Abstract
Variation in female reproductive traits, such as fertility, fecundity, and fecundability, are heritable in humans, but identifying and functionally characterizing genetic variants associated with these traits have been challenging. Here, we explore the functional significance and evolutionary history of a G/A polymorphism at SNP rs2523393, which is an eQTL for HLA-F and is significantly associated with fecundability (the probability of being pregnant within a single menstrual cycle). We replicated the association between the rs2523393 genotype and HLA-F expression by using GTEx data and demonstrate that HLA-F is upregulated in the endometrium during the window of implantation and by progesterone in decidual stromal cells. Next, we show that the rs2523393 A allele creates a GATA2 binding site in a progesterone-responsive distal enhancer that loops to the HLA-F promoter. Remarkably, we found that the A allele is derived in the human lineage and that the G/A polymorphism arose before the divergence of modern and archaic humans and segregates at intermediate to high frequencies across human populations. Remarkably, the derived A allele is has also been identified in a GWAS as a risk allele for multiple sclerosis. These data suggest that the polymorphism is maintained by antagonistic pleiotropy and a reproduction-health tradeoff in human evolution.
Keywords: HLA-F, GATA2, endometrium, decidualization, pregnancy
Introduction
Traits associated with female reproduction are heritable in humans,1, 2 but identifying the genetic bases for these traits had been challenging.1, 2, 3 Several recent studies with large sample sizes or innovative study designs, however, have identified genetic variants associated with menarche,4, 5, 6, 7 menopause,5, 7, 8, 9, 10 gestation length,11 predisposition to preterm birth,11, 12 severe nausea and vomiting during pregnancy,13 and fecundability (the probability of becoming pregnant within a single menstrual cycle).14 Burrows et al.,14 for example, performed an integrated expression quantitative trait locus (eQTL) mapping and association study to identify mid-secretory endometrium eQTLs that influence pregnancy outcomes. Among the eQTLs identified was a G/A polymorphism (rs2523393) that was significantly associated with HLA-F (MIM: 143110) expression and fecundability. Specifically, the G allele of rs2523393 was associated with longer intervals to pregnancy and lower expression of HLA-F in mid-secretory-phase (receptive) endometrium, whereas the A allele was associated with shorter intervals to pregnancy and higher HLA-F expression. The median time to pregnancy, for example, was 2.3, 2.6, and 4.9 months among women with the AA, GA, and GG genotypes, respectively.14
Although the functions of HLA-F are enigmatic, it is thought to regulate immune responses and could play an important role in regulating maternal-fetal immunotolerance during pregnancy.15, 16 HLA-F is highly expressed, for example, in placental villi, on the surface of invasive and migratory extra-villus trophoblasts (EVTs) and decidual stromal cells (DSCs).17, 18, 19 Furthermore, HLA-F expression increases during gestation and reaches a peak at term.17, 20 Like other major histocompatibility complex I (MHC-I) molecules, HLA-F binds natural killer (NK) cell receptors from the leukocyte immunoglobulin-like receptor (LIR) and killer cell immunoglobulin-like receptor (KIR) families, including LIR1 and LIR2,21, 22 KIR2DS4 and KIR3DL2,23 and KIR3DL1 and KIR3DS1.22, 24, 25 Uterine NK (uNK) cells, which are essential for the establishment and maintenance of maternal immunotolerance and spiral artery remodeling, express KIR3DL1 and LIR2, suggesting that HLA-F expressed by EVTs and DSCs mediates interactions with uNK cells during implantation, trophoblast invasion, and establishment of the uteroplacental circulation. HLA-F expression level is also positively correlated with uNK abundance in mid-luteal endometria and is predictive of achieving pregnancy,14, 26 consistent with an important role for HLA-F in the establishment of pregnancy.
Here, we explore the functional and evolutionary history of the rs2523393 G/A polymorphism. We first replicate the association between the rs2523393 genotype and HLA-F expression by using Genotype-Tissue Expression (GTEx) data. We demonstrate that HLA-F expression increases during the menstrual cycle and is upregulated by progesterone during the differentiation of endometrial stromal fibroblasts (ESFs) into DSCs. Next, we show that rs2523393 is located within a cAMP/progesterone responsive enhancer that makes long range regulatory interactions with the promoter of HLA-F, and that the A allele creates a new binding site for the progesterone receptor (PGR) co-factor GATA2. Remarkably the G/A polymorphism arose before the divergence of modern and archaic humans, is segregating at intermediate to high frequencies across human populations, and associated with a predisposition to several diseases. These data suggest the G/A polymorphism is maintained by antagonistic pleiotropy and that there is a reproduction-health tradeoff in human evolution.
Material and Methods
rs2523393 Is an eQTL for HLA-F
We replicated the association between the G/A polymorphism at rs2523393 and HLA-F expression levels by using data from GTEx Analysis Release V6 (dbGaP: phs000424.v7.p2) for 35 tissues, including the 101 uterus samples.27, 28 We also used GTEx data to identify other genes for which rs2523393 was an eQTL. In brief, we queried the GTEx Portal by using the “single tissue eQTLs search form” for SNP rs2523393.
HLA-F Expression
Using previously generated microarray expression data, we examined the expression of HLA-F in the endometrium across the menstrual cycle (GEO: GSE488829) and in endometrial samples from fertile women (n = 5), women who had implantation failure after in vitro fertilization (n = 5), and woman with recurrent spontaneous abortions (n = 5) (GEO: GSE2678730). These microarray datasets were analyzed with the GEO2R analysis package, which implements the GEOquery31 and limma R32 packages from the Bioconductor project to quantify differential gene expression. We also examined HLA-F expression in RNA-sequencing (RNA-seq) data previously generated from ESFs treated with control media or differentiated (decidualized) with 100 nM medroxyprogesterone acetate (MPA) and 1 mM 8-bromo-cAMP (all from Sigma-Aldrich) into DSCs.34, 35 We used previously generated RNA-seq data to characterize the expression level of HLA-F in human ESFs, DSCs, and DSCs treated with a small interfering RNA (siRNA) to knock down PGR expression.36
Isolation of Primary Human ESFs
Human endometrial samples were obtained from six healthy, reproductive-age volunteers with regular menstrual cycles and no history of gynecological malignancies under a human subject protocol approved by the institutional review board of Baylor College of Medicine. After receipt of informed consent from participants, an endometrial biopsy was performed during the follicular phase of the menstrual cycle, and endometrial stromal cells were isolated by enzymatic digestion and filtration, as described previously.37, 38 ESFs were cultured in DMEM/F12 media with 10% fetal bovine serum (FBS) (see below for greater details about specific culture conditions for different experiments), and all experiments were carried out within four cell passages. For both the primary and the hTERT immortalized ESFs, progesterone and cAMP lasted 2–3 days, which is sufficient to induce decidualization and differentiation into DSCs39 yet short enough to maintain specific needs of experiments (such as transient transfection).
GATA2 ChIP-Seq
ESFs were cultured in 150 mm culture dishes with ESF growth media (DMEM/F12 containing 10% FBS and 1% antibiotic-antimycotic). Cells were grown to approximately 90% confluency and started on treatment with decidual media (Opti-MEM media containing 2% charcoal-stripped FBS, 1% antibiotic-antimycotic, 1 μM MPA, 10 nM E2, and 50 μM cAMP). Decidual media were changed every 48 hr. After 72 hr of treatment, cells were fixed. Cells were fixed for 15 min with 1/10 volume of freshly prepared formaldehyde solution (11% formaldehyde, 0.1 M NaCl, 1 mM EDTA, and 50 mM HEPES). The fixation was stopped by the addition of 1/20 volume 2.5 M glycine for 5 min. Fixed DSCs were collected and pelleted at 800 × g for 10 min at 4°C. Cell pellets were washed twice with cold PBS-Igepal (0.5% Igepal and 1 mM PMSF). DSCs from six individuals were pooled before genomic DNA isolation. GATA2 immunoprecipitation and DNA library generation were performed by Active Motif. Chromatin immunoprecipitation (ChIP) and input DNA were amplified with the Illumina ChIP-Seq DNA Sample Prep Kit. In brief, DNA ends were polished and 59-phosphorylated with T4 DNA polymerase, Klenow polymerase, and T4 polynucleotide kinase. 3′-adenine was added to blunt ends with the Klenow fragment (3′-5′ exo minus), Illumina genomic adapters were ligated, and the sample was size fractionated to 175–225 bp on a 2% agarose gel. After amplification for 18 cycles with Phusion polymerase, the resulting DNA libraries were tested by RT-qPCR at the same specific genomic regions as the original ChIP DNA for quality assessment of the amplification reactions. 36 nt sequence reads were identified by the FastTrack Sequencing Services (with Illumina’s HiSeq). Reads were mapped to the human genome (UCSC Genome Browser hg19) with the Burrows-Wheeler Aligner. Only tags that mapped uniquely, had no more than two mismatches, and passed quality-control filtering were used in the subsequent analysis. The number of unique alignments without duplicate reads was totaled, and MACS analysis was performed at a p value of 1 × 10−7. Raw and processed data are available under accession numbers SRA: SRX3503584 and GEO: GSE108409.
