Abstract
Bacteria use several different secretion systems to deliver toxic EndoU ribonucleases into neighboring cells. Here, we present the first structure of a prokaryotic EndoU toxin in complex with its cognate immunity protein. The contact‐dependent growth inhibition toxin CdiA‐CTSTECO31 from Escherichia coli STEC_O31 adopts the eukaryotic EndoU fold and shares greatest structural homology with the nuclease domain of coronavirus Nsp15. The toxin contains a canonical His‐His‐Lys catalytic triad in the same arrangement as eukaryotic EndoU domains, but lacks the uridylate‐specific ribonuclease activity that characterizes the superfamily. Comparative sequence analysis indicates that bacterial EndoU domains segregate into at least three major clades based on structural variations in the N‐terminal subdomain. Representative EndoU nucleases from clades I and II degrade tRNA molecules with little specificity. In contrast, CdiA‐CTSTECO31 and other clade III toxins are specific anticodon nucleases that cleave tRNAGlu between nucleotides C37 and m2A38. These findings suggest that the EndoU fold is a versatile scaffold for the evolution of novel substrate specificities. Such functional plasticity may account for the widespread use of EndoU effectors by diverse inter‐bacterial toxin delivery systems.
EndoU ribonucleases are commonly used as weapons in inter‐bacterial conflict. We present the firststructure of a bacterial EndoU toxin domain. The CdiA‐CTSTECO31 toxin shares the same fold and catalytic triad as RNA processing enzymes from Xenopus laevis and human SARS virus. Unlike the eukaryotic EndoU ribonucleases, CdiA‐CTSTECO31 is exquisitely specific for the anticodon loop of tRNAGlu. These findings suggest that the EndoU fold is a versatile scaffold for the evolution of novel substrate specificities.
Introduction
Bacteria are social organisms that engage in cooperative and antagonistic relationships with their neighbors. Many of these interactions are mediated by small signaling molecules, such as quorum sensing auto‐inducers, which coordinate group activities like biofilm formation and virulence gene expression (Lopez et al., 2010; Ng and Bassler, 2009). Diffusible inhibitory factors, such as antibiotics and bacteriocins, are exploited as weapons to eliminate rivals (Cascales et al., 2007, Ghequire and De Mot, 2014). Research over the past decade has revealed that bacteria also antagonize their neighbors through direct delivery of protein toxins (Willett et al., 2015; Souza et al., 2015; Whitney et al., 2017; Vassallo et al., 2017). This phenomenon was first discovered as “contact‐dependent growth inhibition” (CDI) between different strains of Escherichia coli (Aoki et al., 2005). CDI is mediated by a subfamily of type V secretion systems (T5SS) that are widely distributed throughout proteobacteria, fusobacteria and negativicutes (Aoki et al., 2010; Zhang et al., 2012; Willett et al., 2015; Jones et al., 2017). T5SS/CDI+ bacteria use CdiB transporters to export and present filamentous CdiA proteins on the cell surface. CdiA proteins carry a variety of polymorphic C‐terminal effector domains (CdiA‐CT), which are transferred directly into neighboring bacteria upon binding specific receptors (Aoki et al., 2008; Beck et al., 2016; Ruhe et al., 2017). T5SS/CDI loci also encode immunity proteins that neutralize CdiA‐CT toxin activity and protect the cell against self‐inhibition. After the discovery of CDI, type VI secretion systems (T6SS) were also found to mediate proximity‐dependent inter‐bacterial competition (Hood et al., 2010; MacIntyre et al., 2010). The T6SS is a bacteriophage‐like contractile apparatus that injects protein effectors directly into nearby target bacteria (Basler et al., 2012). In contrast to CdiA effectors, which carry a single toxin domain, the T6SS apparatus supports simultaneous delivery of multiple toxins with distinct activities (Russell et al., 2014). Other secretion systems were first predicted to mediate inter‐bacterial competition based on genetic linkage to toxin/immunity gene pairs. Zhang et al. predicted that Neisseria species export MafB toxins through type II secretion systems (T2SS), and that Gram‐positive bacteria use type VII secretion systems (T7SS) to deploy LXG/WXG100/ESAT‐6 effectors (Zhang et al., 2012). Those predictions were subsequently supported by experimental work showing that both systems produce toxins and mediate inter‐cellular competition (Holberger et al., 2012; Jamet et al., 2015; Cao et al., 2016; Ohr et al., 2017; Whitney et al., 2017). Further experimental studies uncovered additional polymorphic toxin delivery systems. Bacillus and Listeria use large cell‐wall associated YD‐repeat proteins to exchange polymorphic toxin domains (Koskiniemi et al., 2013). Xanthomonas species deliver toxins though a specialized type IV secretion system (Souza et al., 2015), and a type I secretion system mediates contact‐dependent competition in Caulobacter crescentus (Garcia‐Bayona et al., 2017). Most recently, polymorphic lipoprotein toxins were shown to inhibit cell growth when transferred between strains of Myxococcus xanthus through outer membrane exchange (Dey et al., 2016; Vassallo et al., 2017). Collectively, these observations indicate that inter‐cellular toxin exchange is a fundamental and ubiquitous facet of prokaryotic biology.
Toxin delivery mechanisms are manifold because the architecture of the bacterial cell envelope varies considerably between phyla. Thus, systems capable of breaching the Gram‐negative envelope are ineffective against Gram‐positive bacteria and vice versa. Nonetheless, divergent secretion systems from unrelated species often deploy closely related toxins. For example, uropathogenic E. coli isolates use T5SS/CdiA to deliver Ntox28 RNase domains into target bacteria (Beck et al., 2016). Ntox28 toxins also form the C‐terminal domains of YD‐peptide repeat proteins and T7SS effectors from Gram‐positive bacteria (Zhang et al., 2012; Diner et al., 2012). The Ntox28 toxins of Gram‐negative and Gram‐positive bacteria share similar tRNA anticodon nuclease activities, and their cognate immunity proteins are clearly homologous (Johnson et al., 2016). This sporadic distribution across unrelated species strongly suggests that toxin‐immunity gene pairs are spread through horizontal gene transfer (Zhang et al., 2012; Poole et al., 2011; Ruhe et al., 2016). In this model, newly acquired toxin‐immunity sequences are integrated into resident secretion system loci through genetic recombination (Aoki et al., 2010; Poole et al., 2011; Zhang et al., 2012; Arenas et al., 2013; Unterweger et al., 2014). The modular organization of antibacterial effectors facilitates this process. Effector proteins are typically composites in which variable C‐terminal toxins are fused to conserved N‐terminal elements that guide export through a specific secretion system (Zhang et al., 2012; Koskiniemi et al., 2013). Thus, polymorphic toxins are versatile payloads that can be delivered through several different secretion pathways.
EndoU (Endoribonuclease specific for uridylate) RNase domains are among the most commonly deployed toxins in bacterial competition (Zhang et al., 2012). Aravind and colleagues first predicted that these toxins are related in structure and activity to eukaryotic and viral RNA processing enzymes (Zhang et al., 2012; Zhang et al., 2011). XendoU is the founding member of the EndoU superfamily and was isolated from Xenopus laevis as a uridylate specific endonuclease that releases small nucleolar RNAs from introns (Caffarelli et al., 1994; Caffarelli et al., 1997; Gioia et al., 2005; Laneve et al., 2003). RNA viruses of the order Nidovirales also encode EndoU nucleases. Nidoviral EndoU (NendoU) domains form the C‐termini of nonstructural proteins (Nsp) Nsp15 in coronaviruses and Nsp11 in arteriviruses (Snijder et al., 2003; Nedialkova et al., 2009). These endonucleases are essential for viral replication and have recently been shown to interfere with innate immune responses (Bhardwaj et al., 2004; Ivanov et al., 2004; Kindler et al., 2017; Deng et al., 2017). XendoU and NendoU nucleases have the same core built from two α/β subdomains and share a common catalytic triad that emanates from the N‐terminal subdomain (Joseph et al., 2007; Renzi et al., 2006; Ricagno et al., 2006; Xu et al., 2006; Shi et al., 2016; Zhang et al., 2017). Though unrelated in sequence and fold, EndoU and eukaryotic RNase A nucleases have remarkably similar active sites and use the same catalytic mechanism to produce cyclic 2′‐3′‐phosphodiester and 5′‐hydroxyl termini (Laneve et al., 2003; Ivanov et al., 2004; Renzi et al., 2006; Ricagno et al., 2006; Nedialkova et al., 2009). Two bacterial EndoU toxins have been shown to have RNase activity (Holberger et al., 2012; Jamet et al., 2015), but there is currently no high‐resolution structural information for the prokaryotic enzymes. Here, we present the first structure of a bacterial EndoU toxin‐immunity protein complex from the T5SS/CDI system of E. coli STEC_O31. The CdiA‐CTSTECO31 nuclease domain adopts the EndoU fold and contains a canonical catalytic triad, but unexpectedly it lacks the characteristic uridylate specificity of the superfamily. Instead, CdiA‐CTSTECO31 is an anticodon loop nuclease that preferentially cleaves after a cytidylate residue in tRNAUUC Glu. This unique specificity is correlated with a distinct architecture that differs from previously characterized prokaryotic EndoU toxins.
Results
Structure of the CdiA‐CT/CdiISTECO31 complex
We co‐expressed the CdiA‐CTSTECO31 toxin together with His6‐tagged CdiISTECO31 immunity protein and purified the complex by Ni2+‐affinity chromatography. Limited proteolysis with subtilisin was used to generate a complex that is suitable for crystallization (Fig. S1). This treatment removed the N‐terminal “translocation” domain of CdiA‐CTSTECO31. This domain does not affect toxin activity, but is required to transfer the CdiA‐CT into the cytoplasm of target bacteria (Willett et al., 2015). The truncated CdiA‐CTSTECO31 and full‐length CdiISTECO31 crystallized in space group P65 with one complex in the asymmetric unit. The structure was determined at 2.0 Å by the single wavelength anomalous dispersion approach using selenomethionine (SeMet)‐labeled proteins (Table 1). The final model contains residues Lys181 – Lys323 of CdiA‐CTSTECO31 and CdiISTECO31 residues Met1 – Pro120. The C‐terminal domain of CdiA‐CTSTECO31 adopts a V‐shaped structure comprised of two α/β subdomains (Fig. 1A). The N‐terminal subdomain consists of a β‐hairpin (β1 and β2) followed by helix α1 and an antiparallel β‐sheet. The sheet is composed of three major strands (β3, β4 and β6) and a shorter β5 strand that together with β6 flanks β4. The C‐terminal subdomain begins at helix α2 and continues as an antiparallel β‐sheet (β7, β8 and β9). The N‐terminal tail crosses over to the adjacent subdomain, bringing the N‐ and C‐termini into close proximity. The CdiISTECO31 immunity protein is composed of a core antiparallel β‐sheet (β1′, β2′, β3′ and β4′) surrounded by helices α1′, α2′, α3′ and α4′ (Fig. 1A). The β‐sheet is located topologically between helices α2′ and α3′. The immunity protein also contains three short sections (G1′, G2′ and G3′) that adopt 310‐helical geometry (Fig. 1A).
Table 1.
Data processing and refinement statistics
| Processing | |
| Wavelength (Å) | 0.9793 |
| Resolution range (Å)a | 30.0 – 2.00 (2.03 – 2.00) |
| Space group | P65 |
| Unit cell parameters (Å) | a = 89.43 b = 89.43 c = 76.15 |
| Unique reflections | 23,397 (1,192) |
| Multiplicity | 6.3 (5.7) |
| Completeness (%) | 99.2 (99.8) |
| <I >/< σI> | 17.03 (2.21) |
| Wilson B factor (Å2) | 21.0 |
| Rmerge b | 0.117 (0.839) |
| CC1/2 | 0.689 |
| CC* | 0.903 |
| Refinement | |
| Resolution (Å) | 30.00 ‐ 2.00 |
| Reflections work/test set | 22,180/1,140 |
| Rwork/Rfree | 0.1674/0.2076 |
| Average B factor (Å2) (No of atoms) | |
| macromolecules | 30.4 (2,125) |
| solvent | 34.3 (194) |
| Rmsd bond lengths (Å) | 0.014 |
| Rmsd bond angles (°) | 1.487 |
| Ramachandran favoredc (%) | 98.1 |
| Ramachandran outliers (%) | 0 |
| Clashscorec | 0.72 |
Values in parentheses correspond to the highest resolution shell.
R merge = ΣhΣj|Ihj–<Ih>|/ΣhΣjIhj, where Ihj is the intensity of observation j of reflection h.
R = Σh|Fo|–|Fc|/Σh|Fo| for all reflections, where Fo and Fc are observed and calculated structure factors respectively. R free is calculated analogously for the test reflections, randomly selected and excluded from the refinement.
