Abstract
Insular wildlife populations provide opportunities to examine biological questions in systems that are relatively closed and potentially tractable, striking examples being the long-term studies of ecology and evolution in the red deer and feral sheep populations on the Hebridean islands of Rum and St Kilda. In the case of parasitology, Understanding of parasitic infections insular wildlife populations in conjunction with knowledge of their origins has the potential to add a fresh perspective to disease control in humans and domestic animals. In the case of parasitology, understanding infections of insular wildlife populations, in conjunction with knowledge of their origins, has the potential to add a fresh perspective to disease control in humans and domestic animals. With this in mind, gross and molecular examination for the presence of cyclophyllidean tapeworms was performed on the viscera and rectal contents of 17 preserved specimens of Apodemus sylvaticus field mice and on the naturally voided faeces of a further four mice on the remote archipelago of St Kilda. Molecular speciation of hexacanth embryos extracted from the faeces of two mice, using nucleotide sequence analysis of the ribosomal cytochrome c-oxidase subunit-1, confirmed infection with Hymenolepis hibernia. Phylogenetic analysis showed that these were genetically distinct from Hymenolepis diminuta, previously reported in the insular A. sylvaticus mice, and from other published H. hibernia haplotypes. There was insufficient hymenolepidid tapeworm phylogeographic variation to resolve the origins of the co-evolved St Kilda mice, primarily due to a lack of published H. hibernia Cox-1 sequence data across the parasite's geographical range. Nevertheless, the Maximum Likelihood haplotype tree shows the potential for molecular parasitology to resolve a host-parasite relationship once more data become available. Morphological diagnostic features of zoonotic H. hibernia eggs are also described.
Keywords: Hymenolepis hibernia cyclophyllidean tapeworm, Apodemus sylvaticus field mouse, Host parasite relationship, Maximum likelihood tree, Phylogeographic variation
Graphical abstract
Highlights
-
•
First identification of Hymenolepis hibernia in Apodemus sylvaticus hirtensis mice.
-
•
Molecular speciation of hexocanth eggs.
-
•
Morphological description of Hymenolepis hibernia eggs.
-
•
Maximum likelihood hymenolepidid tapeworm haplotype tree.
-
•
Consideration of parasite origins and those of co-evolved hosts.
1. Introduction
St Kilda is an uninhabited archipelago lying in the north Atlantic Ocean about 40 miles west of the Outer Hebrides, and comprising the islands of Soay, Hirta, Dun and Boreray. Hirta, the largest of these islands, with about 670 ha of exposed hill and moorland, was continuously inhabited for thousands of years. The islanders kept sheep and cattle to supplement their diet of seabirds and cereal crops and for wool to trade and pay the landlord, along with large numbers of dogs. In 1930 the human population of St Kilda was evacuated along with their animals. In 1932, 107 feral Soay sheep were relocated onto Hirta from the smaller neighbouring island of Soay, to maintain the pasture. These sheep have been left unmanaged since then, and the only other terrestrial mammals present on St Kilda have been mice and visiting humans.
Two years after the 1930 evacuation of St Kilda, the resident population of house mice (Mus musculus) became extinct (Harrisson and Moy-Thomas, 1933). Without the human inhabitants, these commensal animals were unable to compete with the less strongly anthropopilous field mice (Apodemus sylvaticus), which have survived on the islands of Hirta and Dun (Morton-Boyd, 1956; Berry, 1969; Berry and Tricker, 1969). Neither of these species would have survived the Last Glacial Maximum about 24 thousand years ago (Johnsen et al., 1992) in this location, hence both would have been introduced more recently, and presumably inadvertently, by humans (Corbet, 1961). The source of these animals has been a matter of considerable interest to zoologists and conservationists, with some evidence for introductions from Scandinavia that have been attributed to Vikings. However there may have been earlier and later introductions from mainland Britain or elsewhere (Berry, 1969; Jones et al., 2010; Herman et al., 2017). At various times, both the St Kilda house mouse and the field mouse have been considered as distinct species or subspecies, due to their size and pelage colour, however genetic evidence does not support any substantial differentiation from populations elsewhere in the British Isles or northern and western Europe (Jones et al., 2010; Herman et al., 2017) and this distinction consequently seems unwarranted. The St Kilda field mice survive on seeds, invertebrates and scavenged carcasses of sheep and seabirds (Berry, 1969). Previous parasitological studies have found that they are infected with adult Hymenolepis diminuta (Morton-Boyd, 1959) and larval metacestodes of Taenia taeniaeformis (referred to as Taenia crassicolis or cysticercus fasciolaris) (Waterston, 1906; Morton-Boyd, 1959).
