SUMMARY
Universally conserved NusG/Spt5 factors reduce RNA polymerase pausing and arrest. In a widely accepted model, these proteins bridge the RNA polymerase clamp and lobe domains across the DNA channel, inhibiting the clamp opening to promote pause-free RNA synthesis. However, recent structures of paused transcription elongation complexes show that the clamp does not open and suggest alternative mechanisms of antipausing. Among these mechanisms, direct contacts of NusG/Spt5 proteins with the nontemplate DNA in the transcription bubble have been proposed to prevent unproductive DNA conformations and thus inhibit arrest. We used Escherichia coli RfaH, whose interactions with DNA are best characterized, to test this idea. We report that RfaH stabilizes the upstream edge of the transcription bubble, favoring forward translocation, and protects the upstream duplex DNA from exonuclease cleavage. Modeling suggests that RfaH loops the nontemplate DNA around its surface and restricts the upstream DNA duplex mobility. Strikingly, we show that RfaH-induced DNA protection and antipausing activity can be mimicked by shortening the nontemplate strand in elongation complexes assembled on synthetic scaffolds. We propose that remodeling of the nontemplate DNA controls recruitment of regulatory factors and R-loop formation during transcription elongation across all life.
Keywords: Non-template DNA strand, NusG, RfaH, RNA polymerase, transcription elongation
Graphical Abstract
Introduction
In all three domains of life, transcription elongation factors from NusG/Spt5 family coordinate RNA synthesis with co-transcriptional RNA processing. These proteins are composed of an N-terminal domain (NTD) and at least one C-terminal domain (CTD) bearing a Kyrpides, Ouzounis, Woese (KOW) motif (Kyrpides et al., 1996). The two domains, which are nearly superimposable across this family, are connected by a flexible linker and can make simultaneous interactions with the elongating RNA polymerase (RNAP) and other cellular partners, coupling transcription to RNA processing, modification, and translation (Tomar & Artsimovitch, 2013). While KOW domains interact with a diverse set of structurally distinct proteins, the NTDs bind to the same site on the transcription elongation complex (EC) and modulate RNA synthesis in the absence of any auxiliary factors (Tomar and Artsimovitch, 2013 and references therein).
Escherichia coli RfaH, an operon-specific paralog of NusG, is the best-characterized member of this family of regulators. RfaH is recruited to RNAP paused at an ops element, an exemplar of a consensus pause signal (Larson et al., 2014, Vvedenskaya et al., 2014), via specific interactions of the NTD with the nontemplate (NT) DNA strand exposed on the RNAP surface (Belogurov et al., 2007). Following recruitment in the leader region, RfaH remains bound to RNAP throughout transcription of the entire operon (Belogurov et al., 2009) and abrogates Rho-dependent termination, in part by inhibiting pausing (Sevostyanova et al., 2011). E. coli NusG and Spt5 proteins from yeast and archaea share the ability to reduce RNAP pausing and arrest (Crickard et al., 2016, Artsimovitch & Landick, 2000, Hirtreiter et al., 2010).
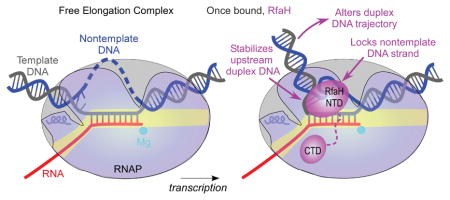
Studies of RfaH identified key contacts with RNAP thought to underpin antipausing activity in all NusG-like proteins. In RfaH-NTD, a patch of hydrophobic residues mediates high-affinity interactions with the tip of the β′ clamp helices (CH), a part of the β′ clamp domain, and a separate HTTT motif interacts with the β gate loop (GL; Fig. 1A), a part of the β lobe domain (Belogurov et al., 2010). The β′ clamp and β lobe form two pincers of the crab-claw-shaped RNAP (Zhang et al., 1999) that lie on two sides of the main channel, which contains the active site and the nucleic acids in the EC (Kang et al., 2017, Vassylyev et al., 2007). The clamp domain makes up much of the β′ pincer and undergoes swinging motions that open the channel to permit entry of nucleic acids during initiation or close the channel around the nucleic acids to enable processive elongation (Chakraborty et al., 2012, Feklistov et al., 2017, Gnatt et al., 2001). In a long-standing model of RNAP pausing, the clamp was hypothesized to open during the EC isomerization into off-pathway states (Zhang & Landick, 2016), suggesting that RfaH may inhibit pausing by bridging the RNAP pincers across the main channel (Sevostyanova et al., 2011), akin to processivity clamps in DNA polymerases.
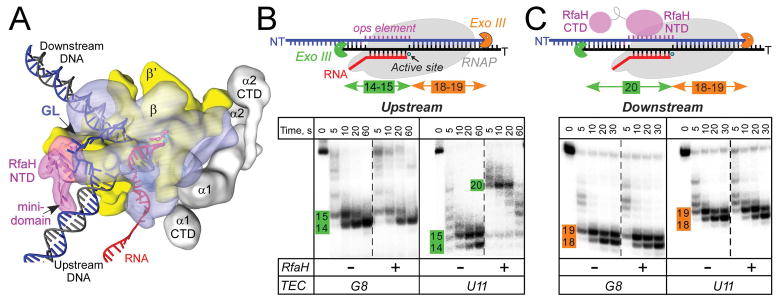
Figure 1.
RfaH interactions with the EC.
A. A model of the RfaH-bound EC. RNAP α (grey), β (blue), and β′ (yellow) subunits and RfaH NTD (magenta) are depicted by simplified molecular surfaces; β (with the GL labeled) and RfaH NTD are rendered semi-transparent. Nucleic acids are shown as cartoons, two Mg2+ ions in the active site – as cyan spheres, and an incoming NTP – as red sticks.
B and C. Footprinting of the upstream (B) and downstream (C) RNAP boundary in G8 and U11 ops ECs. Exo III was added to ECs assembled on synthetic scaffolds in which RNAP is halted at G8 and U11 positions in the ops element (NT sequence G1GCGGTAG8CGT11G); the probed DNA strand (T, panel B; NT, panel C) was 5′-end labeled with [γ32P]-ATP. RfaH was present at 100 nM where indicated. Aliquots were quenched at the indicated times (0 represents an untreated DNA control) and analyzed on a 12 % denaturing gel; a representative of three independent experiments is shown. Numbers indicate the distance from the RNAP active site.
Subsequent structural analyses and molecular modeling revealed that the NTD-RNAP interactions are broadly conserved among bacterial, yeast, mammalian and archaeal factors (Klein et al., 2011, Martinez-Rucobo et al., 2011, Turtola & Belogurov, 2016, Yakhnin et al., 2016, Bernecky et al., 2017, Ehara et al., 2017) and that the clamp opens a paused bacterial EC (Weixlbaumer et al., 2013). Together, these data supported a ubiquitous antipausing mechanism shared by all NusG homologs.