As defined by Molprobity (Davis et al., 2004).
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Figure 1.
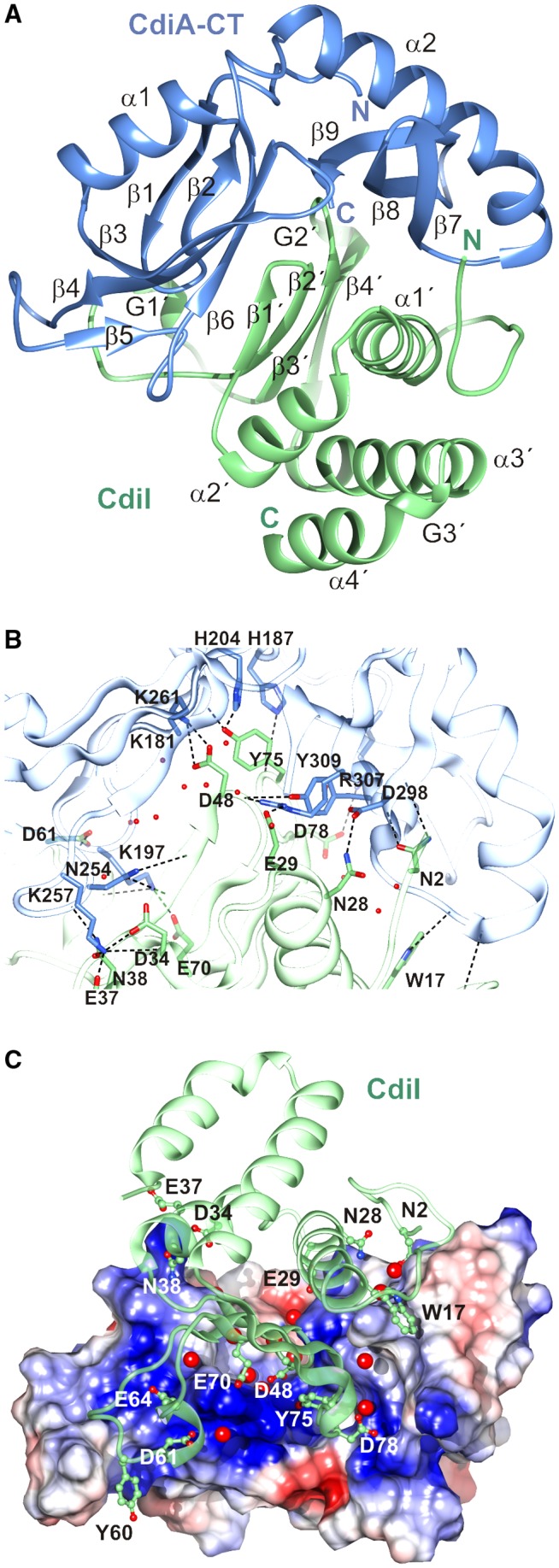
Structure of the CdiA‐CT/CdiISTECO31 complex. A. The CdiA‐CT/CdiISTECO31 complex is depicted in cartoon with the toxin domain colored blue and the immunity protein colored green. Secondary structure elements are labeled with CdiISTECO31 elements denoted by a prime (′) symbol. B. The toxin‐immunity protein interface is depicted with selected side‐chains forming direct hydrogen bonds (black dashed lines) shown in a stick representation. Water molecules that mediate interactions are shown as red spheres. C. Charge complementarity at the toxin‐immunity protein interface. The electrostatic potential of the toxin surface was calculated using Coulomb's law with Chimera (Pettersen et al., 2004). Potentials range from –10 kcal/mol*e (red) to +10 kcal/mol*e (blue). Water molecules that mediate interactions are shown as red spheres. [Colour figure can be viewed at http://wileyonlinelibrary.com]
CdiISTECO31 binds between the two subdomains of the toxin domain, with its β‐sheet inserting into the toxin cleft (Fig. 1A). The N‐terminus and helix α1′ of CdiISTECO31 interact with the C‐terminal subdomain of the toxin, and the long G1′ loop contacts the N‐terminal subdomain (Fig. 1A). The interface buries approximately 1,600 Å2, corresponding to ~20% of the total solvent‐accessible surface area as determined by PISA (Krissinel and Henrick, 2007). The interaction network is elaborate and features numerous direct contacts and more than 20 water‐mediated hydrogen bonds (Fig. 1B, Tables 2 and S1). Moreover, the toxin cleft is strongly electropositive, and several anionic residues from CdiISTECO31 form salt‐bridges (Fig. 1C). Asp48 and Asp78 of CdiISTECO31 form salt‐bridges with toxin residues Lys261 and Lys181 respectively. Arg307 and Tyr309 from the toxin also converge on the main‐chain carbonyl of CdiISTECO31 Asp48. CdiISTEO31 Tyr75 makes direct H‐bond contacts with His187, Gly202 and His204 (Fig. 1B and Table 2). The side‐chains of Trp17, Thr18, Asn28 and Glu29 emanate from CdiISTECO31 helix α1′ to make contacts with toxin residues Gly293, Gln295, Asp298 and Arg307 respectively (Figs. 1B and C). Additionally, CdiISTECO31 Trp17 makes van der Waals contacts in a pocket formed by toxin residues Gln295 and Gln312 (Fig. 1C). The N‐terminal subdomain of the toxin interacts primarily with CdiISTECO31 helix α2′ and the long G1′ loop that links β2′ to β3′. Toxin residue Lys257 is the focal point of interactions with side‐chains of Asp34, Glu37 and Asn38 from immunity helix α2′ (Fig. 1B and C). Residues Asp61 and Ser63 from the G1′ loop form direct H‐bonds with toxin residue Lys197 (Fig. 1B and Table 2), and CdiISTECO31 Tyr60 packs against residues Ala243, Ala244 and Gly245, which form the β‐turn connecting toxin strands β4 and β5 (Fig. 1C).
Table 2.
Direct H‐bonds and salt‐bridges at the CdiA‐CT/CdiISTECO31 interface
| CdiA‐CT atom | CdiI atom | distance (Å) |
|---|---|---|
| Lys197 (NZ) | Ser63 (O) | 3.23 |
| Lys257 (NZ) | Glu37 (OE1) | 2.81 |
| Lys197 (NZ) | Glu70 (OE2) | 2.54 |
| Lys257 (NZ) | Asp34 (O) | 3.16 |
| Lys257 (NZ) | Asp34 (OD1) | 2.85 |
| Lys261 (NZ) | Asp48 (OD1) | 3.30 |
| Lys261 (NZ) | Asp48 (OD2) | 2.76 |
| Lys197 (N) | Asp61 (OD2) | 3.26 |
| Lys181 (NZ) | Asp78 (OD2) | 3.49 |
| Lys257 (NZ) | Asn38 (OD1) | 2.77 |
| His187 (NE2) | Tyr75 (O) | 2.84 |
| His204 (NE2) | Tyr75 (OH) | 2.69 |
| Gly202 (N) | Tyr75 (OH) | 2.70 |
| Gln295 (NE2) | Thr18 (OG1) | 3.25 |
| Asp298 (N) | Asn2 (OD1) | 2.80 |
| Asn254 (ND2) | Ile46 (O) | 2.96 |
| Asp298 (O) | Asn2 (ND2) | 3.11 |
| Asp298 (OD2) | Asn28 (ND2) | 2.85 |
| Asn254 (O) | Asn38 (ND2) | 3.01 |
| Arg307 (NH1) | Glu29 (OE1) | 2.96 |
| Arg307 (NE) | Asp48 (O) | 3.38 |
| Arg307 (NH2) | Asp48 (O) | 2.74 |
| Tyr309 (OH) | Asp48 (O) | 3.01 |
| Asn292 (O) | Tyr10 (N) | 3.10 |
| Gly293 (O) | Trp17 (NE1) | 2.92 |
| Asn254 (OD1) | Ile46 (N) | 2.75 |
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
CdiA‐CTSTECO31 contains an EndoU ribonuclease domain
Consistent with its annotation as an EndoU toxin, the DALI server returned eukaryotic EndoU domains as the closest structural homologs of CdiA‐CTSTECO31 (Holm and Rosenstrom, 2010). Coronaviral NendoU domains were the top hits in the search: coronavirus 229E Nsp15 (PDB:4S1T), Z = 6.1, rmsd 3.6 Å over 100 Cα atoms; SARS Nsp15 (PDB:2RHB (Bhardwaj et al., 2008)) Z = 6.0, rmsd 3.6 Å over 101 Cα atoms; SARS Nsp15 (PDB:2H85 (Ricagno et al., 2006)), Z = 6.0, rmsd 3.5 Å over 100 Cα atoms, and murine hepatitis virus Nsp15 (PDB:2GTH (Xu et al., 2006)), Z = 5.5, rmsd 3.8 Å for 101 Cα atoms. The NendoU domains and CdiA‐CTSTECO31 toxin have essentially identical topologies, though some secondary structure elements vary in length and relative position (Fig. 2A and B). Notably, NendoU domains harbor the two catalytic His residues within helix α8 and the adjacent loop, which corresponds to loop β2‐α1 that follows the β1‐β2 hairpin of the toxin domain (Figs. 2A and B). CdiA‐CTSTECO31 also contains additional residues that form strand β5 and contribute to an elongated helix α2 (Fig. 2B). XendoU (PDB:2C1W (Renzi et al., 2006)) is more distantly related to the toxin at Z = 5.2, rmsd 3.1 Å over 108 Cα atoms. As with Nsp15, the XendoU active‐site loop aligns poorly with the β1‐β2 hairpin of the toxin (Fig. 2C and D). NendoU domains from arteriviral Nsp11 proteins show the lowest structural similarity to the toxin. Nsp11 from porcine reproductive and respiratory virus (PDB:5DA1) was recovered at Z = 4.5 with rmsd 3.7 Å over 84 Cα atoms. Notably, sequence homology between CdiA‐CTSTECO31 and the eukaryotic EndoU domains is low, ranging from ~9% identity with XendoU up to about 15% for the coronavirus NendoU domains (Fig. 2B and D). The structural homology search also recovered a handful of pseudopilin proteins from T2SSs. However, structural similarity with pseudopilins is limited to the C‐terminal subdomain of the toxin.
Figure 2.
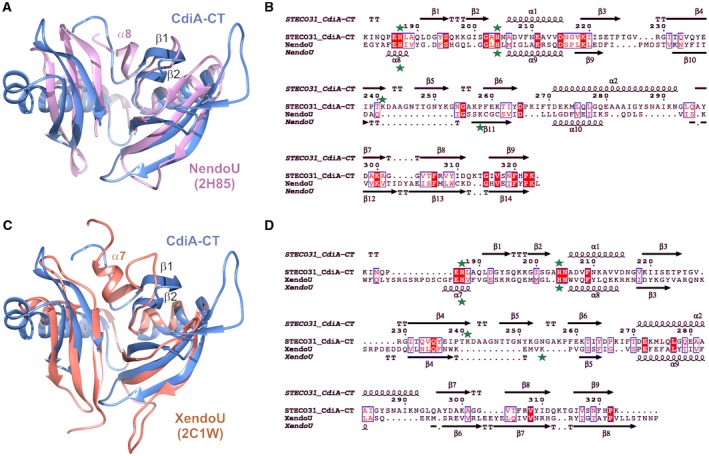
The CdiA‐CTSTECO31 toxin is a prokaryotic EndoU domain. A. CdiA‐CTSTECO31 (blue) and the C‐terminal domain of SARS Nsp15 (PDB: 2H85, pink) were aligned using secondary structure matching (SSM) superposition calculated in Coot (Emsley and Cowtan, 2004). The β1‐β2 hairpin of CdiA‐CTSTECO31 and helix α8 in the NendoU active‐site loop are indicated. B. The sequences of the CdiA‐CTSTECO31 and SARS Nsp15 C‐terminal domains were aligned based on structure using DALI (Holm and Rosenstrom, 2010). The resulting sequence alignment was rendered using Espript (Robert and Gouet, 2014). Identical residues are highlighted in red, and similar residues are shown in red font. The predicted catalytic triad residues are marked with green stars. C. CdiA‐CTSTECO31 (blue) and the C‐terminal domain of XendoU (PDB: 2C1W, coral) were aligned by SSM superposition. The β1‐β2 hairpin of CdiA‐CTSTECO31 and helix α7 in the XendoU active‐site loop are indicated. D. CdiA‐CTSTECO31 and XendoU sequences were aligned as described in panel B. [Colour figure can be viewed at http://wileyonlinelibrary.com]
Two prokaryotic EndoU toxins have been characterized to date. BC0920 from Bacillus cereus ATCC 14579 is a putative T7SS effector that degrades rRNA and tRNA in vivo (Holberger et al., 2012). MafB1 from Neisseria meningitidis 8013 cleaves RNA after uridylate nucleotides in vitro (Jamet et al., 2015). To determine whether CdiA‐CTSTECO31 possesses similar activity, we expressed the toxin in E. coli and examined RNA for degradation. Although the toxin inhibited cell growth (Fig. S2A), there was no obvious RNase activity in the intoxicated cells (Fig. S2B). Because several CDI toxins cleave specific tRNAs (Nikolakakis et al., 2012; Willett et al., 2015; Jones et al., 2017; Michalska et al., 2017), we used Northern blot analysis to identify possible tRNA substrates. This screen revealed efficient cleavage of tRNAUUC Glu in CdiA‐CTSTECO31 intoxicated cells, but not in cells expressing other toxins (Fig. 3). We also detected modest nuclease activity against tRNAGUC Asp, tRNAUUG Gln, tRNACCC Gly, tRNAUCC Gly, tRNAGCC Gly, tRNAUUU Lys, tRNAGGU Thr and tRNACCA Trp (Fig. 3). These results indicate that CdiA‐CTSTECO31 exhibits a tRNase activity that is novel for the EndoU superfamily.
