Abstract
Background & Aims
Several studies have shown that signaling via the interleukin 23 (IL23) receptor is required for development of colitis. We studied the roles of IL23, dietary factors, alterations to the microbiota, and T cells in development and progression of colitis in mice.
Methods
All mice were maintained on lab diet 5053, unless otherwise noted. We generated mice that express IL23 in CX3CR1-positive myeloid cells (R23FR mice) upon cyclic administration of tamoxifen dissolved in diet 2019. Diet 2019 and 5053 have minor differences in the overall composition of protein, fat, fiber, minerals, and vitamins. CX3CR1CreER mice (FR mice) were used as controls. Some mice were given antibiotics and others were raised in a germ-free environment. Intestinal tissues were collected and analyzed by histology and flow cytometry. Feces were collected and analyzed by 16S rDNA sequencing. Feces from C57/Bl6, R23FR, or FR mice were fed to FR and R23FR germ-free mice in microbiota transplant experiments. We also performed studies with R23FR/Rag−/−, R23FR/Mu−/−, and R23FR/Tcrd−/− mice. R23FR mice were given injections of antibodies against CD4 or CD8 to deplete T cells. Mesenteric lymph nodes and large intestine CD4+ cells from R23FR or FR mice in remission from colitis were transferred into Rag−/− mice. CD4+ cells were isolated from donor R23FR mice and recipient Rag−/− mice, and T-cell receptor sequences were determined.
Results
Expression of IL23 led to development of a relapsing–remitting colitis that was dependent on the microbiota and CD4+ T cells. The relapses were caused by switching from the conventional diet used in our facility (diet 5053) to the diet 2019, and were not dependent on tamoxifen after the first cycle. The switch in the diet modified the microbiota, but did not alter levels of IL23 in intestinal tissues, compared to mice that remained on the conventional diet. Mesenteric lymph nodes and large intestine CD4+ cells from R23FR mice in remission, but not from FR mice, induced colitis after transfer into Rag−/− mice, but only when these mice were placed on the diet 2019. The CD4+ T-cell receptor repertoire of Rag−/− mice with colitis (fed the 2019 diet) was less diverse than that from donor mice and Rag−/− mice without colitis (fed the 5053 diet), due to expansion of dominant T-cell clones.
Conclusions
We developed mice that express IL23 in CX3CR1-positive myeloid cells (R23FR mice) and found they are more susceptible to diet-induced colitis than mice that do not express IL23. The R23FR mice have a population of CD4+ T cells that becomes activated in response to dietary changes and alterations to the intestinal microbiota. The results indicate that alterations in the diet, intestinal microbiota, and IL23 signaling can contribute to pathogenesis of inflammatory bowel disease.
Keywords: cytokine, immune response, microbiome, inflammatory bowel disease model
INTRODUCTION
Crohn’s disease (CD) and ulcerative colitis (UC) are two distinct phenotypic patterns of inflammatory bowel disease (IBD) affecting over 1.5 million people in Europe and almost one million people in North American 1. At the turn of the 21st century, IBD has become a global disease with newly industrialized countries now facing rising incidence, analogous to trends seen in the western world during the latter part of the 20th century2. IBD is associated with morbidity, mortality, and substantial costs to the health-care system. Both CD and UC are characterized by periods of asymptomatic remission interrupted by episodes of symptomatic disease flares or exacerbations. While its exact cause is unknown, IBD seems to be due to a combination of genetic predisposition and environmental factors 3–6. Genome-wide association studies identified polymorphisms in several genes, including in the Interleukin-23 (IL23) receptor (IL23R)7, 8. IL23 is a heterodimeric cytokine formed by the IL23-specific p19 subunit and the IL12p40 subunit. IL23 interacts with cells that co-express the IL23R subunit and the shared IL-12R β1 chain9. Several experimental models support a role for IL23 in colitis. Loss-of-function studies show that IL23p19 is essential for chronic colitis development in IL10−/− spontaneous colitis model10, CD45RBhiCD4+ T-cell transfer models11, Helicobacter hepaticus-driven colitis12, anti-CD40-induced acute innate colitis model13, 14, and chemically induced colitis15. Other studies show that IL23R expression by CD4+ T cells is required for disease development in murine T-cell transfer colitis model16 and that IL23R expression by innate lymphoid cells (ILCs) is important for colitis development in an anti-CD40 antibody-induced acute innate colitis model14. Although mounting evidence suggests that IL23 is relevant in IBD pathogenesis, no direct demonstration that increased IL23 signaling causes colitis exists to date.
Despite many advances, it is still not clear what environmental factors trigger development of IBD, nor it is known what factors cause flares of UC and CD. Besides genetic factors, smoking, diet, microbiota, and stress, appear to contribute to IBD development or aggravation17. Dietary changes have been proposed as a key factor in the increasing incidence of IBD in developing nations. Several large longitudinal studies have pointed to a lower risk of IBD among people who consume more fruits and vegetables, and a higher risk in people who consume less of these and more animal fats and sugar18. Consumption of specific foods has also been associated with CD and UC flares 18. In mice, the contribution of specific genes and the microbiota to colitis has been extensively analyzed. Mice bearing specific gene alterations develop colitis, but a significant number of them do not develop colitis when raised in germ-free (GF) conditions, suggesting a critical role for genes and the microbiota in promoting disease 4, 19. Other animal studies suggest a critical role of dietary components in the onset and severity of colitis 20–22. Yet, the development of relapsing-remitting disease models has not been reported. Because of this limitation it has been difficult to evaluate the contribution of diet and microbiota changes to disease initiation and progression.
To examine the contribution of IL23, the microbiota and diet to development of colitis, we created a novel mouse model in which IL23 is conditionally expressed by fractalkine chemokine receptor positive (CX3CR1+) cells. CX3CR1+ macrophages and dendritic cells (DC) are the main cells expressing IL23 in the gut upon exposure to bacterial antigens 23, 24. Our results show that CX3CR1+derived IL23 expression triggers development of a colitis that is dependent on the microbiota and the diet, with diet-driven cycles of active disease (relapse/flares) followed by remission. The development of colitis in this model is dependent on the generation of a CD4+ T cell response to the gut microbiota that is elicited by changes in the diet. Colitis-inducing CD4+ T cells are found in the mesenteric lymph nodes (mLN) and large intestine (LI) during remission and are able to trigger disease when transferred to lymphopenic mice, but only upon diet modification. Collectively, our experiments reveal a critical role for IL23 in generation of a CD4+ T cell population that is sensitive to modification of intestinal bacterial flora subsequent t to a specific dietary manipulation. The demonstration of a critical role for the diet in eliciting disease in genetically prone organism strongly supports the hypothesis of multi-causality in the etiopathogenesis of IBD.
METHODS
Mice
C57BL/6, Rag1−/−, Tcrd−/− and muMt−/− mice were purchased from The Jackson laboratory (Bar Harbor, ME). Mice were maintained under specific pathogen-free (SPF) conditions at the Icahn School of Medicine at Mount Sinai. The generation of IL-23 conditional knock-in mice (R23 mice and R23FR mice) is detailed in Supplementary Figure 1. All the germ-free mice were bred in-house and housed in standard flexible film isolators in our GF animal facility. All animal experiments in this study were approved by the Institutional Animal Care and Use Committee of Icahn School of Medicine at Mount Sinai, and were performed in accordance with the approved guidelines for animal experimentation at the Icahn School of Medicine at Mount Sinai.
Diet Treatment
All mice were raised on the basal diet 5053, which was purchased from LabDiet (St. Louis, MO). The basal diet 2019 was purchased from Envigo (Madison, WI). Tamoxifen (500mg/kg) (Sigma) was added to the Envigo diet 2019. R23FR mice and control FR mice were fed with tamoxifen diet during the indicated times shown as Figure 1A. After each cycle of TAM treatment, animals were switched back to the basal diet 5053.
