Abstract
Polymeric supramolecular assemblies that can effectively transport proteins across an incompatible solvent interface are described. We show that electrostatics and ligand-protein interactions can be used to selectively transport proteins from an aqueous phase to organic phase. These transported proteins have been shown to maintain their secondary structure and function. This approach opens up new possibilities for application of supramolecular assemblies in sensing, diagnostics and catalysis.
Table of Contents (TOC)
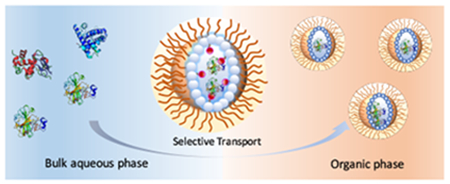
Transporting molecules across incompatible interfaces is a significant challenge, especially for macromolecules. A striking example of an interfacial barrier is the cellular membrane, where an organized presentation of hydrophilic and hydrophobic functional groups provides a formidable barrier for molecular transport.1 While small ions or molecules can cross the membrane through ion channels or passive diffusion, globular proteins with large hydrophilic surfaces offer no easy access.2,3 Nonetheless, cells do transport proteins when necessary for inter-cellular communication, often using nanoscopic vesicular compartments called exosomes.4,5 Inspired by these cell-derived vesicles, we became interested in exploring the possibility of transporting proteins into a nanoscopic compartment across a solvent interface. Indeed, small molecule surfactants mostly and occasionally polymers have been explored as a means of loading proteins into reverse micelles.6–9 Loading selectivity with such systems is limited or non-existent, even in an electrostatics context, presumably due to the low stability of small molecule based micelles.10 While transporting proteins across interfaces has many implications, selective transport, while retaining structure and function, could be transformative in applications such as sensing, delivery, and diagnostics. Supramolecular assemblies have shown great potential in these areas11–13 and supramolecular protein transport would be an important advance. Here we report a simple polymeric platform that selectively transports water-soluble proteins from an aqueous phase to the water-pool of a reverse micelle in an apolar organic phase based on complementary electrostatic interactions or specific ligand-protein binding interactions (Figure 1).
Figure 1.
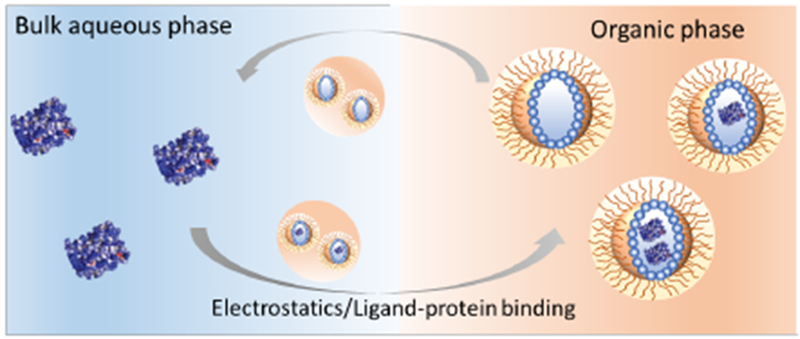
Schematic representation of reverse micelle driven protein transportation.
For the initial proof-of-concept, we synthesized a polystyrene-based amphiphilic anionic random copolymer P1 (Mn= 11 kDa, Đ=1.09) (Figure 2a) and a cationic polymer, P2, (Figure 2b) using nitroxide-mediated polymerization (see SI for details). By distributing these polymers in toluene along with two equivalents of water per carboxylate or quaternary ammonium moiety, assemblies with a fairly homogeneous size distribution of 50 nm for P1 and 37 nm for P2 were observed, as discerned by both dynamic light scattering (DLS) (Figure 2c) and transmission electron microscopy (TEM) (Figure 2d, 2e, S1).
Figure 2.
