Abstract
A new cyclic hexapeptide, nocardiotide A (1), together with three known compounds—tryptophan (2), kynurenic acid (3), and 4-amino-3-methoxy benzoic acid (4)—were isolated and identified from the broth culture of Nocardiopsis sp. UR67 strain associated with the marine sponge Callyspongia sp. from the Red Sea. The structure elucidation of the isolated compounds were determined based on detailed spectroscopic data including 1D and 2D nuclear magnetic resonance (NMR) experimental analyses in combination with high resolution electrospray ionization mass spectrometry (HR-ESI-MS), while the absolute stereochemistry of all amino acids components of nocardiotide A (1) was deduced using Marfey’s method. Additionally, ten known metabolites were dereplicated using HR-ESI-MS analysis. Nocardiotide A (1) displayed significant cytotoxic effects towards the murine CT26 colon carcinoma, human HeLa cervix carcinoma, and human MM.1S multiple myeloma cell lines. The results obtained revealed sponge-associated Nocardiopsis as a substantial source of lead natural products with pronounced pharmacological activities.
Keywords: Nocardiopsis, cyclic hexapeptide, cytotoxicity, marine actinomycetes, sponges
1. Introduction
Actinomycetes are a diverse group of aerobic Gram-positive microorganisms withhigh guanine-cytosine DNA content [1]. They belong to the phylum Actinobacteria, which is one of the largest bacterial phyla, distributed in both terrestrial and marine ecosystems [2,3]. About 70% of all naturally derived drugs in clinical use originate from Actinobacteria as they contain biologically active secondary metabolites accounting for their clinical use, mainly as antibacterial, antifungal, antiviral, and cytotoxic drugs [4,5,6]. The genus Nocardiopsis was first described by Mayer in 1976 [7] as belonging to the family Nocardiopsaceae and as morphologically similar to members of the genera Actinomadura and Nocardia [7,8]. By reviewing the literature on the genus Nocardiopsis [9,10], it has been clearly demonstrated that it is a prolific producer of a wide variety of bioactive compounds, mainly cyclic peptides [11,12], polyketides [13,14], macrolides [15], alkaloids [16], diketopiperazines [17,18], α and γ-pyrones [19,20], naphthoquinones [21], phenazines [22], and phenoxazine derivatives [23], which contributes to a broad spectrum of biological activities, mainly as cytotoxic [21], anticancer [22], antitumor [24], antibacterial [11], antifungal [25], immunemodulatory [15],and protein kinase inhibitory [26].
Cancer still remains one of the most serious challenges to human health. Despite intense efforts to develop treatments, effective—particularly highly selective—agents are still not available for many cancer types. Therefore, it is necessary to continue the discovery of new classes of molecules with cytotoxic activity. One strategy to treat cancer is to find compounds with new scaffolds that have increased chances of possessing novel binding modes or even addressing novel targets. Consequently, this current investigation is a continuation of our efforts to seek new, effective cytotoxic agents from actinomycetes—associated with marine sponges, specifically, the Nocardiopsis sp. UR67 strain—and to evaluate their cytotoxic biological activities.
2. Results and Discussion
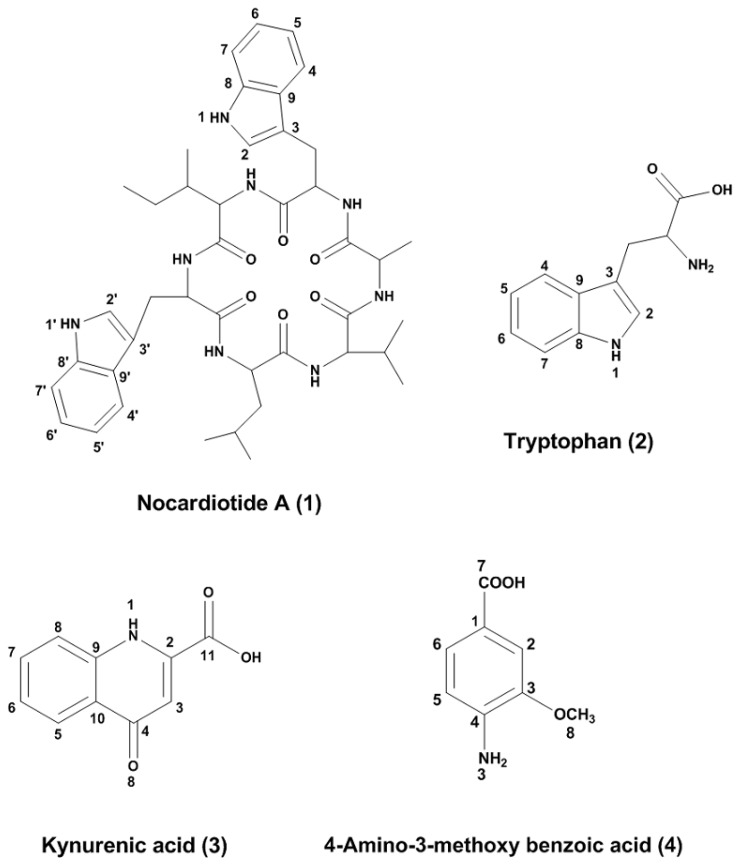
Nocardiopsis sp. UR67 was cultivated from the sponge Callyspongia sp. (family Callyspongiidae) that was collected from the Red Sea (Ras Mohamed, Sinai, Egypt; (GPS: 27°47.655′ N; 34°12.904′ W) in August 2008. ISP2 liquid broth with calcium alginate beads [27] of Nocardiopsis sp. UR67 was extracted with ethyl acetate, and the obtained organic extract was fractionated on Sephadex LH20. This was followed by purification using semi-preparative reversed phase high performance liquid chromatography (HPLC) to yield a new cyclic hexapeptide nocardiotide A (1), along with three known compounds—tryptophan (2), kynurenic acid (3), which was isolated for the first time from microbial origins, and 4-amino-3-methoxy benzoic acid (4) (Figure 1).
Figure 1.
Structures of the isolated compounds.