However, recent findings put this model in doubt and suggest that NusG-like proteins may utilize other contacts with the EC to reduce pausing. First, the bridging contacts to GL do not appear to be universally important; E. coli NusG does not require GL for antipausing (NandyMazumdar et al., 2016) and antibacktracking (Turtola & Belogurov, 2016) activities. Second, in cryo-EM structures of the hairpin-stabilized paused E. coli ECs (Guo et al., 2018, Kang et al., 2018a), the clamp does not open, and instead undergoes a minor rotation termed “swiveling”. Third, the evidence in support of the key roles of direct NTD-DNA contacts has been accumulating. E. coli RfaH (Artsimovitch & Landick, 2002), Bacillus subtilis NusG (Yakhnin et al., 2016) and Saccharomyces cerevisiae Spt5 (Crickard et al., 2016) have been shown to directly contact NT DNA. We proposed that NusG-DNA contacts compensate for missing contacts with GL (NandyMazumdar et al., 2016), providing alternative means to stabilize the NT DNA-GL-NTD network of interactions. We hypothesized that excessive flexibility of NT DNA could be detrimental, and that a factor that restrains NT DNA may promote processive elongation (NandyMazumdar et al., 2016); a similar “bubble-chaperone” model was proposed for yeast Spt5 (Crickard et al., 2016). Finally, interactions of NusG proteins with the upstream DNA duplex and the fork junction may facilitate forward translocation to reduce pausing and arrest. Footprinting and crosslinking data suggest that E. coli (Turtola & Belogurov, 2016) and Thermus thermophilus (Sevostyanova & Artsimovitch, 2010) NusGs promote DNA reannealing behind the RNAP. Biochemical analysis demonstrated interactions of S. cerevisiae (Crickard et al., 2016) and Methanocaldococcus jannaschii (Guo et al., 2015) Spt5 with the upstream DNA. A recent structure of yeast Komagataella pastoris EC revealed that Spt4/5 encircles one turn of the duplex DNA behind the RNAP in a “DNA exit tunnel” (Ehara et al., 2017). In this work, we set out to investigate the contribution of DNA interactions to RfaH effects on the EC. Our results show that RfaH stabilizes the upstream fork junction and restricts the mobility of NT DNA. The congruity of their binding sites and structures suggests that NusG proteins from all three domains of life may utilize similar interactions to control transcription.
Results
RfaH induces protection of the upstream DNA from exonuclease digestion
We first asked whether RfaH interacts with the duplex DNA to promote strand reannealing at the rear end of the transcription bubble. We utilized footprinting with Exo III, an exonuclease that digests DNA in a 3′→5′ direction and has been used extensively in the analysis of ECs (Kireeva & Kashlev, 2009). We prepared ECs on nucleic-acid scaffolds containing the 12-nt ops DNA element in the NT strand (Supporting Information Fig. S1); the template (T) strand and the nascent RNA were end-labeled with γ32P-ATP (Fig. 1B). Throughout the manuscript, we assign the positions of RNA 3′ ends relative to the ops element; U11 corresponds to the position at which RNAP pauses during elongation through the ops signal (Belogurov et al., 2007).
We compared ECs in which the 3′ end of the RNA was at G8 and U11. We have previously shown that RfaH is recruited to U11 EC assembled on a scaffold or during elongation (Artsimovitch & Landick, 2002), but not to G8 EC (see below). As expected based on previous reports (Nedialkov & Burton, 2013), in the absence of RfaH, RNAP protected 14 nts of T DNA upstream from the active site from Exo III digestion (Fig. 1B). Since the RNA:DNA hybrid is 9 bp long (Kang et al., 2017), RNAP alone protects 5 bp of the duplex upstream from the bubble. In the presence of RfaH, the upstream boundary of U11 EC was extended by up to 7 nt, but remained stationary in G8 EC. This protection was observed at 10 nM RfaH (Supporting Information Fig. S2), at or below its cellular concentration (Schmidt et al., 2016).
An upstream shift of the footprint may be indicative of backtracking, reverse translocation of RNAP accompanied by the nascent RNA threading into the secondary channel. Although ops is an example of a backtracked pause (Artsimovitch & Landick, 2000), we considered this explanation unlikely because (i) RfaH-NTD inhibits pyrophosphorolysis, a result consistent with an enhancement of forward translocation (Svetlov et al., 2007) and (ii) the upstream segment of the nascent RNA in U11 scaffold EC is not complementary to the T DNA, a structure expected to inhibit reverse translocation. To exclude a possibility that the full-length RfaH promotes backtracking even in mismatched ECs, we probed their downstream boundaries (Fig. 1C). We found that RfaH did not alter the downstream Exo III protection in G8 and U11 ECs (Fig. 1C), confirming that the ECs in which the RNA cannot reanneal with the T strand are resistant to backtracking.
To test RfaH effects on RNAP translocation in complexes which could backtrack, we walked G8 ECs to the U11 position in one-nucleotide steps and tested their sensitivity to GreB-induced cleavage (Supporting Information Fig. S3). We found that G8 and C9 ECs were resistant to cleavage, both in the absence and in the presence of RfaH. C10 EC was backtracked by 2 nt in the absence of RfaH but became resistant to Gre cleavage in the presence of RfaH. Similarly, RfaH counteracted backtracking in U11 EC. We conclude that RNAP backtracks while approaching the ops pause site and that RfaH, once bound, inhibits backtracking.
The expanded protection is independent of the RfaH-CTD
What could be the source of an expanded footprint? One possibility is that the observed protection is due to RfaH itself. Two RfaH regions could mediate contacts with the upstream DNA, the KOW-CTD (residues 112–162) and the β-hairpin mini-domain (residues 32–49). The upstream DNA binds to the yeast Spt5 KOW domain (Ehara et al., 2017) and can be modeled to interact with the RfaH mini-domain (Fig. 1A); alanine substitution of Lys42 abolishes RfaH antipausing activity, whereas R43A substitution modestly reduces it (Belogurov et al., 2010). To test the contribution of these two elements to RfaH/TEC interactions, we carried out Exo III probing in the presence of RfaH-NTD or the K42A variant. We found that both proteins protected the upstream DNA from Exo III cleavage similarly to WT RfaH (Supporting Information Fig. S4). Thus, the RfaH-NTD is sufficient for the observed protection and the substitution of Lys42, the only residue in the mini-domain shown to be critical for antipausing activity (Belogurov et al., 2010), does not alter Exo III accessibility.
RfaH-bound ECs are not scrunched
DNA scrunching would also lead to an enlarged footprint. Scrunching has been observed in initiation (Winkelman & Gourse, 2017) and elongation (Strobel & Roberts, 2014, Zhilina et al., 2012) complexes in which the σ-DNA contacts stabilize a scrunched intermediate. During initial rounds of RNA synthesis, the front end of RNAP moves forward as nucleotides are added to the chain, but the rear end is held in place by σ-DNA contacts and the unwound DNA is pushed into the active site channel (Winkelman & Gourse, 2017). These interactions must be broken to allow escape from a promoter or from σ-dependent pause induced by a -10-like sequence. Stress accumulated during scrunching can also be relieved by release of a short abortive RNA (Winkelman & Gourse, 2017) or by backtracking, followed by cleavage of the nascent RNA and restart (Strobel & Roberts, 2014, Zhilina et al., 2012). Like σ, RfaH binds to the β′CH and the NT strand (Sevostyanova et al., 2008) and induces a pause downstream of the ops element (Belogurov et al., 2010), suggesting that it could stabilize a scrunched intermediate.
In RfaH-bound ECs, 5–7 additional bases are protected from Exo cleavage (Fig. 1B), suggesting scrunching of 5+ nucleotides. We used two approaches to test this idea. First, we assembled ECs in which NT DNA was nicked 2 nt upstream of the active site (Supporting Information Fig. S5). While the RfaH-binding region is preserved in this EC, potential steric compaction should be relieved. In initiation complexes, the scrunched NT strand is extruded into solution (Winkelman & Gourse, 2017) and might not create strain; indeed, NT nicks do not affect promoter escape (Samanta & Martin, 2013). In contrast, in the RfaH-bound EC, the NT strand is expected to be constrained by contacts to the RfaH-NTD and perhaps to GL. We found that RfaH protected the upstream DNA from Exo III cleavage in complexes assembled with either intact or nicked NT strand (Supporting Information Fig. S5), suggesting either that (i) the RfaH-bound EC is not scrunched or (ii) scrunching does not induce steric stress that can be alleviated by a break in the NT DNA.