Figure 3.
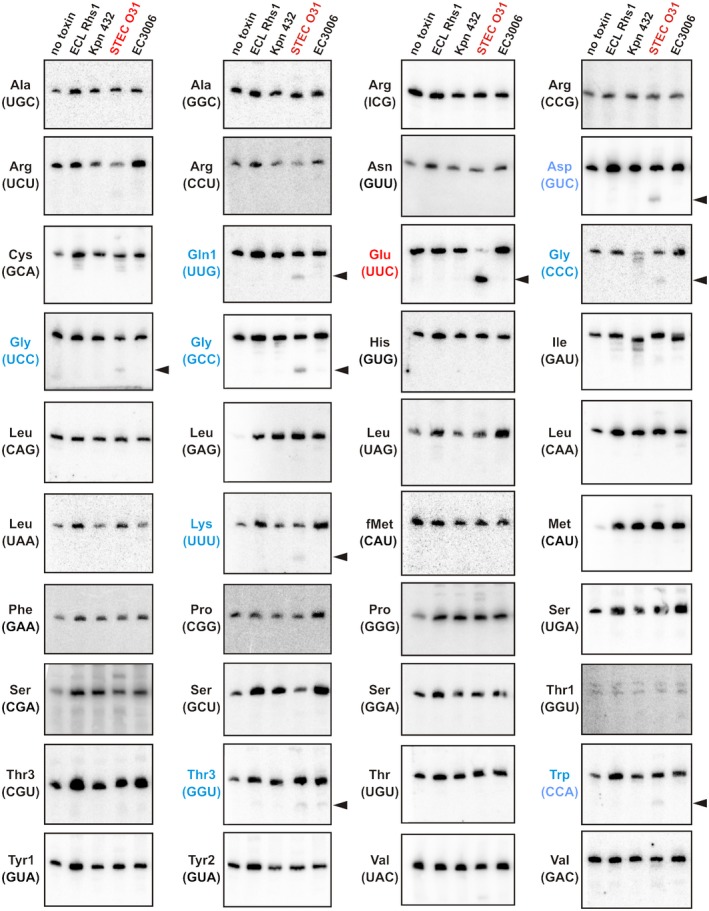
CdiA‐CTSTECO31 is a specific tRNase. Northern blot analyses of RNA isolated from intoxicated E. coli cells. The C‐terminal toxin domains of Rhs1 from E. cloacae ATCC 13047 (ECL Rhs1: YP_003612075.1), CdiA from K. pneumoniae 342 (Kpn 342: ACI08381.1), CdiA from E. coli STEC_O31 (STEC O31: EJK94116.1) and CdiA E. coli 3006 (EC3006: EKI34460.1) were expressed in E. coli, and total RNA was isolated for Northern blot hybridization using radiolabeled probes to the indicated tRNAs. Anticodon sequences are shown in parentheses for specific isoacceptors. Arrows to the left of the blot indicate cleavage products detected in CdiA‐CTSTECO31 intoxicated cells. [Colour figure can be viewed at http://wileyonlinelibrary.com]
CdiISTECO31 homologs and the specificity of immunity
A DALI server search for structural homologs of CdiISTECO31 returned NMB0503 from N. meningitidis MC58 (PDB ID:4Q7O, (Tan et al., 2015)) with Z = 13.4 and rmsd 2.6 Å over 111 Cα atoms (Fig. 4A). Although it shares only ~16% sequence identity with CdiISTECO31 (Fig. 4B), NMB0503 is probably an immunity protein because the upstream NMB0502 reading frame encodes a C‐terminal EndoU domain that is 39% identical to CdiA‐CTSTECO31 (Fig. 4C). Moreover, NMB0502 has the same tRNase activity as CdiA‐CTSTECO31 when expressed in E. coli (Fig. S3). The structures of the two immunity proteins deviate in three regions: (i) NMB0503 contains an N‐terminal helix (α0′) that is absent from CdiISTECO31; (ii) the loop connecting β2′ to β3′ is extended in NMB0503 and contains an additional 310 helix and (iii) CdiISTECO31 contains a 310 helix (G2′) in the loop connecting β3′ to β4′ (Fig. 4B). Given that these secondary structure elements interact with the toxin domain in the CdiA‐CT/CdiISTECO31 structure (Fig. 4A), the discrepancies may reflect the toxin‐free state of NMB0503 (Tan et al., 2015). However, NMB0502 diverges from CdiA‐CTSTECO31 in regions that interact directly with CdiISTECO31 (Fig. 4C), suggesting that these differences are critical for specific binding interactions between toxins and cognate immunity proteins. This conclusion is supported by alignments of closely related EndoU toxins and immunity proteins, which show that each pair possesses a unique set of residues contributing to the binding interface (Fig. S4).
Figure 4.
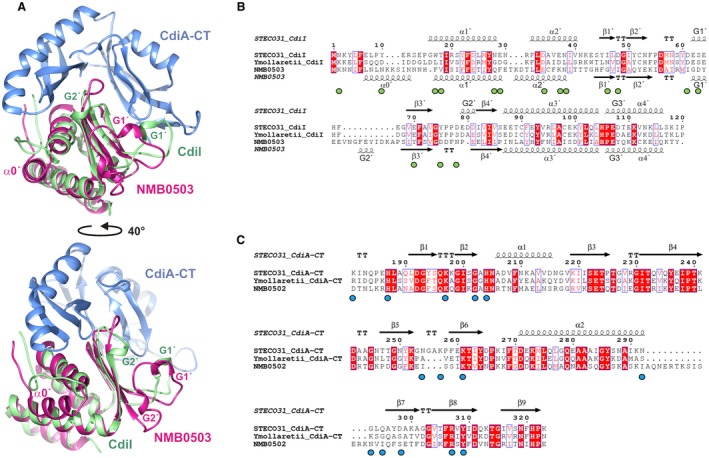
Structure‐based alignment of CdiISTECO31 and the NMB0503 immunity protein of N. meningitidis MC58. A. CdiISTECO31 (green) and NMB0503 (PDB: 4Q7O, magenta) were aligned by SSM superposition. The CdiA‐CTSTECO31 toxin domain (blue) is included to illustrate the toxin‐immunity protein binding interface. The bottom view is rotated 40° with respect to the top view. B. CdiISTECO31 and NMB0503 sequences were aligned based on structure using DALI. The CdiIYmo43969 immunity protein from Y. mollaretii ATCC 43968 was aligned using Clustal‐Omega. The resulting alignment was rendered using Espript (Robert and Gouet, 2014) with identical residues highlighted in red, and similar residues shown in red font. Green circles below the alignment indicate CdiISTECO31 residues that form direct H‐bonds with the EndoU domain of CdiA‐CTSTECO31. C. EndoU toxins from E. coli STEC_O31, Y. mollaretii ATCC 43969 and N. meningitidis MC58 were aligned using Clustal‐Omega. The resulting alignment was rendered using Espript (Robert and Gouet, 2014) with identical residues highlighted in red, and similar residues shown in red font. Blue circles below the alignment indicate CdiA‐CTSTECO31 residues that form direct H‐bonds with CdiISTECO31 immunity protein. [Colour figure can be viewed at http://wileyonlinelibrary.com]
To explore the specificity of immunity proteins, we tested whether EndoU toxins can be neutralized by near‐cognate CdiI. We first fused the cdiA‐CT/cdiI STECO31 module to the cdiA EC93 gene of E. coli EC93, thereby generating a plasmid‐borne CDI system capable of delivering CdiA‐CTSTECO31 toxin into E. coli target cells. Inhibitor cells expressing the chimeric CDISTECO31 system significantly outcompeted E. coli target bacteria when seeded at a 1:1 ratio in co‐culture (Fig. 5A). Northern blot analysis revealed cleaved tRNAUUC Glu in the mixed culture (Fig. 5C). Because inhibitor cells are immune to the toxin, the latter result suggests that most of the tRNAUUC Glu is degraded in target bacteria. Target cells expressing CdiISTECO31 were protected from both growth inhibition and tRNase activities (Fig. 5A and C). By contrast, targets expressing near‐cognate NMB0503 were inhibited to the same extent as cells with no immunity gene (Fig. 5A), and Northern blot showed that NMB0503 failed to neutralize tRNase activity (Fig. 5C). We then tested whether a more closely related immunity protein blocks CdiA‐CTSTECO31. The T5SS/CDI locus of Yersinia mollaretii ATCC 43969 encodes an EndoU toxin and immunity protein that are ~55% and ~58% identical to CdiA‐CTSTECO31 and CdiISTECO31 respectively (Fig. 4B and C). Despite this homology, cdiI Ymo43969 expression did not protect target cells from CDISTECO31 mediated growth inhibition (Fig. 5A and C). To confirm that the cdiI Ymo43969 construct produces functional immunity protein, we tested it against inhibitor cells that express a CdiAEC93‐CTYmo43969 fusion protein. The chimeric CDIYmo43969 system is less potent than CDISTECO31 (compare Fig. 5B to A), but toxic nuclease activity was detected in co‐cultures with non‐immune target cells (Fig. 5D). As expected, target cells that express CdiIYmo43969 were immune to CDIYmo43969 mediated growth inhibition and nuclease activity, but near‐cognate CdiISTECO31 failed to protect target bacteria (Figs. 5B and 5D). Surprisingly, we found that NMB0503‐expressing target cells were not killed in co‐culture with CDIYmo43969 inhibitors (Fig. 5B). Nevertheless, cleaved tRNAUUC Glu accumulated in this latter co‐culture (Fig. 5D), demonstrating that NMB0503 immunity protein does not completely block tRNase activity. Presumably, the NMB0503 expressing target cells retained enough full‐length tRNA to support growth. Taken together, these results indicate that each EndoU toxin‐immunity protein interaction is distinct.
Figure 5.