2.1. Metabolomic Analysis
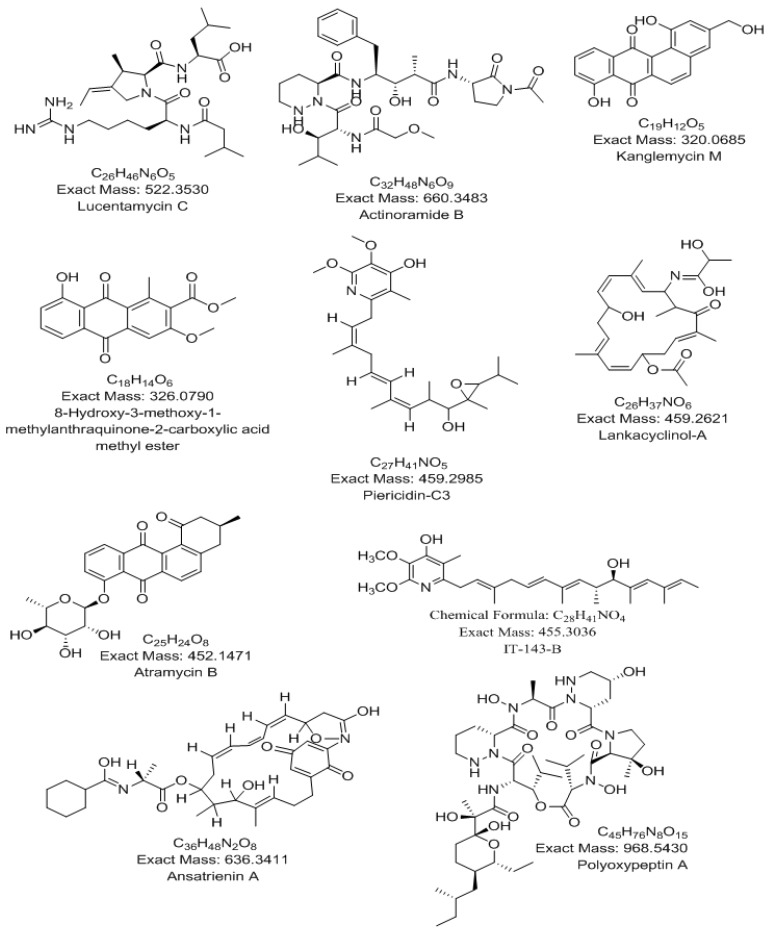
HPLC high resolution electrospray ionization mass spectrometry (HPLC-HR-ESIMS) analysis for dereplication purpose was used for identification of the metabolites from the ethyl acetate extract obtained from the culture broth of Nocardiopsis sp. UR67. The dereplication study of the metabolites (Figure 2, Table 1) against the Dictionary of Natural Products (DNP) database, AntiMarin, and METLIN databases led to the characterization of the following natural products: cytotoxic peptide lucentamycin C [28], immunosuppressant kanglemycin M [29], 8-hydroxy-3-methoxy-1-methyl-anthraquinone-2-carboxylic acid [30], antimicrobial, antitumor and insecticidal piericidin-C3 [31], sotetracenone-type antitumor atramycin B [32], piericidin group antibiotic IT-143-B [33], antibiotic lankacyclinol-A [34], antifungal polyketide ansatrienin A [35], actinoramide B [36] and, finally, a potent apoptosis inducer polyoxypeptin A [37].
Figure 2.
Dereplicated metabolites from metabolomic analysis of Nocardiopsis sp. UR67.
Table 1.
The dereplication results of the ethyl acetate fraction.
| Polarity | m/z | Rt (min.) | Formula | Name | Source |
|---|---|---|---|---|---|
| [M + H]+ | 327.0866 | 2.91 | C18H14O6 | 8-Hydroxy-3-methoxy-1-methylanthraquinone-2-carboxylic acid | Streptomyces sp. |
| [M − H]− | 967.5399 | 3.53 | C45H76N8O15 | Polyoxypeptin A | Streptomyces sp. MK498-98 F14 |
| [M − H]− | 635.3316 | 3.81 | C36H48N2O8 | Ansatrienin A | Streptomyces collinus |
| [M + H]+ | 321.0760 | 4.6 | C19H12O5 | Kanglemycin M | Nocardiamediterranei var. kanglensis 1747-64 |
| [M + H]+ | 661.3568 | 6.41 | C32H48N6O9 | Actinoramide B | Streptomyces ballenaensis and Streptomyces bangulaensis |
| [M + H]+ | 460.2697 | 6.95 | C26H37NO6 | Lankacyclinol-A | Streptomyces rochei var. volubilis |
| [M − H]− | 451.1392 | 7.24 | C25H24O8 | Atramycin B | Streptomyces atratus |
| [M − H]− | 458.2906 | 7.56 | C27H41NO5 | Piericidin-C3 | Streptomyces pactum |
| [M + H]+ | 456.3108 | 7.98 | C28H41NO4 | IT-143-B | Streptomyces species |
| [M + H]+ | 523.3601 | 11.23 | C26H46N6O5 | Lucentamycin C | Nocardiopsis lucentensis |
2.2. Structure Elucidation
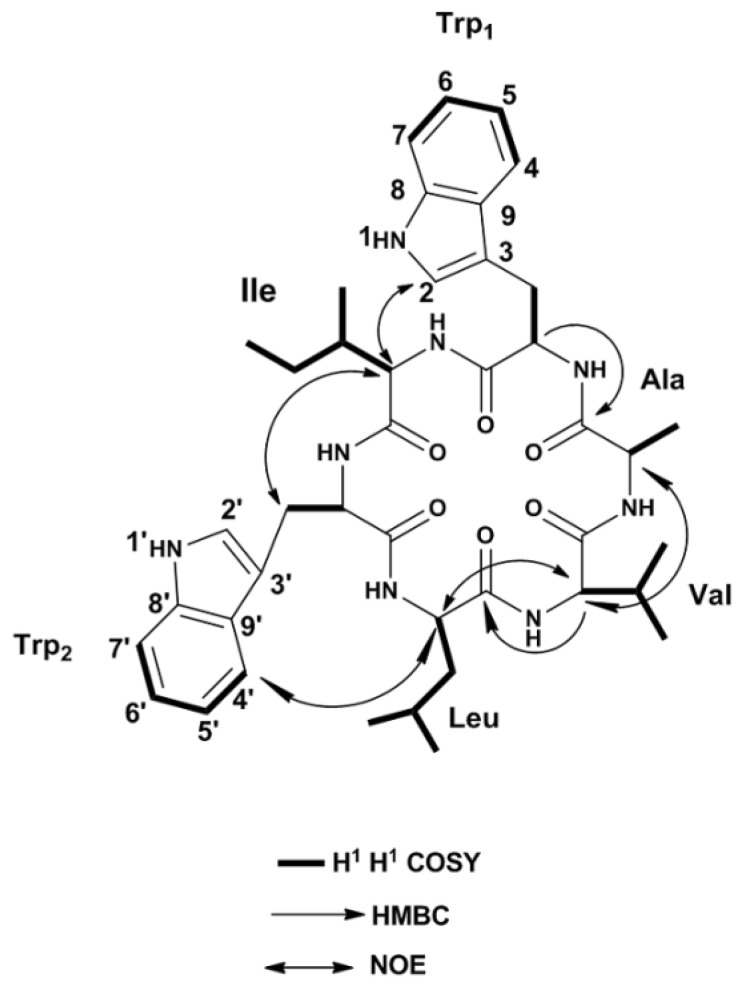
Nocardiotide A (1) was obtained as a pale yellow powder with a molecular formula of C42H56N8O6 determined by HR-ESI-MS analysis (m/z 791.931 [M + Na]+, calcd. for C42H56N8O6Na), indicating 19 degrees of unsaturation. The peptidic nature of nocardiotide A (1) was recognized from the 1H and 13C nuclear magnetic resonance (NMR) spectral data (Table 2). The 1H-NMR spectrum (Figures S1–S3) revealed the presence of six α-amino acid hydrogen resonances (δH 3.35–4.37). Additionally, the 13C NMR spectrum (Figure S4) contained six amide carbonyl signals resonating between δC 171 and 179 ppm and six α-amino acid carbon signals between δC 41 and 60 ppm, thus corroborating the presence of six amino acid moieties [38,39]. The 1H-NMR and COSY spectra (Figures S1 and S7) showed two distinct aromatic spin systems (δH 6.85–7.55), and the 13C-NMR and 13C-DEPT-135 spectrum (Figures S4 and S5) displayed ten methines and six quaternary carbons consistent with two tryptophan moieties. One tryptophan (Trp1) was assigned at δH 4.37 (dd, J = 3.8, 9.9 Hz, H1-α), 3.30 (dd, J = 14.7, 3.8 Hz, H1-β), 3.05 (dd, J = 14.7, 9.98 Hz, H1-β′), 7.03 (s, H1-2), 7.55 (dt, J = 7.84, 0.9 Hz, H1-4), 6.88 (td, overlapped, H1-5), 6.94 (td, overlapped, H1-6), 7.18 (dt, J = 8.11, 0.9 Hz, H1-7), and their corresponding carbons signals were assigned at δC 179.06 (CO1), 56.90 (C1-α), 29.19 (C1-β), 124.65 (C1-2), 