RfaH stabilizes the upstream edge of the transcription bubble
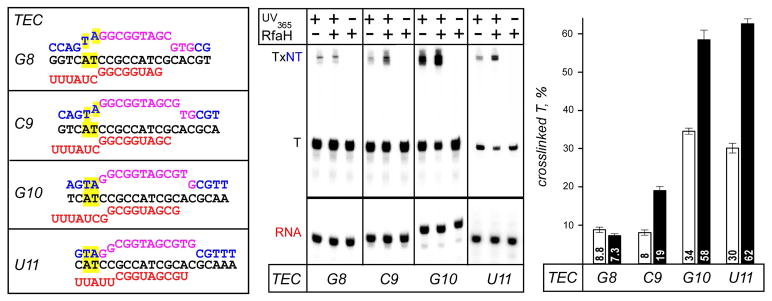
The resistance to Exo III, which is specific for the double-stranded DNA, could be explained by the duplex DNA distortion at the rear edge of the EC. To probe the upstream fork junction (the boundary between the transcription bubble and the upstream duplex) directly, we used crosslinking with 8-methoxypsoralen (8-MP). 8-MP specifically intercalates into double-stranded 5′-TA-3′ motifs and introduces a T-T inter-strand crosslink upon exposure to UV light, and has been used to map the transcription bubble in the E. coli EC (Turtola & Belogurov, 2016). We assembled G8 EC on a scaffold with a TA motif positioned nine nucleotides upstream of the RNA 3′ end (Fig. 2), with 5′-labeled T DNA and RNA. We then walked this complex in one-nucleotide steps to U11 and monitored the inter-strand crosslinking efficiency in the absence and the presence of RfaH. In G8 EC, the efficiency of DNA:DNA crosslinking was below 10%, as expected because the TA motif is located inside the transcription bubble. Upon formation of C9 EC, crosslinking remained low in the absence of RfaH, but was increased ~2-fold (from 8% to 19%) upon addition of RfaH; an analogous but smaller effect was observed for E. coli NusG (Turtola & Belogurov, 2016). Further extension of RNA to G10 and U11 led to increased crosslinking (~30% efficiency), as anticipated upon duplex formation, and RfaH effects remained the same (~2-fold enhancement) in both ECs. These results show that the upstream boundary of the transcription bubble is located at the expected position in ECs moving through the ops element. RfaH does not induce bubble expansion; on the contrary, it appears to stabilize the upstream fork junction, favoring forward translocation (Fig. 2 and Supporting Information Fig. S3). Interestingly, RfaH-mediated enhancement of 8-MP crosslinking is evident at C9, two nucleotides ahead of U11, indicating that pausing at the ops site (at U11) is not absolutely required for RfaH recruitment as long as RNAP moves slowly or not at all, as is the case in scaffold complexes.
Figure 2.
Probing the upstream fork junction by crosslinking with 8-MP. G8 and U11 ECs containing the TA intercalation site (highlighted in yellow) upstream from the ops element were assembled with [γ32P]-ATP labeled T DNA and RNA; C9 and G10 ECs were made by walking from G8. ECs were supplemented with 100 nM RfaH (where indicated) and illuminated with the 365 nm UV light. The products were analyzed on 12 % gels; U11 ECs were run on a different gel. The crosslinked T strand (TxNT) migrates much slower that free T DNA. Fractions of the crosslinked T DNA were determined in the absence (white bars) and presence (black bars) of RfaH. Error bars indicate the SDs of triplicate measurements.
The GL-RfaH interactions are required for upstream protection
Together, these results argue that RfaH alters RNAP contacts with DNA. To elucidate the basis for the observed protection, we used RNAP variants with changes in four elements that, based on the available data, could alter DNA accessibility directly or allosterically. First, the β′ lid loop (LL) acts in concert with NusG to surround the upstream DNA and inhibit backtracking (Turtola & Belogurov, 2016). RfaH is expected to establish a similar crosstalk with the LL that may restrict the DNA accessibility to Exo III directly. Second, pause-resistant substitutions in RNAP are insensitive to RfaH and mimic its effect on elongation (Svetlov et al., 2007), hypothetically by promoting conformational changes in the EC similarly to RfaH. β′ F773V substitution in the bridge helix, a quintessential example of this class (Malinen et al., 2014), could be expected to hinder Exo III digestion. Third, tightening of the clamp may inhibit Exo cleavage by stabilizing RNAP-DNA interactions. The clamp position is linked to movements of the β flap domain, which forms one side of the RNA exit channel and the β flap tip (FT) is expected to clash with the approaching Exo III (see below). Repositioning of the β flap upon the pause RNA hairpin formation is accompanied by the clamp swiveling (Kang et al., 2018a), whereas RfaH exerts an opposite effect (Hein et al., 2014). Finally, the GL, whose interactions with the RfaH HTTT motif are essential for antipausing activity (Sevostyanova et al., 2011), could stabilize DNA in the RfaH-bound EC through a network of NTD/NT DNA/GL contacts (NandyMazumdar et al., 2016).
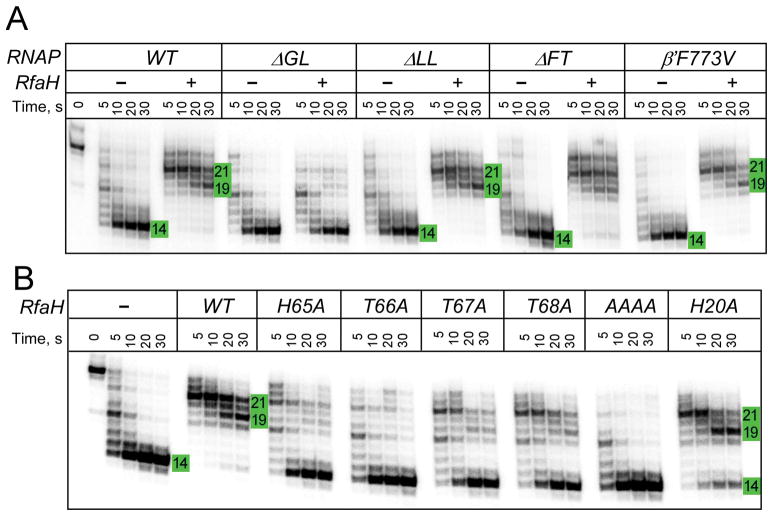
To evaluate if any of these elements contributes to the upstream DNA protection, we tested RNAPs with the F773V substitution and deletions of the LL, FT, and GL elements by Exo III footprinting (Fig. 3A). Our results indicate that all variant enzymes displayed similar Exo III boundaries in the absence of RfaH. Strikingly, the GL deletion eliminated the upstream protection by RfaH, whereas other changes had no effect. This observation suggests that the antipausing activity and the DNA accessibility, both of which are altered upon the GL deletion, are functionally linked.
Figure 3.
Protection of the upstream DNA by RfaH and RNAP variants.
A. U11 ECs were assembled with [γ32P]-ATP labeled T strand with the WT or altered RNAP. Exo III was added following incubation with RfaH (or storage buffer) and the reactions were analyzed as above.
B. The WT U11 ECs assembled as in A were probed with Exo III in the presence of selected RfaH variants. AAAA is a quadruple mutant in which HTTT residues 65–68 were replaced with four alanines.
To confirm that the effect of ΔGL is due to the disruption of the GL-NTD contacts, as opposed to contacts with DNA, we used RfaH variants with substitutions in the HTTT motif. We found that alanine substitution of each single HTTT residue strongly reduced protection, and the quadruple substitution completely eliminated it (Fig. 3B). By comparison, alanine substitution of His20, a residue that interacts with DNA, increased susceptibility to cleavage only slightly. Although the deletion of GL and HTTT substitutions do not significantly alter RfaH binding to the EC (Belogurov et al., 2010, Sevostyanova et al., 2011), we performed these experiments at 100 nM RfaH, a ten-fold excess over the WT RfaH concentration that confers protection (Supporting Information Fig. S2), to compensate for possible minor differences in affinity. We conclude that the upstream DNA protection is due to allosteric effects of stabilizing RfaH-GL contacts.