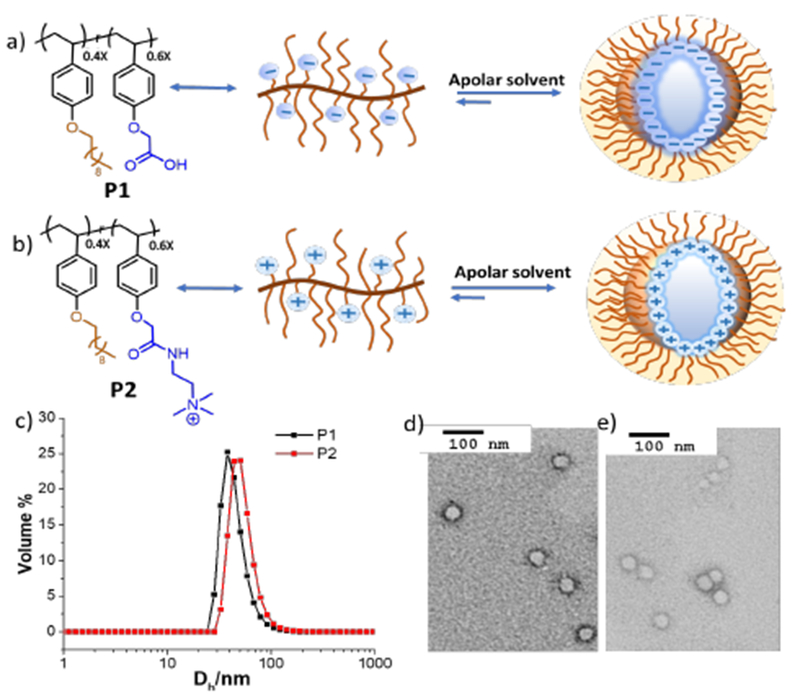
Structural features of polymeric reverse micelles. Molecular structure of polymer P1 (Mn= 11 kDa, Đ= 1.09) a) and P2 b) (Mn= 12 kDa, Đ= 1.15), c) DLS profile of P1 and P2 in toluene, TEM of P1(d) and P2(e).
The key premise for the work here is that these self-assembled structures would bind to complementarily charged proteins in the aqueous phase, and ferry them across the interface to the interior of the reverse micelles in toluene. To test this possibility, porcine liver esterase (plE, MW = 16.8 kDa), a negatively charged enzyme at pH 8.0 (pI = 5.3), was used as the model protein. Upon equilibrating a plE solution with cationic P2 (1 mg/mL) reverse micelles, presence of proteins in both phases was detected using matrix assisted laser desorption/ionization mass spectrometry (MALDI-MS). Figures 3a and 3b show the MS of the aqueous and organic phases, respectively, before equilibration. After equilibration, we were gratified to observe a peak corresponding to plE in the organic phase (Figure 3c), suggesting that plE was successfully transported into interior of the reverse micelles.
Figure 3.
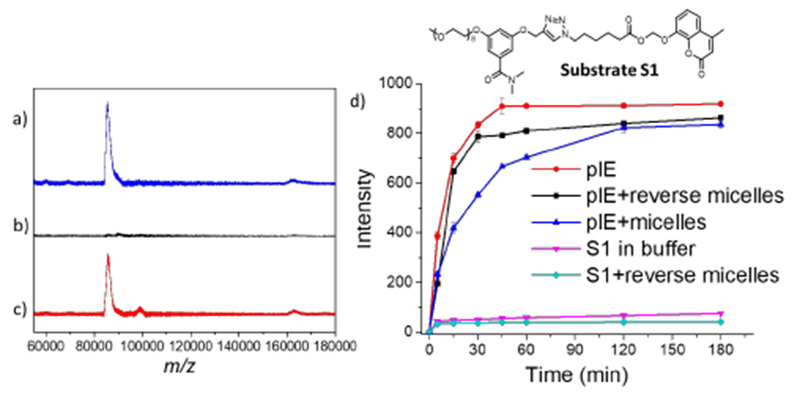
MALDI-MS analysis of a) aqueous phase before equilibration, b) organic phase before equilibration, and c) organic phase after equilibration. d) Activity of esterase (based on substrate cleavage) inside reverse micelles compared with esterase activity in bulk aqueous phase.
To quantify the extent of protein that was encapsulated within the reverse micelles, we analyzed the organic phase for proteins using the bicinchoninic acid (BCA) assay. This analysis showed that 1 mg of polymer is capable of transporting and binding to 0.05 mg of plE, an equivalent of 5 wt% loading capacity. This estimate is consistent with the corresponding SDS-PAGE analysis of the liberated proteins (Figures S6 and S7). This capacity compares more favorably than that with liposomes.14 A more compelling analysis is to identify whether the extracted enzyme remains active in the reverse micelle. To investigate this possibility, we designed an amphiphilic coumarin-based profluorophore S1 as a substrate for plE. The alkylated ether of this substrate is non-fluorescent. When the ester bond of S1 is cleaved by the enzyme, the resultant hemiacetal rapidly degrades to generate umbelliferone, a fluorescent coumarin molecule (see SI for details).
