maximum intensity to generate a series of Z-projection images of 16-mm-thick optical sections. At every 250-mm interval from the crush site, the number of CTB-labeled axons was counted and the width of the nerve was measured in each optical section. Both numbers were used to calculate the number of axons per micrometer of nerve width, which was then averaged over all optical sections. Sad, the total number of axons extending distance d in a nerve with a radius of r, was estimated by summing over all optical sections with a thickness of t (16 μ): Sad = pr2 x (average axons/mm)/t.
Analysis of RGC transduction rate and survival rate
Fixed whole-mount retinae were first radially cut into petal shape and then stained. The procedure and buffers used were the same with those used for DRG cryosections except the percentage of Triton X-100 in the blocking buffer was 1%. Fluorescent images were acquired with a 20x objective on a Zeiss LSM510 confocal microscope.
For quantification of RGC transduction rate, uninjured retinae (no optic nerve crush) were taken from transcardially fixed mice 2 weeks after intravitreal AAV2-Lin28a-Flag injection. The retinae were stained with anti-tubulin β3 (TUJ1, 1:500, BioLegend) and anti-Flag (1:500, Cell Signaling Technology) primary antibodies, followed by corresponding Alexa Fluor conjugated secondary antibodies (1:500, Thermo Fisher Scientific). Six fields under 20x objective were randomly obtained from the peripheral regions of each retina. For each mouse, RGC transduction rate was represented by the ratio of total Flag+/TUJ1+ cells to total TUJ1+ cells. Only cells in the ganglion cell layer were counted.
For quantification of RGC survival rate, mice injected with AAV2-GFP or AAV2-Lin28a-Flag were transcardially perfused 2 or 4 weeks after optic nerve crush and both retinae of each mouse were collected. Whole-mount retinae were sequentially stained with anti-tubulin β3 antibody (TUJ1, 1:500, BioLegend) and Alexa Fluor 488 conjugated secondary antibody (1:500, Thermo Fisher Scientific). Six or eight fields under 20x objective were randomly taken from the peripheral regions of each retina. For each mouse, RGC survival rate was calculated by dividing the total number of TUJ1+ cells in all eight fields in the injured retina by that in the uninjured retina. Only cells in the ganglion cell layer were counted.
Immunohistochemistry of retinal cryosections
The procedure and buffers used were the same with those used for DRG cryosections. Sections of 12-μ thick were stained with anti-tubulin β3 (TUJ1, 1:500, Biolegend) and anti-pS6 Ser235/236 (1:500, Cell Signaling Technology) antibodies, followed by corresponding Alexa Fluor conjugated secondary antibodies (1:500, Thermo Fisher Scientific). Fluorescent images were acquired using the inverted fluorescent microscope mentioned above. Three to five non-adjacent retinal sections from each mouse were used for analysis. For each mouse, the percentage of pS6+ RGCs was calculated by dividing the number of pS6+/TUJ1+ cells by the number of TUJ1+ cells. Only cells in the ganglion cell layer were counted.
Analysis of backward turning of RGC axons
For each optic nerve, a single Z-projection image was obtained by Z-projecting all optical slices with maximum intensity. The trajectory near the axonal tip of the most identifiable 10–30 axons in each nerve were traced. Backward turning was defined when an axonal tip faced backward (the angle between the final direction of the axonal tip and the anterograde longitudinal axis of the optic nerve was wider than 90 degrees). Backward turning rate was calculated by dividing the number of backward turning axons by the number of total axonal tips traced. Three mice were used in each condition.
Western blot analysis
Proteins were extracted from DRG or retinal tissues, or CAD cells using the RIPA buffer containing protease inhibitor cocktail (SigmaAldrich) and phosphatase inhibitor cocktail (Sigma-Aldrich). The extracted proteins were then separated by 4%–12% gradient SDSPAGE gel electrophoresis and transferred onto polyvinylidene fluoride membranes. After being blocked with TBST containing 5% blotting-grade blocker (Bio-Rad), blots were incubated overnight with primary antibodies against target proteins at 4C, followed by corresponding HRP-linked secondary antibodies (Cell Signaling Technology) for 1 hr at room temperature. All antibodies were diluted with blocking buffer. Blots were washed with TBST for 15 min for 4 times after each antibody incubation. Bands from 3 independent experiments were analyzed with the ImageJ software (NIH). Band density was first normalized to the loading control, b-actin, and then normalized to the control group. Primary antibodies against Lin28a (1:1000), Lin28b (1:1000), hLin28b (1:1000), pS6 Ser235/236 (1:2000), pAkt Ser473 (1:2000), pGSK3β Ser9 (1:1000) were purchased from Cell Signaling Technology. Anti-b-actin primary antibody (1:10000) was from Sigma-Aldrich.