112.29 (C1-3), 119.55 (C1-4), 119.44 (C1-5), 122.05 (C1-6), 112.04 (C1-7), 137.98 (C1-8), 128.92 (C1-9). The other tryptophan residue (Trp2) was assigned at δH 3.35 (t, J = 8.04 Hz, H2-α), 2.82 (td, J = 8.64, 0.9 Hz, H2-β), 6.96 (s, H2-2′), 7.45 (dt, J = 7.92, 0.9 Hz, H2-4′), 6.90 (td, overlapped, H2-5′), 6.98 (td, overlapped, H2-6′), 7.22 (dt, J = 8.17, 0.9 Hz, H2-7′), while their corresponding carbons signals were detected at δC 173.24 (CO2), 41.56 (C2-α), 26.21 (C2-β), 123.33 (C2-2′), 113.27 (C2-3′), 119.22 (C2-4′), 119.44 (C2-5′), 122.28 (C2-6′), 112.20 (C2-7′), 138.17 (C2-8′), 128.80 (C2-9′). These assignments were further confirmed by heteronuclear single quantum coherence (HSQC) and heteronuclear multiple bond correlation (HMBC) correlations (Figure 3) [40]. In addition to these two tryptophan moieties, the 13C-NMR, 13C-DEPT-135, and HSQC spectra (Figures S4–S6), displayed seven methyl, two methylene, and seven methine carbons. A spin system characteristic for isoleucine was observed at δH 4.20 (d, J = 3.9 Hz, H-α), 2.03 (m, H-β), 0.84 (d, J = 7.0 Hz, H-γ), 1.28 (m, H-γ′), and 0.85 (d, J = 7.4 Hz, H-δ), while their corresponding carbon signals were observed at δC 58.94 (C-α), 36.93 (C-β), 12.14 (C-γ), 27.49 (C-γ′), and 14.65 (C-δ), respectively. The previous assignments were corroborated using HSQC experiment (Figure S6). In addition, HMBC correlation (Figure S10) from α-proton of isoleucine (δH 4.20, d, J = 3.9 Hz) to its own amide-type carbonyl at δC 173.42 was detected. Furthermore, an alanine spin system was displayed at δH 4.31 (q, J = 7.2 Hz, H-α), and 1.17 (d, J = 7.2 Hz, H-β), and their corresponding carbons were observed at δC 49.77 (C-α) and 18.05 (C-β), respectively. The α and β protons of alaninemoiety (δH 4.31 and 1.17, respectively) showed a strong HMBC correlations with their amide-type carbonyl at δC 173.93. Additionally, a spin system for a leucine residue was observed at δH 3.81 (t, J = 7.3 Hz, H-α), 1.59 (m, H-β), 1.62 (m, H-γ), 0.92 (d, J = 6.2 Hz, H-δ), and 0.93 (d, J = 6.2 Hz, H-δ′), and their corresponding carbons were assigned at δC 52.97 (C-α), 41.95 (C-β), 25.60 (C-γ), 22.56 (C-δ), and 22.85 (C-δ′), respectively, using HSQC experiment correlation. Moreover, the α and β protons of leucine residue exhibited HMBC correlations with their amide-type carbonyl at δC 171.56. Finally, a valine moiety was detected from the spin system at δH 4.22 (d, J = 7.7 Hz, H-α), 1.97 (m, H-β), 0.88 (d, J = 3.1 Hz, H-γ), and 0.89 (d, J = 3.1 Hz, H-γ′), and their corresponding carbons were displayed at δC 60.76 (C-α), 31.75 (C-β), 19.79 (C-γ), and 18.95 (C-γ′), respectively. The amide-type carbonyl at δC 175.33 was attributed to the valine residue, which could be confirmed from the strong HMBC correlations observed amongst α and β protons of valine moiety [40]. Detailed analysis of the 1D (1H, 13C and DEPT-135) and 2D (HSQC, HMBC and NOE) NMR spectroscopic data (Table 2) revealed that nocardiotide A (1) was a hexapeptide containing Ile, Leu, Val, Ala, and two Trp residues. The amino acid sequence was elucidated to be Ile-Trp1-Ala-Val-Leu-Trp2 on the basis of the following HMBC correlations (Figures S10 and S11): α-Trp1 (δH 4.37)/Ala-CO (δC 173.9), α-Val (δH 4.22)/Leu-CO (δC 171.5) and the following NOE correlations (Figures S8 and S9): Trp1H-2(δH 7.02)/α-Ile (δH 4.20), α-Ala (δH 4.31)/α-Val (δH 4.22), α-Val (δH 4.22)/α-Leu (δH 3.81), α-Ile (δH 4.20)/β,β′ Trp2 (δH 2.82) and Trp2 H-4′ (δH 7.45)/α-Leu (δH 3.81) (Table 2, Figure 3) [38,41].
Table 2.
NMR-spectroscopic data of nocardiotide A (1) in MeOD-d4 (1H: 600 MHz; 13C: 150 MHz, δ in ppm, J in Hz).