The substrate itself was quite stable in PBS buffer as well as after equilibration with toluene solution containing P2. In the presence of plE, however, a rapid hydrolysis of S1 to generate the fluorescent umbelliferone was observed (Figure 3d). We then analyzed the possibility of this reaction in toluene in the presence of reverse micelles loaded with plE. Interestingly, the hydrolysis rate was found to be quite similar to that of the free enzyme, suggesting that the plE activity is maintained inside the reverse micelles. As another control experiment, we mixed plE and P2 in aqueous phase and found that the activity of the enzyme was slightly lower, suggesting that interactions between P2 and plE have little effect on plE activity.
When considering the pathway by which these polymers could transport proteins across the interface, we equilibrated the reverse micelle assemblies of polymers P1 and P2 with water. UV-visible absorption spectra of both phases indicate that these polymers fully remain in the apolar phase (Figure S8). While these polymers were initially assembled as micelles in the aqueous phase and equilibrated with toluene (Figure S9), the exclusive presence of these polymers in the aqueous phase shows that these assemblies are kinetically trapped in the solvent that they are initially assembled. Overall, these results suggest protein molecules can exchange between phases, but only remain in the apolar phase if they have favorable interactions with the reverse micelle interior.
Following these observations, we were interested in exploring the applicability of this approach to other non-enzymatic proteins. Green fluorescent protein (GFP) was chosen, not only because it can be readily monitored using fluorescence, but also because the fluorescence itself is a good indicator of whether the protein maintains its tertiary structure. Wild-type GFP (pi 6.2) has a net charge of −7 at pH 7.4, we treated this GFP (−7) solution with P2 reverse micelle solution in toluene. We were gratified to find that the emission spectrum of the organic phase clearly showed the presence of GFP. MALDI-MS peak with a m/z of 28,432 Da further confirmed the presence of GFP in the organic phase (Figure S10). However, there was no discernible change in the emission intensity of the aqueous phase (Figure 4b) using the anionic reverse micelle P1, suggesting that this transport is indeed due to electrostatic complementarity.
Figure 4.
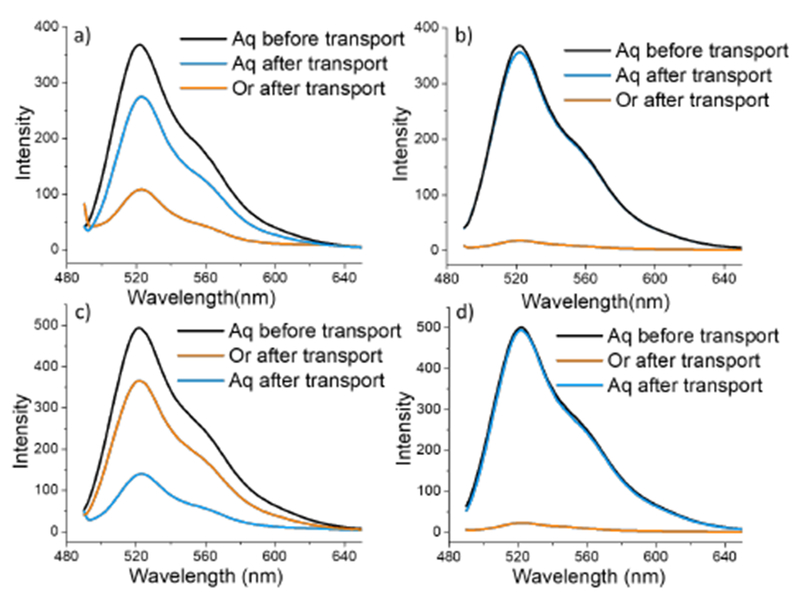
Emission spectrum of GFP before and after transport, a) GFP (−7) transport by P2, b) GFP (−7) transported by P1, c) GFP (+15) transported by P1, d) GFP (+15) transported by P2.
To further test this idea, we utilized the so-called supercharged GFP (+15).15 Indeed here, the anionic polymeric reverse micelle from P1 is able to transport the protein across the interface, while the cationic reverse micelle from P2 does not affect the protein in the aqueous phase (Figures 4c and 4d). The results from these studies show that: (i) transport of proteins across the interface is due to electrostatic complementarity, not due to spurious differences in inherent binding abilities of P1 and P2; (ii) the tertiary structure of the proteins can be preserved upon transport across the interface as indicated by the roughly equal emission intensities before and after equilibration; (iii) at similar polymer and protein concentrations, the extent of protein extraction in GFP (+15) is considerably higher than GFP (−7), showing that binding affinity can influence the extent of proteins transported across the interface.