The present study was undertaken to investigate the possibility that the St Kilda field mouse might be a definitive host for Taenia hydatigena, accounting for the high prevalence of cysticerci tenuicollis previously identified in the St Kilda Soay sheep (Torgerson et al., 1995), but this highly speculative hypothesis was not supported. Nevertheless, the study provided an opportunity for the post-mortem examination of the intestinal tracts from a series of preserved specimens of St Kilda field mice, showing the presence of adult cyclophyllidean tapeworms. In addition, examination of mouse faeces revealed the presence of taeniid eggs and permitted molecular confirmation of their species identity. The serendipitous and novel identification of Hymenolepis hibernia provided an opportunity to investigate the phylogenetic origins of the parasite, with reference to those of its wildlife host and to consideration of its zoonotic potential (Nkouawa et al., 2016).
2. Materials and methods
2.1. Mouse samples used to identify adult tapeworm infections
Viscera from carcasses of 17 St Kilda field mice, preserved in the vertebrate collections of the National Museums of Scotland, were examined. These had been collected between June 2011 and June 2012 from locations throughout Hirta and kept frozen at −20 °C, albeit with occasional cycles of thawing and re-freezing. Any tapeworms or tapeworm segments that were found were recovered and transferred to 70% ethanol. The rectal contents were collected for coprological examination. Naturally-voided faecal samples were also obtained opportunistically from an additional four locations in the Village Bay area on the island of Hirta.
2.2. Parasitological examination of faeces
Faecal samples were suspended in water. Sedimented material was then processed through a series of sieves. Material deposited on 53 μm and 30 μm sieves was retained and examined microscopically for the presence of cyclophyllidean cestode eggs. Eggs were picked and transferred to 70% ethanol.
2.3. Genomic DNA isolation, PCR amplification and sequence analysis of mitochondrial cytochrome c oxidase subunit-1 (Cox-1)
Aliquots of about 100 cyclophyllidean cestode eggs with hexacanth onchospheres were lysed in single 0.2 ml tubes containing 50 μl of proteinase K lysis buffer and stored at −80 °C as previously described (Redman et al., 2008). 1 μl of a 1:5 dilution of neat lysate was used as PCR template and the same dilutions of lysate buffer made in parallel were used as negative controls. A fragment of 396 bp of the mitochondrial cytochrome c oxidase subunit-1 (Cox-1), was amplified using forward (CeCox-For- TTTTTTGGGCATCCTGAGGTTTAT) and reverse primers (CeCox-Rev- TAAAGAAAGAACATAATGAAAATG) (Lavikainen et al., 2008). PCR reaction conditions consisted of 1x thermopol reaction buffer (NEB BioLab), 2 mM MgSO4, 200 μM dNTPs, 0.2 μM forward and reverse primers and 1U of Phusion high fidelity DNA polymerase (Finenzyme). The thermo-cycling parameters consisted of an initial 98 °C for 30 s followed by 40 cycles of 98 °C for 10 s, 54 °C for 30 s and 72 °C for 2 min with a single final extension cycle of 72 °C for 7 min. DNA templates for direct sequencing of the Cox-1 region were cleaned using QIAquick PCR Purification Kit (Cat No./ID: 28104) following the manufacturers’ protocols. Amplicons were sequenced from both ends using the same primers used for PCR amplification for each region with BigDye Terminator Cycle Sequencing (Applied Biosystems). Sequences of Cox-1 were edited to remove primers and poor quality sequence on both ends using Geneious Pro 5.4 software (Kearse et al., 2012). Sequences showing 100% base pair similarity were grouped into haplotypes using the CD-HIT Suite software (Huang et al., 2010). Haplotype and GenBank reference sequences for Hymenolepis spp. were imported into MEGA 6 (Tamura et al., 2013) and a Maximum Likelihood tree was inferred, using the Hasegawa-Kishino-Yano (1985) nucleotide subsitution model with gamma distribution of rates across sites (HKY+G), as selected with the Bayesian Information Criterion. Model parameters were estimated from the data and branch support was estimated from 1000 bootstrap pseudoreplicates of the data. The Maximum Likelihood tree was rooted using a closely related outgroup sequence from H. contortus (GenBank accession no. AF070785).