Quantitative real-time PCR
Total RNA from cells or tissues was first isolated with the miRNeasy mini kit (QIAGEN), and then reversely transcribed to first strand cDNA using Transcriptor first strand cDNA synthesis kit (Roche). For mRNAs, anchored oligo(dT)18 primer was used. Gene-specific stem-loop primers were used for miRNAs following procedures described in a previous study (Chen et al., 2005). To detect the level of mRNAs or miRNAs, 10 ng first strand cDNA was amplified with gene-specific primers and LightCycler 480 SYBR green I master (Roche) using the LightCycler 480 II (Roche). All experiments were done in triplicate. Relative levels of mRNAs or miRNAs were determined using the ddCt method and normalized to control. Gapdh and Rnu6b were used as the endogenous controls for mRNAs and miRNAs, respectively. All primers used are listed in Table S1.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyses were done with GraphPad Prism 7 and the significance level was set as p < 0.05. Data are presented as mean ± SEM unless specifically stated. For comparisons between two groups, if the data were normalized to the control group and presented as relative levels, one sample t test with hypothetical value set as 1 was used to determine the statistical significance; otherwise regular two-tailed Student’s t test was used. For comparisons among three or more groups, one-way ANOVA followed by Tukey’s multiple comparison test was used to determine the statistical significance. All statistical details of experiments, including the statistical tests used, exact value of n, definition of n, can be found in figure legends.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-β-actin | Sigma-Aldrich | Cat#1978; RRID: AB_476692 |
| Mouse monoclonal anti-tubulin β3 (TUJ1) | Biolegend | Cat#801202; RRID: AB_10063408 |
| Chicken polyclonal anti-GFP | Thermo Fisher Scientific | Cat#A10262; RRID: AB_2534023 |
| Rabbit polyclonal anti-Lin28a | Cell Signaling Technology | Cat#3978S; RRID: AB_2297060 |
| Rabbit polyclonal anti-Lin28b (mouse preferred) | Cell Signaling Technology | Cat#5422S; RRID: AB_10697489 |
| Rabbit polyclonal anti-hLin28b | Cell Signaling Technology | Cat#4196S; RRID: AB_2135047 |
| Rabbit monoclonal anti-pS6 (Ser235/236) | Cell Signaling Technology | Cat#4858S; RRID: AB_916156 |
| Rabbit monoclonal anti-pAkt (Ser473) | Cell Signaling Technology | Cat#4060S; RRID: AB_2315049 |
| Rabbit monoclonal anti-pGSK3β (Ser9) | Cell Signaling Technology | Cat#5558S; RRID: AB_10013750 |
| Rabbit monoclonal anti-DYKDDDDK tag (Flag) | Cell Signaling Technology | Cat#14793S; RRID: AB_2572291 |
| HRP-linked horse anti-rabbit IgG | Cell Signaling Technology | Cat#7074S; RRID: AB_2099233 |
| HRP-linked horse anti-mouse IgG | Cell Signaling Technology | Cat#7076S; RRID: AB_330924 |
| Alexa Fluor 488-conjugated goat anti-mouse IgG (H+L) cross-adsorbed |
Thermo Fisher Scientific | Cat#A-11001; RRID: AB_2534069 |
| Alexa Fluor 568-conjugated goat anti-mouse IgG (H+L) cross-adsorbed |
Thermo Fisher Scientific | Cat#A-11004; RRID: AB_2534072 |
| Alexa Fluor 647-conjugated goat anti-mouse IgG (H+L) cross-adsorbed |
Thermo Fisher Scientific | Cat#A-21235; RRID: AB_2535804 |
| Alexa Fluor 488-conjugated goat anti-rabbit IgG (H+L) cross-adsorbed |
Thermo Fisher Scientific | Cat#A-11008; RRID: AB_143165 |
| Alexa Fluor 488-conjugated goat anti-chicken IgY (H+L) | Thermo Fisher Scientific | Cat#A-11039; RRID: AB_2534096 |
| Bacterial and Virus Strains | ||
| DH5α competent cells | Thermo Fisher Scientific | Cat#18265–017 |
| AAV2-GFP | SignaGen Laboratories | Cat#SL100812 |
| AAV2-Lin28a-Flag (codon optimized) | SignaGen Laboratories | N/A |