While electrostatic complementarity can be utilized to simplify protein mixtures and enable identification of the presence of specific proteins, this ability will be greatly enhanced if proteins can be transported across an interface in response to a specific ligand-protein interaction. To investigate this possibility, we used bovine carbonic anhydrase (bCA) as the model protein, because aryl sulfonamides are well-established as small molecule ligands for this protein.16 The design hypothesis here is that if this ligand was installed in the polymeric reverse micelles, it should be able to selectively transport bCA to the organic phase due to specific binding.
For this purpose, we designed a zwitterionic amphiphilic polymer P3 (Figure 5a), containing 40% decyl chain as the hydrophobic moiety, 40% zwitterionic sulphobetain group as the hydrophilic moiety and 20% benzene sulfonamide as the ligand moiety. P3 forms a similar-sized assembly in apolar solvents (Figure S3). When P3 was equilibrated with tetramethylrhodamine-5-isothiocyanate (TRITC)-labeled bCA, we observed a strong emission peak in organic phase, indicating the transportation of TRITC-bCA conjugates. Concurrently, there is a dramatic decrease in the fluorescence intensity in the aqueous phase, indicating bCA is successfully transported across the interface.
Figure 5.
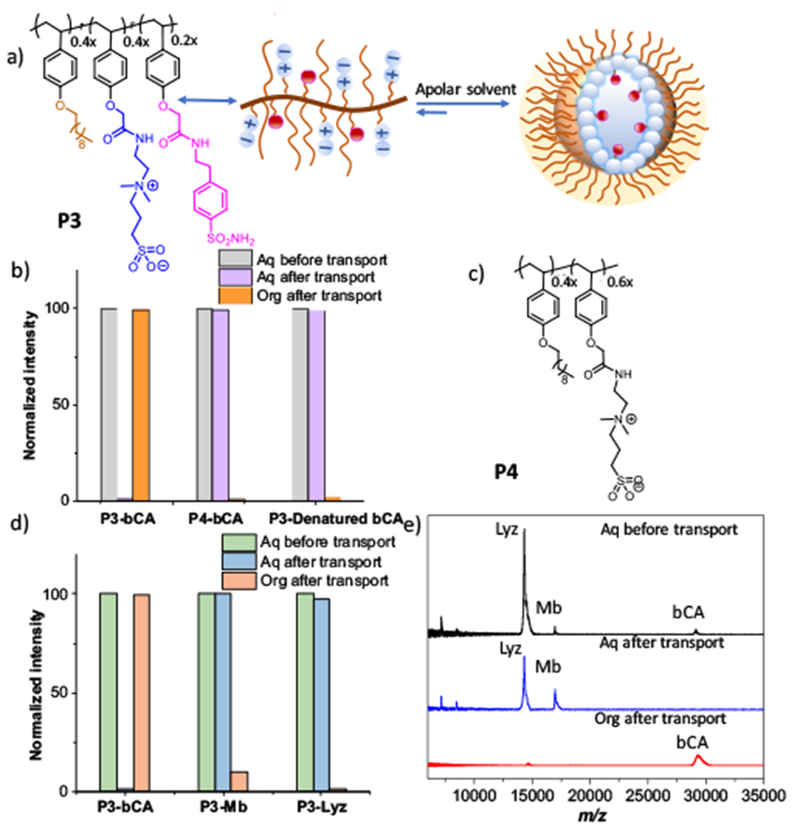
a) Molecular structure of P3, b) Fluorescence change of aqueous/organic phase using P3 or P4 for bCA transport, c) Molecular structure of P4, d) Fluorescence change of aqueous/organic phase of P3 to transport bCA, Myb or Lyz, e) MALDI-MS analysis of protein mixture before and after transportation.
To investigate whether this is driven by the ligand-protein binding, we designed a structurally similar amphiphilic polymer, P4, which forms reverse micelles but lacks the sulfonamide functional group. No change is observed in the emission spectrum of both organic and aqueous phases, when using P4 as the transporter for TRITC-bCA (Figure 5b). To further test whether the specific ligand-protein interaction is responsible for the observed transport across the interface, we designed another control experiment where the structure of the protein was disrupted with acetonitrile and heat. The denatured bCA should not be able to bind the sulfonamide ligands and thus would not be transported into the organic phase. Indeed, no fluorescence changes in the aqueous or organic phase are observed, showing that no bCA was transported into the organic phase (Figure 5b). These results confirm that transportation occurs only when bCA’s native structure is maintained in such a way to preserve its ability to bind the sulfonamide ligand. Overall, these results suggest that specific ligand-protein interactions can be utilized to bind and transport proteins across the solvent interface.