Modeling the effects of DNA conformation on Exo III accessibility
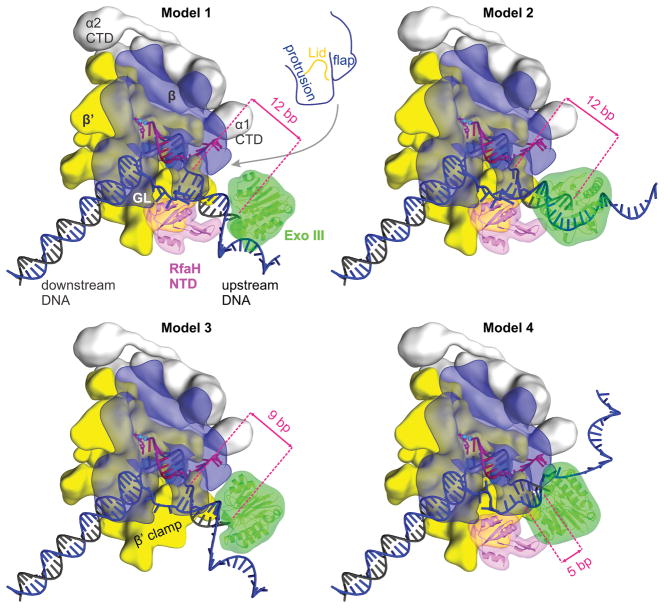
The above results exclude scrunching and backtracking as plausible explanations for the upstream DNA protection. We hypothesized that RfaH may restrict the upstream DNA duplex conformation, thereby hindering Exo III access, in several ways. First, a five amino-acid loop of RfaH/NusG faces the upstream fork junction and may alter the trajectory of the upstream DNA (Turtola & Belogurov, 2016). Second, the β-hairpin mini-domain could contact the upstream DNA duplex to restrict its conformation. Third, the single-stranded NT DNA loops around RfaH (Zuber et al., 2018, Kang et al., 2018b), thereby effectively shortening the NT DNA tether between the upstream and downstream DNA and constraining them in an orthogonal orientation. We refer to a change in the NT DNA trajectory by RfaH as a NT DNA lock model.
To evaluate these possibilities, we built models in which Exo III is approaching the EC (with or without the bound RfaH) from behind (Fig. 4). A detailed description of the models, along with an alternative view (Fig. S6), pdb files, and 3D PDF files are provided in Supporting Information. Modeling revealed that Exo III does not clash with any EC components when positioned 12 bp upstream of the transcription bubble if the upstream DNA is orthogonal to the downstream DNA and lines the β′ clamp (Model 1; Fig. 4). However, tilting the upstream DNA towards the β protrusion brings Exo III in contact with the protrusion (Model 2; Fig. 4 and Supporting Information Fig. S6). Given that RfaH protects 12 upstream bp (Figs. 1A and 3), these results suggest that RfaH restricts the upstream DNA in a protrusion-proximal conformation in addition to restricting its orthogonal orientation relative to the downstream DNA.
Figure 4.
Modeling Exo III approach to the EC.
Models of the Exo III digesting the upstream duplex DNA in different ECs (see the main text and for details). The EC components, RfaH, and Exo III are depicted as in Figure 1. Numbers indicate distances from the Exo III active site to the upstream fork junction. Supplementary Information Fig. S6 shows alternative views of the models, which can also be viewed as 3D PDB models (see Supplementary Information).
Next, we considered a possibility that RfaH effect on the DNA trajectory in the EC is mediated solely by the NT DNA looping. If this were the case, the upstream duplex DNA would be restricted in the orthogonal orientation by physically shortening the NT DNA. By in silico modeling, we estimated that minimum of five single-stranded nucleotides is needed to tether the orthogonal upstream and the downstream DNA without compromising the duplex structures (Model 3; Fig. 4). In a resulting model, Exo III is positioned 9 bp from the bubble.
The observation that the upstream protection is largely absent when RfaH-GL contacts are compromised (Fig. 3) strongly argues that the NT DNA lock makes the major contribution to restricting the upstream DNA flexibility by RfaH. To structurally evaluate effects of the loss of RfaH/NT DNA contacts, we constructed a fourth model in which Exo III is positioned five bp upstream from the transcription bubble (Model 4), as observed in ECs without RfaH or RfaH-GL contacts (Fig. 3). We threaded ten nucleotides of the single-stranded NT DNA so that it does not clash with RfaH, but did not attempt to loop the NT strand around the RfaH surface implicated in interactions with ops (Belogurov et al., 2010). The resulting model (Model 4; Fig. 4) shows that a conformation in which Exo III leaves only five bp of the upstream DNA undigested is spatially feasible if the NT DNA is not required to loop around the RfaH surface.
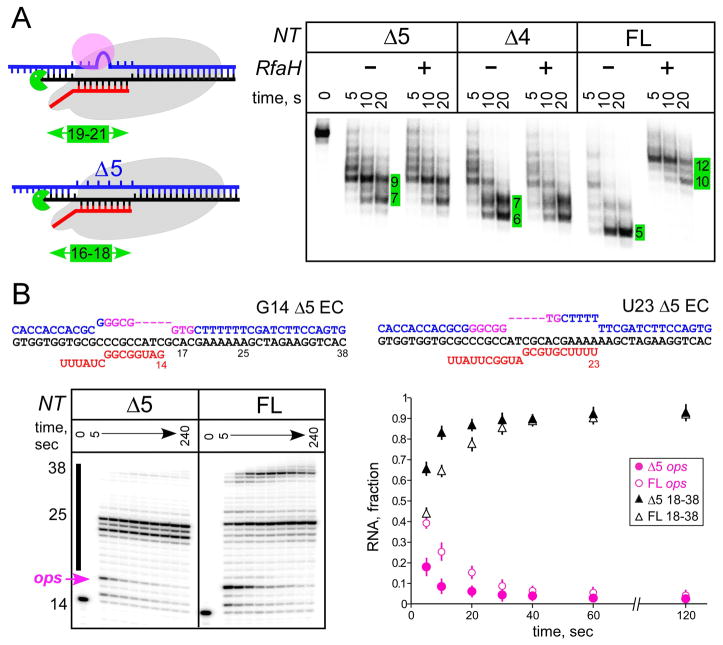
Shortening the NT DNA mimics RfaH effects on the EC
In the EC, the transcription bubble is 10 nt, and the T DNA strand is held tightly within the 9-bp RNA:DNA hybrid (Kang et al., 2017). By contrast, the NT strand is very flexible and may potentially assume conformations incompatible with processive transcription (NandyMazumdar et al., 2016). Modeling suggests that up to five NT DNA nucleotides can be deleted without compromising the EC structure (Model 3; Fig. 4), altering the upstream duplex DNA trajectory. To test if RfaH-mediated Exo III protection could be mimicked by physically shortening the NT DNA, we assembled scaffold ECs in which four or five NT DNA nucleotides were deleted (Fig. 5A) while the T DNA and RNA strands were identical to those in U11 EC (Fig. 4). These deletions remove the ops bases that interact with RfaH (Zuber et al., 2018, Kang et al., 2018b), explaining the lack of RfaH effect on these ECs (Fig. 5A). In support of the DNA lock model, four- and five-nt deletions protected seven and nine bp of upstream DNA from cleavage, respectively (Fig. 5A). This result is in agreement with model 3 (Fig. 4) where Exo III stops nine bp upstream of the transcription bubble when the NT strand is 5 nt long, matching the five-nt deletion. Strikingly, forthcoming X-ray, NMR, and cryo-EM structures of RfaH bound to the ops DNA and to a complete opsEC reveal that the NT DNA forms a short hairpin in which the ops bases are presented for RfaH recognition (Kang et al., 2018b, Zuber et al., 2018).
Figure 5.
Constraining NT DNA inhibits Exo III cleavage and promotes elongation.
A. U11 ECs were assembled on scaffolds with the full-length (FL) NT DNA or NT DNAs containing deletions of four or five nucleotides. Exo III probing was carried out in the presence and in the absence of RfaH. Numbers indicate the distance from the bubble, to facilitate comparison with Fig. 4.
B. ECs were assembled on scaffolds with the FL or Δ5 NT DNA and [γ32P]-ATP labeled G14 RNA, and incubated with 10 μM GTP, 150 μM ATP, CTP, UTP for 5–240 sec before quenching. RNAs were analyzed on 12% gel. RNA fractions were determined from 3 replicates (±SD).