| AAV2-shPten | Vigene Biosciences | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Type I Collagense | Thermo Fisher Scientific | Cat#17018–017 |
| Dispase II | Thermo Fisher Scientific | Cat#17105–041 |
| MEM | Thermo Fisher Scientific | Cat#11090–081 |
| DMEM/F12 (1:1) | Thermo Fisher Scientific | Cat#11330–032 |
| Opti-MEM reduced serum medium | Thermo Fisher Scientific | Cat#31985–070 |
| GlutaMAX-I | Thermo Fisher Scientific | Cat#A12860–01 |
| HBSS | Thermo Fisher Scientific | Cat#13175–095 |
| Penicillin-Streptomycin | Thermo Fisher Scientific | Cat#15140–122 |
| Fetal bovine serum | GE Healthcare Life Sciences | Cat#SH30071.03HI |
| Poly-D-lysine | Sigma-Aldrich | Cat#P6407 |
| Laminin | Thermo Fisher Scientific | Cat#23017–015 |
| 5-Fluoro-2′-deoxyuridine | Sigma-Aldrich | Cat#F0503 |
| Uridine | Sigma-Aldrich | Cat#U3750 |
| Lipofectamine 2000 | Thermo Fisher Scientific | Cat#11668–027 |
| Doxycycline diet | Bio-Serv | Cat#F3893 |
| Fluoroshield histology mounting medium | Sigma-Aldrich | Cat#F6182 |
| Alexa Fluor 594-conjugated cholera toxin subunit B | Thermo Fisher Scientific | Cat#C-22842 |
| Goat serum | Sigma-Aldrich | Cat#G9023 |
| Tetrahydrofuran | Sigma-Aldrich | Cat#186562 |
| Triton X-100 | Sigma-Aldrich | Cat#T9284 |
| Benzyl alcohol | Sigma-Aldrich | Cat#305197 |
| Benzyl benzoate | Sigma-Aldrich | Cat#B6630 |
| RIPA buffer | Sigma-Aldrich | Cat#R0278 |
| Protease inhibitor cocktail | Sigma-Aldrich | Cat#11836153001 |
| Phosphatase inhibitor cocktail | Sigma-Aldrich | Cat#4906845001 |
| Blotting-grade blocker | Bio-Rad | Cat#1706404 |
| EcoRI-HF restriction endonuclease | New England Biolabs | Cat#R3101S |
| NotI-HF restriction endonuclease | New England Biolabs | Cat#R3189S |
| BamHI-HF restriction endonuclease | New England Biolabs | Cat#R3136S |
| EcoRV-HF restriction endonuclease | New England Biolabs | Cat#R3195S |
| T4 DNA ligase | New England Biolabs | Cat#M0202S |
| Critical Commercial Assays | ||
| Mouse neuron nucleofector kit | Lonza | Cat#VPG-1001 |
| miRNeasy mini kit | QIAGEN | Cat#217004 |
| Transcriptor first strand cDNA synthesis kit | Roche | Cat#04897030001 |
| LightCycler 480 SYBR green I master | Roche | Cat#04707516001 |
| Endofree plasmid maxi kit | QIAGEN | Cat#12362 |
| Experimental Models: Cell Lines | ||
| CAD (Cath.-a-differentiated) cells | Gift from Dr. William Snider | N/A |
| Experimental Models: Organisms/Strains | ||
| CF-1 mice | Charles River Laboratories | Strain#023 |
| CD-1 IGS mice | Charles River Laboratories | Strain#022 |
| C57BL/6J mice | The Jackson Laboratory | Stock#000664 |
| hLin28b mice | Gift from Dr. George Daley | Stock#023911 |
| M2rtTA mice | Gift from Dr. Angelika Doetzlhofer | Stock#006965 |
| Oligonucleotides | ||
| For all primers, see Table S1 | This paper | N/A |
| siRNA targeting Lin28a (for target sequence, see Table S2) |
Dharmacon | Cat#L-051530–01-0005 |
| siRNA targeting Lin28b (for target sequence, see Table S2) |
Dharmacon | Cat#L-063393–01-0005 |
| Non-targeting siRNA control (for target sequence, see Table S2) |
Dharmacon | Cat#L-001810–10-20 |
| Fluorescent siRNA control | Sigma-Aldrich | Cat#SIC003 |
| let-7a microRNA mimic | Dharmacon | Cat#C-310503–07-0005 |
| let-7b microRNA mimic | Dharmacon | Cat#C-310504–05-0005 |
| Non-mammalian targeting microRNA control (cel-miR-67) |
Dharmacon | Cat#CN-001000–01-05 |
| let-7 microRNA family inhibitor (for target sequence, see Table S2) |
QIAGEN | Cat#YFI0450006 |
| Recombinant DNA | ||
| pCMV-GFP | Addgene | Cat#11153 |
| pMSCV-mLin28A | Addgene | Cat#26357 |
| pCMV-Lin28a | This paper | N/A |
| Lin28a-Flag open reading frame (codon optimized) | Integrated DNA Technologies | N/A |