GATA2 Knockdown and Expression Profiling
Three ESF subcultures were transfected with siRNA and treated with decidual media (see above) for 3 days for DSC differentiation. Total RNA was extracted with the QIAGEN RNeasy RNA Isolation Kit (QIAGEN). The RNA from three replicates (wells) was pooled for each treatment per cell line. The integrity of all RNA samples was tested with the 2100 Bioanalyzer (Agilent Technologies). The concentration of RNA was quantified on the Nanodrop Spectrophotometer (Nanodrop Technologies). Samples with 260/280 ratios greater than 1.8 were used for microarray hybridization. Microarrays were performed by the Genomic and RNA Profiling Core of the Baylor College of Medicine with Affymetrix Human Genome U133 Plus 2.0 arrays (Affymetrix). Microarray CEL files were analyzed by dChip with the PM-MM model and quantile normalization. Combat was used to normalize differences and for batch correction. Two-sided t tests and fold changes were used to define differentially expressed genes. Genes with p = 0.05 and an absolute fold change of 1.4 were considered significant. Raw and processed data are available under accession numbers SRA: SRX3503584 and GEO: GSE108409.
Luciferase Assays
A 1,000 bp region centered on rs2523393 (chr6: 29,705,520–29,706,519; hg19) was synthesized with either the A or G allele (GenScript) and cloned into the pGL3-Basic luciferase vector (Promega). A pGL3-Basic plasmid without the 1 kb rs2523393 insert was used as a negative control. GATA2 and PGR expression vectors were also obtained from GenScript. ESFs (ATCC CRL-4003) immortalized with telomerase were maintained in phenol-red-free DMEM (GIBCO) supplemented with 10% charcoal-stripped FBS (CSFBS; GIBCO), 1× insulin-transferrin-selenium (GIBCO), 1% sodium pyruvate (GIBCO), and 1% l-glutamine (GIBCO). Confluent ESFs in 96-well plates in 80 μL of Opti-MEM (GIBCO) were transfected with 100 ng of the luciferase plasmid, 100 ng of GATA2 and/or PGR as needed, and 10 ng of pRL-null with 0.1 μL of PLUS reagent (Invitrogen) and 0.25 μL of Lipofectamine LTX (Invitrogen) in 20 μL of Opti-MEM. The cells incubated in the transfection mixture for 6 hr, and the medium was replaced with the phenol-red-free maintenance medium overnight. Cell decidualization was then induced by incubation in the decidualization medium: DMEM with phenol red (GIBCO), 2% CSFBS (GIBCO), 1% sodium pyruvate (GIBCO), 0.5 mM 8-Br-cAMP (Sigma), and 1 μM of the progesterone analog MPA (Sigma) for 48 hr. Decidualization controls were incubated in the decidualization control medium (phenol-red-free DMEM, 2% CSFBS, and 1% sodium pyruvate; GIBCO) instead for 48 hr. After decidualization, we began Dual Luciferase Reporter Assays (Promega) by incubating the cells for 15 min in 20 μL of 1× passive lysis buffer. Luciferase and Renilla activity were then measured with the GloMax-Multi+ Detection System (Promega). We standardized luciferase activity values by the Renilla activity values and background activity values by measuring luminescence from the pGL3-Basic plasmid with no insert. Each luciferase experiment was replicated in at least four independent experiments.
LCL Promoter in Capture Hi-C Data
We also took advantage of the fact that rs2523393 was an eQTL for HLA-F in Epstein-Barr virus (EBV)-transformed lymphoblastoid cell lines (LCLs) to determine whether this locus interacts with the HLA-F promoter in capture Hi-C data generated from LCLs. We used the Capture HiC Plotter (CHiCP),40 a web-based tool for the integrative and interactive visualization of promoter capture Hi-C (PCHi-C) datasets, to identify long-range interactions in the high-resolution capture Hi-C dataset of Mifsud et al. from the LCL GM12878.41
Evolutionary Analyses of the rs2523393 G/A Polymorphism
To reconstruct the evolutionary history of the G/A polymorphism, we used a region spanning 50 bp upstream and downstream of rs2523393 from hg19 (chr6: 32,814,778–32,814,878) as a query sequence to BLAT search the chimpanzee (CHIMP2.1.4), gorilla (gorGor3.1), orangutan (PPYG2), gibbon, (Nleu1.0), rhesus monkey (MMUL_1), hamadryas baboon (Pham_1.0), olive baboon (Panu_2.0), vervet monkey (ChlSab1.0), marmoset (C_jacchus3.2.1), Bolivian squirrel monkey (SalBol1.0), tarsier (tarSyr1), mouse lemur (micMur1), and galago (OtoGar3) genomes. For all other non-human species, we used the same 101 bp region as a query for SRA-BLAST against high-throughput sequencing reads deposited in the SRA. The top-scoring 100 reads were assembled into contigs with the “map to reference” option in Geneious v.6.1.2 and the human sequence as a reference. Sequences for the Altai Neanderthal, Denisovan, and Ust-Ishim were obtained from the Ancient Genome Browser. The frequency of the G/A allele across the Human Genome Diversity Project (HGDP) populations was obtained from the Geography of Genetic Variants Browser. We inferred ancestral sequences of the 101 bp region by using the ancestral sequence reconstruction (ASR) module of Datamonkey,42 which implements joint, marginal, and sampled reconstruction methods,43 nucleotide alignment of the 101 bp, the best-fitting nucleotide substitution model (HKY85), a general discrete model of site-to-site rate variation with three rate classes, and the phylogeny shown in Figure 6A. The phylogeny was retrieved from TimeTree.44 All three ASR methods reconstructed the same sequence for the ancestral human sequence at 1.0 support. Tajima’s D and Fu and Li’s F data for the CEU (Utah residents with northern and western European ancestry from the CEPH collection), YRI (Yoruba in Ibadan, Nigeria), and CHB (Han Chinese in Beijing, China) populations were obtained from the 1000 Genomes Selection Browser 1.0.45 Data from the non-central deviation (NCD) test were extracted from the supplemental materials accompanying the publication describing the method.46
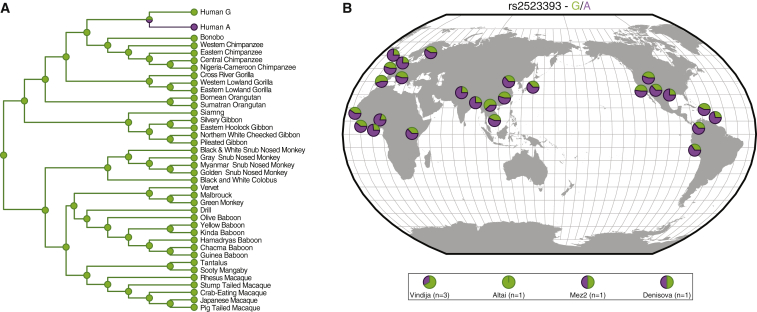
Figure 6.
Evolutionary History of the G/A Polymorphism at rs2523393
(A) rs2523393 genotype across primates and ancestral reconstructions. Genotypes in extant species are shown as circles next to species names at terminal branches. Genotypes at internal nodes based on ancestral reconstruction are shown as circles. A is shown in purple, and G is shown in green.
(B) Distribution of the G and A alleles of rs2523393 across HGDP populations.
Results
rs2523393 Is an eQTL for HLA-F
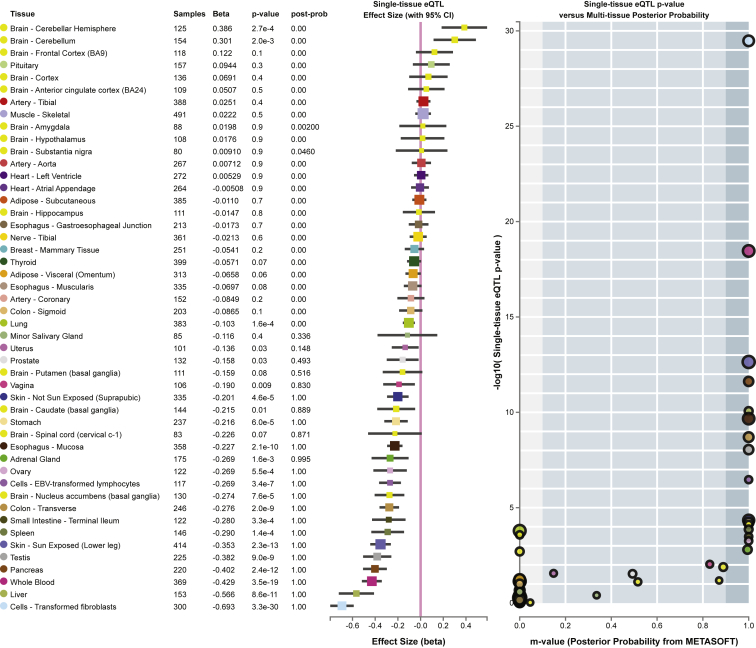
It has been previously shown that the rs2523393 G/A polymorphism is an eQTL for HLA-F in mid-secretory-phase endometrium.14 To replicate this observation in an independent cohort and in additional tissues, we tested whether rs2523393 was correlated with HLA-F expression by using GTEx data. Confirming the previous observation, we found that rs2523393 was an eQTL for HLA-F in the uterus (p = 0.028) as well as 22 other tissues (Figure 1). We also used GTEx data to identify other genes for which rs2523393 was an eQTL and found that it was an eQTL for 15 other genes, including HLA-A (MIM: 142800) and HLA-G (MIM: 142871) (Table S1). The observation that rs2523393 was an eQTL for HLA-A and HLA-G in GTEx data prompted us to explore whether it was an eQTL for these genes in the mid-secretory endometrium data from the original study.14 Indeed, we found that rs2523393 was also an eQTL for HLA-G (p = 0.041), but not HLA-A, in mid-secretory endometrium.