| Aminoacids | δC | δH, mult (J in Hz) | COSY | HMBC | NOESY |
|---|---|---|---|---|---|
| Ile | |||||
| CO | 173.42 | ||||
| α | 58.94 | 4.20, d (4.6) | β | CO, β, γ′, δ | β-Trp2 |
| β | 36.93 | 2.03, m | α, γ, γ′ | γ′, δ | |
| γ | 12.14 | 0.84, d (7.0) | γ′, β | ||
| γ′ | 27.49 | 1.28, m | β, γ, δ | α, β, γ, δ | |
| δ | 14.65 | 0.85, d (7.4) | γ′ | α, β, γ′ | |
| Trp1 | |||||
| CO | 179.06 | ||||
| α | 56.90 | 4.37, dd (3.8, 9.9) | β, β′ | CO, β/β′, C-3, Ala-CO | |
| β | 29.19 | 3.05, dd (14.7, 9.9) | α | CO, α, C-2, C-3, C-8, C-9 | |
| β′ | 29.19 | 3.30, dd (14.7, 3.8) | α | ||
| 2 | 124.65 | 7.03 (s) | β/β′, C-3, C-8, C-9 | α-Ileu | |
| 3 | 112.29 | ||||
| 4 | 119.55 | 7.55, dt (7.84, 0.9) | H-5, H-6, H-7 | C-3, C-6, C-8, C-9 | |
| 5 | 119.44 | 6.88 m | H-4, H-6, H-7 | C-7, C-9 | |
| 6 | 122.05 | 6.94 (m) | H-4, H-5, H-7 | C-4, C-9 | |
| 7 | 112.04 | 7.18, dt (8.11, 0.9) | H-4, H-5, H-6 | C-5, C-9 | |
| 8 | 137.98 | ||||
| 9 | 128.92 | ||||
| Ala | |||||
| CO | 173.93 | ||||
| α | 49.77 | 4.31, q, (7.2) | β | CO, β | α-Val |
| β | 18.05 | 1.17, d, (7.2) | α | CO, α | |
| Val | |||||
| CO | 175.33 | ||||
| α | 60.76 | 4.22, d (7.7) | β | CO, Leu-CO, β, γ′ | α-Leu |
| β | 31.75 | 1.97, m | α, γ, γ′ | CO, α, γ | |
| γ | 19.79 | 0.88, d (3.1) | β | α, β, γ′ | |
| γ′ | 18.95 | 0.9, d (3.1) | β | α, β, γ | |
| Leu | |||||
| CO | 171.56 | ||||
| α | 52.97 | 3.81, t (7.3) | β | CO, β, γ | |
| β | 41.95 | 1.59, m | α, γ | CO, α, γ, δ, δ′ | |
| γ | 25.60 | 1.62, m | β, δ, δ′ | α, β, δ, δ′ | |
| δ | 22.56 | 0.92, d (6.2) | γ | β, γ, δ′ | |
| δ′ | 22.85 | 0.93, d (6.2) | γ | β, γ, δ | |
| Trp2 | |||||
| CO | 173.24 | ||||
| α | 41.56 | 3.35, t (8.04) | β/β’ | CO, β/β′, C-3′ | |
| β/β′ | 26.21 | 2.82, td (8.64, 0.9) | α | α, C-2′, C-3′, C-9′ | α-Ile |
| 2′ | 123.33 | 6.96, s | α, β/β’ | ||
| 3′ | 113.27 | ||||
| 4′ | 119.22 | 7.45, dt (7.92, 0.9) | H-5′, H-6′, H-7′ | C-3′, C-6′, C-8′, C-9′ | |
| 5′ | 119.44 | 6.90, m | H-4′, H-6′, H-7′ | C-7′, C-9′ | |
| 6′ | 122.28 | 6.98, m | H-4′, H-5′, H-7′ | C-4′, C-9′ | |
| 7′ | 112.20 | 7.22, dt (8.17, 0.9) | H-4′, H-’5, H-6′ | C-5′, C-9′ | |
| 8′ | 138.7 | ||||
| 9′ | 128.80 |
Figure 3.
Significant COSY, heteronuclear multiple bond correlation (HMBC), and NOE correlations of nocardiotide A (1).
These six amino acids accounted for18 degrees of unsaturation, indicating that nocardiotide A (1) was a monocyclic hexapeptide. The absolute configurations of the amino acid units in nocardiotide A (1) were determined by acid hydrolysis, followed by chiral derivatization with Marfey’s reagent (1-fluoro-2,4-dinitrophenyl-5-lalanine amide, FDAA). HPLC analysis of the Marfey’s derivatives in comparison to their respective d-and l-authentic reference amino acids revealed the absolute configuration of all amino acids of the new cyclic hexapeptideto beL.
Additionally, three known compounds—tryptophan (2), kynurenic acid (3), and 4-amino-3-methoxy benzoic acid (4) (Figure 1)—were also separated and could be identified by comparing their 1D and 2D NMR spectral analysis (Figures S12–S26) with the published data [42,43]. It is worth mentioning that kynurenic acid (3) was isolated for the first time from microbial origins.
2.3. Biological Activities of the Isolated Compounds
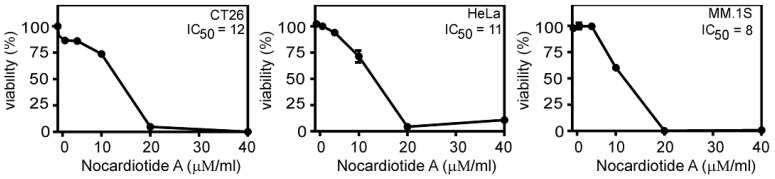
The four aforementioned isolated compounds were examined for their cytotoxicity potential towards the murine CT26 colon carcinoma, the human HeLa cervix carcinoma, and the human MM.1S multiple myeloma cell lines. Nocartiodite A (1) displayedprominent cytotoxic features with IC50 values of 8, 11, and 12 μM/mL against the human MM. 1S multiple myeloma, human HeLa cervix carcinoma, and murine CT26 colon carcinoma, respectively (Figure 4). Tryptophan (2), kynurenic acid (3), and4-amino-3-methoxy benzoic acid (4) did not demonstrate any considerable cell death properties at the examined concentration.
Figure 4.
Nocardiotide A induces cell death in CT26, HeLa, and MM.1S cell lines.
3. Materials and Methods
3.1. General Experimental Procedures
Melting points were measured using Stuart Scientific (SMPI) melting point apparatus and were uncorrected. An ultraviolet lamp (CAMAG, Wilmington, NC, USA) was used for visualization of spots on thin layer chromatograms at 254 and/or 365 nm. 1H (600 MHz) and 13C (150 MHz) NMR spectra were recorded on BrukerAvance III HD 600 instruments (Bruker Biospin, Rheinstetten, Germany) in CD3OD. The samples were degassed by an ultrasonic water bath (Branson 3800 Ultrasonic Cleaner, Branson, Gayton, UK) for 20 min before measurements. Solvent signals of CD3OD (δH 3.3 ppm and δC 49.0 ppm) were considered as the internal reference signals for calibration. Chemical shift values (δ) were recorded in ppm units and coupling constants (J) in Hz. Heteronuclear correlations were measured using HSQC (optimized for 1 JHC = 145 Hz) and HMBC (optimized for n JHC = 8.3 Hz or n JHC = 4.0 Hz) pulse sequences. Positive and negative HR-ESI-MS spectra were obtained using a Synapt G2 HDMS QTOF-mass spectrometer (Waters, Eschborn, Germany). HPLC separations and purifications were performed on the Knauer system (Knauer, Berlin, Germany). This included Smartline S-1000 quaternary pumps coupled with a Smartline S-2600 UV–VIS multiwavelength detector (Knauer, Berlin, Germany), a Knauer dynamic mixing chamber, and using a C18 column (5 µm, 10 mm × 250 mm, Knauer, Berlin, Germany) at ambient temperature with a guard column filled with the same stationary phase. On the other hand, the analytical detection was carried out using an analytical Gemini-NX RP-18 column (5 µm, 4.60 mm × 100 mm; Phenomenex, Aschaffenburg, Germany).
3.2. Sponge Collection
Callyspongia sp. (family Callyspongiidae) was collected at a depth of 10 m in the Red Sea (Ras Mohamed, Sinai, Egypt; (GPS: 27°47.655′ N; 34°12.904′ W) in August 2008. The collected sponge was transferred to plastic bags containing seawater and transported to the laboratory. The sponge was identified by R.W.M. van Soest (University of Amsterdam, Amsterdam, The Netherlands).