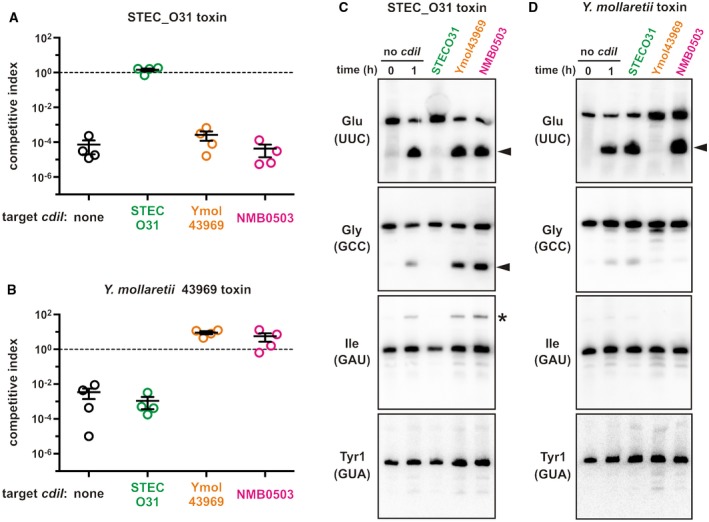
Specificity of CdiI immunity function. E. coli inhibitor strains that deploy CdiA‐CTSTECO31 (panel A) or CdiA‐CTYmo43969 (panel B) were cultured at a 1:1 ratio with E. coli target cells that express the indicated cdiI immunity genes. Viable inhibitor and target bacteria were enumerated as colony forming units (cfu) at t = 0 and after 1 h of co‐culture CDISTECO31 competitions (panel A) and 3 h for CDIYmo43969 competitions (panel B). The competitive index = (cfut = final targets/cfut = final inhibitors)/(cfut = 0 targets/cfut = 0 inhibitors). Competitive indices for four independent experiments are reported together with mean ± standard error. Northern blot analysis of total RNA isolated from CDISTECO31 (panel C) and CDIYmo43969 (panel D) competition co‐cultures. One sample was collected immediately after mixing (t = 0 h) and all other samples were collected after 1 h of co‐culture. Carets indicate cleavage products, and the asterisk indicates an incompletely processed tRNAGAU Ile transcript that accumulates in CdiA‐CTSTECO31 intoxicated cells. [Colour figure can be viewed at http://wileyonlinelibrary.com]
The EndoU nuclease active site
Eukaryotic EndoU nucleases cleave phosphodiester bonds using a His‐His‐Lys catalytic triad (Ivanov et al., 2004; Guarino et al., 2005; Shi et al., 2016; Kang et al., 2007; Nedialkova et al., 2009). The EndoU catalytic mechanism is similar to that of RNase A, which uses two His residues to mediate proton transfers and Lys to stabilize the pentavalent phosphoryl transition‐state (Findlay et al., 1962; Deavin et al., 1966; Cuchillo et al., 2011). Structural superimposition suggests that toxin residues His187 and His204 are equivalent to the catalytic His residues in SARS Nsp15 (His234/His249) and XendoU (His162/His178) (Fig. 6A). Further, CdiA‐CTSTECO31 Lys261 is positioned near Lys289 of Nsp15 and Lys224 of XendoU (Fig. 6A). The EndoU active site also contains conserved residues that contribute to uridylate specificity. Ser293 of SARS Nsp15 (Ser174 in EAV Nsp11, Ser228 in XendoU) is predicted to form a H‐bond with O2 of uracil, and Tyr342 (Tyr216 in EAV Nsp11, Tyr280 in XendoU) is thought to stack onto the pyrimidine ring (Bhardwaj et al., 2008; Nedialkova et al., 2009). The structure overlay suggests that CdiA‐CTSTECO31 residues Thr262 and His321 may also function in substrate discrimination (Fig. 6A). Finally, toxin residue Pro322 superimposes closely onto conserved Pro residues that are required for Nsp15 nuclease activity (Fig. 6A) (Ricagno et al., 2006; Bhardwaj et al., 2008).
Figure 6.
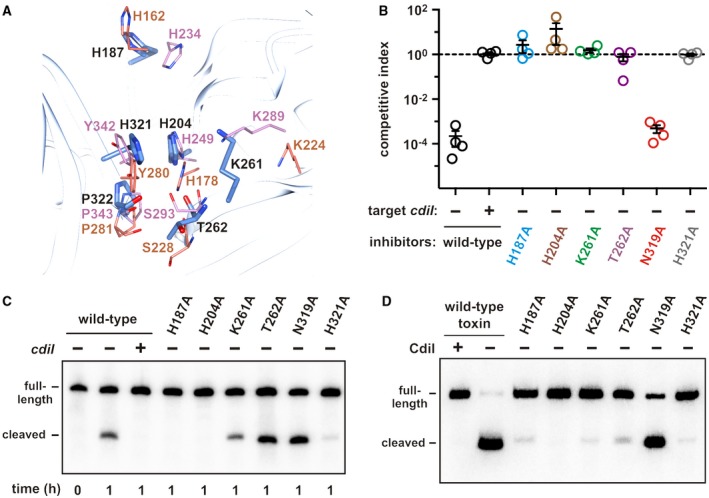
The CdiA‐CTSTECO31 active site. A. Superimposition of XendoU and SARS Nsp15 active‐site residues onto CdiA‐CTSTECO31. Side‐chains from SARS Nsp15 are shown in pink, and those from XendoU are in coral. CdiA‐CTSTECO31 residues are rendered in blue and labeled with black font. B. E. coli inhibitors that deploy the indicated CdiA‐CTSTECO31 variants were cultured at 1:1 ratio with E. coli target cells. Viable inhibitor and target bacteria were enumerated as colony forming units (cfu) at t = 0 and t = 1 h, and the competitive index calculated. Competitive indices for four independent experiments are reported together with mean ± standard error. C. Northern blot analysis of tRNAGlu isolated from the competition co‐cultures in panel B. D. In vitro activities of CdiA‐CTSTECO31 variants. The indicated toxins were purified and incubated with E. coli total RNA for 30 min at 37 °C. Reactions were analyzed by Northern blot using a probe to tRNAUUC Glu. [Colour figure can be viewed at http://wileyonlinelibrary.com]
We probed the CdiA‐CTSTECO31 active site using site‐directed mutagenesis. We first introduced Ala substitutions into the CdiAEC93‐CTSTECO31 chimera and tested inhibition activity in competition co‐cultures. Mutation of His187, His204, Lys261, Thr262 and His321 abrogated growth inhibition (Fig. 6B). In contrast, substitution of Asn319 in the toxin active site had no discernable effect on inhibition activity (Fig. 6B). Immunoblot analysis confirmed that each CdiA variant was produced at the same level as the wild‐type effector (Fig. S5). Furthermore, the mutated effectors were susceptible to degradation with extracellular proteinase K (Fig. S5), indicating that each was exported to the cell surface properly. Although target bacteria were not inhibited by cells deploying the Lys261Ala and Thr262Ala toxin variants (Fig. 6B), we detected tRNase activity in these co‐cultures (Fig. 6C). Presumably, these latter toxins are attenuated enough to allow target cell growth. We then compared the activities of mutant toxins to wild‐type CdiA‐CTSTECO31 using in vitro nuclease assays. Purified wild‐type toxin cleaved tRNAUUC Glu efficiently, but its activity was neutralized when purified CdiISTECO31 immunity protein was included in the reaction (Fig. 6D). The His204Ala substitution appeared to ablate nuclease activity, and toxins carrying His187Ala and Lys261Ala substitutions exhibited little activity in vitro (Fig. 6D). The Thr262Ala and His321Ala substitutions also significantly reduced nuclease activity compared to wild‐type (Fig. 6D). Purified Asn319Ala toxin retained nuclease activity (Fig. 6D), consistent with its near wild‐type function in the competition co‐culture experiments (Fig. 6C). These results suggest that CdiA‐CTSTECO31 uses the same metal‐independent catalytic mechanism as RNase A and eukaryotic EndoU nucleases. Consistent with this model, we found that purified CdiA‐CTSTECO31 retains robust tRNase activity in reactions supplemented with EDTA (Fig. S6). Thus, the CdiA‐CTSTECO31 EndoU domain appears to function as a metal‐independent phosphotransferase.
Prokaryotic EndoU toxins have evolved into three clades
The tRNase activity of CdiA‐CTSTECO31 differs from previously characterized EndoU toxins, suggesting the superfamily may have evolved a broader spectrum of substrate specificities. Analysis of EndoU toxin sequences from predicted T5SS/CdiA effector proteins shows that the domains segregate into at least three major clades. Clade I domains are related to the previously characterized MafB1 toxin from N. meningitidis 8013 (Jamet et al., 2015) (Fig. 7A). Clade I toxins are characterized by greatly abbreviated N‐terminal subdomains that lack the α1, β3, β4 and β5 elements of CdiA‐CTSTECO31 (Fig. S7). Clade I toxins contain additional residues between α2 and β7 in the C‐terminal subdomain and sometimes carry extended C‐terminal tails (Fig. S7). Clade II toxins are similar to BC0920 from B. cereus ATCC 14579 and are predicted to lack helix α1 (Figs. 7A and S7). CdiA‐CTSTECO31, CdiA‐CTYmo43969 and NMB0502 contain clade III nuclease domains (Fig. 7A). All EndoU domains are predicted to carry the two catalytic His residues within a β‐hairpin, though the spacing of these residues varies amongst toxins (Fig. S7). Clades I and III contain the catalytic Lys residue within a conserved K‐(S/T) motif in strand β6 (Fig. S7). The equivalent Lys residues are not easily identified in clade II, though many of these toxins contain irregularly spaced K‐(S/T) motifs within the β5 region (Fig. S7).
Figure 7.
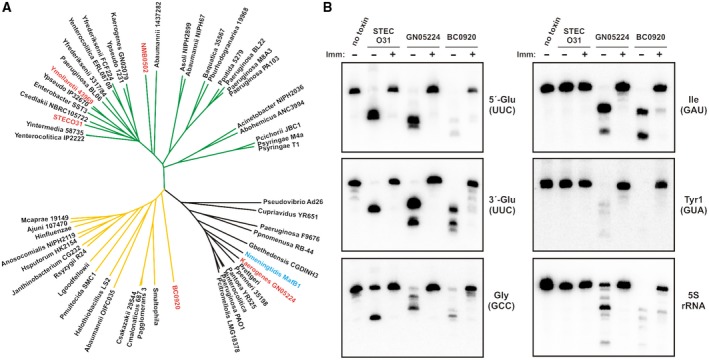
Diversification of prokaryotic EndoU toxin domains. A. EndoU domains from predicted CdiA proteins were aligned using Clustal‐Omega, and the associated unrooted tree rendered using the Tree of Life website. The source multiple sequence alignment is presented in Fig. S7, which also lists the NCBI Refseq accession numbers for each protein. EndoU domains from N. meningitidis MafB1 and B. cereus BC0920 are included for comparison. B. In vitro activities of EndoU toxins. The indicated toxins were purified and incubated with E. coli total RNA in the absence or presence of cognate immunity protein. Reactions were analyzed by Northern blot hybridization with probes to the indicated RNAs. [Colour figure can be viewed at http://wileyonlinelibrary.com]
To directly compare nucleases from each EndoU clade, we purified representative toxins and examined their RNase activities in vitro. For clade I, we chose a predicted CdiA‐CT from Klebsiella aerogenes GN05224, reasoning that it should be functional when fused to CdiAEC93 to generate a chimeric effector. Additionally, the CdiA‐CTGN05224 EndoU domain shares 61% sequence identity with the previously characterized N. meningitidis MafB1 toxin (Jamet et al., 2015). Purified CdiA‐CTGN05224 degrades 5S rRNA and several tRNA species in vitro (Fig. 7B). Similar RNase activity was detected in competition co‐cultures with inhibitor cells that deploy CdiA‐CTGN05224, though 5S rRNA degradation was not observed in vivo (Fig. S8), presumably because it is protected by ribosomal proteins. We also found that CdiA‐CTGN05224 RNase activity is effectively neutralized by CdiIGN05224 immunity protein in vitro (Fig. 7B) and in vivo (Fig. S8). We chose B. cereus BC0920 as a representative clade II toxin and found that its RNase activity is similar to that of CdiA‐CTGN05224 (Fig. 7B). Like the other nucleases, BC0920 activity was suppressed when the reaction was supplemented with purified BC0921 immunity protein (Fig. 7B). Finally, we confirmed the substrate specificity of CdiA‐CTSTECO31 toxin in vitro. Purified CdiA‐CTSTECO31 cleaves tRNAUUC Glu efficiently and shows partial activity on tRNAGCC Gly, but has no discernable activity on tRNAGAU Ile, tRNAGUA Tyr or 5S rRNA (Fig. 7B). Moreover, CdiA‐CTSTECO31 is most likely a specific endonuclease, because it produces stable 5′ and 3′ fragments of tRNAUUC Glu (Fig. 7B).
Finally, we mapped the CdiA‐CTSTECO31 cleavage site within tRNAUUC Glu. The comparable sizes of the 5′ and 3′ fragments suggest that the toxin cleaves tRNAUUC Glu in the anticodon loop. Therefore, we designed an oligonucleotide that anneals downstream of the anticodon for primer extension analysis (Fig. 8A). Analysis of 3′‐fragments generated from in vivo toxin expression revealed cleavage at the 5′‐side of m2A38 (Fig. 8B and C). This same site was observed in tRNAUUC Glu isolated from competition co‐cultures. As expected, these cleavages were not detected in cells that express cdiI STECO31 (Fig. 8B). We also determined the same cleavage site in tRNAUUC Glu digested with purified CdiA‐CTSTECO31 in vitro (Fig. 8B). This latter result confirms that CdiA‐CTSTECO31 is directly responsible for producing the 3′‐fragment, and strongly suggests that the toxin cleaves the phosphodiester linking nucleotides C37 and m2A38 (Fig. 8C, red arrow).
Figure 8.