Next, to test the ligand-protein binding based selectivity associated with this process, we performed another set of control experiments using myoglobin (Mb) and fluorescently labelled lysozyme (Lyz). Since benzene sulfonamide ligands have little to no binding affinity to these proteins, we predicted that Mb and Lyz would remain in the aqueous phase. Indeed, no discernible fluorescence change was observed in both aqueous and organic phases for these two proteins, suggesting that the ligand attached reverse micelles are specific for the target protein bCA (Figure 5d). These experiments were initially done separately due to the possible bleeding of fluorescence emission. Selective transport from a mixture of these proteins by P3 was tested using MS. We were gratified to find that only bCA is transported to the organic phase, while Mb and Lyz remained in the aqueous phase as indicated by the mass spectra before and after equilibration (Figure 5e). These data strongly support the idea that ligand-attached reverse micelle systems are specific for target proteins.
In conclusion, we have demonstrated a set of supramolecular assemblies, based on amphiphilic polymers, that can transport proteins across a solvent interface. We have shown here that: (i) simple electrostatic complementarity in polymeric reverse micelle systems can transport proteins from bulk aqueous phase into the interior of a reverse micelle assembly in the apolar organic phase; (ii) the activity of the transported proteins is retained in the process; (iii) the efficiency of protein binding is dependent on the charge density presented on the protein surface; (iv) the kinetically trapped nature of the assemblies suggest that the polymers do not ferry the proteins, but instead transport likely occurs during the solvent exchange within the interior of the assembly, when these assemblies transiently find themselves at the interface during equilibration, as illustrated in Figure 1; (v) specific ligand-receptor interactions can be used to selectively extract proteins from the aqueous phase. Overall, the most gratifying finding here is that whole proteins can be moved across a solvent interface into the interior of a supramolecular assembly, even though the resident location of the assembly is in an incompatible solvent for the protein. The preliminary findings here have implications in many areas, especially in sensing, diagnostics, and catalysis. For example, these systems can be further developed to detect biomarkers in more complex mixtures of proteins.17–20 Similarly, facile incorporation of active proteins in organic solvents could facilitate enzyme-based catalysis for a broader range of organic substrates.21–25 These constitute examples of future directions for this research in our own laboratories.
Supplementary Material
ACKNOWLEDGMENT
We thank NIH-NCI (CA-169140) for support.
Footnotes
ASSOCIATED CONTENT
Supporting Information
Materials and methods, syntheses and characterization of polymers, and supporting figures. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interests.
REFERENCES
- (1).Yang NJ; Hinner MJ Getting across the cell membrane: an overview for small molecules, peptides, and proteins. Methods Mol. Biol 2015, 1266, 29–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Huang J; Lein M; Gunderson C; Holden MA Direct quantitation of peptide-mediated protein transport across a droplet–interface bilayer. J. Am. Chem. Soc. 2011, 133, 15818–15821. [DOI] [PubMed] [Google Scholar]
- (3).Orozco J; Cortés A; Cheng G; Sattayasamitsathit S; Gao W; Feng X; Shen Y; Wang J Molecularly imprinted polymer-based catalytic micromotors for selective protein transport. J. Am. Chem. Soc. 2013, 135, 5336–5339. [DOI] [PubMed] [Google Scholar]
- (4).Denzer K; Kleijmeer MJ; Heijnen HF; Stoorvogel W; Geuze HJ Exosome: from internal vesicle of the multivesicular body to intercellular signaling device. J. Cell Sci 2000, 113, 3365–3374. [DOI] [PubMed] [Google Scholar]