| pAAV-Ef1a-EYFP | Gift from Dr. Hongjun Song | N/A |
| pAAV-Lin28a-Flag | This paper | N/A |
| pAAV-shPten | Gift from Dr. David Turner and Dr. Kevin Park |
N/A |
| Software and Algorithms | ||
| LightCycler 480 software, release 1.5 | Roche | Cat#04994884001 |
| AxioVision, release 4.8 | Zeiss | N/A |
| GraphPad Prism 7 | GraphPad Software | N/A |
| LSM 510 software | Zeiss | N/A |
| Office 2016 | Microsoft | N/A |
| ImageJ | NIH | N/A |
| Adobe Illustrator | Adobe | N/A |
| Amaxa Nucelfector II | Lonza | N/A |
| Glass capillaries | World Precision Instruments | Cat#TW100–4 |
| Picospritzer III | Parker Hannifin | Cat#051–0500-900 |
| Flat tweezertrode | BTX | Cat#45–0524 |
| ECM 830 Electro Square Porator | BTX | N/A |
| Dumont #5 fine forceps | Fine Science Tools | Cat#11254–20 |
| LightCycler 480 Instrument II | Roche | Cat#05015243001 |
Highlights.
Upregulation of Lin28a/b is necessary and sufficient for sensory axon regeneration
let-7 microRNAs act downstream of Lin28a/b in regulating sensory axon regeneration
Upregulation of Lin28a in RGCs induces robust and sustained optic nerve regeneration
Lin28a overexpression and Pten knockdown have additive effect on axon regeneration
ACKNOWLEDGMENTS
We thank Dr. George Daley and Dr. Angelika Doetzlhofer for sharing mouse strains. F.-Q.Z. was funded by the NIH (R01NS064288, R01NS085176, R01GM111514, and R01EY027347), the Craig H. Neilson Foundation (259450), and the BrightFocus Foundation (G2017037). S.L. was funded by the NIH (R01NS079432 and R01EY024575), and the Shriners Research Foundation (SHC-86300-PHI, SHC-86200-PHI-16, and SHC-85100).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Ambros V, and Horvitz HR (1984). Heterochronic mutants of the nematode Caenorhabditis elegans. Science 226, 409–416. [DOI] [PubMed] [Google Scholar]
- Amen AM, Ruiz-Garzon CR, Shi J, Subramanian M, Pham DL, and Meffert MK (2017). A Rapid Induction Mechanism for Lin28a in Trophic Responses. Mol. Cell 65, 490–503.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balzer E, Heine C, Jiang Q, Lee VM, and Moss EG (2010). LIN28 alters cell fate succession and acts independently of the let-7 microRNA during neurogliogenesis in vitro. Development 137, 891–900. [DOI] [PubMed] [Google Scholar]
- Belin S, Nawabi H, Wang C, Tang S, Latremoliere A, Warren P, Schorle H, Uncu C, Woolf CJ, He Z, and Steen JA (2015). Injury-induced decline of intrinsic regenerative ability revealed by quantitative proteomics. Neuron 86, 1000–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhuiyan MI, Lee JH, Kim SY, and Cho KO (2013). Expression of exogenous LIN28 contributes to proliferation and survival of mouse primary cortical neurons in vitro. Neuroscience 248, 448–458. [DOI] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. (2005). Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 33, e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y, Sloutsky R, Naegle KM, and Cavalli V (2013). Injury-induced HDAC5 nuclear export is essential for axon regeneration. Cell 155, 894–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erturk A, Becker K, Jahrling N, Mauch CP, Hojer CD, Egen JG, Hellal F, Bradke F, Sheng M, and Dodt HU (2012). Three-dimensional imaging of solvent-cleared organs using 3DISCO. Nat. Protoc 7, 1983–1995. [DOI] [PubMed] [Google Scholar]
- Guo X, Snider WD, and Chen B (2016). GSK3b regulates AKT-induced central nervous system axon regeneration via an eIF2Bε-dependent, mTORC1-independent pathway. eLife 5, e11903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, Joo C, Cho J, Ha M, Han J, and Kim VN (2008). Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol. Cell 32, 276–284. [DOI] [PubMed] [Google Scholar]