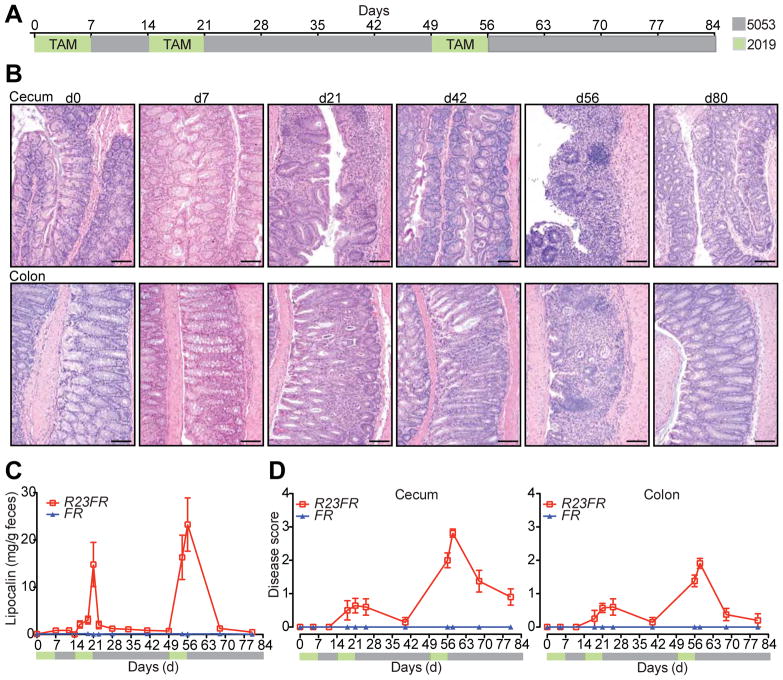
Figure 1.
IL23 Expression by CX3CR1+ Cells Induces Colonic and Cecal Inflammation. (A) Tamoxifen (TAM) in diet 2019 (green) was fed to R23FR and FR mice during the indicated times. After each cycle of TAM treatment, animals were switched to our mouse facility diet 5053 (gray). (B) Representative H&E-stained cecum and colon sections of R23FR mice at different time points. Scale bars, 100 μm. (C) Fecal lipocalin-2 levels in the stools of R23FR and FR mice were measured by ELISA (n= 5–15 per group per time point). (D) Histological scores of the colon and cecum of R23FR and FR mice at different time points (n= 5–15 per group per time point). Error bars represent mean ± SEM.
Histology
Tissues were dissected, fixed in 10% phosphate-buffered formalin, and then processed for paraffin sections. Five-micrometer sections were stained with hematoxylin and eosin (H&E) for histological analyses. See Supplementary Material for more details.
DNA Extraction, 16S rDNA Amplification, and Multiplex Sequencing
DNA was obtained from feces of mice using a bead-beating protocol. Bacterial 16S rRNA genes were amplified using the primers as described in Caporaso et al25. Sample preparation and analysis of 16S rDNA sequence were done as previously described26. See Supplementary Material for details.
T cell Adoptive Transfer
One million CD4+ from mLN and/or large intestine enriched by using MACS-beads (Miltenyi Biotech) were transferred into Rag1−/− recipient mice by intravenous (i.v.) injection. See Supplementary Material for details.
Bulk Sequencing of TCRs
Cell suspensions from the lamina propria and mLN were prepared as described previously27. For details regarding cell isolation and sorting, see Supplementary Material for details. The sorted CD4 T cells were used for TCR sequencing. See Supplementary Material for details.
Statistical Analysis
All statistical analyses were performed with GraphPad Prism 5 software. Differences between groups were analyzed with Student’s t tests or nonparametric Mann-Whitney test. Statistical tests are indicated throughout the Figure legends. Differences were considered significant when p < 0.05 (NS, not significant, * p < 0.05, **p < 0.01, ***p < 0.001), and levels of significance are specified throughout the Figure legends. Data are shown as mean values ± SEM throughout.
RESULTS
IL23 Expression Induces An Inflammatory Disease that Resembles UC in Humans
Although IL23 appears to be relevant in IBD pathogenesis both in human and experimental colitis model, there is no direct evidence that IL23 expression can cause colitis in adult immuno-competent mice. To define the role of IL23 in the intestinal inflammation, we engineered mice in which IL23 expression could be induced by tamoxifen in a subset of myeloid cells, known to express it in the gut (CX3CR1+ cells). This was accomplished by first generating Rosa26-lox-STOP-lox-IL23 mice (R23 mice) (Supplementary Figure 1A). The R23 mice were subsequently mated to CX3CR1CreER mice (FR mice) 28 to generate R23FR mice (Supplementary Figure 1A and B). As expected, TAM treatment promoted Cre-mediated excision of the STOP cassette and expression of IL23 in CX3CR1+ intestinal cells of R23FR mice (Supplementary Figure 1C and D). To examine whether IL23 expression would promote intestinal inflammation, we treated 60–70 days old R23FR and control FR mice with three cycles of TAM (500mg/kg in the diet 2019, Harlan Teklad). After each TAM cycle, animals were fed the conventional diet used in our mouse facility (diet 5053, Lab Diet) (Figure 1A). Histological evaluation of the LI of R23FR mice at d7 did not present inflammatory infiltrates (Figure 1B). However, by d21 we observed the presence of leukocytic infiltrates in the mucosa of LI of R23FR mice (Figure 1B). In the colon, the presence of crypt abscesses was noted, but no erosions were observed (Figure 1B). By day 42, these infiltrates were very mild or absent. By day 56 the LI of R23FR mice showed a severe colitis, with marked leukocytic infiltrate, crypt loss, epithelial damage, and ulcerations (Figure 1B & D). The mucosa was expanded by the presence of a mixed lymphoplasmacytic and histiocytic infiltrates. No granulomata or transmural infiltrates were observed. The mucosal-based colitis with predominant cecal and colonic involvement was strikingly similar to human UC. By d80, the infiltrates were very mild or absent (Figure 1B). Histological analysis of FR treated mice did not show any abnormality at any time points (Supplementary Figure 2). Consistent with histological analyses, the levels of fecal Lipocalin-2 varied during treatment of R23FR mice, but not FR mice (Figure 1C). The small intestine of the R23FR mice appeared normal and did not show infiltrates even at the stage of peak disease in the LI (d56) (Supplementary Figure 3). Collectively, these data show that IL23 expression drives development of an intestinal inflammatory disease that resembles UC in humans. Of note, cycles of acute disease and remission, observed in humans29, were also observed in this animal model.
