Abstract
The classification of genetic variants represents a major challenge in the post-genome era by virtue of their extraordinary number and the complexities associated with ascribing a clinical impact, especially for disorders exhibiting exceptional phenotypic, genetic, and allelic heterogeneity. To address this challenge for hearing loss, we have developed the Deafness Variation Database (DVD), a comprehensive, open-access resource that integrates all available genetic, genomic, and clinical data together with expert curation to generate a single classification for each variant in 152 genes implicated in syndromic and non-syndromic deafness. We evaluate 876,139 variants and classify them as pathogenic or likely pathogenic (more than 8,100 variants), benign or likely benign (more than 172,000 variants), or of uncertain significance (more than 695,000 variants); 1,270 variants are re-categorized based on expert curation and in 300 instances, the change is of medical significance and impacts clinical care. We show that more than 96% of coding variants are rare and novel and that pathogenicity is driven by minor allele frequency thresholds, variant effect, and protein domain. The mutational landscape we define shows complex gene-specific variability, making an understanding of these nuances foundational for improved accuracy in variant interpretation in order to enhance clinical decision making and improve our understanding of deafness biology.
Keywords: deafness, genetic variant, variant classification, database, genomic landscape, mutational signature, precision medicine
Introduction
Genomic technologies have revolutionized medicine in the post-genome era by offering the promise of personalized, precision healthcare based on DNA sequencing.1 Prior to and immediately after the completion of the human genome project, the primary bottleneck in advancing precision medicine was generating DNA sequencing and genetic variant data. With the advent of massively parallel sequencing technologies, the bottleneck shifted to clinically meaningful variant interpretation that is comprehensive, easily understandable, free from contradictory categorization, curated by experts, and freely available to the public. Guidelines developed by the American College of Medical Genetics and Genomics (ACMG) aid classification using a structured framework that defines 28 evidence codes by which to score a variant. There are 20 rules for combining codes to reach one of five conclusions that predict variant effect: pathogenic (P), likely pathogenic (LP), variant of uncertain significance (VUS), likely benign (LB), or benign (B).2, 3
The challenging and dynamic process of variant interpretation has spurred the creation of two major variant databases—ClinVar4 and the Human Gene Mutation Database (HGMD)5—to catalog the rapidly increasing volume of reported genetic variants. ClinVar is a freely accessible, public database that archives reports of the relationships between variations and phenotypes with varying degrees of supporting evidence. HGMD, a pay-for-access service, is a comprehensive reference database of published germline mutations that are associated with human inherited diseases based on curation of published literature.6 These databases are invaluable resources but because of their broad all-encompassing design, are not disease specific.
Hearing loss is the most common sensory deficit in humans. It affects an estimated 5% of the world’s population (360 million individuals) and in developed countries is most frequently genetic, segregating in a Mendelian fashion in the case of non-syndromic hearing loss (NSHL) or as a complex genetic disease in the case of age-related hearing loss.7 Its clinical evaluation has been facilitated by the use of comprehensive genetic testing with massively parallel sequencing, which has evolved to become the most informative test in the diagnostic evaluation of the hearing-impaired person. A positive diagnosis is made in more than 40% of persons who undergo this type of testing, and to date more than 6,000 mutations in more than 150 genes have been causally implicated in deafness.8 As the number of genes implicated in NSHL has continued to increase, we sought to provide a freely and continually updated comprehensive database to inform variant classification for deafness.
Called the Deafness Variation Database (DVD, see Web Resources), this resource is collated from major public databases. It provides a single classification for each variant based on collected evidence and is curated by experts in genetic hearing loss to provide a single-source guide to variant interpretation. By capitalizing on the wealth of data the DVD provides to assess the genomic and mutational landscape of deafness, we provide a deeper understanding of hereditary hearing loss and the molecular mechanisms at play.
Material and Methods
Gene Selection
The DVD v.8.1 includes 152 genes and microRNAs known to cause hearing loss-related phenotypes including NSHL, NSHL mimics such as Usher, Perrault, and Pendred syndromes (PDS [MIM: 274600]), and common forms of syndromic hearing loss like Alström (ALMS [MIM: 203800]), branchio-oto-renal (BOR1 [MIM: 113650], BOR2 [MIM: 610896]), Jervell and Lange-Nielsen (JLNS1 [MIM: 220400], JLNS1 [MIM: 612347]), and Wolfram (WFS1 [MIM: 222300], WFS2 [MIM: 604928]) syndromes (Table S1). The genes are curated from the Hereditary Hearing Loss Homepage and published literature after careful review of the supporting evidence including the strength of the genetic data (linkage data, allele frequency and deletriousness of the candidate variant, segregation analysis) and functional data (gene expression in inner ear, in vivo experiments, animal models). The gene list is regularly updated by adding or removing genes based on newly published data.