3. Results
3.1. Gross parasitological tapeworm identification
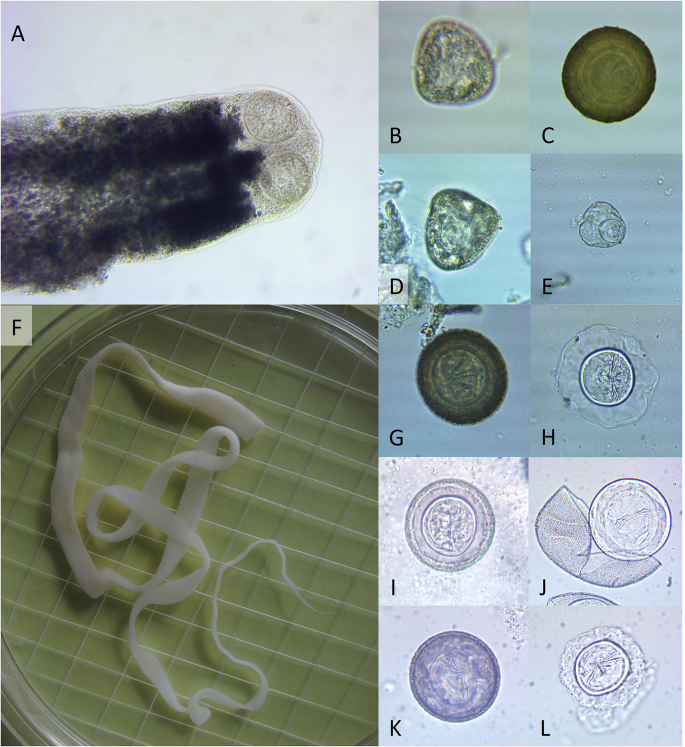
Cyclophyllidean tapeworms or segments were identified in the small intestines of five of the 17 mice that were examined postmortem. These were in poor condition and mostly fragmented, but could be separated into two morphological groups based on their segment dimensions and estimated overall length of about 2 mm wide and 6 cm long, or about 4 mm wide and up to 31 cm long. One of the smaller tapeworms had an intact, unarmed scolex (Fig. 1A), but the scolices were not found for any others. The most intact larger tapeworm, lacking a scolex, is shown in Fig. 1F. Cyclophylllidean tapeworm eggs were only recovered on the 53 μm sieve, from the faeces of each of the five mice in which adult tapeworms were found and from one of the four naturally-voided faecal samples, collected from a cleat (or cleit; an ancient small stone storage hut). Eggs in the faeces of three mice (numbered 5, 8 and 10) in which the smaller tapeworms were identified and in the faeces collected from a cleat (labelled X) were typically anoplocephalid, being approximately pyramidal-shaped, with a maximum width of 62–68 μm (Fig. 1B and D) and containing a similarly shaped onchosphere, about 38 μm across (Fig. 1E). Eggs in the faeces of the three mice (numbered 7, 8 and 9) and in the naturally-voided faeces (lettered X) were near spherical, mean 68 μm in diameter, with a thick, dark outer shell, a granular and non-striated embryophore about 20 μm deep, granular, and a thin onchospheral membrane (Fig. 1C, G, 1I and 1K) surrounding a 36–38 μm spherical hexacanth onchosphere (Fig. 1H and J and 1L). The embryonic hooks were relatively long, and consistently arranged in offset, radiating pairs. Cyclophyllidean co-infections were, therefore, identified in mouse 8 and in the naturally-voided faeces from mouse X. However, the lack of non-overlapping and definitive morphological diagnostic characters for cyclophyllidean eggs and the poor condition of the adult tapeworms prevented identification to generic or specific level.