DNA lock promotes elongation in the absence of RfaH
The central element of our model is that locking the NT DNA should reduce pausing similarly to, and independently of, RfaH. To test this, we extended RNA in scaffold ops8 EC assembled with the full length or Δ5 NT DNA (Fig. 5B). As observed on standard long templates, RNAP efficiently paused at the ops site (U17 RNA) on the full-length NT scaffold, and a fraction of ECs were paused/arrested at U25, likely due to the lack of downstream duplex ahead of RNAP. On the Δ5 scaffold, the efficiency of pausing at the ops site was reduced 2-fold, an effect that quantitatively matches that of RfaH (Belogurov et al., 2007), and RNAP was arrested at U23 and U25. A block to bubble reannealing behind RNAP is a likely cause of arrest. Together, these results argue that constraining the NT DNA promotes elongation at an adjacent site, but may hinder transcription at other sites; locking the NT DNA by PNAs promotes R-loop formation and inhibits transcription by phage RNAPs (D’Souza et al., 2018).
DNA lock is not restricted to ops EC
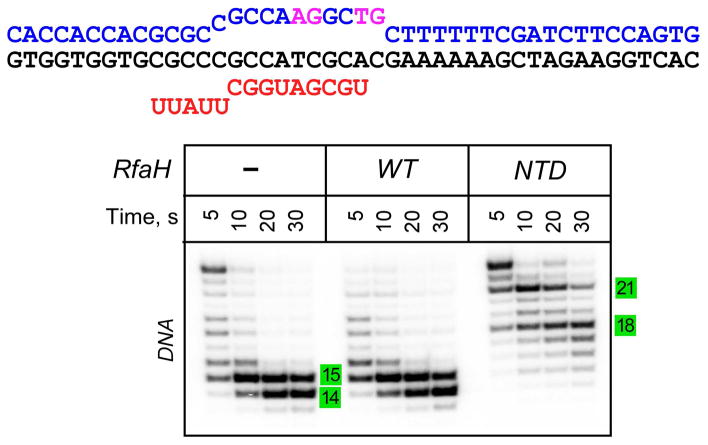
Is the upstream protection dependent on RfaH interactions with ops, and thus restricted to the recruitment step, or is it a general property of RfaH-modified ECs? To answer this question, we probed the Exo sensitivity of a non-ops EC upon binding of the RfaH-NTD. In the isolated RfaH-NTD, the activation step which is dependent on ops is bypassed, allowing sequence-independent recruitment to RNAP (Belogurov et al., 2007). The NTD-bound EC thus mimics a post-recruitment RfaH-modified EC. We assembled an EC in which the ops bases that specifically interact with RfaH (G5 and T6) are mutated but the residues in the RNAP active site (T11 and G12) are preserved (Fig. 6). In this “scrambled ops” EC (Belogurov et al., 2007) the NTD, but not the full-length RfaH, conferred the extended upstream protection against Exo III cleavage. These results suggest that the NTD is able to dictate the DNA trajectory in any and all ECs after recruitment.
Figure 6.
The upstream protection is independent of the ops DNA sequence. The scrambled ops ECs were assembled on a scaffold shown on top; the NT strand residues in magenta match those in ops. The wild-type full-length RfaH does not bind to this EC because contacts with the ops element are required to induce RfaH domain dissociation to expose the RNAP-binding site on the NTD. The isolated NTD binds to any EC and serves and a model of activated, post-recruitment conformation of RfaH. The assembled with the T DNA strand 5′-end labeled with [γ32P]-ATP EC was incubated with RfaH or NTD (at 100 nM) and treated with Exo III. The reactions were analyzed on a 12% urea-acrylamide gel. Numbers indicate the distance from the RNAP active site.
DISCUSSION
NusG/Spt5 proteins bind to the elongating RNAP and favor productive RNA synthesis. While most discussion has focused on similar interactions of the NTD with RNAP, widely assumed to be responsible for the antipausing activity of all members of this family, the role of direct DNA contacts has only recently come to light (Crickard et al., 2016, NandyMazumdar et al., 2016, Yakhnin et al., 2016). Our present results argue that remodeling of the NT DNA and the upstream DNA duplex by RfaH makes a significant contribution to its effects on RNA chain elongation. We hypothesize that the ability to modulate the NT and duplex DNA trajectories is shared by diverse regulators of transcription across all life.
The DNA lock mechanism
Our in silico (Fig. 4) and in vitro (Fig. 5) results support our hypothesis that RfaH-NTD restrains the NT DNA strand and the upstream DNA duplex. The latter effect confers protection of the upstream DNA from Exo III cleavage. Our modeling argues that the orientation of the upstream DNA can modulate Exo III progression (Fig. 4): while the parallel duplex orientation allows for the closest approach to the transcription bubble (5 bp), the upstream DNA that is orthogonal to the downstream DNA offers the longest protection (12 bp). Importantly, RfaH does not impede the Exo III approach unless the upstream DNA is restrained (Model 4; Fig. 4).
By looping the NT DNA around, RfaH effectively shortens it. We observe that physical shortening of the NT DNA by five nucleotides leads to nine bp protection from Exo III (Fig. 5A), a result fully consistent a predicted orthogonal orientations of the upstream and downstream DNA duplexes (Model 3; Fig. 4). The longer protection conferred by RfaH (12 bp) suggests that the upstream DNA is additionally constrained by contacts with RNAP, but the structural basis for this effect is beyond the resolution of our models. One possibility is that, by folding the NT DNA, RfaH precisely positions the junction point between the single-stranded NT DNA and the upstream duplex DNA to bring the latter near the β protrusion.
Controlling the NT DNA trajectory during elongation
Other proteins that interact with the NT DNA would be expected to alter the observed upstream RNAP boundary on the DNA. Indeed, an upstream block to Exo III cleavage was observed in σ-paused ECs (Yarnell & Roberts, 1992, Zhilina et al., 2012). RfaH NTD and σ domain 2 (σ2) make topologically similar contacts to the melted NT DNA and β′CH (Sevostyanova et al., 2008) and delay RNAP escape downstream from their recruitment sites (Artsimovitch & Landick, 2002, Perdue & Roberts, 2011). The isolated σ2 is sufficient to delay escape (Zenkin et al., 2007) and induce upstream protection (Zhilina et al., 2012) in σ-paused complexes, in which the bubble is expanded (Kainz & Roberts, 1992) and the DNA is scrunched (Marr et al., 2001, Zhilina et al., 2012). Although the extended Exo III footprint in σ-paused ECs could be partially due to scrunching (Zhilina et al., 2012), observations that changes in the footprint boundary (6–13 nt) are different between the complexes formed on short and long templates and exceed the number of “scrunchable” nucleotides (~4 nt in both cases) argue that scrunching alone cannot explain this protection. By contrast, RfaH-bound ECs are not scrunched; the upstream fork junction (Fig. 2) and the downstream RNAP boundary (Fig. 1C) are indicative of a relaxed EC. However, as in σ-paused ECs, RfaH protects the upstream DNA from Exo III cleavage by 5–7 nt (Fig. 1B) and the NTD is sufficient for protection (Supporting Information Fig. S3). In the case of RfaH, which makes fewer contacts to the NT strand than σ2 (Zuber et al., 2018, Feklistov & Darst, 2011), protection is dependent on GL (Fig. 3), which possibly supplements the missing contacts by binding to both the NT DNA and RfaH NTD (NandyMazumdar et al., 2016).
NT DNA interactions in other NusG homologs
Initial studies painted a picture in which specific recognition of the ops element arose in evolution to direct RfaH to a few specialized operons in the presence of a large excess of housekeeping NusG. In contrast to E. coli RfaH, which is recruited to the elongating RNAP only at ops sites, E. coli NusG displays no apparent sequence specificity and is able to bind to RNAP across the entire genome (Belogurov et al., 2009). However, studies of other NusGs suggest that the apparent lack of sequence preferences could be a peculiar feature of E. coli NusG. B. subtilis NusG recognizes a specific sequence in the NT DNA and inhibits RNAP escape (Yakhnin et al., 2016), similarly to RfaH effects at the ops site (Artsimovitch & Landick, 2002). Pause-enhancing properties of Mycobacterium bovis (Czyz et al., 2014) and Thermus thermophilus (Sevostyanova & Artsimovitch, 2010) NusGs could also be explained by their sequence-specific contacts with DNA.