Figure 1.
GTEx Multi-tissue eQTL Plot for rs2523393
(Left) Tissue type, sample size, beta value, p value, and posterior probability that rs2523393 in each tissue tested is also an eQTL across other tissues.
(Center) Single-tissue effect size and 95% confidence interval.
(Right) Single-tissue eQTL p value versus multi-tissue posterior probability.
The rs2523393 G/A Polymorphism Occurs within a GATA2 Motif
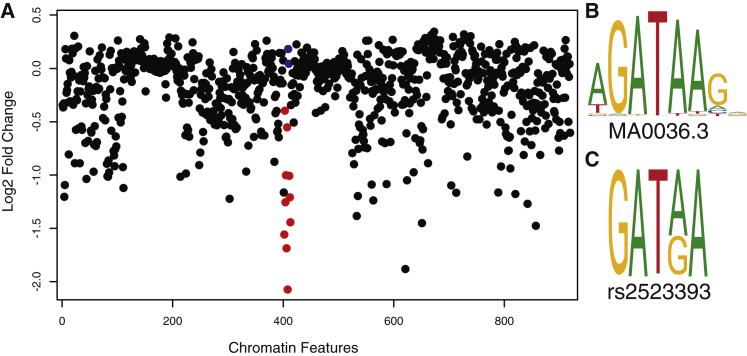
To infer the functional consequences of the G/A polymorphism, we used DeepSea,47 a deep-learning-based algorithm that infers the effects of single-nucleotide substitutions on chromatin features, such as transcription factor binding, DNase I sensitivity, and histone marks. DeepSea predicted that the G allele would have a negative effect on the binding of GATA2 (log2 fold-change effect = −2.07, E-value = 0.035) and GATA3 (log2 fold-change effect = −1.00; E-value = 0.003) (Figure 2A). We next used JASPAR transcription factor binding site (TFBS) motifs48 to identify putative TFBSs in a 36 bp window upstream and downstream of rs2523393. We identified a single TFBS in this window, a GATA motif (matrix ID = MA0036.3, score = 8.70), and found that the G/A polymorphism occurs at an invariant A site in the motif (GATAA). Similar to the DeepSea results, substituting the reference A allele with the alternative G allele is predicted to abolish GATA2 binding (Figures 2B and 2C). These results suggest that the G/A polymorphism alters binding of GATA2, a transcription factor that plays an essential role in mediating the transcriptional response to progesterone in DSCs.49, 50, 51
Figure 2.
The rs2523393 G Allele Is Predicted to Disrupt a GATA2 Binding Site
(A) DeepSea plot showing predicted effects of the G allele on chromatin features. The G allele is predicted to negatively affect GATA2 and GATA3 binding. GATA1 binding is shown in blue, GATA2 and GATA3 binding is shown in red.
(B) JASPAR GATA2 motif.
(C) GATA2 motif at rs2523393.
HLA-F Is Upregulated by Progesterone and GATA2 in DSCs
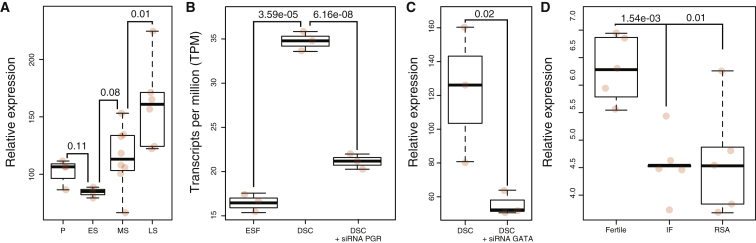
Previous studies have shown that HLA-F is expressed in placental villi, EVTs, and DSCs.17, 18, 19, 52 To determine whether the expression of HLA-F varies in the endometrium across the menstrual cycle, we used previously generated microarray expression data.29 We found that HLA-F expression reached peak expression in the late secretory phase of the menstrual cycle and was significantly differentially expressed throughout the menstrual cycle (Figure 3A). To test whether HLA-F is upregulated by progesterone, we examined its expression in RNA-seq data from ESFs treated with control media or differentiated with cAMP and progesterone into DSCs treated with either a PGR-specific siRNA or a scrambled control siRNA.34, 35, 53 We found that HLA-F was upregulated 2.11-fold (p = 3.59 × 10−5; Figure 3B) in DSCs and that knockdown of PGR downregulated HLA-F 1.67-fold (p = 6.15 × 10−8; Figure 3B). Next, we used a siRNA to knock down the expression of GATA2 in DSCs and assayed global gene expression with an Agilent Human Gene Expression 8x60K Microarray. Consentient with our previous results, we observed a 2.20-fold downregulation of HLA-F (p = 0.02) upon GATA2 knockdown (Figure 3C). We also observed that HLA-F expression was lower in the endometria of women with implantation failure (1.38-fold, p = 1.54 × 10−3) or recurrent spontaneous abortion (1.35-fold, p = 0.011) than in fertile control individuals (Figure 3D). Thus, we conclude that HLA-F expression increases as the menstrual cycle progresses and is upregulated by progesterone, PGR, and GATA2 during the differentiation of ESFs into DSCs and that dysregulation is associated with implantation failure and recurrent spontaneous abortion.
Figure 3.
HLA-F Is Regulated by Upregulated Progesterone and GATA2 during the Menstrual Cycle and in Human DSCs
(A) Relative expression of HLA-F throughout the menstrual cycle. Abbreviations are as follows: P, proliferative phase; ES, early secretory phase; MS, mid-secretory phase; LS, late secretory phase. n = 3–8 (t test).
(B) Expression level of HLA-F in ESFs, DSCs, and DSCs treated with siRNA to knock down PGR expression.
(C) Relative expression level of HLA-F in DSCs and DSCs treated with siRNA to knock down GATA2 expression. n = 3 (t test).
(D) Relative expression level of HLA-F in endometrial fertile women and women with implantation failure (IF) or recurrent spontaneous abortion (RSA). n = 5 (t test).
The rs2523393 A Allele Creates an Enhancer that Loops to HLA-F
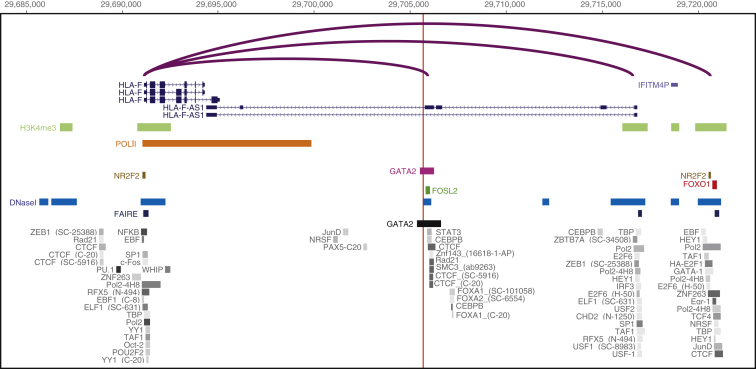
Our observations that rs2523393 is an eQTL for HLA-F and that progesterone and GATA2 upregulate HLA-F in DSCs suggest that rs2523393 could be located within or linked to a progesterone responsive enhancer. To identify such a regulatory element, we generated new GATA2 ChIP-sequencing (ChIP-seq) data from DSCs and used previously published ChIP-seq data from DSCs for the transcription factors PGR,35, 36, 54 FOXO1,54 FOSL2,54 and NR2F2 (COUP-TFII),37 H3K27ac (which marks active enhancers),35 and H3K4me3 (which marks active promoters),35 as well as DNase I hypersensitive site sequencing (DNaseI-seq) and formaldehyde-assisted isolation of regulatory elements sequencing (FAIRE-seq), to identify regions of open chromatin.35 We also took advantage of the fact that rs2523393 is an eQTL for HLA-F in multiple tissues, including EBV-transformed LCLs, to identify additional TFBSs with ChIP-seq data generated by ENCODE and PCHi-C data generated from LCLs. We found that rs2523393 was located in a region of open chromatin and within a GATA2 ChIP-seq peak in DSCs, 170 bp upstream of a FOSL2 ChIP-seq peak in DSCs, and nearby a cluster of TFBSs, including a binding site for GATA2, in ENCODE data (Figure 4). Finally, we found that this region looped to the HLA-F promoter located ∼14.5 kb upstream of rs2523393, suggesting that this region is an enhancer for HLA-F. (This region also looped to the HLA-G promoter.)