Diet Change Induces Development of Relapse that was not Caused by Increased IL23 Expression
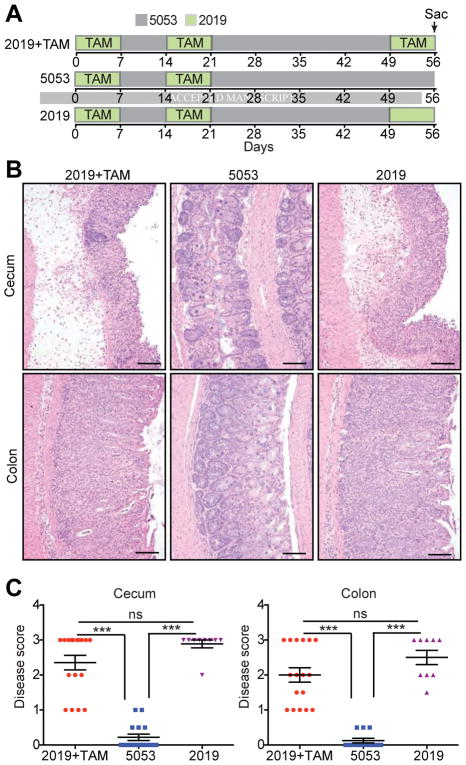
One of the striking aspects of this model was the development of relapse upon administration of a third cycle of TAM. To investigate the factors contributing to development of relapse we first investigated if the levels of transgenic IL23 fluctuated in the intestine as function of TAM treatment. To do so, we performed qPCR analysis of IL23 mRNA expression. Although IL23p19 mRNA expression levels increased at d7 in the CX3CR1+ (YFP+) cells and in CX3CR1− (YFP− CD11c+/CD11b+) myeloid populations compared with non-treated R23FR mice (d0), the transgenic-specific 2A mRNA expression levels increased only in the CX3CR1+ (YFP+) cells at this time point (Supplementary Figure 4). Subsequent treatments (d56) did not increase IL23p19 mRNA expression further (Supplementary Figure 4). In addition, we probed if repeated injection of recombinant mouse IL23 into R23FR mice starting at d49 elicited disease (Supplementary Figure 5A). None of the mice developed disease at day 56(Supplementary Figure 5B and C). The results suggested that the relapses were not function of increased levels of IL23. We also investigated if the relapse was due to the anti-estrogen receptor activities of TAM (Figure 2). Animals were treated with 2 cycles of TAM and subsequently treated with diet 2019 with TAM (Group 1), diet 5053 without TAM (Group 2), or diet 2019 without TAM (Group 3) (Figure 2A). As expected, animals in Group 1 had marked colitis (Figure 2B–2C), as shown in Figure 1. Animals in Group 2 did not develop colitis (Figure 2B–2C). Surprisingly, animals in Group 3, which did not receive TAM, but were fed with the diet in which TAM was dissolved (diet 2019), developed marked colitis (Figure 2B–2C). Diet 2019 promoted development of colitis independent of the levels of IL23, as measured by the levels of IL23 mRNA expression at d49 and d56 (Supplementary Figure 6). Subsequent experiments showed that development of colitis required a first cycle of TAM, as administration of diet 2019 without TAM to R23FR mice using the 3 cycles paradigm did not elicit disease (Supplementary Figure 7). Furthermore intraperitoneal injection of TAM into R23FR mice on the basal diet 5053 did not induce colitis (Supplementary Figure 8A and B), but intraperitoneal injection of TAM together with diet switch (from diet 5053 to 2019) into R23FR mice promoted colitis (Supplementary Figure 8C and D). Collectively, these results indicate that: 1) the induction of disease is dependent on the initial treatment of animals with TAM, 2) the relapses were not a function of increased IL23 expression nor a function of estrogen receptor-blocking properties of TAM, and 3) the relapses were caused by the diet switch (from diet 5053 to diet 2019).
Figure 2.
Change in Diet Causes Colitis Relapse in R23FR Mice. (A) Schematic representation of the diet switch experiments. Animals were treated with 2 cycles of TAM and subsequently treated with diet 2019 with TAM, diet 5053 without TAM, or diet 2019 without TAM. (B) Representative H&E stained cecum and colon sections of R23FR mice at d56 from each group. Scale bars, 100 μm. (C) Histological scores of the colon and cecum of R23FR mice at d56. *** p<0.001, ns, not significant; by nonparametric Mann-Whitney test.
Commensal Bacteria Are Required for IL23 Induced Colitis
Many genetically susceptible animals do not develop colitis when raised in germ-free conditions 4. To test if microbes were required in our model, we developed a gnotobiotic colony of R23FR mice and FR mice, and treated them with the same protocol used for SPF mice (Figure 1). In striking contrast to the SPF R23FR mice, none of the GF R23FR mice showed inflammation in the LI at d56 (Supplementary Figure 9A). Next we asked if colonization of GF mice with commensal bacterial would be sufficient to cause disease. We found that introduction of C57BL/6 microbiota into GF FR mice did not cause disease, but that it caused colitis in GF R23FR mice (Supplementary Figure 9B). We then performed experiments to test whether different donor microbiota (from Figure 2 group 1, R23FR and FR mice at day 56) could induce disease (Supplementary Figure 10A). Transfer of diseased SPF R23FR microbiota into GF R23FR recipient promoted disease, but this flora was not sufficient to cause disease when transferred into GF FR mice, that did not express transgenic IL23 (Supplementary Figure 10B). No disease was observed in GF FR mice that received FR SPF microbiota (Supplementary Figure 10B). Transplant of the microbiota of diseased SPF R23FR mice into FR mice did not cause disease suggesting that the R23FR microbiota at peak disease is not sufficient to induce disease and that development of colitis in this setting requires co-expression of IL23. Together, these results indicate the microbiota is a key factor contributing to IL23-induced colitis.
Development of Relapse is Dependent on the Diet-modified Microbiota
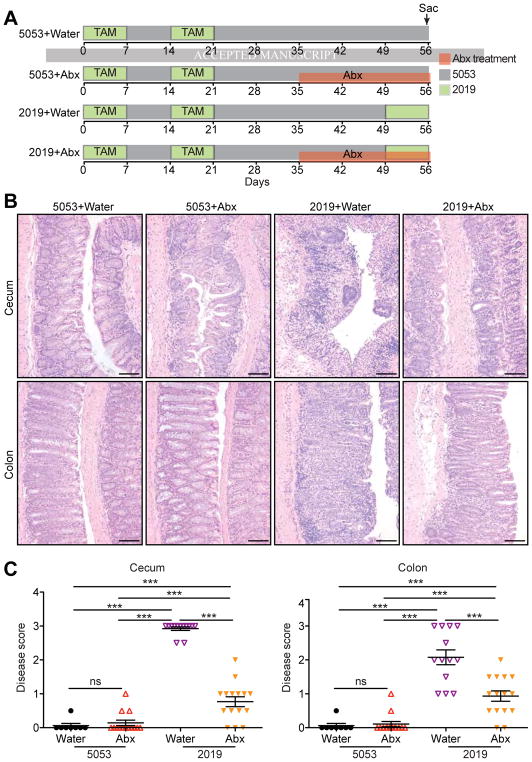
Having established a critical requirement for the microbiota in disease development, we tested next if the induction of relapse induced by the diet switch required microbes. To do so, we treated R23FR mice with 2 cycles of TAM and then with an antibiotic cocktail (ampicillin metronidazole, neomycin, vancomycin) from d35 to d56 (Figure 3A). All Antibiotic treatment significantly reduced colitis severity in the R23FR mice that received the diet 2019 from days 49–56 (Figure 3B–3C). These results further implicate the microbiota in the development of the diet-induced relapse.
Figure 3.
Microbiota is Required for Colitis Relapse. (A) Schematic representation of the experimental design. R23FR mice after treatment with 2 cycles of TAM were treated with an antibiotic cocktail (ampicillin metronidazole, neomycin, vancomycin) or water from d35 to d56 with different diets. (B) Representative H&E stained sections of cecum and colon of R23FR mice at d56 from each group. Scale bars, 100 μm. (C) Histological scores of the colon and cecum of R23FR mice at d56. *** p<0.001, ns, not significant; by nonparametric Mann-Whitney test.