3.3. Isolation, Fermentation, and Extract Preparation of Nocardiopsis sp. UR67
Sponge specimens were rinsed in sterile seawater, cut into pieces of ca. 1 cm3, and then thoroughly homogenized in a sterile mortar with 10 volumes of sterile seawater. The supernatant was diluted in a tenfold series (10−1, 10−2, 10−3) and subsequently plated out on agar plates. Four different media—M1, ISP medium 2, Oligotrophic medium (OLIGO), and Marine Agar (MA)—were used for the isolation of actinobacteria. All media were supplemented with 0.2 µm pore size filtered cycloheximide (100 µg/mL), nystatin (25 µg/mL), and nalidixic acid (25 µg/mL) to facilitate the isolation of slow-growing actinobacteria; cycloheximide and nystatininhibit fungal growth, while nalidixic acid inhibits many fast-growing Gram-negative bacteria [44]. All media contained DifcoBacto agar (18 g/L) and were prepared in 1 L artificial sea water (NaCl 234.7 g, MgCl2·6 H2O 106.4 g, Na2SO4 39.2 g, CaCl2 11.0 g, NaHCO3 1.92 g, KCl 6.64 g, KBr 0.96 g, H3BO3 0.26 g, SrCl2 0.24 g, NaF 0.03 g, and ddH2O to 10.0 L). The inoculated plates were incubated at 30 °C for 6–8 weeks. Distinct colony morphotypes were picked and restreaked until visually free of contaminants. The isolates were maintained on plates for short-term storage and long-term strain collections. Nocardiopsis sp. UR67 was fermented in 10 Erlenmeyer flasks (2 L), each containing 1 L of ISP 2 (International Streptomyces Project) medium in artificial sea water and incubated at 30 °C for 10 days with shaking at 150 rpm. After fermentation and filtration, the supernatant was extracted with ethyl acetate (3 × 500 mL) to give the organic extract for subsequent compound isolation.
3.4. LC-HR/MS Analysis
Ethyl acetate extract of 1 mg/mL in MeOH was analyzed on an Accela HPLC (Thermo Scientific, Karlsruhe, Germany) coupled to a UV detector at 280 and 360 nm and an Exactive-Orbitrap high resolution mass spectrometer (Thermo Fisher Scientific, Karlsruhe, Germany). The HPLC column was an ACE (ACE, Mainz, Germany) C18, 75 mm × 3.0 mm, 5 μm column. The mobile phase consisted of purified water (A) and acetonitrile (B) with 0.1% formic acid in each solvent. The gradient program started with 10% B linearly increased to 100% B at a flow rate of 300 µL/min for 30 min and remained isocratic for 5 min before linearly decreasing back to 10% B in 1 min. The column was then re-equilibrated with 10% B for 9 min before the next injection. The total analysis time for each sample was 45 min. The injection volume was 10 µL, and the tray temperature was maintained at 12 °C. High resolution mass spectrometry was carried out in both positive and negative ESI ionization modes with a spray voltage at 4.5 kV and capillary temperature at 320 °C. The mass range was set from m/z 150–1500. Both negative and positive ionization switch modes were used to include the highest number of metabolites from the investigated bacterial fractions subjected to LC–HR-ESIMS analysis. The dereplication was achieved for each m/z ion peak with metabolites recorded in the customized databases based on established parameters (m/z threshold of ±3 ppm and retention time) [45], which provided a high level of confidence in metabolites identity; consequently, the number of the remaining unknown metabolites in each bacterial fraction was refined.
3.5. Metabolites Isolation
Theethyl acetateextract (5 g) was fractionated on a silica gel (250 g, 15–25 μm, 120 cm × 2.5 cm, Merck, Darmstadt, Germany) column and eluted with a DCM/MeOH gradient from (90:10%) to 100% methanol. The effluents were collected in fractions (50 mL each). Similar fractions monitored by TLC were combined and concentrated to yield six raw fractions. Fraction 2 (850 mg) was further fractionated bySephadex LH-20 column chromatography (50 g, 32–64 µm, 120 cm × 2 cm, Merck, Darmstadt, Germany) eluted with MeOH to yield five subfractions. Subfractions II and IV (compounds-rich subfractions) were further purified by semi-preparative HPLC using H2O/acetonitrile(90:10%) initially for 10 min, then by a linear gradient to 100% acetonitrilewithin 60 min, which was then followed by an isocratic elution at 100% acetonitrilefor a further 10 min with a flow rate of 2.0 mL/min using a C18 column (5 µm, 10 mm × 250 mm, Knauer, Berlin, Germany) to yield compound 1 (4 mg; Rt = 14.3 min) and compound 2 (3 mg; Rt = 16.5 min) from subfraction II as well as compound 3 (1 mg; Rt = 17.1 min) and compound 4 (2 mg; Rt = 17.9 min) from subfraction IV.
3.6. Marfey’s Analysis
The absolute configurations of the amino acids in compound 1 were elucidated by Marfey’s derivatization and compared to the corresponding standard amino acids each with D and L configurations (Sigma, Darmstadt, Germany) by HPLC. Compound 1 (1 mg) was initially hydrolyzed with 6 M HCl (2 mL) in a water bath at 100 °C for 24 h. The hydrolysate was cooled to room temperature, dried using a vacuum evaporator and dissolved in 100 μL of water. The Marfey’s derivatization was carried out by adding 100 μL of 1% Marfey’s reagent (1-fluoro-2,4-dinitrophenyl-5-l-alanine amid) dissolved in acetone and 20 μL of 1 M NaHCO3 (H2O) to 50 μL of the hydrolysate of compound 1 as well as 50 mM standard amino acid, respectively, and incubated at 40 °C for 1 h with frequent shaking. The reaction was stopped by adding 10 μL of 2 M HCl after cooling. The Marfey’s derivatization products were finally dried and prepared in MeOH for further HPLC analysis. The HPLC chromatography was carried out on Gemini-NX RP-C18 column by eluting with H2O/acetonitrile (95:5%) for the first 5 min, linearly gradient to 100% acetonitrile for 30 min, and staying at 100% actonitrile for a further 10 min with a flow rate at 1 mL/min and UV detection at 340 nm. The configuration was eventually determined with the observation of the same retention times compared to the standard enantiomeric amino acids [46,47,48]. Retention times (min) of authentic amino acids were as follows: l-Val (25.4), d-Val (27.3), l-Leu (21.9), d-Leu (22.2), l-Ala (18.7), d-Ala (20.3), l-Trp (27.3), d-Trp (29.6). A better resolution of the l-Ile, d-Ile derivatives was achieved using a linear gradient of acetonitrile in 0.1% (v/v) aqueous TFA (30–45% acetonitrile over 50 min): l-Ile (30.7), d-Ile (38.5).
3.7. Cytotoxic Activity
The cytotoxicity of the isolated compounds was evaluated in cell lines using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. CT26, HeLa, and MM.1S cells were maintained in RPMI medium (Merck, Darmstadt, Germany), supplemented with 10% fetal bovine serum (FBS), and grown at 37 °C and 5% CO2. HeLa cells (2 × 104 per well) were plated in 96-well tissue culture plates in 100 μ Lcell culture medium. The following day, cells were stimulated overnight in triplicates with the reagents of interest. Cell viability was assessed by crystal violet staining. In case of the CT26 and MM.1S cell lines, cells were seeded in 96-well plates (7× 104cells per well) and were challenged the same day overnight with the reagents of interest; the cytotoxic effect was evaluated using the MTT assay [49]. To normalize cell viability values, each plate included a triplicate of cells treated with the compound carrier DMSO to define 100% viable cells as well as a triplicate of cells incubated with a cytotoxic mixture (200 ng/ML Tumor Necrosis Factor TNF, 200 ng/mL CD95L (Fas ligand), 200 ng/mL TRAIL (TNF-related apoptosis-inducing ligand), 25 µg/mL CHX (Cycloheximide), 1% (w/v) sodium azide) to define maximal cell death and thus 0% viability. All other viability values were normalized according to the averages of these triplicates and analyzed by the Graph Pad Prism 5 software (La Jolla, CA, USA).