Figure 4.
The rs2523393 G/A Polymorphism Is Located in a Distal Enhancer of HLA-F
Location of the rs2523393 polymorphism (vertical red line) with respect to histone modifications that characterize promoters (H3K4me4 ChIP-seq) and enhancers (H3K27ac ChIP-seq), open chromatin (DNaseI-seq and FAIRE-seq), and GATA2 and NR2F2 ChIP-seq binding sites in human DSCs. ENCODE TFBSs are shown in gray, and intrachromosomal loop (PCHiC) data from LCLs is shown in purple.
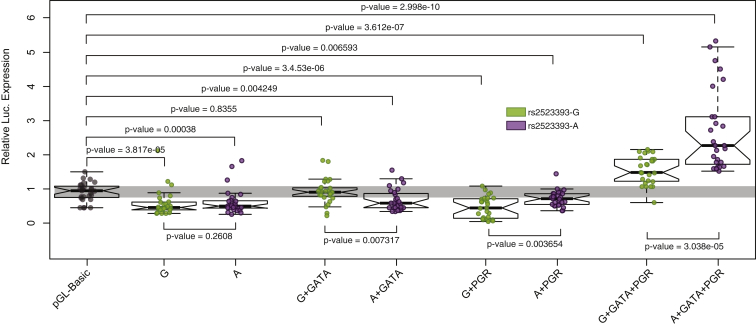
To test whether this locus has regulatory potential, we synthesized a 1,000 bp region spanning 500 bp upstream and downstream of rs2523393 (Figure 4) with either the reference A allele or the alternate G allele and cloned them into the pGL3-Basic luciferase reporter vector, which lacks an enhancer but encodes a minimal promoter. Next, we transiently transfected either the pGL3-Basic-rs2523393A or the pGL3-Basic-rs2523393G luciferase reporter along with the pRL-null internal control vector into DSCs and quantified luciferase and Renilla activity by using a dual luciferase assay. We found that luciferase activity was significantly lower in DSCs transfected with either pGL3-Basic-rs2523393A (Wilcox test p = 3.80 × 10−4) or pGL3-Basic-rs2523393G (Wilcox test p = 3.82 × 10−5) than in DSCs transfected with empty-vector pGL3-Basic controls (Figure 5). Next, we co-transfected pGL3-Basic-rs2523393A or pGL3-Basic-rs2523393G along with either a GATA2 expression vector or a PGR expression vector. Although co-transfection of neither the GATA2 expression vector nor the PGR expression vector enhanced luciferase expression above that of empty-vector controls, there were significant differences between alleles (Figure 5). Finally, we co-transfected pGL3-Basic-rs2523393A or pGL3-Basic-rs2523393G with both GATA2 and PGR expression vectors. Consistent with cooperative interaction between GATA2 and PGR, we observed significantly more luciferase expression from both the pGL3-Basic-rs2523393A (Wilcox test p = 3.00 × 10−10) and pGL3-Basic-rs2523393G (Wilcox test p = 3.61 × 10−7) vectors than from empty-vector controls. We also observed that the pGL3-Basic-rs2523393A vector drove significantly higher luciferase expression than the pGL3-Basic-rs2523393G vector (Wilcox test p = 3.65 × 10−3). Thus, we conclude that rs2523393 is located within a GATA2 binding site in a progesterone-responsive enhancer and that the G/A polymorphism affects enhancer function.
Figure 5.
The rs2523393 G/A Polymorphism Alters the Function of a Progesterone Response Enhancer
Luciferase assay results testing the regulatory potential of the G (pGL3Basic-rs2523393G) and A (pGL3Basic-rs2523393A) alleles in DSCs treated with cAMP and MPA for 48 hr. Data are shown as luciferase activity from the pGL3Basic-rs2523393G or pGL3Basic-rs2523393A reporter in relation to Renilla activity (pRL-null and empty-vector [pGL3Basic] controls). Abbreviations are as follows: +GATA, relative luciferase activity from pGL3Basic-rs2523393G or pGL3Basic-rs2523393A in DSCs co-transfected with GATA; +PGR, relative luciferase activity from pGL3Basic-rs2523393G or pGL3Basic-rs2523393A in DSCs co-transfected with PGR; +GATA+PGR, relative luciferase activity from pGL3Basic-rs2523393G or pGL3Basic-rs2523393A in DSCs co-transfected with GATA and PGR. n = 14; p values from Wilcox test.
The rs2523393 A Allele Is Derived in Humans
To reconstruct the evolutionary history of the G/A polymorphism, we identified a region spanning 50 bp upstream and downstream of rs2523393 from 37 primates, including species from each of the major primate lineages, multiple sub-species of African apes (Homininae), and modern and archaic humans (Altai Neanderthal and Denisova). Next, we used maximum-likelihood methods to reconstruct ancestral sequences for this 101 bp region. We found that the G allele was ancestral in primates and that the A variant was found only in Neanderthal, Denisovan, and modern human populations (Figure 6A). Next, we examined the frequency of these alleles across the HGDP populations and found that the derived and ancestral alleles segregated at intermediate to high frequencies in nearly all human populations (Figure 6B). These results indicate that the derived A variant at rs2523393, which creates a new GATA2 binding site in an enhancer of HLA-F expression, arose before the population split between archaic and modern humans between 550,000 and 765,000 years ago.55
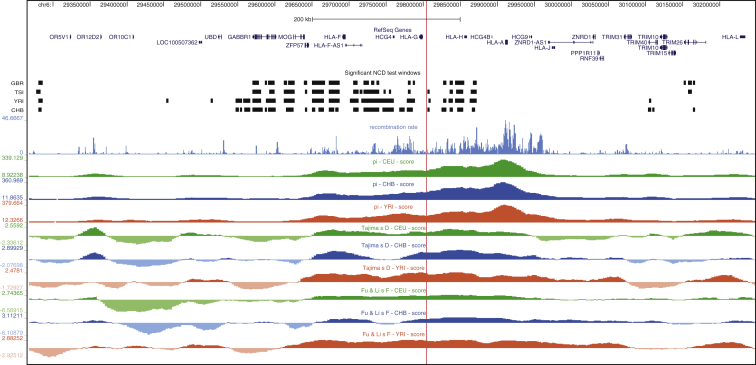
We previously found that a derived allele in an enhancer of TAP2 also arose before the divergence of archaic and modern humans and has signatures of long-term balancing selection, suggesting that the rs2523393 G/A polymorphism might also be the target of balancing selection. Long-term balancing selection leaves characteristic signatures in the genome, such as an increased local density of polymorphisms and an excess of alleles segregating at intermediate frequencies, which can be detected by methods such as Tajima’s D, Fu and Li’s F, and NCD tests.46 Consistent with the action of balancing selection, we found that the region encoding HLA-F has high nucleotide diversity and positive Tajima’s D and Fu and Li’s F values and was among the human genomic regions identified from the NCD test as under balancing selection (Figure 7). Indeed, the rs2523393 polymorphism is only 2 kb away from one of the significant regions (p < 0.0001) identified by the NCD test (Figure 7), suggesting that it is a target of balancing selection.
Figure 7.
Genomic Signatures of Balancing Selection at the HLA-F Locus
Recombination rate scores, nucleotide diversity (pi), Tajima’s D, and Fu and Li’s F statistics for 1000 Genomes CEU (Utah residents with northern and western European ancestry from the CEPH collection; green), CHB (Han Chinese in Beijing, China; blue), and YRI (Yoruba in Ibadan, Nigeria; red) populations are shown at the HLA-F encoding region of chromosome 6. The locations of windows with significant evidence of balancing selection from the NCD test are shown above for 1000 Genomes GBR (British in England and Scotland), TSI (Toscani in Italia), YRI, and CHB populations. The location of rs2523393 is shown with a vertical red line.
rs2523393 G/A Variants Are Associated with Disease
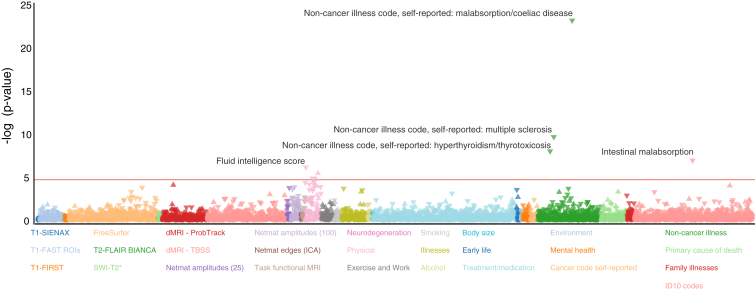
Our observation that the rs2523393 G/A polymorphism shows evidence of balancing selection suggests that processes such as frequency-dependent or antagonistic pleiotropy could maintain the polymorphism.56 Intriguingly, a previous genome-wide association study (GWAS) found that the rs2523393 A allele is associated with multiple sclerosis (MIM: 126200) with an odds ratio of 1.28 [1.18–1.39] (p = 1.04 × 10−17),57 suggesting that antagonistic pleiotropy maintains both alleles. To explore whether other diseases are associated with rs2523393, we took advantage of the UK Biobank GWAS results of ∼2,000 phenotypes (Figure 8). We found that the rs2523393 G allele is most significantly associated with malabsorption and celiac disease (p = 8.8 × 10−24, beta = −0.0016; MIM: 212750), multiple sclerosis (p = 2.7 × 10−10, beta = −0.0009; MIM: 126200), and hyperthyroidism and thyrotoxicosis (p = 1.2 × 10−8, beta = −0.0012; MIM: 188580).