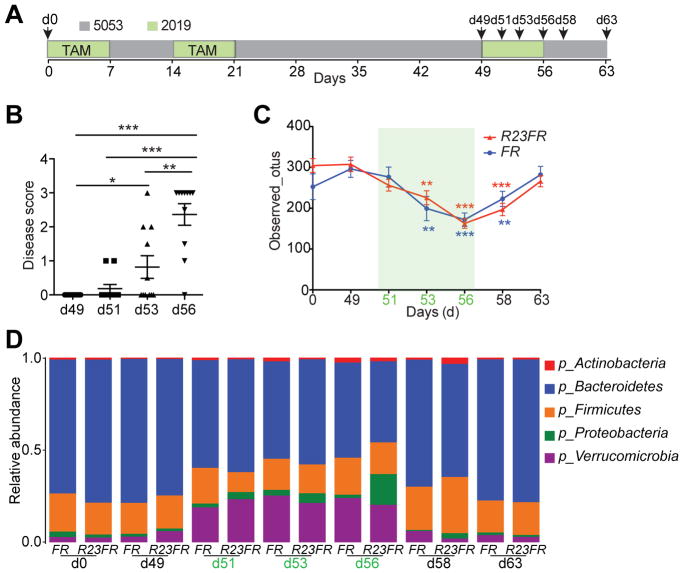
To examine if the diet changes promoted changes in the composition of the microbiota we performed a longitudinal study with R23FR and FR mice. We collected fecal samples for microbiota analysis at different time points (Figure 4A), and evaluated disease by histology (Figure 4B). Cecum inflammation was not observed in animals at d49. Diet switch promoted colitis in 2/11(~18%) of the animals at d51, 5/11(~ 46%) of the animals at d53 and 10/11 (~91%) of the animals at d56 (Figure 4B). There was an increase not only in penetrance but also in severity of the disease during this time (Figure 4B). Analysis of the microbiome during this period revealed that there was a significant reduction in alpha diversity (a measure of overall microbiome diversity) in both groups as early as 4 days after the diet switch (d53). A very pronounced difference was observed at d56 (Figure 4C). Further analysis showed that there were no significant taxa differences between R23FR and FR mice at d49 (prior to treatment), indicating that changes in diversity were not due to pre-treatment differences in the microbiome (Figure 4D). However, by d51, two days after diet switch, there was a significant expansion of Verrucomicrobia and a decrease of Bacteroidetes compared with day 49 in both R23FR and FR groups (Figure 4D and Supplementary Figure 11). Similar changes were found at d53 and d56 (Figure 4D and Supplementary Figure 11). At d56 there was a significant difference in the relative abundance of Proteobacteria in R23FR mice compared to FR mice, which coincided with peak inflammatory disease (Figure 4D and Supplementary Figure 11). Re-conversion to the 5053 diet (d58 and d63) resulted in rapid restoration of the microbiota to pre-treatment levels (d49) (Figure 4D). Overall, our results indicate that development of flares caused by diet switch is dependent on the intestinal microbiota, and that a diet switch can rapidly change the composition of the microbiota.
Figure 4.
Diet Switch Changes the Composition of the Intestinal Microbiota. (A) Schematic representation of the experimental design. The fecal samples were collected for microbiota analysis at different time points (d0, d49, d51, d53 and d56, d58 and d63) from R23FR and FR mice. (B) Histological scores of the colon and cecum of R23FR mice at d49, d51, d53 and d56. *p<0.05, **p<0.01, *** p<0.001; by nonparametric Mann-Whitney test. (C) Alpha diversity (estimated as number of observed OTUs) at a sequencing depth of 12,000 seqs/sample. Asterisks (colored according to the different genotyping group) indicate a statistically significant difference in the number of observed OTUs compared to OTUs at day 49 within each group. N=8–15 per time point for each group. **p<0.01, *** p<0.001; by nonparametric Mann-Whitney test. (D) Bacterial phylum-level community composition in the stool of R23FR and FR mice at each time point.
Lymphocytes Are Required for Intestinal Inflammation
To examine the number and type of leukocytes present in the LI during disease we performed flow cytometry in cells obtained from the cecum of R23FR and FR mice. The overall number of leukocytes (as estimated by the number of CD45+ cells) present in the cecum of R23FR mice was not different from that of controls (FR mice) by the end of the first cycle of treatment (d7) (Supplementary Figure 12A). The number of leukocytes rose significantly in the cecum of R23FR mice at the end of the second cycle (d21), but fell afterwards. By d42, the number of leukocytes was similar to that of FR mice. At the end of the third cycle, the number of leukocytes in the cecum of R23FR mice was, again, markedly increased (Supplementary Figure 12A). These results are consistent with the histological results presented before (Figure 1B & 1D). Next we performed a phenotypic analysis of the leukocyte subsets present in the lamina propria of FR and R23FR mice at d56 (Supplementary Figure 12B and C). We found that the relative and absolute number of B cells, CD4+ and CD8+ T cells was dramatically increased in R23FR compared to FR mice (Supplementary Figure 12B and C) in overall analyses. These results indicate that expression of IL23 induced markedly accumulation of B cells and T cells in the lamina propria.
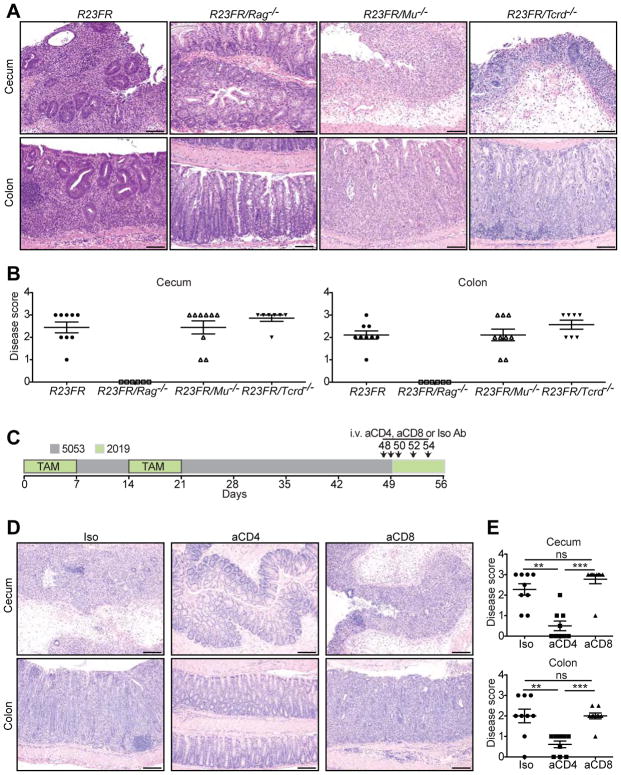
Given the significant changes in the numbers of lymphocytes observed in the LI of the R23FR mice and relevance of these cells to pathogenesis in experimental models, we tested next whether they were required for disease. To do so, we crossed R23FR mice with Rag−/− mice, which lack B and T cells, to generate R23FR/Rag−/− mice and subjected them to the protocol described in Figure 2A. Disease was completely absent in R23FR/Rag−/− mice, as determined by histological analysis of the LI (Figure 5A & 5B). These results indicated that lymphocytes were required for disease. To discriminate between the major lymphocyte populations causing disease, we crossed R23FR mice into muMt−/− mice, which lack B cells. By the end of third cycle of TAM treatment, we observed similar histological scores for R23FR and R23FR/muMt−/− mice (Figure 5A & 5B), suggesting that B cells were not required for disease.
Figure 5.
T Cells Are Required for Intestinal Inflammation in R23FR Mice. (A) Representative H&E stained sections from R23FR, R23FR/Rag−/−, R23FR/Mu−/− and R23FR/Tcrd−/− mice at d56. Scale bars, 100 μm. (B) Histological scores of the colon and cecum of R23FR, R23FR/Rag−/−, R23FR/Mu−/− and R23FR/Tcrd−/− mice at d56. Notice that disease was completely absent in R23FR/Rag−/− mice but not in R23FR/Mu−/− and R23FR/Tcrd−/− mice. (C) Schematic representation depicts experimental design for cell depletion experiments. (D) Representative H&E-stained colon and cecum sections (left) and histological scores (right) of R23FR mice after treatment with anti-CD4 and anti-CD8 antibodies. Depletion of CD4+, but not CD8+ T cells significantly reduced disease severity. Scale bars, 100 μm. **p<0.01, *** p<0.001, ns, not significant; by nonparametric Mann-Whitney test.