Annotation Collection
Data for the DVD are collected, combined, filtered, and analyzed using a custom-built internal computational pipeline we have developed called Kafeen. The pipeline was built using the Ruby programming language and integrates open-source and freely available bioinformatics utilities including BCFtools and BEDtools.9, 10 Variants are collected and annotated from multiple data sources including the 1000 Genomes Project (phase 3.5a),11 Exome Sequencing Project (ESP) (ESP6500SI-V2-SSA137 release) (see Web Resources), Exome Aggregation Consortium (ExAC) (v.0.3),12 HGMD (2015.2 release),5 ClinVar (2016-03-02 release),4 dbSNP 146,13 and our manual curations (Figure 1A). Additional annotations for pathogenicity prediction algorithms are collected from dbNSFP (v.3.0a).14, 15 Transcripts on which the variant has the most deleterious impact are selected. When all transcripts are equally impacted (e.g., the variant is missense in all of them), then the canonical transcript is selected. All tab-delimited files are converted to VCF. All VCF sources are further left-aligned, normalized, quality filtered, and de-duplicated before input to the pipeline. The pipeline is extensible to incorporate additional variant and annotation sources as they become available. Copy number variants (CNVs) are not included in the DVD.
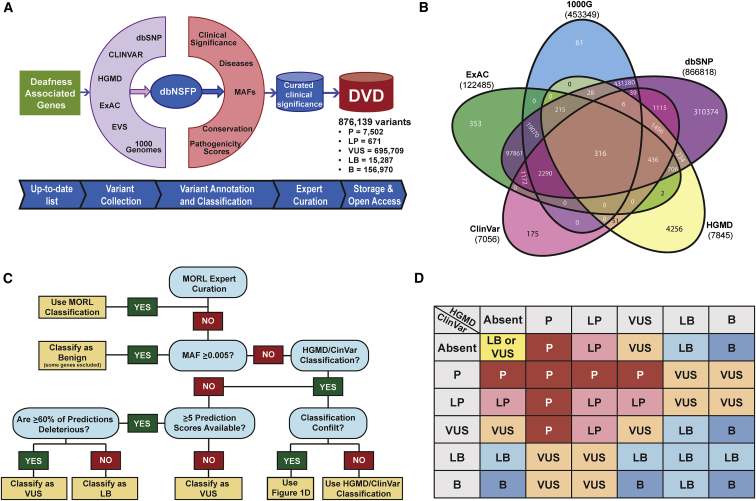
Figure 1.
The Deafness Variation Database
(A) Kafeen, a custom internal pipeline, gathers data for the DVD by collecting variants and annotations from multiple data sources. Deleteriousness predictions collected from dbNSFP and MAF data are extracted from our local database, EVS, 1000 Genomes, and ExAC to inform the DVD classification. A comparison between DVD versus ClinVar and HGMD classifications captures all changes that result in medically significant differences (defined as up-grading a variant to P/LP or down-grading a variant from P/LP), each of which is manually curated to ensure that the DVD reclassification is appropriate.
(B) Venn diagram showing number of variants collated from major population-scale MAF databases and the count of variants that are shared among them and those that are database specific.
(C) Decision tree for Kafeen classification.
(D) Decision matrix detailing Kafeen logic regarding variants classified in ClinVar and HGMD.
Scoring System and Interpretation
Available annotations are utilized to make an informed interpretation about the pathogenicity of each variant. MAF data from 1000 Genomes, ESP, ExAC, and our in-house database are used to determine whether a variant is too common to be considered pathogenic (Figure 1B). When considering multiple MAF annotations for a variant across databases and populations, we select the highest population-specific MAF to use in our computational evaluation. As a general rule, we use a MAF threshold of 0.5%,16 with the exception of select variants in specific genes (i.e., GJB2 [MIM: 121011]) (Table S2). A minimum of 400 alleles in the population with the highest MAF is required to use this classification threshold.