- Hochedlinger K, Yamada Y, Beard C, and Jaenisch R (2005). Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell 121, 465–477. [DOI] [PubMed] [Google Scholar]
- Hou P, Li Y, Zhang X, Liu C, Guan J, Li H, Zhao T, Ye J, Yang W, Liu K, et al. (2013). Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science 341, 651–654. [DOI] [PubMed] [Google Scholar]
- Huang YW, Ruiz CR, Eyler EC, Lin K, and Meffert MK (2012). Dual regulation of miRNA biogenesis generates target specificity in neurotrophininduced protein synthesis. Cell 148, 933–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur EM, Yang IH, Kim DH, Byun J, Saijilafu, Xu WL, Nicovich PR, Cheong R, Levchenko A, Thakor N, and Zhou FQ (2011). Engineering neuronal growth cones to promote axon regeneration over inhibitory molecules. Proc. Natl. Acad. Sci. USA 108, 5057–5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowski MP, McIlwrath SL, Jing X, Cornuet PK, Salerno KM, Koerber HR, and Albers KM (2009). Sox11 transcription factor modulates peripheral nerve regeneration in adult mice. Brain Res 1256, 43–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang JJ, Liu CM, Zhang BY, Wang XW, Zhang M, Saijilafu, Zhang SR, Hall P, Hu YW, and Zhou FQ (2015). MicroRNA-26a supports mammalian axon regeneration in vivo by suppressing GSK3b expression. Cell Death Dis 6, e1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing X, Wang T, Huang S, Glorioso JC, and Albers KM (2012). The transcription factor Sox11 promotes nerve regeneration through activation of the regeneration-associated gene Sprr1a. Exp. Neurol 233, 221–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar MS, Lu J, Mercer KL, Golub TR, and Jacks T (2007). Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat. Genet 39, 673–677. [DOI] [PubMed] [Google Scholar]
- Leibinger M, Andreadaki A, Golla R, Levin E, Hilla AM, Diekmann H, and Fischer D (2017). Boosting CNS axon regeneration by harnessing antagonistic effects of GSK3 activity. Proc. Natl. Acad. Sci. USA 114, E5454–E5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Wang X, Gu Y, Chen C, Wang Y, Liu J, Hu W, Yu B, Wang Y, Ding F, et al. (2015). Let-7 microRNAs regenerate peripheral nerve regeneration by targeting nerve growth factor. Mol. Ther 23, 423–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Sako Y, Imai A, Nishiyama T, Thompson K, Kubo M, Hiwatashi Y, Kabeya Y, Karlson D, Wu SH, et al. (2017). A Lin28 homologue reprograms differentiated cells to stem cells in the moss Physcomitrella patens. Nat. Commun 8, 14242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Salgueiro Y, Beckerman SR, Lemmon VP, Tsoulfas P, and Park KK (2013). Three-dimensional evaluation of retinal ganglion cell axon regeneration and pathfinding in whole mouse tissue after injury. Exp. Neurol 247, 653–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore DL, Blackmore MG, Hu Y, Kaestner KH, Bixby JL, Lemmon VP, and Goldberg JL (2009). KLF family members regulate intrinsic axon regeneration ability. Science 326, 298–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss EG, Lee RC, and Ambros V (1997). The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin4 RNA. Cell 88, 637–646. [DOI] [PubMed] [Google Scholar]