CD4+ T Cells Are the Main Effector Cells Promoting Colitis Upon Diet Switch
Since our results strongly suggested that T cells were the effector cells promoting colitis, we examined next which type of T cells accounted for the disease in the R23FR mice. To do so we crossed R23FR mice into Tcrd−/− mice that lack gamma-delta T cells. R23FR/Tcrd−/− mice had a disease that was indistinguishable from that observed in R23FR/Tcrd+/+ mice (Figure 5A & 5B), suggesting that TCRαβ T cells, not TCR gamma-delta cells, were responsible for disease in the R23FR mice. To define which TCRαβ T cells were responsible for disease, we used antibodies to deplete CD4+ or CD8+ T cells (Figure 5C and Supplementary Figure 13). Depletion of CD4+, but not CD8+ T cells significantly reduced disease severity (Figure 5D) suggesting that CD4+ T cells had a critical effector role in this model.
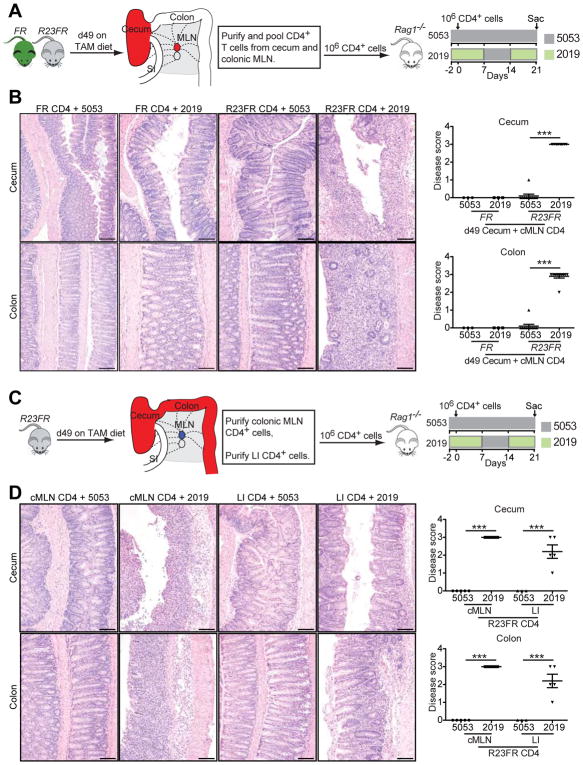
To investigate if the CD4+ T cells obtained from R23FR mice could induce disease when transferred into Rag−/− mice (Figure 6A), we transferred MACS purified mLN and colonic CD4+ T cells from d49 R23FR and FR mice to Rag−/− mice and treated them with different diets for 21 days (Figure 6A). Transfer of control FR CD4+ T cells did not elicit colitis in Rag−/− mice, regardless of the diet regimen (Figure 6B). Transfer of R23FR CD4+ T cells to Rag−/− mice that were fed with diet 5053 also did not promote disease. However Rag−/− mice that received R23FR CD4+ T cells and were fed with 2 cycles of 2019 diet developed severe colitis (Figure 6B). Of note, there were comparable frequencies of CD4+ T cells in the blood of the recipients after adoptive transfer of CD4+ T cells (Supplementary Figure 14). Next we examined if both mLN and colonic CD4+ T cells could cause disease after transfer into Rag−/− mice. To do so, we purified R23FR d49 CD4+ T cells from mLN or LI separately and transferred them into Rag−/− mice (Figure 6C). Both mLN and colonic CD4+ T cells caused severe colitis in reconstituted Rag−/− mice after a diet switch (Figure 6D). These results indicate that CD4+ T cells capable of inducing disease can be found in both mLN and LI of R23FR mice, but can only induce disease upon a diet-switch.
Figure 6.
IL23-induced CD4+ T Cells Drive Inflammation in Adoptively Transferred Mice. (A) Experimental setup for adoptive transfer of pooled CD4+ T cells from cecum and mLN of R23FR or FR mice into Rag−/− mice. (B) Representative H&E staining (left) and histological scores (right) of the colon and cecum of Rag−/− mice that received R23FR and FR mice CD4+ T cells fed with different diets. Rag−/− mice that received R23FR CD4+ T cells, but not FR CD4+ T cells, and were fed with 2 cycles of 2019 diet developed severe colitis. Scale bars, 100 μm. *** p<0.001; by nonparametric Mann-Whitney test. (C) Experimental setup for adoptive transfer of CD4+ T cells from large intestine (LI) or colonic mLN (cMLN) of R23FR mice into Rag−/− mice fed with different diets. (D) Representative H&E stained sections (left) and histological scores (right) of the colon and cecum of Rag−/− mice that received mLN or intestinal CD4+ T cells fed with different diets. Scale bars, 100 μm. Both mLN and colonic CD4+ T cells from R23FR mice caused severe colitis in reconstituted Rag−/− mice after a diet switch. *p<0.05, *** p<0.001; by nonparametric Mann-Whitney test.
Differential Expansion of T cell Clones in Adoptively Transferred Mice Fed with Distinct Diets
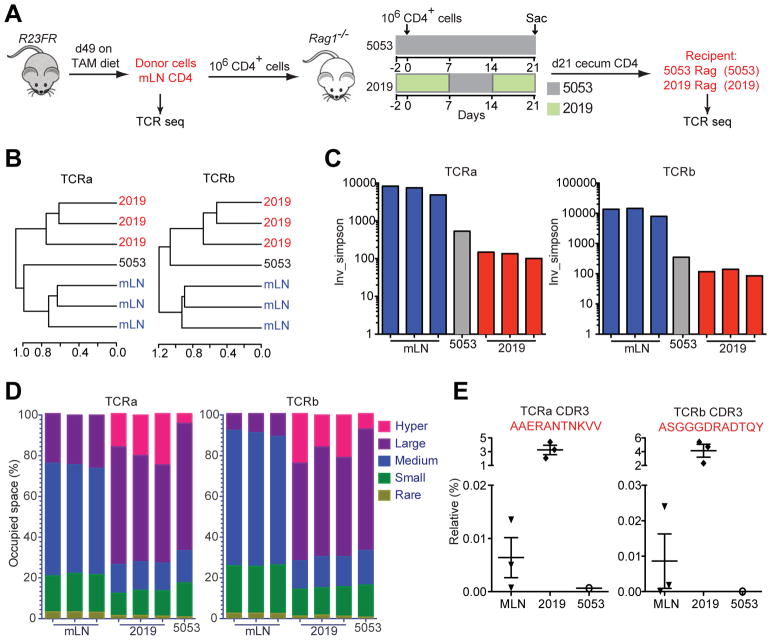
The T cells in the gut specifically recognize antigens by virtue of their heterodimeric αβ T-cell receptor (TCR). Once T cells encounter their cognate antigen, they proliferate to produce clonal copies of themselves. In the homeostatic stage, the intestinal mucosal T cell repertoire is shaped by environmental antigens. Under chronic intestinal inflammation, the TCR repertoire is thought to be more profoundly shaped by immune response against lumen antigens30. To examine if there was selective expansion of T cell clones upon diet switch, we sorted CD4+ T cells from donor R23FR mice, transferred them to recipient Rag−/− mice treated with different diets (Figure 7A), and sequenced both alpha and beta TCR chains from T cells recovered from the cecum at the end of treatment (Figure 7A). We clustered the sample populations on the basis of the complementary determining region 3 (CDR3) profiles, using hierarchical clustering. As expected, all donor cells from mLN for both alpha and beta chains clustered together (Figure 7B). Cells recovered from the cecum of three individual recipient Rag−/− mice treated with diet 2019 grouped together and apart from the cells recovered from recipient Rag−/− mice treated with diet 5053, and from mLN donor cells (Figure 7B). In addition, analysis of inverse Simpson’s diversity index revealed that the both alpha and beta chains of T cells from Rag−/− mice treated with 2019 diet had lower diversity compared to those of donor cells (Figure 7C). The decreased diversity of TCR repertoire in the cells from Rag−/− mice treated with diet 2019 was associated with expansion of T cell clones illustrated by an increase in the number of hyper-expanded clones (comprising >1% of all TCR sequences, Supplementary Table 1) of both TCRα and TCRβ (Figure 7D and Supplementary Table 2 and 3). These hyper-expanded clones present in the cecum of recipient Rag−/− mice treated with diet 2019 take up ~25% of the total TCR repertoire, while they take up less than 7% in Rag−/− mice treated with 5053 diet (Figure 7D). More importantly, a few of the dominant clones expanded in all three individual recipient Rag−/− mice treated with 2019 diet (Figure 7E and Supplementary Table 2 and 3). These results indicate that the TCR repertoire in the diseased recipient Rag−/− mice had reduced diversity associated with the expansion of dominant T cell clones.