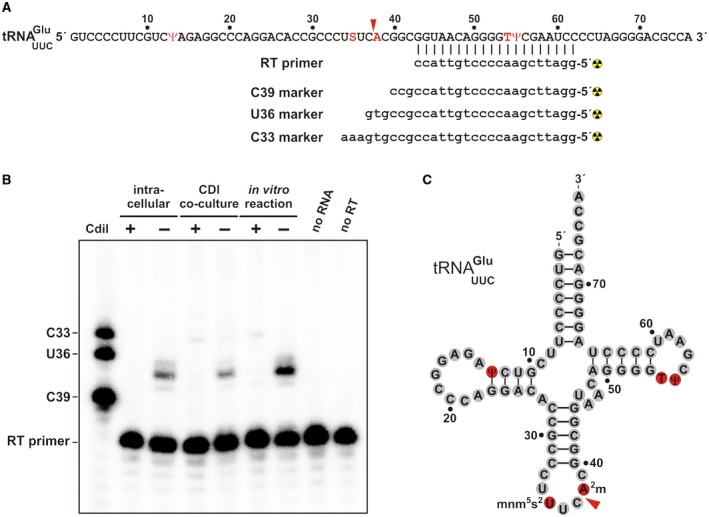
CdiA‐CTSTECO31 is an anticodon nuclease. A. tRNAUUC Glu sequence showing the hybridized reverse transcriptase (RT) primer and oligonucleotide standards used to map the CdiA‐CTSTECO31 cleavage site. B. Primer extension analysis. RNA was isolated competition co‐cultures and cells intoxicated by intracellular CdiA‐CTSTECO31 expression. Samples from in vitro nuclease assays were also analyzed. The neutralizing effect of CdiISTECO31 was tested where indicated. The reverse transcriptase (RT) primer was radiolabeled, hybridized to tRNAUUC Glu, and extended with reverse transcriptase. Reactions and radiolabeled marker oligonucleotides were run on a 50% urea, 15% polyacrylamide gel and visualized by phosphorimaging. C. Position of CdiA‐CTSTECO31 cleavage within the tRNAUUC Glu anticodon loop. Modified nucleotides are highlighted in red. [Colour figure can be viewed at http://wileyonlinelibrary.com]
Discussion
Aravind and colleagues first identified the link between eukaryotic EndoU RNases and a broadly distributed family of antibacterial toxins (Zhang et al., 2012; Zhang et al., 2011). The structure of CdiA‐CTSTECO31 confirms those predictions, and mutational analysis suggests that this toxin uses the same catalytic mechanism as eukaryotic EndoU nucleases. However, there are also significant differences between the prokaryotic and eukaryotic nucleases. Purified XendoU and Nsp15 are only active in the presence of millimolar Mn2+ or Ca2+ (Laneve et al., 2003; Schwarz and Blower, 2014; Ivanov et al., 2004; Bhardwaj et al., 2004), whereas the bacterial toxins examined here have no metal requirement. There is no direct role for divalent metal in the EndoU catalytic mechanism (Ricagno et al., 2006; Renzi et al., 2006), and like the bacterial toxins, arteriviral Nsp11 is active in the absence of metal (Nedialkova et al., 2009). These observations suggest that metals may promote XendoU and Nsp15 nuclease activity through allostery. This model is consistent with data showing that SARS Nsp15 undergoes conformational changes upon titration with Mn2+ (Bhardwaj et al., 2004). Allosteric regulation could be mediated by the additional structural elements found in eukaryotic EndoU enzymes. Bacterial EndoU domains are about half the size of XendoU, which contains ~150 additional N‐terminal residues that form an extensive α‐helical cradle that surrounds and supports the nuclease core (Renzi et al., 2006). In this model, conformational changes in the supporting cradle would reshape the active‐site cleft and/or alter the configuration of the catalytic triad. The supporting structures in NendoU domains are abbreviated, but Nsp11 and Nsp15 contain additional N‐terminal domains that mediate oligomerization (Bhardwaj et al., 2008; Ricagno et al., 2006). Oligomerization is critical for Nsp15 activity because the active‐site loop is stabilized by the β10‐β11 “supporting loop”, which is in turn conformationally constrained by the neighboring protomer (Joseph et al., 2007; Guarino et al., 2005). In monomeric Nsp15, the active‐site loop is untethered and migrates into the substrate‐binding groove, where it presumably blocks the interaction with substrate RNA. Similarly, one of the active‐site loops is disordered in crystal structures of homodimeric Nsp11, again raising the possibility that RNase activity is regulated through multimerization (Shi et al., 2016; Zhang et al., 2017; Joseph et al., 2007). These findings suggest that eukaryotic EndoU proteins must be carefully controlled because unrestrained RNase activity is lethal. Such allosteric control should not be required of prokaryotic EndoU nucleases, because these enzymes are used to inhibit bacterial cell growth. Nevertheless, the potent toxic effects of EndoU nucleases must be suppressed in the producing cell, thus providing the selective pressure to evolve immunity proteins that protect against self‐intoxication.
The narrow substrate specificity of CdiA‐CTSTECO31 is surprising given that all previously characterized EndoU enzymes cleave RNAs after uridylate. The determinants governing EndoU nuclease specificity have been examined most extensively for viral NendoU proteins. Ser293 in Nsp15 (Ser228 in XendoU) is predicted to form specific H‐bond contacts with uracil, and Ala substitutions at this position relax specificity and increase the rate of cleavage at cytidylate nucleotides (Ricagno et al., 2006; Bhardwaj et al., 2008; Nedialkova et al., 2009). The corresponding residue in CdiA‐CTSTECO31 is Thr262, which is able to form the same H‐bond contacts with uracil as Ser. Moreover, Thr residues at this position have been shown to promote uridylate specificity in other NendoU‐containing proteins (Bhardwaj et al., 2008; Nedialkova et al., 2009). Nevertheless, CdiA‐CTSTECO31 does not recognize uridylate and instead cleaves tRNAUUC Glu between nucleotides C37 and m2A38. The in vivo toxin activity screen suggests that the C37‐A38 motif is a positive recognition determinant, because tRNAGUC Asp and tRNAGly isoacceptors share this element and are subject to partial cleavage. However, the toxin probably monitors the overall structure of the anticodon loop, because it discriminates against the tRNAGUC Asp anticodon, which differs from tRNAUUC Glu only at nucleotide 35. All of the clade III toxins examined here have anticodon nuclease activity, suggesting that their shared architecture determines substrate specificity. Clade III domains are characterized by helix α1, which is apparently absent from the other clades. Helix α1 supports the β1‐β2‐hairpin in the CdiA‐CTSTECO31 structure, and therefore it is conceivable that this structural element shapes the active‐site cleft. We note that clade III also contains toxins from Pseudomonas and Acinetobacter species that appear to lack strand β5 in the N‐terminal subdomain (see Fig. S7). It will be of interest to determine whether these enzymes share anticodon nuclease activity with CdiA‐CTSTECO31 or perhaps exhibit distinct substrate specificities.
The EndoU superfamily is one of the most widely used toxins in inter‐bacterial competition. Aravind and colleagues initially identified EndoU effectors associated with T2SS/MafB, T5SS, T6SS and T7SS delivery platforms (Zhang et al., 2012; Holberger et al., 2012; Jamet et al., 2015). More recent database searches indicate that EndoU domains are deployed by several other systems – some of which have been previously described and others that appear to be novel. We identified bacterial EndoU domains fused to the C‐termini of PrsW, SpvB and MuF‐like phage‐head morphogenesis proteins (Table S2), all of which have been proposed to deliver polymorphic toxins (Zhang et al., 2012). Intriguingly, EndoU nuclease are also fused to VgrG‐like proteins in Gram‐positive Paenibacillus species (Table S2). VgrG is a critical structural component of T6SS and often carries C‐terminal toxin domains (Russell et al., 2014; Ho et al., 2014). However, T6SSs have only been characterized in Gram‐negative bacteria, and Paenibacillus genomes contain no recognizable T6SS core genes. Nevertheless, these VgrG proteins are encoded together with DUF4280 (PAAR‐like) and pentapeptide repeat adaptor proteins that are similar to those found in T6SS effector operons from Gram‐negative bacteria. We also found a PAAR‐like protein with a C‐terminal EndoU domain in Clostridium sp. ASBs410 (Table S2). Together, these observations strongly suggest that Gram‐positive bacteria possess phage‐like secretion systems that are functionally analogous to T6SS. In addition to these recognizable toxin delivery systems, we identified EndoU‐containing proteins with unusual N‐terminal domains and several others that contain no other conserved domains whatsoever (Table S2). Some of these proteins carry N‐terminal secretion signal sequences, and nearly all are encoded together with probable immunity proteins. We speculate that these latter uncharacterized proteins are also used for inter‐bacterial toxin delivery. Finally, we identified a handful of extraordinarily large (1.5 to 2.9 MDa) proteins from Actinobacteria that carry internal EndoU domains. These proteins typically contain one or more APH aminoglycoside phosphotransferase domains together with a variety of RNA/DNA‐binding modules (Table S3). The function of these proteins is not known, but they appear unlikely to mediate inter‐cellular competition given the central location of the EndoU domain and the absence of linked immunity genes. The unusual domain assemblage suggests that these enormous proteins could process nucleic acids, or perhaps synthesize secondary metabolites or antibiotics in a manner similar to polyketide synthases. Regardless of their precise activities, these latter proteins suggest that EndoU domains have also been coopted to perform non‐inhibitory functions in bacteria.
Experimental Procedures
Plasmid constructions
Plasmid constructs and oligonucleotides used in this study are listed in Tables 3 and S4 respectively. DNA fragments encoding CdiA‐CT/CdiI proteins from E. coli STEC_O31 and Y. mollaretii ATCC 43969 were synthesized by Genscript (Piscataway, NJ) and supplied in plasmid pUC57. The fragment from E. coli STEC_O31 (Genbank: AFEX01000038) encodes residues Val2931 – Lys3253 of CdiASTECO31 (locus tag: ECSTECO31_4009) and CdiISTECO31 (ECSTECO31_4008). The initiation codon for ECSTECO31_4008 is not annotated correctly, and CdiISTECO31 contains 121 residues. The fragment from Y. mollaretii ATCC 43969 (NCBI reference: NZ_AALD02000043.1) encodes residues Val2694 – Asn2963 of CdiAYmo43969 (YMOLL0001_RS03095) and its putative CdiIYmo43969 immunity protein (YMOLL0001_RS0219640). A fragment encoding Val1 – Lys360 of CdiA‐CTGN05224 (YA39_RS24095) and CdiIGN05224 (YA39_RS16570) from K. aerogenes GN05224 (NCBI reference: NZ_LDBZ01000036.1) was synthesized by GenArt Gene Synthesis (ThermoFisher) and provided in plasmid pMA‐T. The cdiA‐CT/cdiI STECO31 module was amplified with primers 204F40 and 204R49 and introduced into plasmid pMCSG58 using ligation‐independent cloning (LIC) (Eschenfeldt et al., 2013; Eschenfeldt et al., 2010; Eschenfeldt et al., 2009). All of the resulting clones contained frame‐shift mutations in the toxin coding region, suggesting that the toxin was not completely neutralized by CdiISTECO31. Therefore, cdiI STECO31 was amplified with primer pair 204CdiIF/04R49 and introduced into plasmid pMCSG88 as described above. Vector pMCSG88 was generated by digesting pMCSG58 with HindIII/NcoI, followed by ligation to plasmid pMCSG76 (Eschenfeldt et al., 2013). The resulting pMCSG88 construct contains the LIC site with the Clo DF13 origin of replication and a spectinomycin‐resistance marker. The pMCSG58‐CdiA‐CT/CdiISTECO31 and pMCSG88‐CdiISTECO31 expression vectors were then introduced into E. coli BL21(DE3) to generate an over‐expression strain for large‐scale protein purification and crystallography.
Table 3.