Figure 8.
PheWAS Plot of Phenotypes Associated with rs2523393 in the UK Biobank
Associations are shown for 3,870 ICD10 (10th revision of the International Statistical Classification of Diseases and Related Health Problems) codes, ICD10 code groups are shown in color, associations with p < 0.05 are shown are triangles, and those with p ≥ 0.05 are shown as circles. The most statistically significant phenotype associations are labeled. The horizontal red line indicates the false-discovery-rate threshold.
Discussion
The mechanisms that underlie maternal tolerance of the antigenically distinct fetus are complex58 and have been the subject of intense study since Medawar proposed that tolerance is achieved by maternal immunosuppression and immaturity of fetal antigens.59 It is now clear that rather than being suppressed at the maternal-fetal interface, the immune system plays an active role in establishing a permissive environment for implantation, placentation, and gestation. Among the diverse immune cells that contribute to the successful establishment and maintenance of pregnancy are maternal regulatory T cells and γδ T cells,60, 61, 62 uNK cells,63, 64 uterine dendritic cells,65 uterine macrophage,66 and DSCs themselves.67, 68, 69 DSCs, for example, recruit uNK cells to the endometrium via IL-15 expression,70, 71, 72, 73, 74, 75, 76, 77, 78 stimulate the migration of macrophages into the endometrium via CSF1 expression,70, 71, 72, 79, 80 and suppress cytokine secretion by allogenic CD4+ T cells via PD-1 ligand interactions,68 which are all essential for establishing maternal-fetal immunotolerance. These data indicate that DSCs directly regulate local immune responses through multiple mechanisms, potentially also through HLA-F.
MHC genes play an important role in the rejection of non-self-tissues but also contribute to maternal tolerance of the fetus.1, 2, 3, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97 Although the precise functions of HLA-F are unclear, our observations that its expression level in the endometrium during the window of implantation is associated with fecundability14 and that it is upregulated by progesterone implicate it in the establishment of pregnancy. HLA-F binds LIR and KIR NK cell receptors,21, 22, 23, 24, 25 suggesting that HLA-F expressed by DSCs directly signals to uNK cells, which are essential for the establishment of maternal immunotolerance and spiral artery remodeling. Consistent with this hypothesis, HLA-F expression is positively correlated with uNK abundance in mid-luteal endometria and is predictive of achieving pregnancy.14, 26
Our observation that the G/A polymorphism is shared among modern humans, Neanderthals, and Denisovans indicates that it is relatively ancient and shows genetic signatures characteristic of balancing selection. We found that although the derived A allele creates a new GATA2 binding site and augments the function of a progesterone responsive enhancer of HLA-F, a previous GWAS found that it is also associated with multiple sclerosis,57 and the G polymorphism is also associated with malabsorption and celiac disease, multiple sclerosis, and hyperthyroidism and thyrotoxicosis in the UK Biobank. These data are consistent with maintenance of the polymorphism through antagonistic pleiotropy. We have previously shown that another fecundability-associated variant that switches a repressor into an enhancer of endometrial TAP2 (MIM: 170261) expression is also shared among modern humans, Neanderthals, and Denisovans.98 Remarkably, whereas the ancestral T allele in the TAP2 cis-regulatory element is associated with shorter time to pregnancy and ulcerative colitis,40 the derived C allele is associated with longer time to pregnancy and Crohn disease (MIM: 266600),41 suggesting that these alleles are also maintained by antagonistic pleiotropy. Collectively, these data raise the intriguing possibility that there is a reproduction-health tradeoff in recent human evolution.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
This work was funded by a Burroughs Wellcome Preterm Birth Initiative grant (1013760), an NIH National Institute of General Medical Sciences Graduate Training Grant (T32GM007197), and the March of Dimes Prematurity Research Center at the University of Chicago, Northwestern University, and Duke University (22-FY18-809).
Published: September 20, 2018
Footnotes
Supplemental Data include one table and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.08.009.
Accession Numbers
The accession numbers for the ChIP-seq and microarray data reported in this paper are SRA: SRX3503584 and GEO: GSE108409, respectively.
Web Resources
1000 Genomes Selection Browser 1.0, http://www.hsb.upf.edu
Capture HiC Plotter (CHiCP), https://www.chicp.org
Datamonkey, http://www.datamonkey.org
Geography of Genetic Variants Browser, http://popgen.uchicago.edu/ggv/
GTEx Portal, http://www.gtexportal.org/home/eqtls/bySnp
OMIM, http://www.omim.org
Oxford Brain Imaging Genetics (BIG) Server, http://big.stats.ox.ac.uk
TimeTree, http://www.timetree.org
UCSC Genome Browser, https://genome.ucsc.edu/
Supplemental Data
References
- 1.Kosova G., Abney M., Ober C. Colloquium papers: Heritability of reproductive fitness traits in a human population. Proc. Natl. Acad. Sci. USA. 2010;107(Suppl 1):1772–1778. doi: 10.1073/pnas.0906196106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Christensen K., Kohler H.P., Basso O., Olsen J., Vaupel J.W., Rodgers J.L. The correlation of fecundability among twins: Evidence of a genetic effect on fertility? Epidemiology. 2003;14:60–64. doi: 10.1097/00001648-200301000-00015. [DOI] [PubMed] [Google Scholar]
- 3.Tropf F.C., Lee S.H., Verweij R.M., Stulp G., van der Most P.J., de Vlaming R., Bakshi A., Briley D.A., Rahal C., Hellpap R. Hidden heritability due to heterogeneity across seven populations. Nat. Hum. Behav. 2017;1:757–765. doi: 10.1038/s41562-017-0195-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perry J.R.B., Day F., Elks C.E., Sulem P., Thompson D.J., Ferreira T., He C., Chasman D.I., Esko T., Thorleifsson G., Australian Ovarian Cancer Study. GENICA Network. kConFab. LifeLines Cohort Study. InterAct Consortium. Early Growth Genetics (EGG) Consortium Parent-of-origin-specific allelic associations among 106 genomic loci for age at menarche. Nature. 2014;514:92–97. doi: 10.1038/nature13545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He C., Kraft P., Chen C., Buring J.E., Paré G., Hankinson S.E., Chanock S.J., Ridker P.M., Hunter D.J., Chasman D.I. Genome-wide association studies identify loci associated with age at menarche and age at natural menopause. Nat. Genet. 2009;41:724–728. doi: 10.1038/ng.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elks C.E., Perry J.R., Sulem P., Chasman D.I., Franceschini N., He C., Lunetta K.L., Visser J.A., Byrne E.M., Cousminer D.L., GIANT Consortium Thirty new loci for age at menarche identified by a meta-analysis of genome-wide association studies. Nat. Genet. 2010;42:1077–1085. doi: 10.1038/ng.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mbarek H., Steinberg S., Nyholt D.R., Gordon S.D., Miller M.B., McRae A.F., Hottenga J.J., Day F.R., Willemsen G., de Geus E.J. Identification of common genetic variants influencing spontaneous dizygotic twinning and female fertility. Am. J. Hum. Genet. 2016;98:898–908. doi: 10.1016/j.ajhg.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stolk L., Perry J.R.B., Chasman D.I., He C., Mangino M., Sulem P., Barbalic M., Broer L., Byrne E.M., Ernst F., LifeLines Cohort Study Meta-analyses identify 13 loci associated with age at menopause and highlight DNA repair and immune pathways. Nat. Genet. 2012;44:260–268. doi: 10.1038/ng.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen C.T.L., Liu C.-T., Chen G.K., Andrews J.S., Arnold A.M., Dreyfus J., Franceschini N., Garcia M.E., Kerr K.F., Li G. Meta-analysis of loci associated with age at natural menopause in African-American women. Hum. Mol. Genet. 