Figure 7.
T Cell Receptor Clonality Analysis. (A) Experimental setup for TCR sequencing analysis. Note that donor cells from mLN of R23FR mice were obtained at remission stage (d49). (B) Unsupervised hierarchical clustering of TCR repertoire from donor mLN cells, cecum CD4+ T cells recovered from recipient Rag−/− mice treated with diets 5013 and 2019. N=3 individual mice in both donor group and 2019 group; n=1 of pooled 10 mice for group 5053. (C) Inverse Simpson’s diversity index was used to assess the diversity of TCR repertoires for each sample. (D) The percent of clonal space occupied by clones of a given type (classified by size, Supplementary Table 1). (E) Frequencies of selected TCRα and TCRβ clonotypes in different samples.
DISCUSSION
Here we show that low-level expression of IL23 by CX3CR1+ cells triggered development of a colitis that was dependent on the microbiota and the diet, and that had a striking resemblance to human disease, with cycles of active disease (relapse or flares) followed by remission29. Expression of IL23 resulted in the generation of a memory T cell population that was impacted by the microbiota, and had a skewed T cell repertoire. The colitogenic capacity of these T cells depended on the presence of a specific microbiota, which fluctuated in accordance to the diet.
Although IL23 appears to be relevant in IBD pathogenesis both in human and experimental colitis models, there is no direct evidence to date that IL23 expression can cause colitis in adult immunocompetent mice. Studies employing gene-modified mice have shown that mice expressing one of the IL23 subunits (p19) in multiple tissues develop a multiorgan inflammatory disease that results in early lethality31. Dysregulated expression of IL23 (both subunits) in the intestinal epithelium of transgenic mice induces severe intestinal inflammation in the small intestine, but not LI, and causes neonatal death32. Systemic exposure of adult animals to IL23 via hydrodynamic delivery of plasmids encoding the IL23 subunits results in rapid onset of psoriatic-like lesions in the ears, arthropathies in the paws and spine 33, 34, adenomatous tumors in the proximal duodenum 35, but not colitis. Here we show that IL23 expression can lead to development of colitis; but that it requires simultaneous diet-driven changes in the microbiota to cause disease. These results add to the clinical evidence that targeting IL23 through p40 (ustekinumab)36 or more specifically anti-p19 antibodies (brazikumab and risankizumab)37, 38 are effective in IBD.
The hypothesis that diet is an important factor contributing to the onset and progression of IBD, has remained formally unproven to date. Our results indicate that diet is a critical factor affecting development of IL23-driven experimental IBD, and suggest that it does so by modifying the microbiota. Dietary constituents have been shown to affect the immune response and the inflammatory status, in part by modulating the microbiota 21, 39. Various dietary factors influence the growth of different bacterial populations and/or their functions in the gut. However, most studies to date have focused on how specific diet components (e.g. fat, carbohydrate, protein, fiber, et al.) promote or inhibit inflammation in experimental models 39. Analysis of the overall composition of the diets used in our experiments did not show significant differences in the content of fat (2019 9% vs 5053 6.3%), protein (2019 19% vs 5053 20%), fiber (2019 2.6% vs 5053 6%), minerals or vitamins, so it is unclear what components are critical for the changes in the microbiota, but it is clear from our studies that colitis can not develop in the absence of the microbiota and that interference with the microbiota via antibiotic treatment prevents development of flares associated with the diet switch.
Our studies showed a marked reduction in alpha diversity as function of the diet change. These changes were already clear at d53 in both control and R23FR mice, and more pronounced at d56. Reduced diversity generally results in a reduced capacity of the microbial community to adapt to environmental pressures, and is likely to impact negatively on the microbial functional capacity 40. The taxa composition was similar between the groups at days 51 and 53, but was different from that of the same animals before the diet switch. The most evident change induced by the diet switch was an increased abundance of Proteobacteria and Verrucomicrobia in both groups. By d56, R23FR mice had an increased abundance of Verrucomicrobia and of Proteobacteria and a decrease in the abundance of Bacteroidetes compared to FR mice. The early increased abundance of Verrucomicrobia may have facilitated destruction of the mucus layer and adhesion of other bacterial species to the epithelium41. These changes, however, were not sufficient to promote colitis in FR mice, suggesting that the expression of IL23 in the R23FR mice was critical for disease induction.
Our studies showed that the colitis induced by IL23 is dependent on lymphocytes, and that the other IL23R+ cell populations are not sufficient to drive disease. Studies employing cell-depleting antibodies revealed a critical role for CD4+ T cells in pathogenesis, which was confirmed by adoptive transfer of these cells into Rag−/− mice. However, transfer of IL23 induced CD4+ T cells was not sufficient to cause colitis. These cells could only elicit disease in response to an environmental modification (diet switch/microbiota changes). Thus, our findings provide the first evidence that IL23 expression results in the generation of a colitogenic CD4+ T cell population induced by a dietary switch. In this context, we note that changes in the microbiota induced by diet have been hypothesized to contribute to the development of IBD in humans42, 43. Our results suggest that increased IL23 signaling caused by intrinsic genetic defects or infection, may predispose the generation of pathogenic T cells. The nature of the antigenic determinant eliciting the activation of the CD4+ T cell response in R23FR mice is not clear, but it appears to be derived from the bacteria, rather than from the diet. A food antigen would have triggered disease in the germ-free conditions, which was not observed here. However, factors other than bacterial antigens may also contribute to the overall immune response. As immune cells are sensitive to the action of bacterial metabolites, it is possible that in addition to bacterial antigens, they may be also responding to bacterial or host derived metabolites.
We propose that naive T cells become primed against bacterial antigens during the first cycle of TAM treatment. Expression of IL23 and a concomitant change in the microbiota would result in generation of effector T cells. Effector T cells would migrate back to the intestine but would not respond with an inflammatory response since diet and microbiota would have reverted by then. These T cells would convert into memory cells and persist in the colon and mLN for extended periods of time, only to be activated upon diet-driven microbiota changes. The presence of microbiota-reactive T cells would not be detrimental on its own. Reactivity to intestinal bacteria is a normal property of the human CD4+ T cells present in the periphery and intestine44. Such auto reactive cells may occur as a function of previous pathogen exposure. Indeed, break in tolerance to commensals has been shown in mice previously exposed to infection with T. gondii45.