3.8. Compounds Characterization
3.8.1. (Nocardiotide A) (1)
Pale yellow solid (4 mg; Rt = 14.3 min) UV (EtOH) λ max 232, 305 nm; its 1H NMR (CD3OD, 600 MHz) and 13C NMR (CD3OD, 150 MHz) data aredetailed anddisplayed in Table 2.
3.8.2. (Tryptophan) (2)
Pale yellow crystalline solid (3 mg; Rt = 16.5 min) (m.p. 283–285 °C); 1H NMR (CD3OD, 600 MHz): δ 7.20 (1H, s, H-2), 7.61 (1H, dt, J = 8.0, 1.0 Hz, H-4), 7.06 (1H, td, J = 7.6, 1.0 Hz, H-5), 7.14 (1H, td, J = 7.6, 1.0 Hz, H-6), 7.38 (1H, dt, J = 8.0, 0.96 Hz, H-7), 3.50 (1H, dd, J = 4.47, 15.3 Hz, H-10a), 3.33 (obscured by solvent, H-10b), 4.23 (1H, dd, J = 4.8, 8.1 Hz, H11); 13C NMR (CD3OD, 150 MHz): δ 125.4 (CH, C-2), 108.0 (C, C-3), 117.0 (CH, C-4), 120.3 (CH, C-5), 123.0 (CH, C-6), 112.6 (CH, C-7), 138.4 (C, C-8), 128.4 (C, C-9), 27.7 (CH2, C-10), 54.7 (CH, C-11), 171.9 (C, C-12). The physical and spectral data were in accordance with those reported in the literature [42].
3.8.3. (Kynurenicacid) (3)
White amorphous powder; (1 mg; Rt = 17.1 min) 1H NMR (CD3OD, 600 MHz): δ 7.01 (1H, s, H-3), 8.26 (1H, ddd, J = 8.5, 1.4, 0.6 Hz, H-5), 7.47 (1H, td, J = 7.7, 1.4 Hz, H-6), 7.78 (1H, td, J = 7.7, 1.0 Hz, H-7), 7.88 (1H, ddd, J = 8.5, 1.0, 0.6 Hz, H-8); 13C NMR (CD3OD, 150 MHz): δ 143.3 (C, C-2), 110.3 (CH, C-3), 181.1 (C, C-4), 126.0 (CH, C-5), 126.0 (CH, C-6), 134.3 (CH, C-7), 120.4 (CH, C-8), 141.3 (C, C-9), 126.5 (C, C-10), 164.8 (C, C-11). The physical and spectral data were in accordance with those reported in the literature [43]. This is the first isolation of this compound from microbial origins.
3.8.4. (4-Amino3-methoxy benzoic acid) (4)
Pale yellow crystalline powder (2 mg; Rt = 17.9 min) (m.p. 185–187 °C); 1H NMR (CD3OD, 600 MHz): δ 7.61 (1H, d, J = 1.2 Hz, H-2), 6.79 (1H, d, J = 9.8 Hz, H-5), 7.53 (1H, dd, J = 9.8, 1.2 Hz, H-6); 13C NMR (CD3OD, 150 MHz): δ 127.4 (C, C-1), 112.5 (CH, C-2), 146.8 (C, C-3), 149.3 (C, C-4), 113.9 (CH, C-5), 123.1 (CH, C-6). The physical and spectral data were in accordance with those reported in the literature.
4. Conclusions
In continuation of our interest to isolate and identify new antiproliferative agents from natural sources, the chemical characterization of Nocardiopsis sp.UR67—an actinomycete associated with the sponge (Callyspongia sp.) previously collected from the Red Sea—was conducted alongside with evaluation of the cytotoxic properties of the attained compounds versus the murine CT26 colon carcinoma, the human HeLa cervix carcinoma, and the human MM.1S multiple myeloma cell lines. Ten known metabolites were identified by dereplication using LC-HR-ESI-MS techniques. Additionally, four compounds were isolated and characterized for the first time from the broth culture of Nocardiopsis sp. UR67. Most importantly, one new cyclic hexapeptide—nocardiotide A—was identified, along with tryptophan, kynurenic acid, and 4-amino-3-methoxy benzoic acid. Among them, only nocardiotide A demonstrated significant cytotoxic property.
Acknowledgments
We thank Arwa A. Makki for the fruitful discussion.
Supplementary Materials
The following are available online at http://www.mdpi.com/1660-3397/16/9/290/s1, Figures S1–S11: 1H-NMR, 13C-NMR, COSY, HSQC, NOESY and HMBC spectra of 1, Figures S12–S17: 1H-NMR, 13C-NMR, COSY, HSQC and HMBC spectra of 2, Figures S18–S21: 1H-NMR, 13C-NMR, COSY and HMBC spectra of 3, Figures S22–S26: 1H-NMR, 13C-NMR, COSY, HSQC and HMBC spectra of 4.
Author Contributions
Conceptualization, U.R.A.; Formal analysis, A.H.I., M.A.F. and U.R.A.; Investigation, E.Z.A., S.Y.D., M.A.A., D.H.; Supervision, M.S.K., M.A.F., S.Y.D. and U.R.A.; Writing—original draft, A.H.I., D.H., E.Z.A. and M.A.A.; Writing—review & editing, T.A.M.G., H.W. and U.R.A.