Fig. 1.
Examples of cyclophyllidean tapeworms and eggs recovered from Apodemus sylvaticus viscera and faeces. A: Unarmed scolex of an intact tapeworm - mouse 5. B: Anoplocephalid tapeworm egg - mouse 8. C: Hymenolepidid egg cropped without changing dimensions from the same image as B, for comparison - mouse 8. D: Anoplocephalid tapeworm egg - mouse X (faeces from a cleat). E: Anoplocephalid onchosphere released after squashing an egg under a cover slip - mouse X (faeces from a cleat). F: Tapeworm from mouse 8 (scolex not intact). G: hymenolepidid tapeworm egg - mouse X (faeces from a cleat). H: Hexacanth onchosphere surrounded by an onchospheral membrane and inner zone of the embryophore, which has swollen, having been released from the egg shell by squashing under a cover slip - mouse 9. I: Hymenolepidid tapeworm egg - mouse 7. J: Hexacanth onchosphere surrounded by an intact onchospheral membrane and inner and outer zones of the embryophore, being released from a cracked egg shell by squashing under a cover slip - mouse 7. K: Hymenolepidid tapeworm egg - mouse 8. L: Hexacanth onchosphere surrounded by an onchospheral membrane and inner zone of the embryophore, which has swollen, having been released from the egg shell by squashing under a cover slip - mouse 8.
3.2. Confirmation of H. hibernia in St Kilda mice by analysis of mt-Cox-1 sequences
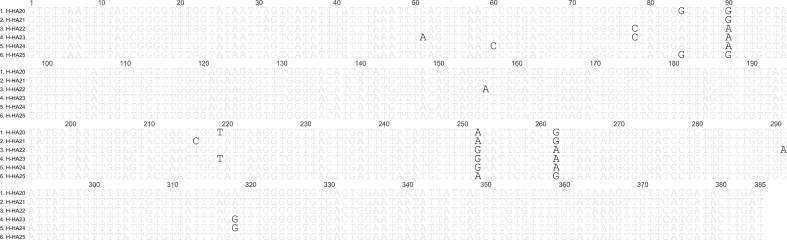
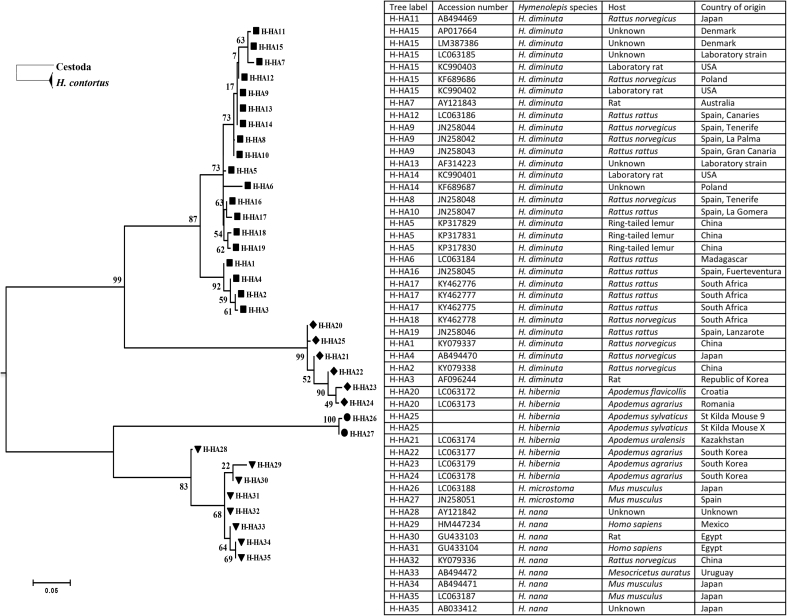
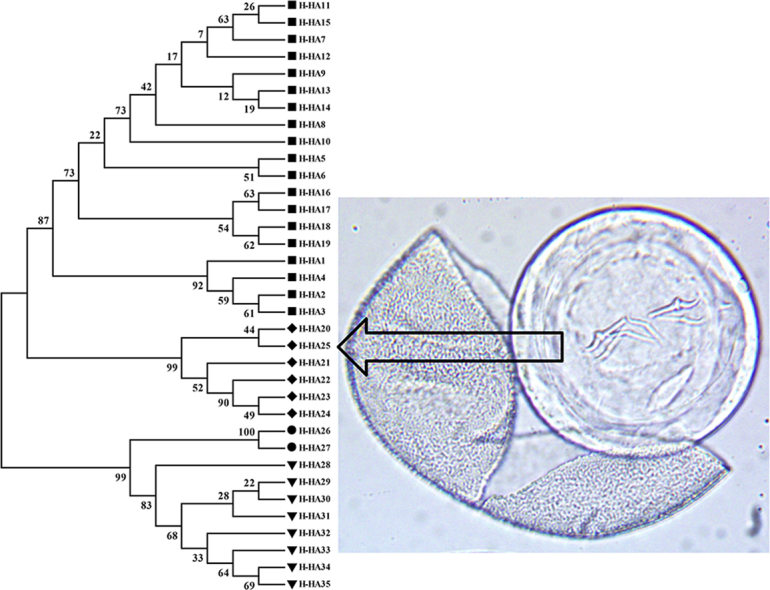