- (5).Skog J; Wurdinger T; Van Rijn S; Meijer D; Gainche L; Sena-Esteves M; Curry WT Jr; Carter RS; Krichevsky AM; Breakefield XO Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Cöklen KE; Hatton TA Protein extraction using reverse micelles. Biotechnol. Prog. 1985, 1, 69–74. [DOI] [PubMed] [Google Scholar]
- (7).) Oldfield C Biotechnol. Genet. Enzymes in water-in-oil microemulsions (‘Reversed Micelles’): Principles and applications. Eng. Rev 1994, 12, 255–327. [DOI] [PubMed] [Google Scholar]
- (8).Leser ME; Mrkoci K; Luisi PL Reverse micelles in protein separation: The use of silica for the back-transfer process. Biotechnol. Bioeng. 1993, 41, 489–492. [DOI] [PubMed] [Google Scholar]
- (9).Du X; Song N; Yang YW; Wu G; Ma J; and Gao H Reverse micelles based on β-cyclodextrin-incorporated amphiphilic polyurethane copolymers for protein delivery. Polym. Chem. 2014, 5, 5300–5309. [Google Scholar]
- (10).Matzke SF; Creagh AL; Haynes CA; Prausnitz JM; Blanch HW Mechanisms of protein solubilization in reverse micelles.Biotechnol. Bioeng. 1992, 40, 91–102. [DOI] [PubMed] [Google Scholar]
- (11).Yu G; Jie K; Huang F Supramolecular amphiphiles based on host–guest molecular recognition motifs. Chem. Rev. 2015, 115, 7240–7303. [DOI] [PubMed] [Google Scholar]
- (12).Ma X; Zhao Y Biomedical applications of supramolecular systems based on host–guest interactions. Chem. Rev. 2015, 115, 7794–7839. [DOI] [PubMed] [Google Scholar]
- (13).Cui H; Xu B Supramolecular medicine. Chem. Soc. Rev 2017, 46, 6430–6432. [DOI] [PubMed] [Google Scholar]
- (14).Colletier J-P; Chaize B; Winterhalter M; Fournier D Protein encapsulation in liposomes: efficiency depends on interactions between protein and phospholipid bilayer. BMC Biotechnol. 2002, 2, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Lawrence MS; Phillips KJ; Liu DR Supercharging proteins can impart unusual resilience. J. Am. Chem. Soc. 2007, 129, 10110–10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Alterio V; Di Fiore A; D’Ambrosio K; Supuran CT; De Simone G Multiple binding modes of inhibitors to carbonic anhydrases: how to design specific drugs targeting 15 different isoforms? Chem. Rev. 2012, 112, 4421–4468. [DOI] [PubMed] [Google Scholar]
- (17).Whitcombe MJ; Chianella I; Larcombe L; Piletsky SA; Noble J; Porter R; Horgan A The rational development of molecularly imprinted polymer-based sensors for protein detection. Chem. Soc. Rev. 2011, 40, 1547–1571. [DOI] [PubMed] [Google Scholar]
- (18).Rodthongkum N; Ramireddy R; Thayumanavan S; Richard WV Selective enrichment and sensitive detection of peptide and protein biomarkers in human serum using polymeric reverse micelles and MALDI-MS. Analyst 2012, 137, 1024–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Grzelakowski M; Onaca O; Rigler P; Kumar M; Meier W Immobilized protein–polymer nanoreactors. Small 2009, 5, 2545–2548. [DOI] [PubMed] [Google Scholar]
- (20).Santra S; Zhang P; Wang K; Tapec R; Tan W Conjugation of biomolecules with luminophore-doped silica nanoparticles for photostable biomarkers. Anal. Chem. 2001, 73, 4988–4993. [DOI] [PubMed] [Google Scholar]
- (21).Nam J-M; Thaxton CS; Mirkin CA Nanoparticle-based bio-bar codes for the ultrasensitive detection of proteins. Science. 2003, 301, 1884–1886. [DOI] [PubMed] [Google Scholar]
- (22).Zhang H; Piacham T; Drew M; Patek M; Mosbach K; Ye L Molecularly imprinted nanoreactors for regioselective huisgen 1, 3-dipolar cycloaddition reaction. J. Am. Chem. Soc. 2006, 128, 4178–4179. [DOI] [PubMed] [Google Scholar]
- (23).Vriezema DM; Comellas Aragonès M; Elemans JAAW; Cornelissen JJLM; Rowan AE; Nolte RJM Self-assembled nanoreactors. Chem. Rev. 2005, 105, 1445–1490. [DOI] [PubMed] [Google Scholar]
- (24).Bruns N; Tiller JC Amphiphilic network as nanoreactor for enzymes in organic solvents. Nano Lett. 2005, 5, 45–48. [DOI] [PubMed] [Google Scholar]
- (25).Broz P; Driamov S; Ziegler J; Ben-Haim N; Marsch S; Meier W; Hunziker P Toward intelligent nanosize bioreactors: a pH-switchable, channel-equipped, functional polymer nanocontainer. Nano Lett. 2006, 6, 2349–2353. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