- Nam Y, Chen C, Gregory RI, Chou JJ, and Sliz P (2011). Molecular basis for interaction of let-7 microRNAs with Lin28. Cell 147, 1080–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MA, Thomson JM, and Hammond SM (2008). Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA 14, 1539–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LH, Robinton DA, Seligson MT, Wu L, Li L, Rakheja D, Comerford SA, Ramezani S, Sun X, Parikh MS, et al. (2014). Lin28b is sufficient to drive liver cancer and necessary for its maintenance in murine models. Cancer Cell 26, 248–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norsworthy MW, Bei F, Kawaguchi R, Wang Q, Tran NM, Li Y, Brommer B, Zhang Y, Wang C, Sanes JR, et al. (2017). Sox11 Expression Promotes Regeneration of Some Retinal Ganglion Cell Types but Kills Others. Neuron 94, 1112–1120.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KK, Liu K, Hu Y, Smith PD, Wang C, Cai B, Xu B, Connolly L, Kramvis I, Sahin M, and He Z (2008). Promoting axon regeneration in the adult CNS by modulation of the PTEN/mTOR pathway. Science 322, 963–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernet V, and Schwab ME (2014). Lost in the jungle: new hurdles for optic nerve axon regeneration. Trends Neurosci 37, 381–387. [DOI] [PubMed] [Google Scholar]
- Pernet V, Joly S, Jordi N, Dalkara D, Guzik-Kornacka A, Flannery JG, and Schwab ME (2013). Misguidance and modulation of axonal regeneration by Stat3 and Rho/ROCK signaling in the transparent optic nerve. Cell Death Dis 4, e734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planello AC, Ji J, Sharma V, Singhania R, Mbabaali F, Muller F, Alfaro JA, Bock C, De Carvalho DD, and Batada NN (2014). Aberrant DNA methylation reprogramming during induced pluripotent stem cell generation is dependent on the choice of reprogramming factors. Cell Regen. (Lond.) 3, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R, Fausett BV, and Goldman D (2010). Ascl1a regulates Muller glia dedifferentiation and retinal regeneration through a Lin-28-dependent, let-7 microRNA signalling pathway. Nat. Cell Biol 12, 1101–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, and Wulczyn FG (2008). A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat. Cell Biol 10, 987–993. [DOI] [PubMed] [Google Scholar]
- Saijilafu H, Hur EM, and Zhou FQ (2011). Genetic dissection of axon regeneration via in vivo electroporation of adult mouse sensory neurons. Nat. Commun 2, 543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijilafu H, Hur EM, Liu CM, Jiao Z, Xu WL, and Zhou FQ (2013). PI3K-GSK3 signalling regulates mammalian axon regeneration by inducing the expression of Smad1. Nat. Commun 4, 2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinoda G, Shyh-Chang N, Soysa TY, Zhu H, Seligson MT, Shah SP, Abo-Sido N, Yabuuchi A, Hagan JP, Gregory RI, et al. (2013). Fetal deficiency of lin28 programs life-long aberrations in growth and glucose metabolism. Stem Cells 31, 1563–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang N, Zhu H, Yvanka de Soysa T, Shinoda G, Seligson MT, Tsanov KM, Nguyen L, Asara JM, Cantley LC, and Daley GQ (2013). Lin28 enhances tissue repair by reprogramming cellular metabolism. Cell 155, 778–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DS, and Skene JH (1997). A transcription-dependent switch controls competence of adult neurons for distinct modes of axon growth. J. Neurosci 17, 646–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, and Yamanaka S (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676. [DOI] [PubMed] [Google Scholar]