385 bp mt-Cox-1 fragments of cyclophyllidean DNA were amplified and sequenced from eggs recovered from mouse 9 and faeces X (despite repeated attempts, DNA did not amplify from the eggs containing hexacanth onchospheres recovered from mice 7 and 8). The fragments were aligned with 34 sequences of four Hymenolepis species (H. Hibernia, H. diminuta, Hymenolepis nana and Hymenolepis microstoma) published in the NCBI Genbank database. The St Kilda sequences were identical and comparison with the previously published H. hibernia sequences revealed that they formed a distinct haplotype, characterised by 12 intraspecific SNPs within the 385 bp fragment of the mt-Cox-1 locus (Fig. 2). The 31 H. diminuta, 6 H. hibernia, 2 H. microstoma and 9 H. nana mt-Cox-1 sequences from Genbank comprise of 19, 5, 2 and 8 unique haplotypes, respectively. In the Maximum Likelihood tree inferred from the 35 haplotypes, including the new one from St Kilda, there are four well-supported monophyletic clades, comprising the H. diminuta, H. hibernia, H. microstoma and H. nana sequences (Fig. 3). The two rodent-origin species, H. diminuta and H. hibernia, and the two human-origin species, H. microstoma and H. nana, form distinct clades in the phylogenetic tree. The H. hibernia clade is highly supported (bootstrap percentage 100) and includes the new haplotype from the St Kilda mice (H-HA25). The St Kilda H. hibernia sequence is nevertheless distinct and clusters with an eastern European haplotype (H-HA20), separately from haplotypes of distant origins in Asia (H-HA21, 22, 23 and 24).
Fig. 2.
The Sanger sequence of 385 bp of mt-Cox-1 fragments of cyclophyllidean DNA generated from eggs recovered from mouse 9 and faeces X (6. H-HA25), aligned with 5 corresponding H. hibernia sequences published in the NCBI Genbank database. There are 12 intraspecific SNPs at positions 5, 60, 78, 84, 90, 156, 216, 219, 252, 262, 291 and 318. GeneBank submission ID: 2151861.
Fig. 3.