Identification of DNA elements recognized by the NTD is challenging. The role of ops in RfaH recruitment is not limited to direct interactions with RfaH. In the ops-paused EC, no more than six out of 12 ops nucleotides could interact with RfaH, and G5 and T6 mediate most contacts with the NTD (Zuber et al., 2018, Kang et al., 2018b). However, substitutions of almost every ops base confer strong defects on RfaH function in vivo, including those of T11 and G12 located in the RNAP active site and inaccessible to RfaH (Zuber et al., 2018). Our results that the effects of T11 and G12 mutations, which inhibit pausing, can be suppressed in vitro by reducing NTP concentration to promote pausing (Zuber et al., 2018) support a model in which pausing at the ops site extends a time window for RfaH recruitment to the moving RNAP. Finally, transient contacts between RfaH and the ops-paused EC trigger RfaH domain dissociation, which exposes the β′CH binding site on the NTD and allows RfaH recruitment to the EC (Belogurov et al., 2007). This activation step is unique to RfaH, in which the NTD and CTD tightly interact to stabilize the autoinhibited state (Shi et al., 2017). NusG could travel with RNAP, scanning the NT DNA. Given the proximity of the NT DNA to the NTDs, it is hard to imagine how NTD-DNA interactions can be avoided and that all sequences will be recognized equally. Preferred sequences, possibly able to form a secondary structure that could promote base flipping to augment the information content of a short NT DNA sequence exposed on the EC surface (Zuber et al., 2018), would be expected to delay RNAP at a specific site. Consistently, the isolated RfaH-NTD in which the activation state is bypassed increases RNAP pausing at the ops site as efficiently as does the full-length RfaH (Belogurov et al., 2007). Systematic analysis of NusG interactions with their cognate ECs would be required to determine if their hypothetical interactions with the NT DNA are specific.
Diverse roles of the NT DNA in RNA synthesis
The NT strand has been shown to play many roles in regulating gene expression, from controlling properties of promoter complexes to serving as a recruitment platform for diverse protein factors, which include AID deaminase during immunoglobulin class switching (Pavri et al., 2010), UvrD during transcription-coupled repair (Epshtein et al., 2014), and C37 during RNAP III termination (Arimbasseri & Maraia, 2015). We propose that some of these proteins constrain NT DNA as part of their mechanisms. Our hypothesis is based on protection against exonuclease cleavage in complexes assembled on short linear templates. What role could folding of the NT strand play in a physiological context? We envision several possibilities. First, specific protein recruitment to a NT signal during elongation could be challenging because each short DNA segment is exposed on the RNAP surface only transiently. Formation of a secondary structure would augment the information content of the signal and could transiently slow the EC, thus aiding recruitment. Second, the NT strand could be trapped in a nonproductive conformation, hindering rapid elongation. By guiding NT DNA along the optimal path, a regulator could support pause-free transcription. Third, holding the NT strand in place could modulate DNA reannealing behind RNAP, in turn controlling translocation, R-loop formation, and the trajectory of upstream DNA duplex that interacts with Spt5 (Crickard et al., 2016, Ehara et al., 2017) and perhaps other accessory factors. While relative contributions of NTD interactions with the DNA and RNAP may vary, the available data suggest that many if not all NusG-like proteins are able to restrict the mobility of the transcription bubble to modify RNAP into a pause- and arrest-resistant state.
EXPERIMENTAL PROCEDURES
Reagents and proteins
All general reagents were obtained from Sigma Aldrich (St. Louis, MO) and Fisher (Pittsburgh, PA); NTPs, from GE Healthcare (Piscataway, NJ); [γ32P]-ATP and [α32P]-GTP, from Perkin Elmer (Boston, MA); PCR reagents, restriction and modification enzymes, from NEB (Ipswich, MA) and Roche (Indianapolis, IN). Oligonucleotides were obtained from Integrated DNA Technologies (Coralville, IA) and Sigma Aldrich. DNA purification kits were from Qiagen (Valencia, CA). E. coli RNAP variants (Svetlov & Artsimovitch, 2015), RfaH variants (Belogurov et al., 2007), and GreB (Vassylyeva et al., 2007) were purified as described previously.
Exonuclease footprinting
Scaffolds were assembled from oligonucleotides listed in Supplementary Table S1. In all scaffolds, the RNA primer was end-labeled with [γ32P]-ATP using T4 polynucleotide kinase (PNK; NEB). For the upstream RNAP edge mapping, T DNA oligonucleotide was also labeled; for the downstream edge mapping, NT DNA oligonucleotide was labeled. Following labeling, oligonucleotides were purified using QIAquick Nucleotide Removal Kit (Qiagen). To assemble a scaffold, RNA and T DNA oligonucleotides were combined in PNK buffer and annealed in a PCR machine as follows: 5 min at 45 °C; 2 min each at 42, 39, 36, 33, 30, and 27 °C, 10 min at 25 °C. 12 pmoles of T/RNA hybrid were mixed with 14 pmoles of His-tagged core RNAP in 30 μl of transcription buffer (TB) [20 mM Tris-Cl, 5% Glycerol, 40 mM KCl (upstream mapping) or 120 mM KCl (downstream mapping), 5 mM MgCl2, 10 mM β-mercaptoethanol, pH 7.9], and incubated at 37 °C for 10 min. 15 μl of His-Select® HF Nickel Affinity Gel (Sigma Aldrich) was washed once in TB and incubated with 20 μg Bovine Serum Albumin in a 40-μl volume for 15 min at 37 °C, followed by a single wash step in TB-40 or TB-120 (for the upstream and downstream mapping, respectively). The T/RNA/RNAP complex was mixed with the Affinity Gel for 15 min at 37 °C on a thermomixer (Eppendorf) at 900 rpm, and washed twice with TB. 30 pmoles of the NT oligonucleotide were added, followed by incubation for 20 min at 37 °C, one 5-min incubation with TB-1000 in a thermomixer, and five washes with TB-40/120. The assembled ECs were eluted from beads with 90 mM imidazole in a 15-μl volume, purified through a Durapore (PVDF) 0.45 μm Centrifugal Filter Unit (Merck Millipore), and resuspended in TB-40/120. The EC was divided in two aliquots; one was incubated with 100 nM RfaH and the other – with storage buffer for 3 min at 37°C. For each time point, 5 μl EC were mixed with 5 μl of Exo III (NEB, 40 U) and incubated at 21 °C. At times indicated in figure legends, the reactions were quenched with an equal volume of Stop buffer (8 M Urea, 20 mM EDTA, 1 × TBE, 0.5 % Brilliant Blue R, 0.5 % Xylene Cyanol FF).
Psoralen crosslinking
G8 and U11 scaffolds were assembled as described for the upstream Exo III mapping; C9 and G10 ECs were obtained by walking in the presence of a cognate NTP. The ECs eluted from beads were re-suspended in TB-40 supplemented with 6.3 % DMSO and 0.92 mM 8-MP and incubated for 2 min at 37 °C, followed by addition of 100 nM RfaH (or storage buffer) and a 3-min incubation at 37 °C. Complexes were then exposed to 365 nm UV light (8W Model UVLMS-38; UVP, LLC) for 20 min on ice. The reactions were quenched as above.
Gre-mediated cleavage
R40 oligonucleotide was 5′-end labeled with [γ32P]-ATP and scaffolds were assembled as described for the upstream Exo III mapping. Aliquots of the assembled EC were incubated with 5 μM NTP substrates to extend the nascent G14 RNA to C15, G16 and U17 for 5 min at 37°C. Following three washes with TB-40, complexes were incubated with 50 nM of RfaH or storage buffer for 3 min at 37°C. GreB (or storage buffer) was then added to 500 nM for 5 min at 37°C, and the reactions were quenched as above.