A fundamental property of memory T cells is the ability to proliferate rapidly and elicit effector functions in response to secondary challenge. We suggest that IL23 expression favors development of a memory T cell population that can become pathogenic upon diet switch. As such, IL23 could contribute to a break in tolerance to antigens present in the commensal microbiota. Indeed, a break in tolerance to commensal antigens is thought to induce chronic intestinal inflammation in humans46. Increased expansion and skewing of T cells has been reported in patients with IBD30, 47 and in murine T-cell transfer colitis model48. The clonal expansion that we observed here in adoptive transferred Rag−/− mice fed with diet 2019 suggest that these expanded clones are pathogenic, but further studies are required to test this hypothesis.
In conclusion, we propose that IL23 has a critical role in the generation of a CD4+ T cell population that reacts to bacterial shifts elicited by dietary manipulation. Our studies open new routes for the identification of dietary factors leading to expansion of the colitis-inducing microbiota and to the delineation of the immune mechanisms associated with the marked epithelial cell destruction and inflammation. The demonstration of a critical role for the diet in eliciting disease in genetically prone organism strongly supports the important role of diet, microbiome changes and IL23 in the pathogenesis of IBD.
Supplementary Material
Acknowledgments
Funding:
This work was supported by grants from the National Institutes of Health (R01 DK110352) and from the Bacchetta Foundation (SUCCESS) grant to S.A.L.. L.C. was supported by a Research Fellowship Award (327362) from the Crohn’s & Colitis Foundation of America (CCFA).
We thank all members of the Lira Lab for their support. We thank the germ-free animal facility of the Icahn School of Medicine at Mount Sinai for expert support. We thank Claudia Canasto-Chibuque for colony maintenance and Dr. Kevin Kelley and the Mouse Genetics and Gene Targeting CoRE Facility for assistance in generation of gene-modified mice. We thank Drs. Peter S. Heeger and Nocholas Chun for technical assistance and comments. We also thank Mr. Alan J. Soto (Biorepository and Pathology CoRE) for technical support with histology.
Abbreviations
- CD
Crohn’s disease
- CDR3
complementary determining region 3
- DC
dendritic cells
- GF
germ-free
- IBD
inflammatory bowel diseases
- IL23
Interleukin-23
- ILC
innate lymphoid cell
- Lcn2
Lipocalin 2
- LI
large intestine
- mLN
mesenteric lymph node
- SPF
specific pathogen-free
- TAM
tamoxifen
- TCR
T-cell receptor
- UC
ulcerative colitis
Footnotes
Conflicts of interest
Both R.S. and A.J. are co-founders of Girihlet Inc., which has licensed the TCR sequencing technology from Mount Sinai, with the goal of developing it as a commercial product. JF.C. has served as consultant, advisory board member or speaker for AbbVie, Amgen, Boehringer-Ingelheim, Celgene Corporation, Celltrion, Enterome, Ferring, Genentech, Janssen and Janssen, Lilly, Medimmune, Merck & Co., Pfizer, PPM Services, Protagonist, Second Genome, Seres, Shire, Takeda, Theradiag, Theravance Biopharma. Stock options: Intestinal Biotech Development, Genfit. JF.C. has research Grants from AbbVie, Takeda, Janssen. The remaining authors disclose no conflicts.
Author contributions
L.C. and Z.H. designed study, did experiments, analyzed data and wrote the manuscript; A.C.I., and S.N.M.F. contributed with pathological analyses; M.D. contributed to histology; J.J.F. and J.C.C. assisted with microbiota analyses; J.J.L., R.S. and A.J. contributed to TCR sequencing and analyses; JF.C. contributed with data analyses; G.C.F and S.A.L designed study, analyzed data and wrote the manuscript. All authors reviewed the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Author names in bold designate shared co-first authorship.
- 1.Tontini GE, Vecchi M, Pastorelli L, et al. Differential diagnosis in inflammatory bowel disease colitis: state of the art and future perspectives. World J Gastroenterol. 2015;21:21–46. doi: 10.3748/wjg.v21.i1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ng SC, Shi HY, Hamidi N, et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet. 2017;390:2769–2778. doi: 10.1016/S0140-6736(17)32448-0. [DOI] [PubMed] [Google Scholar]
- 3.Gkouskou KK, Deligianni C, Tsatsanis C, et al. The gut microbiota in mouse models of inflammatory bowel disease. Frontiers in Cellular and Infection Microbiology. 2014:4. doi: 10.3389/fcimb.2014.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kamada N, Seo SU, Chen GY, et al. Role of the gut microbiota in immunity and inflammatory disease. Nature Reviews Immunology. 2013;13:321–335. doi: 10.1038/nri3430. [DOI] [PubMed] [Google Scholar]
- 5.Nishikawa J, Kudo T, Sakata S, et al. Diversity of mucosa-associated microbiota in active and inactive ulcerative colitis. Scandinavian Journal of Gastroenterology. 2009;44:180–186. doi: 10.1080/00365520802433231. [DOI] [PubMed] [Google Scholar]
- 6.Noor SO, Ridgway K, Scovell L, et al. Ulcerative colitis and irritable bowel patients exhibit distinct abnormalities of the gut microbiota. Bmc Gastroenterology. 2010:10. doi: 10.1186/1471-230X-10-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burton PR, Clayton DG, Cardon LR, et al. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duerr RH, Taylor KD, Brant SR, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parham C, Chirica M, Timans J, et al. A receptor for the heterodimeric cytokine IL-23 is composed of IL-12R beta 1 and a novel cytokine receptor subunit, IL-23R. Journal of Immunology. 2002;168:5699–5708. doi: 10.4049/jimmunol.168.11.5699. [DOI] [PubMed] [Google Scholar]
- 10.Yen D, Cheung J, Scheerens H, et al. IL-23 is essential for T cell-mediated colitis and promotes inflammation via IL-17 and IL-6. Journal of Clinical Investigation. 2006;116:1310–1316. doi: 10.1172/JCI21404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hue S, Ahern P, Buonocore S, et al. Interleukin-23 drives innate and T cell-mediated intestinal inflammation. Journal of Experimental Medicine. 2006;203:2473–2483. doi: 10.1084/jem.20061099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kullberg MC, Jankovic D, Feng CG, et al. IL-23 plays a key role in Helicobacter hepaticus-induced T cell-dependent colitis. Journal of Experimental Medicine. 2006;203:2485–2494. doi: 10.1084/jem.20061082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buonocore S, Ahern PP, Uhlig HH, et al. Innate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature. 2010;464:1371–1375. doi: 10.1038/nature08949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eken A, Singh AK, Treuting PM, et al. IL-23R(+) innate lymphoid cells induce colitis via interleukin-22-dependent mechanism. Mucosal Immunology. 2014;7:143–154. doi: 10.1038/mi.2013.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cox JH, Kljavin NM, Ota N, et al. Opposing consequences of IL-23 signaling mediated by innate and adaptive cells in chemically induced colitis in mice. Mucosal Immunology. 2012;5:99–109. doi: 10.1038/mi.2011.54. [DOI] [PubMed] [Google Scholar]
- 16.Ahern PP, Schiering C, Buonocore S, et al. Interleukin-23 Drives Intestinal Inflammation through Direct Activity on T Cells. Immunity. 2010;33:279–288. doi: 10.1016/j.immuni.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ananthakrishnan AN. Epidemiology and risk factors for IBD. Nat Rev Gastroenterol Hepatol. 2015;12:205–17. doi: 10.1038/nrgastro.2015.34. [DOI] [PubMed] [Google Scholar]