Maximum Likelihood tree for the 35 haplotypes identified from 39 mt-cox-1 sequences of Hymenolepis species. Of these, 19, 5, 2 and 8 haplotypes are identified in the Genbank database as H. diminuta, H. hibernia, H. microstoma and H. nana, respectively, and one haplotype (H-HA25) was identified here from the faeces of St Kilda mice 9 and X. Branches with bootstrap values (1000 replications) represented at the base of the nodes. The phylogeny is rooted with mt-cox-1 sequence of parasitic nematode H. contortus.
4. Discussion
The tapeworm species H. diminuta was previously reported in the A. sylvaticus mouse population from St Kilda (Morton-Boyd, 1959). However, species identification of helminth parasites by morphology can be challenging, not least in the case of Hymenolepis sensu lato (Makarikov and Tkach, 2013; Binkienė et al., 2018). Biochemical, growth and behavioural studies suggest that H. hibernia and H. diminuta are genetically distinct, with H. hibernia being adapted to Palaearctic mice from the genus Apodemus, whereas H. diminuta is a rat tapeworm (Montgomery et al., 1987). The molecular evidence provided by our study supports the suggestion (Montgomery et al., 1987) that H. diminuta and H. hibernia are genetically distinct. It also suggests that the former species may previously have been wrongly identified as the latter on St Kilda, and potentially elsewhere, as a result of their overlapping morphological characteristics. Similar genetic differentiation has been shown between H. diminuta and a cryptic species, closely related to H. hibernia, that was recovered from a Tibetan woman (Nkouawa et al., 2016).
The presence of H. hibernia in field mice from St Kilda is of potential interest with regard to the origins of both the parasite itself and its co-evolved host (Wilson et al., 1998). The presence of a glacial refugium for A. sylvaticus in northern Europe was previously predicted from the distribution of mitochondrial genetic variation in its nematode parasite Heligmosomoides polygyrus, and this was subsequently confirmed from genetic variation in A. sylvaticus itself (Nieberding et al., 2004, 2005; Herman et al., 2017). However, there was insufficient resolution in the Maximum Likelihood tree (Fig. 2) for this approach to be effective in the present case, which may to some extent reflect the limited variation present in the 385 bp mt-Cox-1 fragment, although it is most likely due to insufficient sampling across its geographical range. Nevertheless, genetic variation in H. hibernia may become sufficiently informative in the future, once more molecular data become available. The sensitivity of the approach could be improved by cloning and sequencing to identify all of the haplotypes present in a mixed pool of eggs, albeit the number of haplotypes present in clonally reproducing cyclophyllidean tapeworms would be predicted to be small. Other approaches such as the use of polymorphic microsatellite markers (Pajeulo et al., 2015), or of next generation sequencing methods to describe the phylogenetic origins of multiple parasite species derived from the same host (Avramenko et al., 2015) might also provide future direction.
Molecular speciation methods have proved to be sensitive and unambiguous in situations where complementary phenotypic characterisation has already been undertaken (Wimmer et al., 2004). Precise species identification is important in the study of parasite life histories and epidemiology that is required to define control strategies. Hymenolepis senso latu including the rodent-origin species H. hibernia (Nkouawa et al., 2016) and H. diminuta (Hancke and Suarez, 2017) are zoonotic, hence informed preventive management is necessary. The basic life history of hymenolepidid tapeworms involves incidental human and definitive rodent hosts being infected when they ingest cysticercoid metacestodes with beetle intermediate hosts, although autoinfection with eggs can also occur in humans. It is therefore important that further studies are made of hymenolepidid tapeworm phylogeographic variation and epidemiology.
Acknowledgments
We are grateful for the support of the St Kilda Soay Sheep Project in facilitating our work. The work was undertaken as a R(D)SVS BVM&S undergraduate Student Research Project (PB), which was formally approved by the University of Edinburgh Veterinary Ethics in Research Committee. Work at the Roslin Institute uses facilities funded by the UK Biotechnology and Biological Sciences Research Council (BBSRC). NS and UC work under a BBSRC Responsive Mode Strategic LoLa (RA3176. Building Upon the Genome: using Haemonchus contortus genomic resources to develop novel interventions to control endemic GI parasites.)