2014;23:3327–3342. doi: 10.1093/hmg/ddu041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruth K.S., Beaumont R.N., Tyrrell J., Jones S.E., Tuke M.A., Yaghootkar H., Wood A.R., Freathy R.M., Weedon M.N., Frayling T.M., Murray A. Genetic evidence that lower circulating FSH levels lengthen menstrual cycle, increase age at menopause and impact female reproductive health. Hum. Reprod. 2016;31:473–481. doi: 10.1093/humrep/dev318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang G., Feenstra B., Bacelis J., Liu X., Muglia L.M., Juodakis J., Miller D.E., Litterman N., Jiang P.-P., Russell L. Genetic associations with gestational duration and spontaneous preterm birth. N. Engl. J. Med. 2017;377:1156–1167. doi: 10.1056/NEJMoa1612665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rappoport N., Toung J., Hadley D., Wong R.J., Fujioka K., Reuter J., Abbott C.W., Oh S., Hu D., Eng C. A genome-wide association study identifies only two ancestry specific variants associated with spontaneous preterm birth. Sci. Rep. 2018;8:226. doi: 10.1038/s41598-017-18246-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fejzo M.S., Sazonova O.V., Sathirapongsasuti J.F., Hallgrímsdóttir I.B., Vacic V., MacGibbon K.W., Schoenberg F.P., Mancuso N., Slamon D.J., Mullin P.M., 23andMe Research Team Placenta and appetite genes GDF15 and IGFBP7 are associated with hyperemesis gravidarum. Nat. Commun. 2018;9:1178. doi: 10.1038/s41467-018-03258-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burrows C.K., Kosova G., Herman C., Patterson K., Hartmann K.E., Velez Edwards D.R., Stephenson M.D., Lynch V.J., Ober C. Expression quantitative trait locus mapping studies in mid-secretory phase endometrial cells identifies HLA-F and TAP2 as fecundability-associated genes. PLoS Genet. 2016;12:e1005858. doi: 10.1371/journal.pgen.1005858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishitani A., Sageshima N., Hatake K. The involvement of HLA-E and -F in pregnancy. J. Reprod. Immunol. 2006;69:101–113. doi: 10.1016/j.jri.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 16.Persson G., Melsted W.N., Nilsson L.L., Hviid T.V.F. HLA class Ib in pregnancy and pregnancy-related disorders. Immunogenetics. 2017;69:581–595. doi: 10.1007/s00251-017-0988-4. [DOI] [PubMed] [Google Scholar]
- 17.Hackmon R., Pinnaduwage L., Zhang J., Lye S.J., Geraghty D.E., Dunk C.E. Definitive class I human leukocyte antigen expression in gestational placentation: HLA-F, HLA-E, HLA-C, and HLA-G in extravillous trophoblast invasion on placentation, pregnancy, and parturition. Am. J. Reprod. Immunol. 2017;77:e12643. doi: 10.1111/aji.12643. [DOI] [PubMed] [Google Scholar]
- 18.Ishitani A., Sageshima N., Lee N., Dorofeeva N., Hatake K., Marquardt H., Geraghty D.E. Protein expression and peptide binding suggest unique and interacting functional roles for HLA-E, F, and G in maternal-placental immune recognition. J. Immunol. 2003;171:1376–1384. doi: 10.4049/jimmunol.171.3.1376. [DOI] [PubMed] [Google Scholar]
- 19.Nagamatsu T., Fujii T., Matsumoto J., Yamashita T., Kozuma S., Taketani Y. Human leukocyte antigen F protein is expressed in the extra-villous trophoblasts but not on the cell surface of them. Am. J. Reprod. Immunol. 2006;56:172–177. doi: 10.1111/j.1600-0897.2006.00414.x. [DOI] [PubMed] [Google Scholar]
- 20.Shobu T., Sageshima N., Tokui H., Omura M., Saito K., Nagatsuka Y., Nakanishi M., Hayashi Y., Hatake K., Ishitani A. The surface expression of HLA-F on decidual trophoblasts increases from mid to term gestation. J. Reprod. Immunol. 2006;72:18–32. doi: 10.1016/j.jri.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 21.Lepin E.J., Bastin J.M., Allan D.S., Roncador G., Braud V.M., Mason D.Y., van der Merwe P.A., McMichael A.J., Bell J.I., Powis S.H., O’Callaghan C.A. Functional characterization of HLA-F and binding of HLA-F tetramers to ILT2 and ILT4 receptors. Eur. J. Immunol. 2000;30:3552–3561. doi: 10.1002/1521-4141(200012)30:12<3552::AID-IMMU3552>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 22.Dulberger C.L., McMurtrey C.P., Hölzemer A., Neu K.E., Liu V., Steinbach A.M., Garcia-Beltran W.F., Sulak M., Jabri B., Lynch V.J. Human leukocyte antigen F presents peptides and regulates immunity through interactions with NK cell receptors. Immunity. 2017;46:1018–1029.e7. doi: 10.1016/j.immuni.2017.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goodridge J.P., Burian A., Lee N., Geraghty D.E. HLA-F and MHC class I open conformers are ligands for NK cell Ig-like receptors. J. Immunol. 2013;191:3553–3562. doi: 10.4049/jimmunol.1300081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia-Beltran W.F., Hölzemer A., Martrus G., Chung A.W., Pacheco Y., Simoneau C.R., Rucevic M., Lamothe-Molina P.A., Pertel T., Kim T.-E. Open conformers of HLA-F are high-affinity ligands of the activating NK-cell receptor KIR3DS1. Nat. Immunol. 2016;17:1067–1074. doi: 10.1038/ni.3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burian A., Wang K.L., Finton K.A.K., Lee N., Ishitani A., Strong R.K., Geraghty D.E. HLA-F and MHC-I open conformers bind natural killer cell Ig-like receptor KIR3DS1. PLoS ONE. 2016;11:e0163297. doi: 10.1371/journal.pone.0163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kofod L., Lindhard A., Bzorek M., Eriksen J.O., Larsen L.G., Hviid T.V.F. Endometrial immune markers are potential predictors of normal fertility and pregnancy after in vitro fertilization. Am. J. Reprod. Immunol. 2017;78 doi: 10.1111/aji.12684. Published online April 25, 2017. [DOI] [PubMed] [Google Scholar]
- 27.GTEx Consortium Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melé M., Ferreira P.G., Reverter F., DeLuca D.S., Monlong J., Sammeth M., Young T.R., Goldmann J.M., Pervouchine D.D., Sullivan T.J., GTEx Consortium Human genomics. The human transcriptome across tissues and individuals. Science. 2015;348:660–665. doi: 10.1126/science.aaa0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Talbi S., Hamilton A.E., Vo K.C., Tulac S., Overgaard M.T., Dosiou C., Le Shay N., Nezhat C.N., Kempson R., Lessey B.A. Molecular phenotyping of human endometrium distinguishes menstrual cycle phases and underlying biological processes in normo-ovulatory women. Endocrinology. 2006;147:1097–1121. doi: 10.1210/en.2005-1076. [DOI] [PubMed] [Google Scholar]
- 30.Lédée N., Munaut C., Aubert J., Sérazin V., Rahmati M., Chaouat G., Sandra O., Foidart J.M. Specific and extensive endometrial deregulation is present before conception in IVF/ICSI repeated implantation failures (IF) or recurrent miscarriages. J. Pathol. 2011;225:554–564. doi: 10.1002/path.2948. [DOI] [PubMed] [Google Scholar]
- 31.Davis S., Meltzer P.S. GEOquery: A bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23:1846–1847. doi: 10.1093/bioinformatics/btm254. [DOI] [PubMed] [Google Scholar]
- 32.Smyth G.K. limma: Linear models for microarray data. In: Gentleman R., Carey V.J., Huber W., Irizarry R.A., Dudoit S., editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer; 2005. pp. 397–420. [Google Scholar]
- 34.Tamura I., Ohkawa Y., Sato T., Suyama M., Jozaki K., Okada M., Lee L., Maekawa R., Asada H., Sato S. Genome-wide analysis of histone modifications in human endometrial stromal cells. Mol. Endocrinol. 2014;28:1656–1669. doi: 10.1210/me.2014-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lynch V.J., Nnamani M.C., Kapusta A., Brayer K., Plaza S.L., Mazur E.C., Emera D., Sheikh S.Z., Grützner F., Bauersachs S. Ancient transposable elements transformed the uterine regulatory landscape and transcriptome during the evolution of mammalian pregnancy. Cell Rep. 2015;10:551–561. doi: 10.1016/j.celrep.2014.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaya H.S., Hantak A.M., Stubbs L.J., Taylor R.N., Bagchi I.C., Bagchi M.K. Roles of progesterone receptor A and B isoforms during human endometrial decidualization. Mol. Endocrinol. 2015;29:882–895. doi: 10.1210/me.2014-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li X., Large M.J., Creighton C.J., Lanz R.B., Jeong J.-W., Young S.L., Lessey B.A., Palomino W.A., Tsai S.Y., Demayo F.J. COUP-TFII regulates human endometrial stromal genes involved in inflammation. Mol. Endocrinol. 2013;27:2041–2054. doi: 10.1210/me.2013-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ryan I.P., Schriock E.D., Taylor R.N. Isolation, characterization, and comparison of human endometrial and endometriosis cells in vitro. J. Clin. Endocrinol. Metab. 1994;78:642–649. doi: 10.1210/jcem.78.3.8126136. [DOI] [PubMed] [Google Scholar]