Plasmids
| Plasmid | Descriptiona | Reference |
|---|---|---|
| pTrc99a | IPTG‐inducible expression plasmid, AmpR | GE Healthcare |
| pTrc99KX | Derivative of pTrc99a with additional 5′ KpnI, and 3′ SpeI and XhoI restriction sites, AmpR | Koskiniemi et al. (2014) |
| pMCSG58 | LIC overexpression vector containing argU and ileX tRNA genes, AmpR | Eschenfeldt et al. (2013) |
| pMCSG76 | Over‐expression vector with Clo DF13 origin of replication, SpcR StrR | Eschenfeldt et al. (2013) |
| pMCSG88 | LIC overexpression vector containing argU and ileX tRNA genes and Clo DF13 origin of replication, SpcR StrR | This study |
| pDAL8914 | Constitutive expression of chimeric cdiA EC93‐CT GN05224 and cdiI GN05224 genes, CmR | This study |
| pDAL8924 | pTrc99KX::cdiI GN05224, AmpR | This study |
| pDAL8956 | pET28A(+)::cdiA‐CT//cdiI GN05224, KanR | This study |
| pMCSG58‐APC200204 | pMCSG58::cdiA‐CT/cdiI STECO31, AmpR | This study |
| pMCSG88‐APC111471 | pMCSG88::cdiI STECO31, SpcR StrR | This study |
| pCH450 | pACYC184 derivative with E. coli araBAD promoter for arabinose‐inducible expression, TetR | Hayes and Sauer (2003) |
| pCH2408 | Constitutive expression of chimeric cdiA EC93‐CT STECO31 and cdiI STECO31 genes, CmR | This study |
| pCH4066 | pTrc99KX::cdiI Ymo43969, AmpR | This study |
| pCH4496 | pTrc99KX::cdiI STECO31, AmpR | This study |
| pCH6278 | pUC57::cdiA‐CT/cdiI STECO31, AmpR | Genscript |
| pCH6290 | pUC57::cdiA‐CT/cdiI Ymo43969, AmpR | Genscript |
| pCH6505 | pET21S::cdiA‐CT/cdiI Dd3937, AmpR | Aoki et al. (2010) |
| pCH8102 | pET21::BC0921, AmpR | This study |
| pCH9273 | pET21::BC0920‐CT/BC0921, AmpR | Holberger et al. (2012) |
| pCH10163 | Cosmid pCdiA‐CT/pheS* that carries a kan‐pheS* cassette in place of the E. coli EC93 cdiA‐CT/cdiI coding sequence. Used for allelic exchange and counter‐selection. CmR KanR | Morse et al. (2012) |
| pCH11140 | pCH450::rhsA‐CT/rhsI ECL ‐DAS, TetR | This study |
| pCH11884 | pET21::cdiA‐CT/cdiI STECO31, AmpR | This study |
| pCH12158 | pCH450::cdiA‐CT/cdiI Kpn342 ‐DAS, TetR | This study |
| pCH12599 | pCH450::cdiA‐CT/cdiI EC3006 ‐DAS, TetR | Willett et al. (2015) |
| pCH12847 | Constitutive expression of chimeric cdiA EC93‐CT Ymo43969 and cdiI Ymo43969 genes, CmR | This study |
| pCH12964 | pET21::cdiA‐CT(N319A)/cdiI STECO31, AmpR | This study |
| pCH12966 | pET21::cdiA‐CT(H321A)/cdiI STECO31, AmpR | This study |
| pCH13215 | Constitutive expression of chimeric cdiA EC93‐CT(H321A) STECO31 and cdiI STECO31 genes, CmR | This study |
| pCH13306 | Constitutive expression of chimeric cdiA EC93‐CT(H204A) STECO31 and cdiI STECO31 genes, CmR | This study |
| pCH13307 | Constitutive expression of chimeric cdiA EC93‐CT(N319A) STECO31 and cdiI STECO31 genes, CmR | This study |
| pCH13308 | pCH450::cdiA‐CT/cdiI STECO31 ‐DAS, TetR | This study |
| pCH13309 | Constitutive expression of chimeric cdiA EC93‐CT(T262A) STECO31 and cdiI STECO31 genes, CmR | This study |
| pCH13527 | pCH450::NMB0502/NMB0503‐DAS, TetR | This study |
| pCH13629 | pET21::cdiA‐CT(H204A)/cdiI STECO31, AmpR | This study |
| pCH13638 | pTrc99KX::NMB0503, AmpR | This study |
| pCH13639 | pET21::cdiI STECO31, AmpR | This study |
| pCH13703 | pET21::cdiA‐CT(T262A)/cdiI STECO31, AmpR | This study |
| pCH13871 | Constitutive expression of chimeric cdiA EC93‐CT(H187A) STECO31 and cdiI STECO31 genes, CmR | This study |
| pCH13881 | Constitutive expression of chimeric cdiA EC93‐CT(K261A) STECO31 and cdiI STECO31 genes, CmR | This study |
| pCH14224 | pET21::cdiA‐CT(H187A)/cdiI STECO31, AmpR | This study |
| pCH14226 | pET21::cdiA‐CT(K261A)/cdiI STECO31, AmpR | This study |
| pCH14368 | pET21::cdiI GN05224, AmpR | This study |
Abbreviations: AmpR, ampicillin‐resistance; CmR, chloramphenicol‐resistance; KanR, kanamycin‐resistance; SpcR, spectinomycin‐resistance; StrR, streptomycin‐resistance; and TetR, tetracycline‐resistance.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
The cdiA‐CT/cdiI STECO31 (CH3538/CH3539) and NMB0502/NMB0503 (CH2839/CH2785) modules were amplified and ligated to NcoI/SpeI‐digested plasmid pCH12599 to tag the immunity proteins with C‐terminal ssrA(DAS) degrons. The cdiA‐CT/cdiI STECO31 product was also ligated to pCH6505 to generate plasmid pCH11884 for the purification of CdiA‐CT/CdiISTECO31 complex for biochemical analyses. This latter plasmid was also used as a template for PCR‐based site‐directed mutagenesis using primers CH4016, CH4017, CH4018, CH4019, CH4269 and CH4270. The cdiA‐CT/cdiI GN05224 module was amplified with primers SK724/DL3986 and ligated to pET28a via NcoI/XhoI restrictions sites to generate plasmid pDAL8956. The cdiI STECO31 (CH3854/CH3539), cdiI GN05224 (SK733/DL3986) and BC0921 (CH4398/CH1706) immunity genes were also cloned into pET21S for the purification of His6‐tagged immunity proteins under non‐denaturing conditions. The cdiI STECO31 (CH3854/CH3624), cdiI GN05224 (SK733/SK734), NMB0503 (CH2784/CH2785) and cdiI Ymol43969 (CH3976/CH3977) genes were also cloned into pTrc99a derivatives to test for immunity function in competition co‐cultures.
Plasmid‐borne chimeric CDI systems were constructed by allelic exchange of the counter‐ selectable pheS* marker from plasmid pCH10163 (Morse et al., 2012). Wild‐type and mutant cdiA‐CT/cdiI STECO31 sequences were amplified with primers CH3172/CH3569 and fused to fragments amplified from regions upstream and downstream of the cdiA EC93 gene. The upstream homology fragment was amplified using primer pair CH4100/CH4101 and the downstream fragment with primers CH4102/CH4103. The three products were then fused to each other using overlap‐extension PCR with primers CH4100/CH4103. The final DNA product (100 ng) was electroporated together with plasmid pCH10163 (300 ng) into E. coli DY378 cells (Thomason et al., 2007). Recombinant plasmid clones were selected on yeast extract glucose‐agar supplemented with 33 µg/mL chloramphenicol and 10 mM D/L‐p‐chlorophenylalanine. The same procedure was used to fuse the cdiA‐CT/cdiI Ymol43969 and cdiA‐CT/cdiI GN05224 modules (amplified primers CH3747/CH3748 and SK693/SK694 respectively) to the E. coli EC93 CDI system.
Protein expression and purification for crystallization
E. coli BL21(DE3) cells carrying pMCSG58‐CdiA‐CT/CdiISTECO31 and pMCSG88‐CdiISTECO31 were grown at 37 °C for 6 ‐ 8 h in LB medium supplemented with ampicillin (100 μg/mL) and spectinomycin (50 μg/mL). A portion of the culture (0.5 mL) was diluted into 50 mL of M9 minimal medium supplemented with 0.5% glycerol, 100 μg/mL of ampicillin, 50 μg/mL of spectinomycin, trace minerals and vitamins for overnight culture at 37 °C. Large‐scale cultures were grown to an optical density at 600 nm (OD600) of 0.8 then cooled to 18 °C. SeMet was added to a final concentration of 60 μg/mL together with L‐isoleucine, L‐leucine, L‐lysine, L‐phenylalanine L‐threonine and L‐valine to a final concentration of 100 μg/mL and incubated for 20 min. Protein expression was induced with 0.5 mM isopropyl‐D‐thiogalactopyranoside (IPTG) for overnight culture. Cells were harvested by centrifugation the next day, and the pellets washed and resuspended in 50 mM Tris‐HCl (pH 8.0), 500 mM NaCl, 10 mM 2‐mercaptoethanol (2‐ME), 10% glycerol. Cells were broken with Fast Break reagent (Promega) supplemented with 10 μg/mL lysozyme, 500 U Benzonase Nuclease HC (Novagen) and Complete Protease Inhibitor Cocktail (Roche, Mannheim, Germany). The lysate was clarified by centrifugation at 10,000 × g for 1 h, followed by passage through a 0.22 μm filter prior to loading onto a 5 mL Nickel (II) Sepharose HisTrap column (GE Healthcare Biosciences). The column was washed with 5 volumes of 50 mM Tris‐HCl (pH 8.0), 500 mM NaCl, 20 mM imidazole, 10% glycerol, followed by elution with 250 mM imidazole in the same buffer. Fractions were pooled and loaded onto a Hiload 26/60 Superdex 200 size‐exclusion column equilibrated with 20 mM Tris‐HCl (pH 7.5), 150 mM NaCl and 2 mM dithiothreitol (DTT). Fractions containing two proteins were pooled and concentrated to 14 mg/mL using an Amicon Ultracel 20K concentrator (Millipore).
Crystallization, data collection, structure solution and refinement
The protein sample was treated with subtilisin (20 ng/μL) on ice overnight prior to crystallization trials. Crystallization conditions were screened using the Pi‐PEG Screen HTS (Jena Biosciences GmBh). The complex was crystallized in 10.7% PEG 4000, 50 mM Tris‐HCl (pH 8.0), 8.6% PEG 3000 at 4 °C by sitting‐drop vapor diffusion in 96‐well Crystal Quick plates (Greiner Bio‐one). Prior to flash cooling in liquid nitrogen, the crystals were cryo‐protected in mother liquor supplemented with 17% glycerol. The single‐wavelength anomalous diffraction (SAD) dataset was collected at 100 K near the selenium K‐absorption edge on beamline 19‐ID at the Advanced Photon Source, Argonne National Laboratory. Diffraction images were processed with the HKL3000 suite (Minor et al., 2006). Intensities were converted to structure factor amplitudes in Ctruncate (French and Wilson, 1978; Padilla and Yeates, 2003) from the CCP4 package (Winn et al., 2011). Data collection and processing statistics are presented in Table 1. The structure was solved using the HKL3000 software pipeline (Minor et al., 2006) by the SAD method with Se peak data. The pipeline applied SHELXD for the search of heavy atom sites and SHELXE for initial phases calculations (Sheldrick, 2008). The phases were improved through iterations of MLPHARE (Otwinowski, 1991) and DM (Cowtan, 1994). The initial protein model was built in HKL‐Builder utilizing Buccaneer (Cowtan, 2006). The final model was obtained through alternating manual rebuilding in COOT (Emsley and Cowtan, 2004) and crystallographic refinement in Refmac (Murshudov et al., 1997; Winn et al., 2011). The protocol refinement included optimization of TLS parameters with 5 and 2 groups defined for chains A (CdiA‐CTSTECO31) and I (CdiISTECO31) respectively. The refinement statistics are presented in Table 1. The atomic coordinates and structure factors have been deposited in the Protein Data Bank under accession code 5HKQ.
Protein purification and in vitro nuclease assays
Cultures of E. coli CH2016 harboring expression plasmids were grown to OD600 ~ 0.7, and expression was induced with 1.5 mM IPTG. After incubation for 2 h, cells were harvested and frozen at –80 °C. Cell pellets were re‐suspended in lysis buffer [20 mM sodium phosphate (pH 7.0), 150 mM NaCl, 10 mM 2‐ME, 0.05% Triton X‐100] and broken by two French press passages at 20,000 psi. Cell debris was removed by centrifugation at 16,000 × g at 4 °C. His6‐tagged immunity proteins were purified by Ni2+‐affinity chromatography in lysis buffer and eluted with 250 mM imidazole as described (Nikolakakis et al., 2012). EndoU toxins were first purified in complex with His6‐tagged immunity proteins, then eluted from the column by denaturation in 6 M guanidine‐HCl, 20 mM sodium phosphate (pH 7.0). Purified proteins were dialyzed against 20 mM sodium phosphate (pH 7.0), 150 mM NaCl, 10 mM 2‐ME. Proteins were quantified by absorbance at 280 nm using the following extinction coefficients: CdiA‐CTSTECO31, 25,330 cm–1 M–1; CdiISTECO31, 20,400 cm–1 M–1; CdiA‐CTGN05224, 47,330 cm–1 M–1; CdiIGN05224, 7,450 cm–1 M–1; BC0920, 22,920 cm–1 M–1 and BC0921, 15,470 cm–1 M–1.
In vitro nuclease assays were performed in 20 mM Tris‐HCl (pH 7.5), 150 mM NaCl using E. coli total RNA (0.5 mg mL–1) as a substrate. Reactions were initiated by addition of 2 µM purified toxins, followed by incubation for 30 min at 37 °C. Where indicated, reactions were supplemented with 2 to 6 µM purified immunity protein. Reactions were quenched with denaturing gel‐loading buffer and run on 50% urea‐10% polyacrylamide gels buffered with 1 × Tris‐borate EDTA. Gels were stained with ethidium bromide or electroblotted to positively charged nylon membranes for subsequent Northern blot analyses.