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijppaw.2018.09.007.
Contributor Information
Neil Sargison, Email: neil.sargison@ed.ac.uk.
Jeremy Herman, Email: J.Herman@nms.ac.uk.
Jill Pilkington, Email: J.Pilkington@ed.ac.uk.
Peter Buckland, Email: buckland05@hotmail.com.
Kathryn Watt, Email: Kathryn.Watt@ed.ac.uk.
Alex Chambers, Email: Alex.Chambers@agresearch.co.nz.
Umer Chaudhry, Email: uchaudhr@exseed.ed.ac.uk.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Avramenko R.W., Redman E.M., Lewis R., Yazwinski T.A., Wasmuth J.D., Gilleard J.S. Exploring the gastrointestinal “nemabiome”: deep amplicon sequencing to quantify the species composition of parasitic nematode communities. PLoS One. 2015 doi: 10.1371/journal.pone.0143559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry R.J. History in the evolution of Apodemus sylvaticus (Mammalia) at one edge of its range. J. Zool. 1969;159:311–328. [Google Scholar]
- Berry R.J., Tricker B.J.K. Competition and extinction: the mice of foula, with notes on those of fair isle and St Kilda. J. Zool. 1969;158:247–265. [Google Scholar]
- Binkienė R., Miliūtė A., Stunžėnas V. Molecular data confirm the taxonomic position of Hymenolepis erinacei (Cyclophyllidea: Hymenolepididae) and host switching, with notes on cestodes of Palaearctic hedgehogs (Erinaceidae) J. Helminthol. 2018:1–8. doi: 10.1017/S0022149X18000056. Published online. [DOI] [PubMed] [Google Scholar]
- Corbet G.B. Origin of the British insular races of small mammals and of the ‘Lusitanian’ fauna. Nature. 1961;191:1037–1040. [Google Scholar]
- Hancke D., Suarez O.V. Helminth diversity in synanthropic rodents from an urban ecosystem. EcoHealth. 2017;14(3):603–613. doi: 10.1007/s10393-017-1239-8. [DOI] [PubMed] [Google Scholar]
- Harrisson T.H., Moy-Thomas J.A. The mice of St Kilda, with especial reference to their prospects of extinction and present status. J. Anim. Ecol. 1933;2:109–115. [Google Scholar]
- Hasegawa M., Kishino H., Yano T. Dating of human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- Herman J.S., Jóhannesdóttir F., Jones E.P., McDevitt A.D., Michaux J.R., White T.A., Wójcik J.M., Searle J.B. Post-glacial colonisation of Europe by the wood mouse, Apodemus sylvaticus: evidence of a northern refugium and dispersal with humans. Biol. J. Linn. Soc. 2017;120:313–332. [Google Scholar]
- Huang Y., Niu B.F., Gao Y., Fu L.M., Li W.Z. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics. 2010;26:680–682. doi: 10.1093/bioinformatics/btq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsen S.J., Clausen H.B., Dansgaard W., Fuhrer K., Gundestrup N., Hammer C.U., Iversen P., Jouzel J., Stauffer B., Steffensen J.P. Irregular glacial interstadials recorded in a new Greenland ice core. Nature. 1992;359:311–313. [Google Scholar]
- Jones E.P., Jóhannesdóttir F., Gündüz I., Richards M.B., Searle J.B. The expansion of the house mouse into north-western Europe. J. Zool. 2010;283:257–268. [Google Scholar]
- Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., Thierer T., Ashton B., Meintjes P., Drummond A. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavikainen A., Haukisalm V., Lehtinen M.J., Henttonen H., Oksanen A., Meri S. A phylogeny of members of the family Taeniidae based on the mitochondrial cox1 and nad1 gene data. Parasitology. 2008;135:1457–1467. doi: 10.1017/S003118200800499X. [DOI] [PubMed] [Google Scholar]