- 18.Lewis JD, Abreu MT. Diet as a Trigger or Therapy for Inflammatory Bowel Diseases. Gastroenterology. 2017;152:398–414 e6. doi: 10.1053/j.gastro.2016.10.019. [DOI] [PubMed] [Google Scholar]
- 19.Roulis M, Bongers G, Armaka M, et al. Host and microbiota interactions are critical for development of murine Crohn’s-like ileitis. Mucosal Immunol. 2016;9:787–97. doi: 10.1038/mi.2015.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Devkota S, Wang Y, Musch MW, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature. 2012;487:104–8. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haskey N, Gibson DL. An Examination of Diet for the Maintenance of Remission in Inflammatory Bowel Disease. Nutrients. 2017:9. doi: 10.3390/nu9030259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Llewellyn SR, Britton GJ, Contijoch EJ, et al. Interactions Between Diet and the Intestinal Microbiota Alter Intestinal Permeability and Colitis Severity in Mice. Gastroenterology. 2018;154:1037–1046 e2. doi: 10.1053/j.gastro.2017.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farache J, Zigmond E, Shakhar G, et al. Contributions of dendritic cells and macrophages to intestinal homeostasis and immune defense. Immunology and Cell Biology. 2013;91:232–239. doi: 10.1038/icb.2012.79. [DOI] [PubMed] [Google Scholar]
- 24.Oppmann B, Lesley R, Blom B, et al. Novel p19 protein engages IL-12p40 to form a cytokine, IL-23, with biological activities similar as well as distinct from IL-12. Immunity. 2000;13:715–725. doi: 10.1016/s1074-7613(00)00070-4. [DOI] [PubMed] [Google Scholar]
- 25.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bongers G, Pacer ME, Geraldino TH, et al. Interplay of host microbiota, genetic perturbations, and inflammation promotes local development of intestinal neoplasms in mice. J Exp Med. 2014;211:457–72. doi: 10.1084/jem.20131587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He Z, Chen L, Souto FO, et al. Epithelial-derived IL-33 promotes intestinal tumorigenesis in Apc (Min/+) mice. Sci Rep. 2017;7:5520. doi: 10.1038/s41598-017-05716-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parkhurst CN, Yang G, Ninan I, et al. Microglia Promote Learning-Dependent Synapse Formation through Brain-Derived Neurotrophic Factor. Cell. 2013;155:1596–1609. doi: 10.1016/j.cell.2013.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Solberg IC, Lygren I, Jahnsen J, et al. Clinical course during the first 10 years of ulcerative colitis: results from a population-based inception cohort (IBSEN Study) Scand J Gastroenterol. 2009;44:431–40. doi: 10.1080/00365520802600961. [DOI] [PubMed] [Google Scholar]
- 30.Doorenspleet ME, Westera L, Peters CP, et al. Profoundly Expanded T-cell Clones in the Inflamed and Uninflamed Intestine of Patients With Crohn’s Disease. Journal of Crohns & Colitis. 2017;11:831–839. doi: 10.1093/ecco-jcc/jjx012. [DOI] [PubMed] [Google Scholar]
- 31.Wiekowski MT, Leach MW, Evans EW, et al. Ubiquitous transgenic expression of the IL-23 subunit p19 induces multiorgan inflammation, runting, infertility, and premature death. J Immunol. 2001;166:7563–70. doi: 10.4049/jimmunol.166.12.7563. [DOI] [PubMed] [Google Scholar]
- 32.Chen L, He Z, Slinger E, et al. IL-23 activates innate lymphoid cells to promote neonatal intestinal pathology. Mucosal Immunol. 2015;8:390–402. doi: 10.1038/mi.2014.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adamopoulos IE, Tessmer M, Chao CC, et al. IL-23 Is Critical for Induction of Arthritis, Osteoclast Formation, and Maintenance of Bone Mass. Journal of Immunology. 2011;187:951–959. doi: 10.4049/jimmunol.1003986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sherlock JP, Joyce-Shaikh B, Turner SP, et al. IL-23 induces spondyloarthropathy by acting on ROR-gammat+ CD3+CD4−CD8− entheseal resident T cells. Nat Med. 2012;18:1069–76. doi: 10.1038/nm.2817. [DOI] [PubMed] [Google Scholar]
- 35.Chan IH, Jain R, Tessmer MS, et al. Interleukin-23 is sufficient to induce rapid de novo gut tumorigenesis, independent of carcinogens, through activation of innate lymphoid cells. Mucosal Immunol. 2014;7:842–56. doi: 10.1038/mi.2013.101. [DOI] [PubMed] [Google Scholar]
- 36.Sandborn WJ, Gasink C, Gao LL, et al. Ustekinumab induction and maintenance therapy in refractory Crohn’s disease. N Engl J Med. 2012;367:1519–28. doi: 10.1056/NEJMoa1203572. [DOI] [PubMed] [Google Scholar]
- 37.Feagan BG, Sandborn WJ, D’Haens G, et al. Induction therapy with the selective interleukin-23 inhibitor risankizumab in patients with moderate-to-severe Crohn’s disease: a randomised, double-blind, placebo-controlled phase 2 study. Lancet. 2017;389:1699–1709. doi: 10.1016/S0140-6736(17)30570-6. [DOI] [PubMed] [Google Scholar]
- 38.Sands BE, Chen J, Feagan BG, et al. Efficacy and Safety of MEDI2070, an Antibody Against Interleukin 23, in Patients With Moderate to Severe Crohn’s Disease: A Phase 2a Study. Gastroenterology. 2017;153:77–86 e6. doi: 10.1053/j.gastro.2017.03.049. [DOI] [PubMed] [Google Scholar]
- 39.Rapozo DC, Bernardazzi C, de Souza HS. Diet and microbiota in inflammatory bowel disease: The gut in disharmony. World J Gastroenterol. 2017;23:2124–2140. doi: 10.3748/wjg.v23.i12.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sigall-Boneh R, Levine A, Lomer M, et al. Research Gaps in Diet and Nutrition in Inflammatory Bowel Disease. A Topical Review by D-ECCO Working Group [Dietitians of ECCO] J Crohns Colitis. 2017;11:1407–1419. doi: 10.1093/ecco-jcc/jjx109. [DOI] [PubMed] [Google Scholar]
- 41.Desai MS, Seekatz AM, Koropatkin NM, et al. A Dietary Fiber-Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility. Cell. 2016;167:1339–1353 e21. doi: 10.1016/j.cell.2016.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–63. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dolan KT, Chang EB. Diet, gut microbes, and the pathogenesis of inflammatory bowel diseases. Mol Nutr Food Res. 2017:61. doi: 10.1002/mnfr.201600129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hegazy AN, West NR, Stubbington MJT, et al. Circulating and Tissue-Resident CD4(+) T Cells With Reactivity to Intestinal Microbiota Are Abundant in Healthy Individuals and Function Is Altered During Inflammation. Gastroenterology. 2017;153:1320–1337 e16. doi: 10.1053/j.gastro.2017.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hand TW, Dos Santos LM, Bouladoux N, et al. Acute gastrointestinal infection induces long-lived microbiota-specific T cell responses. Science. 2012;337:1553–6. doi: 10.1126/science.1220961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zechner EL. Inflammatory disease caused by intestinal pathobionts. Curr Opin Microbiol. 2017;35:64–69. doi: 10.1016/j.mib.2017.01.011. [DOI] [PubMed] [Google Scholar]
- 47.Chapman CG, Yamaguchi R, Tamura K, et al. Characterization of T-cell Receptor Repertoire in Inflamed Tissues of Patients with Crohn’s Disease Through Deep Sequencing. Inflammatory Bowel Diseases. 2016;22:1275–1285. doi: 10.1097/MIB.0000000000000752. [DOI] [PubMed] [Google Scholar]
- 48.Matsuda JL, Gapin L, Sydora BC, et al. Systemic activation and antigen-driven oligoclonal expansion of T cells in a mouse model of colitis. J Immunol. 2000;164:2797–806. doi: 10.4049/jimmunol.164.5.2797. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.