- 39.Tierney E.P., Tulac S., Huang S.T.J., Giudice L.C. Activation of the protein kinase A pathway in human endometrial stromal cells reveals sequential categorical gene regulation. Physiol. Genomics. 2003;16:47–66. doi: 10.1152/physiolgenomics.00066.2003. [DOI] [PubMed] [Google Scholar]
- 40.Anderson C.A., Boucher G., Lees C.W., Franke A., D’Amato M., Taylor K.D., Lee J.C., Goyette P., Imielinski M., Latiano A. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat. Genet. 2011;43:246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Franke A., McGovern D.P.B., Barrett J.C., Wang K., Radford-Smith G.L., Ahmad T., Lees C.W., Balschun T., Lee J., Roberts R. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat. Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Delport W., Poon A.F.Y., Frost S.D.W., Kosakovsky Pond S.L. Datamonkey 2010: A suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics. 2010;26:2455–2457. doi: 10.1093/bioinformatics/btq429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kosakovsky Pond S.L., Frost S.D. Not so different after all: A comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol. 2005;22:1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- 44.Hedges S.B., Marin J., Suleski M., Paymer M., Kumar S. Tree of life reveals clock-like speciation and diversification. Mol. Biol. Evol. 2015;32:835–845. doi: 10.1093/molbev/msv037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pybus M., Dall’Olio G.M., Luisi P., Uzkudun M., Carreño-Torres A., Pavlidis P., Laayouni H., Bertranpetit J., Engelken J. 1000 Genomes Selection Browser 1.0: A genome browser dedicated to signatures of natural selection in modern humans. Nucleic Acids Res. 2014;42:D903–D909. doi: 10.1093/nar/gkt1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bitarello B.D., de Filippo C., Teixeira J.C., Schmidt J.M., Kleinert P., Meyer D., Andrés A.M. Signatures of long-term balancing selection in human genomes. Genome Biol. Evol. 2018;10:939–955. doi: 10.1093/gbe/evy054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou J., Troyanskaya O.G. Predicting effects of noncoding variants with deep learning-based sequence model. Nat. Methods. 2015;12:931–934. doi: 10.1038/nmeth.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mathelier A., Fornes O., Arenillas D.J., Chen C.Y., Denay G., Lee J., Shi W., Shyr C., Tan G., Worsley-Hunt R. JASPAR 2016: A major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2016;44(D1):D110–D115. doi: 10.1093/nar/gkv1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.He B., Lanz R.B., Fiskus W., Geng C., Yi P., Hartig S.M., Rajapakshe K., Shou J., Wei L., Shah S.S. GATA2 facilitates steroid receptor coactivator recruitment to the androgen receptor complex. Proc. Natl. Acad. Sci. USA. 2014;111:18261–18266. doi: 10.1073/pnas.1421415111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rubel C.A., Franco H.L., Jeong J.W., Lydon J.P., DeMayo F.J. GATA2 is expressed at critical times in the mouse uterus during pregnancy. Gene Expr. Patterns. 2012;12:196–203. doi: 10.1016/j.gep.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 51.Rubel C.A., Franco H.L., Camper S.A., Lanz R.B., Jeong J.W., Lydon J.P., DeMayo F.J. Gata2 is a master regulator of endometrial function and progesterone signaling. Biol. Reprod. 2011;85(Issue Suppl_1):179. doi: 10.1093/biolreprod/85.s1.179. [DOI] [Google Scholar]
- 52.Blanco O., Tirado I., Muñoz-Fernández R., Abadía-Molina A.C., García-Pacheco J.M., Peña J., Olivares E.G. Human decidual stromal cells express HLA-G: Effects of cytokines and decidualization. Hum. Reprod. 2008;23:144–152. doi: 10.1093/humrep/dem326. [DOI] [PubMed] [Google Scholar]
- 53.Pabona J.M.P., Simmen F.A., Nikiforov M.A., Zhuang D., Shankar K., Velarde M.C., Zelenko Z., Giudice L.C., Simmen R.C.M. Krüppel-like factor 9 and progesterone receptor coregulation of decidualizing endometrial stromal cells: implications for the pathogenesis of endometriosis. J. Clin. Endocrinol. Metab. 2012;97:E376–E392. doi: 10.1210/jc.2011-2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mazur E.C., Vasquez Y.M., Li X., Kommagani R., Jiang L., Chen R., Lanz R.B., Kovanci E., Gibbons W.E., DeMayo F.J. Progesterone receptor transcriptome and cistrome in decidualized human endometrial stromal cells. Endocrinology. 2015;156:2239–2253. doi: 10.1210/en.2014-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meyer M., Arsuaga J.-L., de Filippo C., Nagel S., Aximu-Petri A., Nickel B., Martínez I., Gracia A., Bermúdez de Castro J.M., Carbonell E. Nuclear DNA sequences from the Middle Pleistocene Sima de los Huesos hominins. Nature. 2016;531:504–507. doi: 10.1038/nature17405. [DOI] [PubMed] [Google Scholar]
- 56.Carter A.J.R., Nguyen A.Q. Antagonistic pleiotropy as a widespread mechanism for the maintenance of polymorphic disease alleles. BMC Med. Genet. 2011;12:160. doi: 10.1186/1471-2350-12-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.De Jager P.L., Jia X., Wang J., de Bakker P.I.W., Ottoboni L., Aggarwal N.T., Piccio L., Raychaudhuri S., Tran D., Aubin C., International MS Genetics Consortium Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci. Nat. Genet. 2009;41:776–782. doi: 10.1038/ng.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Erlebacher A. Why isn’t the fetus rejected? Curr. Opin. Immunol. 2001;13:590–593. doi: 10.1016/s0952-7915(00)00264-8. [DOI] [PubMed] [Google Scholar]
- 59.Medawar P.B. Some immunological and endocrinological problems raised by the evolution of viviparity in vertebrates. Symp. Soc. Exp. Biol. 1953;7:320–338. [Google Scholar]
- 60.Erlebacher A. Mechanisms of T cell tolerance towards the allogeneic fetus. Nat. Rev. Immunol. 2013;13:23–33. doi: 10.1038/nri3361. [DOI] [PubMed] [Google Scholar]
- 61.Guerin L.R., Prins J.R., Robertson S.A. Regulatory T-cells and immune tolerance in pregnancy: A new target for infertility treatment? Hum. Reprod. Update. 2009;15:517–535. doi: 10.1093/humupd/dmp004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moffett A., Loke Y.W. The immunological paradox of pregnancy: A reappraisal. Placenta. 2004;25:1–8. doi: 10.1016/S0143-4004(03)00167-X. [DOI] [PubMed] [Google Scholar]
- 63.Tirado-González I., Barrientos G., Freitag N., Otto T., Thijssen V.L., Moschansky P., von Kwiatkowski P., Klapp B.F., Winterhager E., Bauersachs S., Blois S.M. Uterine NK cells are critical in shaping DC immunogenic functions compatible with pregnancy progression. PLoS ONE. 2012;7:e46755. doi: 10.1371/journal.pone.0046755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Felker A.M., Croy B.A. Uterine natural killer cell partnerships in early mouse decidua basalis. J. Leukoc. Biol. 2016;100:645–655. doi: 10.1189/jlb.1HI0515-226R. [DOI] [PubMed] [Google Scholar]
- 65.Tagliani E., Erlebacher A. Dendritic cell function at the maternal-fetal interface. Expert Rev. Clin. Immunol. 2011;7:593–602. doi: 10.1586/eci.11.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nagamatsu T., Schust D.J. The immunomodulatory roles of macrophages at the maternal-fetal interface. Reprod. Sci. 2010;17:209–218. doi: 10.1177/1933719109349962. [DOI] [PubMed] [Google Scholar]
- 67.Nancy P., Tagliani E., Tay C.S., Asp P., Levy D.E., Erlebacher A. Chemokine gene silencing in decidual stromal cells limits T cell access to the maternal-fetal interface. Science. 2012;336:1317–1321. doi: 10.1126/science.1220030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nagamatsu T., Schust D.J., Sugimoto J., Barrier B.F. Human decidual stromal cells suppress cytokine secretion by allogenic CD4+ T cells via PD-1 ligand interactions. Hum. Reprod. 2009;24:3160–3171. doi: 10.1093/humrep/dep308. [DOI] [PubMed] [Google Scholar]
- 69.Komatsu T., Konishi I., Mandai M., Mori T., Hiai H., Fukumoto M. Expression of class I human leukocyte antigen (HLA) and beta2-microglobulin is associated with decidualization of human endometrial stromal cells. Hum. Reprod. 1998;13:2246–2251. doi: 10.1093/humrep/13.8.2246. [DOI] [PubMed] [Google Scholar]