Sample analysis
Samples were heated for 2 min at 95 °C and separated by electrophoresis in denaturing acrylamide (19:1) gels (7 M Urea, 0.5× TBE). The gels were dried and the products were visualized and quantified using a FLA9000 Phosphorimaging System (GE Healthcare), ImageQuant Software, and Microsoft Excel.
Supplementary Material
Acknowledgments
We thank Bob Landick and Rachel Mooney for a gift of pRM933, Matti Turtola for the β′LL deletion enzyme, and Bob Landick and Rich Maraia for comments on the manuscript.
Abbreviations
- CH
clamp helices
- CTD
C-terminal domain
- EC
elongation complex
- FT
flap tip
- GL
gate loop
- LL
lid loop
- NT
non-template
- NTD
N-terminal domain
- RNAP
RNA polymerase
Footnotes
AUTHOR CONTRIBUTIONS:
GAB and IA conceived research; YN, DS, GAB, and IA performed research; YN, DS, GAB, and IA analyzed data; and GAB and IA wrote the paper with input from all authors.
References
- Arimbasseri AG, Maraia RJ. Mechanism of Transcription Termination by RNA Polymerase III Utilizes a Non-template Strand Sequence-Specific Signal Element. Mol Cell. 2015;58:1124–1132. doi: 10.1016/j.molcel.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artsimovitch I, Landick R. Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proc Natl Acad Sci U S A. 2000;97:7090–7095. doi: 10.1073/pnas.97.13.7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artsimovitch I, Landick R. The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell. 2002;109:193–203. doi: 10.1016/s0092-8674(02)00724-9. [DOI] [PubMed] [Google Scholar]
- Belogurov GA, Mooney RA, Svetlov V, Landick R, Artsimovitch I. Functional specialization of transcription elongation factors. Embo J. 2009;28:112–122. doi: 10.1038/emboj.2008.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belogurov GA, Sevostyanova A, Svetlov V, Artsimovitch I. Functional regions of the N-terminal domain of the antiterminator RfaH. Mol Microbiol. 2010;76:286–301. doi: 10.1111/j.1365-2958.2010.07056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belogurov GA, Vassylyeva MN, Svetlov V, Klyuyev S, Grishin NV, Vassylyev DG, Artsimovitch I. Structural basis for converting a general transcription factor into an operon-specific virulence regulator. Mol Cell. 2007;26:117–129. doi: 10.1016/j.molcel.2007.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernecky C, Plitzko JM, Cramer P. Structure of a transcribing RNA polymerase II-DSIF complex reveals a multidentate DNA-RNA clamp. Nat Struct Mol Biol. 2017;24:809–815. doi: 10.1038/nsmb.3465. [DOI] [PubMed] [Google Scholar]
- Chakraborty A, Wang D, Ebright YW, Korlann Y, Kortkhonjia E, Kim T, Chowdhury S, Wigneshweraraj S, Irschik H, Jansen R, Nixon BT, Knight J, Weiss S, Ebright RH. Opening and closing of the bacterial RNA polymerase clamp. Science. 2012;337:591–595. doi: 10.1126/science.1218716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crickard JB, Fu J, Reese JC. Biochemical Analysis of Yeast Suppressor of Ty 4/5 (Spt4/5) Reveals the Importance of Nucleic Acid Interactions in the Prevention of RNA Polymerase II Arrest. J Biol Chem. 2016;291:9853–9870. doi: 10.1074/jbc.M116.716001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czyz A, Mooney RA, Iaconi A, Landick R. Mycobacterial RNA polymerase requires a U-tract at intrinsic terminators and is aided by NusG at suboptimal terminators. MBio. 2014;5:e00931. doi: 10.1128/mBio.00931-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza AD, Belotserkovskii BP, Hanawalt PC. A novel mode for transcription inhibition mediated by PNA-induced R-loops with a model in vitro system. Biochim Biophys Acta. 2018;1861:158–166. doi: 10.1016/j.bbagrm.2017.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehara H, Yokoyama T, Shigematsu H, Yokoyama S, Shirouzu M, Sekine SI. Structure of the complete elongation complex of RNA polymerase II with basal factors. Science. 2017;357:921–924. doi: 10.1126/science.aan8552. [DOI] [PubMed] [Google Scholar]
- Epshtein V, Kamarthapu V, McGary K, Svetlov V, Ueberheide B, Proshkin S, Mironov A, Nudler E. UvrD facilitates DNA repair by pulling RNA polymerase backwards. Nature. 2014;505:372–377. doi: 10.1038/nature12928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feklistov A, Bae B, Hauver J, Lass-Napiorkowska A, Kalesse M, Glaus F, Altmann KH, Heyduk T, Landick R, Darst SA. RNA polymerase motions during promoter melting. Science. 2017;356:863–866. doi: 10.1126/science.aam7858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feklistov A, Darst SA. Structural basis for promoter-10 element recognition by the bacterial RNA polymerase sigma subunit. Cell. 2011;147:1257–1269. doi: 10.1016/j.cell.2011.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science. 2001;292:1876–1882. [Google Scholar]
- Guo G, Gao Y, Zhu Z, Zhao D, Liu Z, Zhou H, Niu L, Teng M. Structural and biochemical insights into the DNA-binding mode of MjSpt4p:Spt5 complex at the exit tunnel of RNAPII. J Struct Biol. 2015;192:418–425. doi: 10.1016/j.jsb.2015.09.023. [DOI] [PubMed] [Google Scholar]
- Guo X, Myasnikov A, Chen J, Crucifix C, Papai G, Takacs M, Schultz P, Weixlbaumer A. Structural basis for NusA stabilized transcriptional pausing. Mol Cell. 2018;69:816–827. doi: 10.1016/j.molcel.2018.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein PP, Kolb KE, Windgassen T, Bellecourt MJ, Darst SA, Mooney RA, Landick R. RNA polymerase pausing and nascent-RNA structure formation are linked through clamp-domain movement. Nat Struct Mol Biol. 2014;21:794–802. doi: 10.1038/nsmb.2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirtreiter A, Damsma GE, Cheung AC, Klose D, Grohmann D, Vojnic E, Martin AC, Cramer P, Werner F. Spt4/5 stimulates transcription elongation through the RNA polymerase clamp coiled-coil motif. Nucleic Acids Res. 2010;38:4040–4051. doi: 10.1093/nar/gkq135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kainz M, Roberts J. Structure of transcription elongation complexes in vivo. Science. 1992;255:838–841. doi: 10.1126/science.1536008. [DOI] [PubMed] [Google Scholar]
- Kang JY, Mishanina TV, Bellecourt MJ, Mooney RA, Darst SA, Landick R. RNA polymerase accommodates a pause RNA hairpin by global conformational rearrangements that prolong pausing. Mol Cell. 2018a;69:802–815. doi: 10.1016/j.molcel.2018.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JY, Mooney RA, Nedialkov Y, Saba J, Mishanina TV, Artsimovitch I, Landick R, Darst SA. Structural basis for transcript elongation control by NusG family universal regulators. Cell. 2018b doi: 10.1016/j.cell.2018.05.017. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JY, Olinares PD, Chen J, Campbell EA, Mustaev A, Chait BT, Gottesman ME, Darst SA. Structural basis of transcription arrest by coliphage HK022 Nun in an Escherichia coli RNA polymerase elongation complex. Elife. 2017:6. doi: 10.7554/eLife.25478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kireeva ML, Kashlev M. Mechanism of sequence-specific pausing of bacterial RNA polymerase. Proc Natl Acad Sci U S A. 2009;106:8900–8905. doi: 10.1073/pnas.0900407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein BJ, Bose D, Baker KJ, Yusoff ZM, Zhang X, Murakami KS. RNA polymerase and transcription elongation factor Spt4/5 complex structure. Proc Natl Acad Sci U S A. 2011;108:546–550. doi: 10.1073/pnas.1013828108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrpides NC, Woese CR, Ouzounis CA. KOW: a novel motif linking a bacterial transcription factor with ribosomal proteins. Trends Biochem Sci. 1996;21:425–426. doi: 10.1016/s0968-0004(96)30036-4. [DOI] [PubMed] [Google Scholar]