Funding
Research in the group of T.A.M.G. is generously funded by the DFG (GU 1233/1-1 and Center for Integrated Protein Science CIPSM). This work was supported by the German Research Foundation (DFG) and the Technical University of Munich (TUM) in the framework of the Open Access Publishing Program.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Yoshida A., Seo Y., Suzuki S., Nishino T., Kobayashi T., Hamada-Sato N., Kogure K., Imada C. Actinomycetal community structures in seawater and freshwater examined by DGGE analysis of 16S rRNA gene fragments. Mar. Biotechnol. 2008;10:554–563. doi: 10.1007/s10126-008-9092-y. [DOI] [PubMed] [Google Scholar]
- 2.Barka E.A., Vatsa P., Sanchez L., Gaveau-Vaillant N., Jacquard C., Klenk H.-P., Clément C., Ouhdouch Y., van Wezel G.P. Taxonomy, physiology, and natural products of Actinobacteria. Microbiol. Mol. Biol. Rev. 2016;80:1–43. doi: 10.1128/MMBR.00019-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abdelmohsen U.R., Balasubramanian S., Oelschlaeger T.A., Grkovic T., Pham N.B., Quinn R.J., Hentschel U. Potential of marine natural products against drug-resistant fungal, viral, and parasitic infections. Lancet Infect. Dis. 2017;17:e30–e41. doi: 10.1016/S1473-3099(16)30323-1. [DOI] [PubMed] [Google Scholar]
- 4.Manivasagan P., Kang K.-H., Sivakumar K., Li-Chan E.C., Oh H.-M., Kim S.-K. Marine actinobacteria: An important source of bioactive natural products. Environ. Toxicol. Pharmacol. 2014;38:172–188. doi: 10.1016/j.etap.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 5.Manivasagan P., Venkatesan J., Sivakumar K., Kim S.-K. Pharmaceutically active secondary metabolites of marine actinobacteria. Microbiol. Res. 2014;169:262–278. doi: 10.1016/j.micres.2013.07.014. [DOI] [PubMed] [Google Scholar]
- 6.Li J.W.-H., Vederas J.C. Drug discovery and natural products: End of an era or an endless frontier? Science. 2009;325:161–165. doi: 10.1126/science.1168243. [DOI] [PubMed] [Google Scholar]
- 7.Meyer J. Nocardiopsis, a new genus of the order Actinomycetales. Int. J. Syst. Evol. Microbiol. 1976;26:487–493. doi: 10.1099/00207713-26-4-487. [DOI] [Google Scholar]
- 8.Rainey F.A., Ward-Rainey N., Kroppenstedt R.M., Stackebrandt E. The genus Nocardiopsis represents a phylogenetically coherent taxon and a distinct actinomycete lineage: Proposal of Nocardiopsaceae fam. nov. Int. J. Syst. Evol. Microbiol. 1996;46:1088–1092. doi: 10.1099/00207713-46-4-1088. [DOI] [PubMed] [Google Scholar]
- 9.Bennur T., Ravi Kumar A., Zinjarde S., Javdekar V. Nocardiopsis species: A potential source of bioactive compounds. J. Appl. Microbiol. 2015;120:1–16. doi: 10.1111/jam.12950. [DOI] [PubMed] [Google Scholar]
- 10.Ibrahim A.H., Desoukey S.Y., Fouad M.A., Kamel M.S., Gulder T.A., Abdelmohsen U.R. Natural Product Potential of the Genus Nocardiopsis. Mar. Drugs. 2018;16:147. doi: 10.3390/md16050147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engelhardt K., Degnes K.F., Kemmler M., Bredholt H., Fjærvik E., Klinkenberg G., Sletta H., Ellingsen T.E., Zotchev S.B. Production of a new thiopeptide antibiotic, TP-1161, by a marine Nocardiopsis species. Appl. Environ. Microbiol. 2010;76:4969–4976. doi: 10.1128/AEM.00741-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shin J., Seo Y., Lee H.-S., Rho J.-R., Mo S.J. A new cyclic peptide from a marine-derived bacterium of the genus Nocardiopsis. J. Nat. Prod. 2003;66:883–884. doi: 10.1021/np030075r. [DOI] [PubMed] [Google Scholar]
- 13.Raju R., Piggott A.M., Quezada M., Capon R.J. Nocardiopsins C and D and nocardiopyroneA: Newpolyketides from an Australian marine-derived Nocardiopsis sp. Tetrahedron. 2013;69:692–698. doi: 10.1016/j.tet.2012.10.104. [DOI] [Google Scholar]
- 14.Dashti Y., Grkovic T., Abdelmohsen U.R., Hentschel U., Quinn R.J. Actinomycete Metabolome Induction/Suppression with N-Acetylglucosamine. J. Nat. Prod. 2017;80:828–836. doi: 10.1021/acs.jnatprod.6b00673. [DOI] [PubMed] [Google Scholar]
- 15.Raju R., Piggott A.M., Conte M., Tnimov Z., Alexandrov K., Capon R.J. Nocardiopsins: New FKBP12-Binding Macrolide Polyketides from an Australian Marine-Derived Actinomycete, Nocardiopsis sp. Chem. Eur. J. 2010;16:3194–3200. doi: 10.1002/chem.200902933. [DOI] [PubMed] [Google Scholar]
- 16.Tian S., Yang Y., Liu K., Xiong Z., Xu L., Zhao L. Antimicrobial metabolites from a novel halophilic actinomycete Nocardiopsis terrae YIM 90022. Nat. Prod. Res. 2014;28:344–346. doi: 10.1080/14786419.2013.858341. [DOI] [PubMed] [Google Scholar]
- 17.Raju R., Piggott A.M., Huang X.-C., Capon R.J. Nocardioazines: A novel bridged diketopiperazine scaffold from a marine-derived bacterium inhibits P-glycoprotein. Org. Lett. 2011;13:2770–2773. doi: 10.1021/ol200904v. [DOI] [PubMed] [Google Scholar]
- 18.Fu P., Liu P., Qu H., Wang Y., Chen D., Wang H., Li J., Zhu W. α-Pyrones and diketopiperazine derivatives from the marine-derived actinomycete Nocardiopsis dassonvillei HR10-5. J. Nat. Prod. 2011;74:2219–2223. doi: 10.1021/np200597m. [DOI] [PubMed] [Google Scholar]
- 19.Kim Y., Ogura H., Akasaka K., Oikawa T., Matsuura N., Imada C., Yasuda H., Igarashi Y. Nocapyrones: α-and γ-Pyrones from a Marine-Derived Nocardiopsis sp. Mar. Drugs. 2014;12:4110–4125. doi: 10.3390/md12074110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim M.C., Kwon O.-W., Park J.-S., Kim S.Y., Kwon H.C. Nocapyrones, H–J, 3, 6-disubstituted α-pyrones from the marine actinomycete Nocardiopsis sp. KMF-001. Chem. Pharm. Bull. 2013;61:511–515. doi: 10.1248/cpb.c12-00956. [DOI] [PubMed] [Google Scholar]
- 21.Ding Z.-G., Zhao J.-Y., Li M.-G., Huang R., Li Q.-M., Cui X.-L., Zhu H.-J., Wen M.-L. Griseusins F and G spiro-naphthoquinones from a tin mine tailings-derived alkalophilic Nocardiopsis species. J. Nat. Prod. 2012;75:1994–1998. doi: 10.1021/np3004936. [DOI] [PubMed] [Google Scholar]
- 22.Gao X., Lu Y., Xing Y., Ma Y., Lu J., Bao W., Wang Y., Xi T. A novel anticancer and antifungusphenazine derivative from a marine actinomycete BM-17. Microbiol. Res. 2012;167:616–622. doi: 10.1016/j.micres.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 23.Lu C., Li Y., Wang H., Wang B., Shen Y. A new phenoxazine derivative isolated from marine sediment actinomycetes, Nocardiopsis sp. 236. Drug Discov. Ther. 2013;7:101–104. [PubMed] [Google Scholar]
- 24.He J., Roemer E., Lange C., Huang X., Maier A., Kelter G., Jiang Y., Xu L.-H., Menzel K.-D., Grabley S., et al. Structure, derivatization, and antitumor activity of new griseusins from Nocardiopsis sp. J. Med. Chem. 2007;50:5168–5175. doi: 10.1021/jm070170n. [DOI] [PubMed] [Google Scholar]