- 70.Dai X.-M., Zong X.-H., Sylvestre V., Stanley E.R. Incomplete restoration of colony-stimulating factor 1 (CSF-1) function in CSF-1-deficient Csf1op/Csf1op mice by transgenic expression of cell surface CSF-1. Blood. 2004;103:1114–1123. doi: 10.1182/blood-2003-08-2739. [DOI] [PubMed] [Google Scholar]
- 71.Ryan G.R., Dai X.M., Dominguez M.G., Tong W., Chuan F., Chisholm O., Russell R.G., Pollard J.W., Stanley E.R. Rescue of the colony-stimulating factor 1 (CSF-1)-nullizygous mouse (Csf1(op)/Csf1(op)) phenotype with a CSF-1 transgene and identification of sites of local CSF-1 synthesis. Blood. 2001;98:74–84. doi: 10.1182/blood.v98.1.74. [DOI] [PubMed] [Google Scholar]
- 72.Ovadia S., Insogna K., Yao G.-Q. The cell-surface isoform of colony stimulating factor 1 (CSF1) restores but does not completely normalize fecundity in CSF1-deficient mice. Biol. Reprod. 2006;74:331–336. doi: 10.1095/biolreprod.105.045047. [DOI] [PubMed] [Google Scholar]
- 73.Ashkar A.A., Black G.P., Wei Q., He H., Liang L., Head J.R., Croy B.A. Assessment of requirements for IL-15 and IFN regulatory factors in uterine NK cell differentiation and function during pregnancy. J. Immunol. 2003;171:2937–2944. doi: 10.4049/jimmunol.171.6.2937. [DOI] [PubMed] [Google Scholar]
- 74.Allavena P., Giardina G., Bianchi G., Mantovani A. IL-15 is chemotactic for natural killer cells and stimulates their adhesion to vascular endothelium. J. Leukoc. Biol. 1997;61:729–735. doi: 10.1002/jlb.61.6.729. [DOI] [PubMed] [Google Scholar]
- 75.Laskarin G., Strbo N., Bogovic Crncic T., Juretic K., Ledee Bataille N., Chaouat G., Rukavina D. Physiological role of IL-15 and IL-18 at the maternal-fetal interface. Chem. Immunol. Allergy. 2005;89:10–25. doi: 10.1159/000087906. [DOI] [PubMed] [Google Scholar]
- 76.Barber E.M., Pollard J.W. The uterine NK cell population requires IL-15 but these cells are not required for pregnancy nor the resolution of a Listeria monocytogenes infection. J. Immunol. 2003;171:37–46. doi: 10.4049/jimmunol.171.1.37. [DOI] [PubMed] [Google Scholar]
- 77.Kitaya K., Yamaguchi T., Honjo H. Central role of interleukin-15 in postovulatory recruitment of peripheral blood CD16(-) natural killer cells into human endometrium. J. Clin. Endocrinol. Metab. 2005;90:2932–2940. doi: 10.1210/jc.2004-2447. [DOI] [PubMed] [Google Scholar]
- 78.Gu L., Tseng S., Horner R.M., Tam C., Loda M., Rollins B.J. Control of TH2 polarization by the chemokine monocyte chemoattractant protein-1. Nature. 2000;404:407–411. doi: 10.1038/35006097. [DOI] [PubMed] [Google Scholar]
- 79.Erlebacher A. Immunology of the maternal-fetal interface. Annu. Rev. Immunol. 2013;31:387–411. doi: 10.1146/annurev-immunol-032712-100003. [DOI] [PubMed] [Google Scholar]
- 80.Svensson J., Jenmalm M.C., Matussek A., Geffers R., Berg G., Ernerudh J. Macrophages at the fetal-maternal interface express markers of alternative activation and are induced by M-CSF and IL-10. J. Immunol. 2011;187:3671–3682. doi: 10.4049/jimmunol.1100130. [DOI] [PubMed] [Google Scholar]
- 81.Fernandez N., Cooper J., Sprinks M., AbdElrahman M., Fiszer D., Kurpisz M., Dealtry G. A critical review of the role of the major histocompatibility complex in fertilization, preimplantation development and feto-maternal interactions. Hum. Reprod. Update. 1999;5:234–248. doi: 10.1093/humupd/5.3.234. [DOI] [PubMed] [Google Scholar]
- 82.Ober C., Elias S., O’Brien E., Kostyu D.D., Hauck W.W., Bombard A. HLA sharing and fertility in Hutterite couples: Evidence for prenatal selection against compatible fetuses. Am. J. Reprod. Immunol. Microbiol. 1988;18:111–115. doi: 10.1111/j.1600-0897.1988.tb00245.x. [DOI] [PubMed] [Google Scholar]
- 83.Ober C.L., Martin A.O., Simpson J.L., Hauck W.W., Amos D.B., Kostyu D.D., Fotino M., Allen F.H., Jr. Shared HLA antigens and reproductive performance among Hutterites. Am. J. Hum. Genet. 1983;35:994–1004. [PMC free article] [PubMed] [Google Scholar]
- 84.Ober C., Hyslop T., Elias S., Weitkamp L.R., Hauck W.W. Human leukocyte antigen matching and fetal loss: Results of a 10 year prospective study. Hum. Reprod. 1998;13:33–38. doi: 10.1093/humrep/13.1.33. [DOI] [PubMed] [Google Scholar]
- 85.Ober C., Elias S., Kostyu D.D., Hauck W.W. Decreased fecundability in Hutterite couples sharing HLA-DR. Am. J. Hum. Genet. 1992;50:6–14. [PMC free article] [PubMed] [Google Scholar]
- 86.Ober C., Aldrich C.L., Chervoneva I., Billstrand C., Rahimov F., Gray H.L., Hyslop T. Variation in the HLA-G promoter region influences miscarriage rates. Am. J. Hum. Genet. 2003;72:1425–1435. doi: 10.1086/375501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tan C.Y., Ho J.F.V., Chong Y.S., Loganath A., Chan Y.H., Ravichandran J., Lee C.G., Chong S.S. Paternal contribution of HLA-G∗0106 significantly increases risk for pre-eclampsia in multigravid pregnancies. Mol. Hum. Reprod. 2008;14:317–324. doi: 10.1093/molehr/gan013. [DOI] [PubMed] [Google Scholar]
- 88.Loisel D.A., Billstrand C., Murray K., Patterson K., Chaiworapongsa T., Romero R., Ober C. The maternal HLA-G 1597ΔC null mutation is associated with increased risk of pre-eclampsia and reduced HLA-G expression during pregnancy in African-American women. Mol. Hum. Reprod. 2013;19:144–152. doi: 10.1093/molehr/gas041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.O’Brien M., McCarthy T., Jenkins D., Paul P., Dausset J., Carosella E.D., Moreau P. Altered HLA-G transcription in pre-eclampsia is associated with allele specific inheritance: possible role of the HLA-G gene in susceptibility to the disease. Cell. Mol. Life Sci. 2001;58:1943–1949. doi: 10.1007/PL00000828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Larsen M.H., Hylenius S., Andersen A.M., Hviid T.V. The 3′-untranslated region of the HLA-G gene in relation to pre-eclampsia: Revisited. Tissue Antigens. 2010;75:253–261. doi: 10.1111/j.1399-0039.2009.01435.x. [DOI] [PubMed] [Google Scholar]
- 91.Moreau P., Contu L., Alba F., Lai S., Simoes R., Orrù S., Carcassi C., Roger M., Rabreau M., Carosella E.D. HLA-G gene polymorphism in human placentas: Possible association of G∗0106 allele with preeclampsia and miscarriage. Biol. Reprod. 2008;79:459–467. doi: 10.1095/biolreprod.108.068874. [DOI] [PubMed] [Google Scholar]
- 92.Yie S.M., Li L.H., Xiao R., Librach C.L. A single base-pair mutation in the 3′-untranslated region of HLA-G mRNA is associated with pre-eclampsia. Mol. Hum. Reprod. 2008;14:649–653. doi: 10.1093/molehr/gan059. [DOI] [PubMed] [Google Scholar]
- 93.Aldrich C.L., Stephenson M.D., Karrison T., Odem R.R., Branch D.W., Scott J.R., Schreiber J.R., Ober C. HLA-G genotypes and pregnancy outcome in couples with unexplained recurrent miscarriage. Mol. Hum. Reprod. 2001;7:1167–1172. doi: 10.1093/molehr/7.12.1167. [DOI] [PubMed] [Google Scholar]
- 94.Pfeiffer K.A., Fimmers R., Engels G., van der Ven H., van der Ven K. The HLA-G genotype is potentially associated with idiopathic recurrent spontaneous abortion. Mol. Hum. Reprod. 2001;7:373–378. doi: 10.1093/molehr/7.4.373. [DOI] [PubMed] [Google Scholar]
- 95.Suryanarayana V., Rao L., Kanakavalli M., Padmalatha V., Raseswari T., Deenadayal M., Singh L. Association between novel HLA-G genotypes and risk of recurrent miscarriages: A case-control study in a South Indian population. Reprod. Sci. 2008;15:817–824. doi: 10.1177/1933719107314061. [DOI] [PubMed] [Google Scholar]
- 96.Fan W., Li S., Huang Z., Chen Q. Relationship between HLA-G polymorphism and susceptibility to recurrent miscarriage: A meta-analysis of non-family-based studies. J. Assist. Reprod. Genet. 2014;31:173–184. doi: 10.1007/s10815-013-0155-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hylenius S., Andersen A.M., Melbye M., Hviid T.V.F. Association between HLA-G genotype and risk of pre-eclampsia: A case-control study using family triads. Mol. Hum. Reprod. 2004;10:237–246. doi: 10.1093/molehr/gah035. [DOI] [PubMed] [Google Scholar]
- 98.Mika K.M., Lynch V.J. An ancient fecundability-associated polymorphism switches a repressor into an enhancer of endometrial TAP2 expression. Am. J. Hum. Genet. 2016;99:1059–1071. doi: 10.1016/j.ajhg.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.