- Makarikov A.A., Tkach V.V. Two new species of Hymenolepis (Cestoda: Hymenolepididae) from Spalacidae and Muridae (Rodentia) from eastern palearctic. Acta Parasitol. 2013;58:37–49. doi: 10.2478/s11686-013-0115-0. [DOI] [PubMed] [Google Scholar]
- Montgomery S.S.J., Montgomery W.I., Dunn T.S. Biochemical, physiological and morphological variation in unarmed hymenolepids (Eucestoda: Cyclophyllidae) Zool. J. Linn. Soc. 1987;91:293–324. [Google Scholar]
- Morton-Boyd J. The St. Kilda field mouse (Apodemus sylvaticus hirtensis barrett-Hamilton), population in the village area, Hirta, may 1955. Oikos. 1956;7(fasc1):110–116. [Google Scholar]
- Morton-Boyd J. Observations on the St. Kilda field-mouse Apodemus sylvaticus hirtensis barrett-Hamilton. J. Zool. 1959;133:47–65. [Google Scholar]
- Nieberding C., Morand S., Libois R., Michaux J.R. A parasite reveals cryptic phylogeographic history of its host. Proc. R. Soc. Lond. Ser. B Biol. Sci. 2004;271:2559–2568. doi: 10.1098/rspb.2004.2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieberding C., Libois R., Douady C.J., Morand S., Michaux J.R. Phylogeography of a nematode (Heligmosomoides polygyrus) in the western Palearctic region: persistence of northern cryptic populations during ice ages? Mol. Ecol. 2005;14:765–779. doi: 10.1111/j.1365-294X.2005.02440.x. [DOI] [PubMed] [Google Scholar]
- Nkouawa A., Haukisalmi V., Li T., Nakao M., Lavikainen A., Chen X., Henttonen H., Ito A. Cryptic diversity in hymenolepidid tapeworms infecting humans. Parasitol. Int. 2016;65:83–86. doi: 10.1016/j.parint.2015.10.009. [DOI] [PubMed] [Google Scholar]
- Pajuelo M.J., Eguiluz M., Dahlstrom E., Requena D., Guzman F., Ramirez M., Sheen P., Frace M., Sammons S., Cama V., Anzick S., Bruno D., Mahanty S., Wilkins P., Nash T., Gonzalez A., Garcia H.H., Gilman R.H., Porcella S., Zimic M. Identification and characterization of microsatellite markers derived from the whole genome analysis of Taenia solium. PLoS Neglected Trop. Dis. 2015;9(12) doi: 10.1371/journal.pntd.0004316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redman E., Packard E., Grillo V., Smith J., Jackson F., Gilleard J.S. Microsatellite analysis reveals marked genetic differentiation between Haemonchus contortus laboratory isolates and provides a rapid system of genetic fingerprinting. Int. J. Parasitol. 2008;38:111–122. doi: 10.1016/j.ijpara.2007.06.008. [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torgerson P., Pilkington J., Gulland M., Gemmell M. Further evidence for the long distance dispersal of taeniid eggs. Int. J. Parasitol. 1995;25:265–267. doi: 10.1016/0020-7519(94)00094-5. [DOI] [PubMed] [Google Scholar]
- Waterson J. Notes on the mice and birds of St Kilda. Ann. Scot. Nat. Hist. 1906;14:199–202. [Google Scholar]
- Wilson K., Eady P., Nevo A.J. del. Origin of an insular population of the wood mouse based on parasitological evidence. J. Wildl. Dis. 1998;34:150–154. doi: 10.7589/0090-3558-34.1.150. [DOI] [PubMed] [Google Scholar]
- Wimmer B., Craig B.H., Pilkington J.G., Pemberton J.M. Non-invasive assessment of parasitic nematode species diversity in wild Soay sheep using molecular markers. Int. J. Parasitol. 2004;34:625–631. doi: 10.1016/j.ijpara.2003.11.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.