- Thornton JE, Chang HM, Piskounova E, and Gregory RI (2012). Lin28mediated control of let-7 microRNA expression by alternative TUTases Zcchc11 (TUT4) and Zcchc6 (TUT7). RNA 18, 1875–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu HC, Schwitalla S, Qian Z, LaPier GS, Yermalovich A, Ku YC, Chen SC, Viswanathan SR, Zhu H, Nishihara R, et al. (2015). LIN28 cooperates with WNT signaling to drive invasive intestinal and colorectal adenocarcinoma in mice and humans. Genes Dev 29, 1074–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbach A, Yermalovich A, Zhang J, Spina CS, Zhu H, Perez-Atayde AR, Shukrun R, Charlton J, Sebire N, Mifsud W, et al. (2014). Lin28 sustains early renal progenitors and induces Wilms tumor. Genes Dev 28, 971–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan SR, and Daley GQ (2010). Lin28: A microRNA regulator with a macro role. Cell 140, 445–449. [DOI] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, and Gregory RI (2008). Selective blockade of microRNA processing by Lin28. Science 320, 97–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan SR, Powers JT, Einhorn W, Hoshida Y, Ng TL, Toffanin S, O’Sullivan M, Lu J, Phillips LA, Lockhart VL, et al. (2009). Lin28 promotes transformation and is associated with advanced human malignancies. Nat. Genet 41, 843–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LD, Rao TN, Rowe RG, Nguyen PT, Sullivan JL, Pearson DS, Doulatov S, Wu L, Lindsley RC, Zhu H, et al. (2015). The role of Lin28b in myeloid and mast cell differentiation and mast cell malignancy. Leukemia 29, 1320–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng YL, An R, Cassin J, Joseph J, Mi R, Wang C, Zhong C, Jin SG, Pfeifer GP, Bellacosa A, et al. (2017). An Intrinsic Epigenetic Barrier for Functional Axon Regeneration. Neuron 94, 337–346.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West JA, Viswanathan SR, Yabuuchi A, Cunniff K, Takeuchi A, Park IH, Sero JE, Zhu H, Perez-Atayde A, Frazier AL, et al. (2009). A role for Lin28 in primordial germ-cell development and germ-cell malignancy. Nature 460, 909–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. (2007). Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920. [DOI] [PubMed] [Google Scholar]
- Yungher BJ, Luo X, Salgueiro Y, Blackmore MG, and Park KK (2015). Viral vector-based improvement of optic nerve regeneration: characterization of individual axons’ growth patterns and synaptogenesis in a visual target. Gene Ther 22, 811–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Yao B, Shin J, Lin L, Kim N, Song Q, Liu S, Su Y, Guo JU, Huang L, et al. (2016). Lin28A Binds Active Promoters and Recruits Tet1 to Regulate Gene Expression. Mol. Cell 61, 153–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ratanasirintrawoot S, Chandrasekaran S, Wu Z, Ficarro SB, Yu C, Ross CA, Cacchiarelli D, Xia Q, Seligson M, et al. (2016). LIN28 Regulates Stem Cell Metabolism and Conversion to Primed Pluripotency. Cell Stem Cell 19, 66–80. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Zhao T, Guan J, Zhang X, Fu Y, Ye J, Zhu J, Meng G, Ge J, Yang S, et al. (2015). A XEN-like State Bridges Somatic Cells to Pluripotency during Chemical Reprogramming. Cell 163, 1678–1691. [DOI] [PubMed] [Google Scholar]
- Zhu H, Shah S, Shyh-Chang N, Shinoda G, Einhorn WS, Viswanathan SR, Takeuchi A, Grasemann C, Rinn JL, Lopez MF, et al. (2010). Lin28a transgenic mice manifest size and puberty phenotypes identified in human genetic association studies. Nat. Genet 42, 626–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Shyh-Chang N, Segre AV, Shinoda G, Shah SP, Einhorn WS, Takeuchi A, Engreitz JM, Hagan JP, Kharas MG, et al. ; DIAGRAM Consortium; MAGIC Investigators (2011). The Lin28/let-7 axis regulates glucose metabolism. Cell 147, 81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Chiu H, Zinovyeva A, Ambros V, Chuang CF, and Chang C (2013). Developmental decline in neuronal regeneration by the progressive change of two intrinsic timers. Science 340, 372–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