- 25.Tian S.-Z., Pu X., Luo G., Zhao L.-X., Xu L.-H., Li W.-J., Luo Y. Isolation and characterization of new p-terphenyls with antifungal, antibacterial, and antioxidant activities from halophilic actinomycete Nocardiopsis gilva YIM 90087. J. Agric. Food Chem. 2013;61:3006–3012. doi: 10.1021/jf400718w. [DOI] [PubMed] [Google Scholar]
- 26.Kase H., Iwahashi K., Matsuda Y. K-252a, a potent inhibitor of protein kinase C from microbial origin. J. Antibiot. 1986;39:1059–1065. doi: 10.7164/antibiotics.39.1059. [DOI] [PubMed] [Google Scholar]
- 27.Grkovic T., Abdelmohsen U.R., Othman E.M., Stopper H., Edrada-Ebel R., Hentschel U., Quinn R.J. Two new antioxidant actinosporin analogues from the calcium alginate beads culture of sponge-associated Actinokineospora sp. strain EG49. Bioorg. Med. Chem. Lett. 2014;24:5089–5092. doi: 10.1016/j.bmcl.2014.08.068. [DOI] [PubMed] [Google Scholar]
- 28.Cho J.Y., Williams P.G., Kwon H.C., Jensen P.R., Fenical W. Lucentamycins A–D, cytotoxic peptides from the marine-derived actinomycete Nocardiopsis lucentensis. J. Nat. Prod. 2007;70:1321–1328. doi: 10.1021/np070101b. [DOI] [PubMed] [Google Scholar]
- 29.Zhou J., Sun C., Wang N., Gao R., Bai S., Zheng H., You X., Li R. Preliminary report on the biological effects of space flight on the producing strain of a new immunosuppressant, Kanglemycin C. J. Ind. Microbiol. Biotechnol. 2006;33:707–712. doi: 10.1007/s10295-006-0118-z. [DOI] [PubMed] [Google Scholar]
- 30.Poumale H.M., Ngadjui B.T., Helmke E., Laatscha H. New anthraquinones from a marine Streptomyces sp.—isolation, structure determination and biological activities. Z. Naturforsch. B. 2006;61:1450–1454. doi: 10.1515/znb-2006-1122. [DOI] [Google Scholar]
- 31.Zhou X., Fenical W. The unique chemistry and biology of the piericidins. J. Antibiot. (Tokyo) 2016;69:582–593. doi: 10.1038/ja.2016.71. [DOI] [PubMed] [Google Scholar]
- 32.Fujioka K., Furihata K., Shimazu A., Hayakawa Y., Seto H. Isolation and characterization of atramycin A and atramycin B, new isotetracenone type antitumor antibiotics. J. Antibiot. (Tokyo) 1991;44:1025–1028. doi: 10.7164/antibiotics.44.1025. [DOI] [PubMed] [Google Scholar]
- 33.Urakawa A., Sasaki T., Yoshida K., Otani T., Lei Y., Yun W. IT-143-A and B, novel piericidin-group antibiotics produced by Streptomyces sp. J. Antibiot. (Tokyo) 1996;49:1052–1055. doi: 10.7164/antibiotics.49.1052. [DOI] [PubMed] [Google Scholar]
- 34.Harada S. Studies on lankacidin-group (T-2636) antibiotics. VI. Chemical structures of lankacidin-group antibiotics. II. Chem. Pharm. Bull. (Tokyo) 1975;23:2201–2210. doi: 10.1248/cpb.23.2201. [DOI] [PubMed] [Google Scholar]
- 35.Patton S.M., Cropp T.A., Reynolds K.A. A novel delta(3),delta(2)-enoyl-CoA isomerase involved in the biosynthesis of the cyclohexanecarboxylic acid-derived moiety of the Polyketideans atrienin A. Biochemistry. 2000;39:7595–7604. doi: 10.1021/bi0005714. [DOI] [PubMed] [Google Scholar]
- 36.Cheng K.C., Cao S., Raveh A., MacArthur R., Dranchak P., Chlipala G., Okoneski M.T., Guha R., Eastman R.T., Yuan J., et al. Actinoramide A Identified as a Potent Antimalarial from Titration-Based Screening of Marine Natural Product Extracts. J. Nat. Prod. 2015;78:2411–2422. doi: 10.1021/acs.jnatprod.5b00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Du Y., Wang Y., Huang T., Tao M., Deng Z., Lin S. Identification and characterization of the biosynthetic gene cluster of polyoxypeptin A, a potent apoptosis inducer. BMC Microbiol. 2014;14:30. doi: 10.1186/1471-2180-14-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wiese J., Abdelmohsen U.R., Motiei A., Humeida U.H., Imhoff J.F. Bacicyclin, a new antibacterial cyclic hexapeptide from Bacillus sp. strain BC028 isolated from Mytilusedulis. Bioorg. Med. Chem. Lett. 2018;28:558–561. doi: 10.1016/j.bmcl.2018.01.062. [DOI] [PubMed] [Google Scholar]
- 39.Ibrahim S.R., Edrada-Ebel R., Mohamed G.A., Youssef D.T., Wray V., Proksch P. Callyaerin G, a new cytotoxic cyclic peptide from the marine sponge Callyspongia aerizusa. Arkivoc. 2008;2008:164. [Google Scholar]
- 40.Wu Z.-C., Li S., Nam S.-J., Liu Z., Zhang C. Nocardiamides A and B, two cyclohexapeptides from the marine-derived actinomycete Nocardiopsis sp. CNX037. J. Nat. Prod. 2013;76:694–701. doi: 10.1021/np400009a. [DOI] [PubMed] [Google Scholar]
- 41.Hsieh P.-W., Chang F.-R., Wu C.-C., Wu K.-Y., Li C.-M., Chen S.-L., Wu Y.-C. New cytotoxic cyclic peptides and dianthramide from Dianthus superbus. J. Nat. Prod. 2004;67:1522–1527. doi: 10.1021/np040036v. [DOI] [PubMed] [Google Scholar]
- 42.Al-Khalil S., Alkofahi A., El-Eisawi D., Al-Shibib A. Transitorine, a new quinoline alkaloid from Ephedra transitoria. J. Nat. Prod. 1998;61:262–263. doi: 10.1021/np9702998. Correction in 1999, 62, 1214. [DOI] [PubMed] [Google Scholar]
- 43.Elsayed Y., Refaat J., Abdelmohsen U.R., Ahmed S., Fouad M.A. Rhodozepinone, a new antitrypanosomal azepino-diindole alkaloid from the marine sponge-derived bacterium Rhodococcus sp. UA13. Med. Chem. Res. 2017;26:2751–2760. doi: 10.1007/s00044-017-1974-y. [DOI] [Google Scholar]
- 44.Webster N.S., Wilson K.J., Blackall L.L., Hill R.T. Phylogenetic diversity of bacteria associated with the marine sponge Rhopaloeidesod orabile. Appl. Environ. Microbiol. 2001;67:434–444. doi: 10.1128/AEM.67.1.434-444.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tawfike A.F., Tate R., Abbott G., Young L., Viegelmann C., Schumacher M., Diederich M., Edrada-Ebel R. Metabolomic Tools to Assess the Chemistry and Bioactivity of Endophytic Aspergillus Strain. Chem. Biodivers. 2017;14:e1700040. doi: 10.1002/cbdv.201700040. [DOI] [PubMed] [Google Scholar]
- 46.Bhushan R., Brückner H. Marfey’s reagent for chiral amino acid analysis: A review. Amino Acids. 2004;27:231–247. doi: 10.1007/s00726-004-0118-0. [DOI] [PubMed] [Google Scholar]
- 47.Kochhar S., Christen P. Amino acid analysis by high-performance liquid chromatography after derivatization with 1-fluoro-2, 4-dinitrophenyl-5-l-alanine amide. Anal. Biochem. 1989;178:17–21. doi: 10.1016/0003-2697(89)90348-5. [DOI] [PubMed] [Google Scholar]
- 48.Marfey P. Determination of d-amino acids. II. Use of a bifunctional reagent, 1,5-difluoro-2,4-dinitrobenzene. Carlsberg Res. Commun. 1984;49:591. doi: 10.1007/BF02908688. [DOI] [Google Scholar]
- 49.Cheng C., Othman E.M., Stopper H., Edrada-Ebel R., Hentschel U., Abdelmohsen U.R. Isolation of Petrocidin A, a New Cytotoxic Cyclic Dipeptide from the Marine Sponge-Derived Bacterium Streptomyces sp. SBT348. Mar. Drugs. 2017;15:383. doi: 10.3390/md15120383. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.