- Larson MH, Mooney RA, Peters JM, Windgassen T, Nayak D, Gross CA, Block SM, Greenleaf WJ, Landick R, Weissman JS. A pause sequence enriched at translation start sites drives transcription dynamics in vivo. Science. 2014;344:1042–1047. doi: 10.1126/science.1251871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinen AM, Nandymazumdar M, Turtola M, Malmi H, Grocholski T, Artsimovitch I, Belogurov GA. CBR antimicrobials alter coupling between the bridge helix and the beta subunit in RNA polymerase. Nat Commun. 2014;5:3408. doi: 10.1038/ncomms4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marr MT, Datwyler SA, Meares CF, Roberts JW. Restructuring of an RNA polymerase holoenzyme elongation complex by lambdoid phage Q proteins. Proc Natl Acad Sci U S A. 2001;98:8972–8978. doi: 10.1073/pnas.161253298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Rucobo FW, Sainsbury S, Cheung AC, Cramer P. Architecture of the RNA polymerase-Spt4/5 complex and basis of universal transcription processivity. Embo J. 2011;30:1302–1310. doi: 10.1038/emboj.2011.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NandyMazumdar M, Nedialkov Y, Svetlov D, Sevostyanova A, Belogurov GA, Artsimovitch I. RNA polymerase gate loop guides the nontemplate DNA strand in transcription complexes. Proc Natl Acad Sci U S A. 2016;113:14994–14999. doi: 10.1073/pnas.1613673114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedialkov YA, Burton ZF. Translocation and fidelity of Escherichia coli RNA polymerase. Transcription. 2013;4:136–143. doi: 10.4161/trns.25527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavri R, Gazumyan A, Jankovic M, Di Virgilio M, Klein I, Ansarah-Sobrinho C, Resch W, Yamane A, Reina San-Martin B, Barreto V, Nieland TJ, Root DE, Casellas R, Nussenzweig MC. Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell. 2010;143:122–133. doi: 10.1016/j.cell.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdue SA, Roberts JW. Sigma(70)-dependent transcription pausing in Escherichia coli. J Mol Biol. 2011;412:782–792. doi: 10.1016/j.jmb.2011.02.011. [DOI] [PubMed] [Google Scholar]
- Samanta S, Martin CT. Insights into the mechanism of initial transcription in Escherichia coli RNA polymerase. J Biol Chem. 2013;288:31993–32003. doi: 10.1074/jbc.M113.497669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Kochanowski K, Vedelaar S, Ahrne E, Volkmer B, Callipo L, Knoops K, Bauer M, Aebersold R, Heinemann M. The quantitative and condition-dependent Escherichia coli proteome. Nat Biotechnol. 2016;34:104–110. doi: 10.1038/nbt.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevostyanova A, Artsimovitch I. Functional analysis of Thermus thermophilus transcription factor NusG. Nucleic Acids Res. 2010;38:7432–7445. doi: 10.1093/nar/gkq623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevostyanova A, Belogurov GA, Mooney RA, Landick R, Artsimovitch I. The beta subunit gate loop is required for RNA polymerase modification by RfaH and NusG. Mol Cell. 2011;43:253–262. doi: 10.1016/j.molcel.2011.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevostyanova A, Svetlov V, Vassylyev DG, Artsimovitch I. The elongation factor RfaH and the initiation factor sigma bind to the same site on the transcription elongation complex. Proc Natl Acad Sci U S A. 2008;105:865–870. doi: 10.1073/pnas.0708432105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi D, Svetlov D, Abagyan R, Artsimovitch I. Flipping states: a few key residues decide the winning conformation of the only universally conserved transcription factor. Nucleic Acids Res. 2017;45:8835–8843. doi: 10.1093/nar/gkx523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel EJ, Roberts JW. Regulation of promoter-proximal transcription elongation: enhanced DNA scrunching drives lambdaQ antiterminator-dependent escape from a sigma70-dependent pause. Nucleic Acids Res. 2014;42:5097–5108. doi: 10.1093/nar/gku147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svetlov V, Artsimovitch I. Purification of bacterial RNA polymerase: tools and protocols. Methods Mol Biol. 2015;1276:13–29. doi: 10.1007/978-1-4939-2392-2_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svetlov V, Belogurov GA, Shabrova E, Vassylyev DG, Artsimovitch I. Allosteric control of the RNA polymerase by the elongation factor RfaH. Nucleic Acids Res. 2007;35:5694–5705. doi: 10.1093/nar/gkm600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomar SK, Artsimovitch I. NusG-Spt5 proteins-Universal tools for transcription modification and communication. Chem Rev. 2013;113:8604–8619. doi: 10.1021/cr400064k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turtola M, Belogurov GA. NusG inhibits RNA polymerase backtracking by stabilizing the minimal transcription bubble. Elife. 2016:e18096. doi: 10.7554/eLife.18096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007;448:157–162. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- Vassylyeva MN, Svetlov V, Dearborn AD, Klyuyev S, Artsimovitch I, Vassylyev DG. The carboxy-terminal coiled-coil of the RNA polymerase beta’-subunit is the main binding site for Gre factors. EMBO Rep. 2007;8:1038–1043. doi: 10.1038/sj.embor.7401079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vvedenskaya IO, Vahedian-Movahed H, Bird JG, Knoblauch JG, Goldman SR, Zhang Y, Ebright RH, Nickels BE. Transcription. Interactions between RNA polymerase and the “core recognition element” counteract pausing. Science. 2014;344:1285–1289. doi: 10.1126/science.1253458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weixlbaumer A, Leon K, Landick R, Darst SA. Structural basis of transcriptional pausing in bacteria. Cell. 2013;152:431–441. doi: 10.1016/j.cell.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkelman JT, Gourse RL. Open complex DNA scrunching: A key to transcription start site selection and promoter escape. Bioessays. 2017:39. doi: 10.1002/bies.201600193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakhnin AV, Murakami KS, Babitzke P. NusG Is a Sequence-specific RNA Polymerase Pause Factor That Binds to the Non-template DNA within the Paused Transcription Bubble. J Biol Chem. 2016;291:5299–5308. doi: 10.1074/jbc.M115.704189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarnell WS, Roberts JW. The phage lambda gene Q transcription antiterminator binds DNA in the late gene promoter as it modifies RNA polymerase. Cell. 1992;69:1181–1189. doi: 10.1016/0092-8674(92)90639-t. [DOI] [PubMed] [Google Scholar]
- Zenkin N, Kulbachinskiy A, Yuzenkova Y, Mustaev A, Bass I, Severinov K, Brodolin K. Region 1.2 of the RNA polymerase sigma subunit controls recognition of the -10 promoter element. Embo J. 2007;26:955–964. doi: 10.1038/sj.emboj.7601555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Campbell EA, Minakhin L, Richter C, Severinov K, Darst SA. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 A resolution. Cell. 1999;98:811–824. doi: 10.1016/s0092-8674(00)81515-9. [DOI] [PubMed] [Google Scholar]
- Zhang J, Landick R. A Two-Way Street: Regulatory Interplay between RNA Polymerase and Nascent RNA Structure. Trends Biochem Sci. 2016;41:293–310. doi: 10.1016/j.tibs.2015.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhilina E, Esyunina D, Brodolin K, Kulbachinskiy A. Structural transitions in the transcription elongation complexes of bacterial RNA polymerase during sigma-dependent pausing. Nucleic Acids Res. 2012;40:3078–3091. doi: 10.1093/nar/gkr1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber PK, Artsimovitch I, Nandymazumdar M, Liu Z, Nedialkov Y, Schweimer K, Rosch P, Knauer SH. The universally-conserved transcription factor RfaH is recruited to a hairpin structure of the non-template DNA strand. eLife. 2018 doi: 10.7554/eLife.36349. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.