In vivo toxin activity and competition co‐cultures
Nuclease activity screens were performed by activating toxins from Enterobacter cloacae ATCC 13047 (Whitney et al., 2014) and E. coli isolates STEC_O31, 3006 and 96.154 inside E. coli X90 cells. Each cognate immunity protein was tagged with a C‐terminal ssrA(DAS) degron as described (McGinness et al., 2006; Poole et al., 2011). E. coli X90 cells carrying these expression constructs were seeded at OD600 ~ 0.05 in LB media supplemented with 25 µg/mL tetracycline and incubated with shaking at 37 °C. After 2 h, the cultures were adjusted to 0.2% L‐arabinose and cultured for an additional 4 h. Cell growth was monitored by measuring the OD600 every 30 min. Culture samples were harvested into an equal volume of ice‐cold methanol after 2 h of induction, and the cells frozen at –80 °C for subsequent RNA extraction and analysis.
Bacterial competitions were conducted in shaking LB broth at 37 °C. Media were seeded with inhibitor and target strains at a 1:1 ratio, and the co‐culture incubated for 1 to 3 h. E. coli EPI100 cells that express chimeric CDI systems were used as inhibitors. Inhibitor strains carried plasmids pCH2408 (wild‐type CDISTECO31), pCH13871 (His187Ala), pCH13306 (His204Ala), pCH13881 (Lys261Ala), pCH13309 (Thr262Ala), pCH13307 (Asn319Ala), pCH13215 (His321Ala), pDAL8914 (CDIGN05224) or pCH12847 (CDIYmo43969). E. coli MC4100 target strains carried plasmids pTrc99A (cdiI–), pCH4496 (cdiI STECO31), pCH4066 (cdiI Ymo43969), pDAL8924 (cdiI GN05224) or pCH13638 (cdiI MC58). Viable inhibitor and target cells were enumerated as colony forming units per mL (CFU mL–1) on selective media at the beginning and end of co‐culture. Competitive indices were calculated as the ratio of target cells to inhibitor cells at the end of co‐culture divided by the initial target to inhibitor cell ratio. Competitive indices are reported for each independent experiment together with the average ± standard error of the mean. Co‐culture samples were also harvested into equal volumes of ice‐cold methanol for RNA extraction.
RNA isolation and analyses
Frozen cell pellets were resuspended in guanidinium isothiocyanate (GITC)‐phenol and total RNA extracted as described (Garza‐Sánchez et al., 2006). RNAs (5 µg) were run on 50% urea‐10% polyacrylamide gels buffered with 1 × Tris‐borate EDTA and electroblotted to positively charged nylon membranes for Northern blot analysis. Blots were hybridized with [32P]‐labeled oligonucleotide probes that are specific for individual E. coli tRNAs (see Table S4) (Garza‐Sánchez et al., 2006; Hayes and Sauer, 2003). Blots were visualized by phosphorimaging using Bio‐Rad Quantity One software. Primer extension analysis was performed as described (Beck et al., 2014) using oligonucleotide CH4275 to map the cleavage site in tRNAUUC Glu. The primer and marker oligonucleotides were 5′‐radiolabeled with [32P] using T4 polynucleotide kinase. The radiolabeled primer was hybridized with RNA samples for 5 min at 50 °C, then extended with Superscript reverse transcriptase at 37 °C for 30 min. Reactions were quenched with denaturing gel‐loading buffer and heated to 95 °C. Primer extension reactions were run on a 50% urea, 15% polyacrylamide gel buffered with 1 × Tris‐borate‐EDTA and visualized on a Bio‐Rad phosphorimager using Quantity One software.
Supporting information
Acknowledgements
We would like to thank Gyorgy Babnigg for help with construct design. This work was supported by National Institutes of Health grants GM102318 (C.W.G., C.S.H., D.A.L. & subcontract to A.J.), GM117373 (C.W.G., D.A.L., C.S.H.), GM094585 and GM115586 (A.J.) and the U. S. Department of Energy, Office of Biological and Environmental Research, under contract DE‐AC02‐06CH11357 (A.J.).
References
- Aoki, S. K. , Diner, E. J. , de Roodenbeke, C. T. , Burgess, B. R. , Poole, S. J. , Braaten, B. A. , et al (2010) A widespread family of polymorphic contact‐dependent toxin delivery systems in bacteria. Nature 468: 439–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki, S. K. , Malinverni, J. C. , Jacoby, K. , Thomas, B. , Pamma, R. , Trinh, B. N. , et al (2008) Contact‐dependent growth inhibition requires the essential outer membrane protein BamA (YaeT) as the receptor and the inner membrane transport protein AcrB. Mol. Microbiol. 70: 323–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki, S. K. , Pamma, R. , Hernday, A. D. , Bickham, J. E. , Braaten, B. A. , and Low, D. A. (2005) Contact‐dependent inhibition of growth in Escherichia coli . Science 309: 1245–1248. [DOI] [PubMed] [Google Scholar]
- Arenas, J. , Schipper, K. , van Ulsen, P. , van der Ende, A. , and Tommassen, J. (2013) Domain exchange at the 3′ end of the gene encoding the fratricide meningococcal two‐partner secretion protein A. BMC Genomics 14: 622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler, M. , Pilhofer, M. , Henderson, G. P. , Jensen, G. J. , and Mekalanos, J. J. (2012) Type VI secretion requires a dynamic contractile phage tail‐like structure. Nature 483: 182–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck, C. M. , Morse, R. P. , Cunningham, D. A. , Iniguez, A. , Low, D. A. , Goulding, C. W. , and Hayes, C. S. (2014) CdiA from Enterobacter cloacae delivers a toxic ribosomal RNase into target bacteria. Structure 22: 707–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck, C. M. , Willett, J. L. , Cunningham, D. A. , Kim, J. J. , Low, D. A. , and Hayes, C. S. (2016) CdiA Effectors from Uropathogenic Escherichia coli Use Heterotrimeric Osmoporins as Receptors to Recognize Target Bacteria. PLoS Pathog 12: e1005925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj, K. , Guarino, L. , and Kao, C. C. (2004) The severe acute respiratory syndrome coronavirus Nsp15 protein is an endoribonuclease that prefers manganese as a cofactor. J Virol 78: 12218–12224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj, K. , Palaninathan, S. , Alcantara, J. M. , Yi, L. L. , Guarino, L. , Sacchettini, J. C. , and Kao, C. C. (2008) Structural and functional analyses of the severe acute respiratory syndrome coronavirus endoribonuclease Nsp15. J Biol Chem 283: 3655–3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffarelli, E. , Arese, M. , Santoro, B. , Fragapane, P. , and Bozzoni, I. (1994) In vitro study of processing of the intron‐encoded U16 small nucleolar RNA in Xenopus laevis . Mol Cell Biol 14: 2966–2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffarelli, E. , Maggi, L. , Fatica, A. , Jiricny, J. , and Bozzoni, I. (1997) A novel Mn++‐dependent ribonuclease that functions in U16 snoRNA processing in X. laevis . Biochem Biophys Res Commun 233: 514–517. [DOI] [PubMed] [Google Scholar]
- Cao, Z. , Casabona, M. G. , Kneuper, H. , Chalmers, J. D. , and Palmer, T. (2016) The type VII secretion system of Staphylococcus aureus secretes a nuclease toxin that targets competitor bacteria. Nat Microbiol 2: 16183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales, E. , Buchanan, S. K. , Duche, D. , Kleanthous, C. , Lloubes, R. , Postle, K. , et al (2007) Colicin biology. Microbiol Mol Biol Rev 71: 158–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowtan, K. (1994) DM: an automated procedure for phase improvement by density modification. Joint CCP4 and ESF‐EACBM newsletter on protein crystallography 31: 34–38. [Google Scholar]
- Cowtan, K. (2006) The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr D Biol Crystallogr 62: 1002–1011. [DOI] [PubMed] [Google Scholar]
- Cuchillo, C. M. , Nogues, M. V. , and Raines, R. T. (2011) Bovine pancreatic ribonuclease: fifty years of the first enzymatic reaction mechanism. Biochemistry 50: 7835–7841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, I. W. , Murray, L. W. , Richardson, J. S. , and Richardson, D. C. (2004) MOLPROBITY: structure validation and all‐atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res 32: W615–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deavin, A. , Mathias, A. P. , and Rabin, B. R. (1966) Mechanism of action of bovine pancreatic ribonuclease. Nature 211: 252–255. [DOI] [PubMed] [Google Scholar]
- Deng, X. , Hackbart, M. , Mettelman, R. C. , O'Brien, A. , Mielech, A. M. , Yi, G. , Kao, C. C. , and Baker, S. C. (2017) Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc Natl Acad Sci USA 114: E4251–E4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey, A. , Vassallo, C. N. , Conklin, A. C. , Pathak, D. T. , Troselj, V. , and Wall, D. (2016) Sibling rivalry in Myxococcus xanthus Is mediated by kin recognition and a polyploid prophage. Journal of bacteriology 198: 994–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diner, E. J. , Beck, C. M. , Webb, J. S. , Low, D. A. , and Hayes, C. S. (2012) Identification of a target cell permissive factor required for contact‐dependent growth inhibition (CDI). Genes Dev 26: 515–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley, P. , and Cowtan, K. (2004) Coot: model‐building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132. [DOI] [PubMed] [Google Scholar]
- Eschenfeldt, W. H. , Lucy, S. , Millard, C. S. , Joachimiak, A. , and Mark, I. D. (2009) A family of LIC vectors for high‐throughput cloning and purification of proteins. Methods Mol Biol 498: 105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eschenfeldt, W. H. , Makowska‐Grzyska, M. , Stols, L. , Donnelly, M. I. , Jedrzejczak, R. , and Joachimiak, A. (2013) New LIC vectors for production of proteins from genes containing rare codons. J Struct Funct Genomics 14: 135–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eschenfeldt, W. H. , Maltseva, N. , Stols, L. , Donnelly, M. I. , Gu, M. , Nocek, B. , et al (2010) Cleavable C‐terminal His‐tag vectors for structure determination. J Struct Funct Genomics 11: 31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findlay, D. , Herries, D. G. , Mathias, A. P. , Rabin, B. R. , and Ross, C. A. (1962) The active site and mechanism of action of bovine pancreatic ribonuclease. 7. The catalytic mechanism. Biochem J 85: 152–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French, S. , and Wilson, K. (1978) On the treatment of negative intensity observations. Acta Crystallogr A A34: 517–525. [Google Scholar]
- Garcia‐Bayona, L. , Guo, M. S. , and Laub, M. T. (2017) Contact‐dependent killing by Caulobacter crescentus via cell surface‐associated, glycine zipper proteins. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garza‐Sánchez, F. , Janssen, B. D. , and Hayes, C. S. (2006) Prolyl‐tRNA(Pro) in the A‐site of SecM‐arrested ribosomes inhibits the recruitment of transfer‐messenger RNA. J Biol Chem 281: 34258–34268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghequire, M. G. , and De Mot, R. (2014) Ribosomally encoded antibacterial proteins and peptides from Pseudomonas. FEMS Microbiol Rev 38: 523–568. [DOI] [PubMed] [Google Scholar]
- Gioia, U. , Laneve, P. , Dlakic, M. , Arceci, M. , Bozzoni, I. , and Caffarelli, E. (2005) Functional characterization of XendoU, the endoribonuclease involved in small nucleolar RNA biosynthesis. J Biol Chem 280: 18996–19002. [DOI] [PubMed] [Google Scholar]
- Guarino, L. A. , Bhardwaj, K. , Dong, W. , Sun, J. , Holzenburg, A. , and Kao, C. (2005) Mutational analysis of the SARS virus Nsp15 endoribonuclease: identification of residues affecting hexamer formation. J Mol Biol 353: 1106–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes, C. S. , and Sauer, R. T. (2003) Cleavage of the A site mRNA codon during ribosome pausing provides a mechanism for translational quality control. Mol. Cell 12: 903–911. [DOI] [PubMed] [Google Scholar]
- Ho, B. T. , Dong, T. G. , and Mekalanos, J. J. (2014) A view to a kill: the bacterial type VI secretion system. Cell Host Microbe 15: 9–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holberger, L. E. , Garza‐Sanchez, F. , Lamoureux, J. , Low, D. A. , and Hayes, C. S. (2012) A novel family of toxin/antitoxin proteins in Bacillus species. FEBS Lett 586: 132–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm, L. , and Rosenstrom, P. (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res 38: W545–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood, R. D. , Singh, P. , Hsu, F. , Guvener, T. , Carl, M. A. , Trinidad, R. R. , et al (2010) A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7: 25–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov, K. A. , Hertzig, T. , Rozanov, M. , Bayer, S. , Thiel, V. , Gorbalenya, A. E. , and Ziebuhr, J. (2004) Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc Natl Acad Sci USA 101: 12694–12699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamet, A. , Jousset, A. B. , Euphrasie, D. , Mukorako, P. , Boucharlat, A. , Ducousso, A. , Charbit, A. , and Nassif, X. (2015) A new family of secreted toxins in pathogenic Neisseria species. PLoS Pathog 11: e1004592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, P. M. , Beck, C. M. , Morse, R. P. , Garza‐Sanchez, F. , Low, D. A. , Hayes, C. S. , and Goulding, C. W. (2016) Unraveling the essential role of CysK in CDI toxin activation. Proc Natl Acad Sci USA 113: 9792–9797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, A. M. , Garza‐Sanchez, F. , So, J. , Hayes, C. S. , and Low, D. A. (2017) Activation of contact‐dependent antibacterial tRNase toxins by translation elongation factors. Proc Natl Acad Sci USA 114: E1951–E1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, A. M. , Low, D. A. , and Hayes, C. S. (2017) Can't you hear me knocking: contact‐dependent competition and cooperation in bacteria. Emerg Top Life Sci 1: 75–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph, J. S. , Saikatendu, K. S. , Subramanian, V. , Neuman, B. W. , Buchmeier, M. J. , Stevens, R. C. , and Kuhn, P. (2007) Crystal structure of a monomeric form of severe acute respiratory syndrome coronavirus endonuclease nsp15 suggests a role for hexamerization as an allosteric switch. J Virol 81: 6700–6708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, H. , Bhardwaj, K. , Li, Y. , Palaninathan, S. , Sacchettini, J. , Guarino, L. , Leibowitz, J. L. , and Kao, C. C. (2007) Biochemical and genetic analyses of murine hepatitis virus Nsp15 endoribonuclease. J Virol 81: 13587–13597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindler, E. , Gil‐Cruz, C. , Spanier, J. , Li, Y. , Wilhelm, J. , Rabouw, H. H. , et al (2017) Early endonuclease‐mediated evasion of RNA sensing ensures efficient coronavirus replication. PLoS Pathog 13: e1006195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskiniemi, S. , Garza‐Sanchez, F. , Sandegren, L. , Webb, J. S. , Braaten, B. A. , Poole, S. J. , et al (2014) Selection of orphan Rhs toxin expression in evolved Salmonella enterica serovar Typhimurium. PLoS Genet 10: e1004255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskiniemi, S. , Lamoureux, J. G , Nikolakakis, K. C , t'Kint de Roodenbeke, C. , Kaplan, M. D , Low, D. A , and Hayes, C. S (2013) Rhs proteins from diverse bacteria mediate intercellular competition. Proc Natl Acad Sci USA 110: 7032–7037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel, E. , and Henrick, K. (2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372: 774–797. [DOI] [PubMed] [Google Scholar]
- Laneve, P. , Altieri, F. , Fiori, M. E. , Scaloni, A. , Bozzoni, I. , and Caffarelli, E. (2003) Purification, cloning, and characterization of XendoU, a novel endoribonuclease involved in processing of intron‐encoded small nucleolar RNAs in Xenopus laevis. J Biol Chem 278: 13026–13032. [DOI] [PubMed] [Google Scholar]
- MacIntyre, D. L. , Miyata, S. T. , Kitaoka, M. , and Pukatzki, S. (2010) The Vibrio cholerae type VI secretion system displays antimicrobial properties. Proc Natl Acad Sci USA 107: 19520–19524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinness, K. E. , Baker, T. A. , and Sauer, R. T. (2006) Engineering controllable protein degradation. Mol. Cell 22: 701–707. [DOI] [PubMed] [Google Scholar]
- Michalska, K. , Gucinski, G. C. , Garza‐Sanchez, F. , Johnson, P. M. , Stols, L. M. , Eschenfeldt, W. H. , et al (2017) Structure of a novel antibacterial toxin that exploits elongation factor Tu to cleave specific transfer RNAs. Nucleic Acids Res 45: 10306–10320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minor, W. , Cymborowski, M. , Otwinowski, Z. , and Chruszcz, M. (2006) HKL‐3000: the integration of data reduction and structure solution–from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr 62: 859–866. [DOI] [PubMed] [Google Scholar]
- Morse, R. P. , Nikolakakis, K. C. , Willett, J. L. , Gerrick, E. , Low, D. A. , Hayes, C. S. , and Goulding, C. W. (2012) Structural basis of toxicity and immunity in contact‐dependent growth inhibition (CDI) systems. Proc Natl Acad Sci USA 109: 21480–21485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov, G. N. , Vagin, A. A. , and Dodson, E. J. (1997) Refinement of macromolecular structures by the maximum‐likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255. [DOI] [PubMed] [Google Scholar]
- Nedialkova, D. D. , Ulferts, R. , van den Born, E. , Lauber, C. , Gorbalenya, A. E. , Ziebuhr, J. , and Snijder, E. J. (2009) Biochemical characterization of arterivirus nonstructural protein 11 reveals the nidovirus‐wide conservation of a replicative endoribonuclease. J Virol 83: 5671–5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, W. L. , and Bassler, B. L. (2009) Bacterial quorum‐sensing network architectures. Annu Rev Genet 43: 197–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolakakis, K. , Amber, S. , Wilbur, J. S. , Diner, E. J. , Aoki, S. K. , Poole, S. J. , et al (2012) The toxin/immunity network of Burkholderia pseudomallei contact‐dependent growth inhibition (CDI) systems. Mol Microbiol 84: 516–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohr, R. J. , Anderson, M. , Shi, M. , Schneewind, O. , and Missiakas, D. (2017) EssD, a nuclease effector of the Staphylococcus aureus ESS pathway. Journal of bacteriology 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski, Z. (1991) Maximum likelihood refinement of heavy atom parameters. In: Proceedings of the CCP4 Study Weekend 25–26 January 1991. Daresbury Laboratory, Warrington, pp. 80–85. [Google Scholar]
- Padilla, J. E. , and Yeates, T. O. (2003) A statistic for local intensity differences: robustness to anisotropy and pseudo‐centering and utility for detecting twinning. Acta Crystallogr D Biol Crystallogr 59: 1124–1130. [DOI] [PubMed] [Google Scholar]
- Pettersen, E. F. , Goddard, T. D. , Huang, C. C. , Couch, G. S. , Greenblatt, D. M. , Meng, E. C. , and Ferrin, T. E. (2004) UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612. [DOI] [PubMed] [Google Scholar]
- Poole, S. J. , Diner, E. J. , Aoki, S. K. , Braaten, B. A. , T'Kint de Roodenbeke, C. , Low, D. A. , and Hayes, C. S. (2011) Identification of functional toxin/immunity genes linked to contact‐dependent growth inhibition (CDI) and rearrangement hotspot (Rhs) systems. PLoS Genet. 7: e1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renzi, F. , Caffarelli, E. , Laneve, P. , Bozzoni, I. , Brunori, M. , and Vallone, B. (2006) The structure of the endoribonuclease XendoU: From small nucleolar RNA processing to severe acute respiratory syndrome coronavirus replication. Proc Natl Acad Sci USA 103: 12365–12370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricagno, S. , Egloff, M. P. , Ulferts, R. , Coutard, B. , Nurizzo, D. , Campanacci, V. , et al (2006) Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family. Proc Natl Acad Sci USA 103: 11892–11897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert, X. , and Gouet, P. (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42: W320–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhe, Z. C. , Nguyen, J. Y. , Chen, A. J. , Leung, N. Y. , Hayes, C. S. , and Low, D. A. (2016) CDI systems are stably maintained by a cell‐contact mediated surveillance mechanism. PLoS Genet 12: e1006145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhe, Z. C. , Nguyen, J. Y. , Xiong, J. , Koskiniemi, S. , Beck, C. M. , Perkins, B. R. , Low, D. A. , and Hayes, C. S. (2017) CdiA effectors use modular receptor‐binding domains to recognize. Target Bacteria. MBio: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, A. B. , Peterson, S. B. , and Mougous, J. D. (2014) Type VI secretion system effectors: poisons with a purpose. Nat Rev Microbiol 12: 137–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz, D. S. , and Blower, M. D. (2014) The calcium‐dependent ribonuclease XendoU promotes ER network formation through local RNA degradation. J Cell Biol 207: 41–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick, G. M. (2008) A short history of SHELX. Acta Crystallogr A 64: 112–122. [DOI] [PubMed] [Google Scholar]
- Shi, Y. , Li, Y. , Lei, Y. , Ye, G. , Shen, Z. , Sun, L. , et al (2016) A dimerization‐dependent mechanism drives the endoribonuclease function of porcine reproductive and respiratory syndrome virus nsp11. J Virol 90: 4579–4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder, E. J. , Bredenbeek, P. J. , Dobbe, J. C. , Thiel, V. , Ziebuhr, J. , Poon, L. L. , et al (2003) Unique and conserved features of genome and proteome of SARS‐coronavirus, an early split‐off from the coronavirus group 2 lineage. J Mol Biol 331: 991–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza, D. P. , Oka, G. U. , Alvarez‐Martinez, C. E. , Bisson‐Filho, A. W. , Dunger, G. , Hobeika, L. , et al (2015) Bacterial killing via a type IV secretion system. Nat Commun 6: 6453. [DOI] [PubMed] [Google Scholar]
- Tan, K. , Johnson, P. M. , Stols, L. , Boubion, B. , Eschenfeldt, W. , Babnigg, G. , et al (2015) The structure of a contact‐dependent growth‐inhibition (CDI) immunity protein from Neisseria meningitidis MC58. Acta Crystallogr F Struct Biol Commun 71: 702–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomason, L. , Court, D. L. , Bubunenko, M. , Costantino, N. , Wilson, H. , Datta, S. , and Oppenheim, A. (2007) Recombineering: genetic engineering in bacteria using homologous recombination. Current protocols in molecular biology / edited by Frederick M. Ausubel … [et al Chapter 1: Unit 1 16. [DOI] [PubMed]
- Unterweger, D. , Miyata, S. T. , Bachmann, V. , Brooks, T. M. , Mullins, T. , Kostiuk, B. , Provenzano, D. , and Pukatzki, S. (2014) The Vibrio cholerae type VI secretion system employs diverse effector modules for intraspecific competition. Nat Commun 5: 3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassallo, C. N. , Cao, P. , Conklin, A. , Finkelstein, H. , Hayes, C. S. , and Wall, D. (2017) Infectious polymorphic toxins delivered by outer membrane exchange discriminate kin in myxobacteria . Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney, J. C. , Beck, C. M. , Goo, Y. A. , Russell, A. B. , Harding, B. N. , De Leon, J. A. , et al (2014) Genetically distinct pathways guide effector export through the type VI secretion system. Mol Microbiol 92: 529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney, J. C. , Peterson, S. B. , Kim, J. , Pazos, M. , Verster, A. J. , Radey, M. C. , et al (2017) A broadly distributed toxin family mediates contact‐dependent antagonism between gram‐positive bacteria. eLife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willett, J. L. , Gucinski, G. C. , Fatherree, J. P. , Low, D. A. , and Hayes, C. S. (2015) Contact‐dependent growth inhibition toxins exploit multiple independent cell‐entry pathways. Proc Natl Acad Sci USA 112: 11341–11346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willett, J. L. , Ruhe, Z. C. , Goulding, C. W. , Low, D. A. , and Hayes, C. S. (2015) Contact‐dependent growth inhibition (CDI) and CdiB/CdiA two‐partner secretion proteins. J Mol Biol 427: 3754–3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn, M. D. , Ballard, C. C. , Cowtan, K. D. , Dodson, E. J. , Emsley, P. , Evans, P. R. , et al (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67: 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X. , Zhai, Y. , Sun, F. , Lou, Z. , Su, D. , Xu, Y. , et al (2006) New antiviral target revealed by the hexameric structure of mouse hepatitis virus nonstructural protein nsp15. J Virol 80: 7909–7917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, D. , de Souza, R. F. , Anantharaman, V. , Iyer, L. M. , and Aravind, L. (2012) Polymorphic toxin systems: Comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol Direct 7: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, D. , Iyer, L. M. , and Aravind, L. (2011) A novel immunity system for bacterial nucleic acid degrading toxins and its recruitment in various eukaryotic and DNA viral systems. Nucleic Acids Res 39: 4532–4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, M. , Li, X. , Deng, Z. , Chen, Z. , Liu, Y. , Gao, Y. , Wu, W. , and Chen, Z. (2017) Structural biology of the arterivirus nsp11 endoribonucleases. J Virol 91: e01309‐e01316. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