We consider a MAF ≥ 0.5 to be inconsistent with P/LP for NSHL and implement this MAF cutoff in our pipeline. Any variant with a MAF ≥ 0.5% is automatically classified as B (Figure 1C), although it remains important to know whether other databases classify common variants as P/LP (Figure 1D). To capture this information, we append an asterisk (∗) to a B classification for common variants that are classified as P/LP by other databases. For example, if a variant has been classified as P in ClinVar but has a MAF ≥ 0.5% in any population, the DVD classification is B∗.
If no population-based MAF meets or exceeds the defined cutoff, the pipeline uses variant classification data from ClinVar and HGMD (Figures 1C and 1D). ClinVar provides user-submitted data, and a single variant can have multiple and conflicting classifications with varying degree of supporting evidence. In such cases, we select the most deleterious ClinVar classification as it carries the highest clinical significance for individuals with hearing loss. When ClinVar and HGMD classifications are concordant, the DVD uses that classification. For ClinVar-HGMD discrepancies, the DVD default classification is based on the level of discordance (Figure 1D). If the variant is reported in only one database, that classification is used for the DVD.
For a variant with MAF < 0.5% that is absent from both ClinVar and HGMD, the DVD relies on functional prediction annotations to classify the variant as either LB or VUS (Figure 1). Our pipeline currently supports two evolutionary conservation algorithms (phyloP17 and GERP++18) and four deleteriousness prediction algorithms (SIFT,19 PolyPhen-2,20 MutationTaster,21 and LRT22). The DVD calculates a composite pathogenicity score (PS), assigning 1 point for each conserved and damaging prediction to make a final prediction of either VUS or LB. When multiple scores for the same prediction or conservation algorithm are provided, DVD selects the most damaging prediction from the set to consider in its algorithms. The LB classification is warranted if a variant has ≤40% pathogenic predictions and at least 5 algorithms make a prediction. In all other instances, the variant is computationally classified as a VUS (Figure 1C).
Classification Metrics
To validate this scoring metric and threshold, we tested all known deafness pathogenic variants with MAF < 0.5% and at least 5 algorithms calls (Figure S1). In calculation of performance of Kafeen variant classification prediction, we make the distinction between two classes of variants: positive as pathogenic classification and negative as benign classification. Within these classes, we consider the subclasses: true positive as predicted (PS ≥ 60%) and classified pathogenic variants; true negative as predicted (PS ≤ 40%) and classified benign variants; false positive as predicted pathogenic but classified benign variants; and false negative as predicted benign but classified pathogenic variants. Then, in calculating the traditional binary classification metrics of sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV): sensitivity = true positive / all positive; specificity = true negative / all negative; PPV = true positive / (true positive + false positive); NPV = true negative / (true negative + false negative).
Implementation, Manual Curation, and Override
The DVD was implemented in our internal NGS pipeline, which we use to generate a clinical report for each subject evaluated with our targeted gene panel OtoSCOPE.8 A multidisciplinary expert panel, including clinicians, geneticists, scientists, bioinformaticians, and genetic counselors, reviews all genetic results in the context of available phenotypic data. When the expert panel does not agree with the variant classification in the DVD, the variant is added to an internal list of manually curated variants with the revised classification. This list is continually updated and integrated back into the DVD to prevent the propagation of an incorrect variant classification. The manually curated list includes pathogenic variants that have been identified exclusively in our screen of more than 5,000 individuals with hearing loss, as these variants were not found in other public databases.8, 23, 24 It also includes known pathogenic variants with MAFs ≥ 0.5% (founder mutations and variants in specific genes, see Table S2), which have been deemed exempt from the MAF restriction.
Versioning the DVD
To keep the DVD up to date, we regularly update it by adding newly discovered deafness-associated genes and the most recent versions of all input data sources. Testing and validation of each new DVD version is performed via a comparison between the newest dataset and the previous dataset to capture all variants that have undergone a major reclassification in pathogenicity (any changes from or to P and LP) resulting in medically significant differences (P/LP versus VUS/LB/B). We evaluate these variants to ensure that the reclassification is appropriate. If upon further review we do not agree with the reclassification, we preserve the previous classification.
Results
The Deafness Variation Database
The DVD classifies and interprets variants in 152 genes and microRNAs implicated in genetic hearing loss. The included genes are associated with a variety of hearing loss-related phenotypes including NSHL, NSHL mimics, and common forms of syndromic hearing loss (Table S1). 876,139 genetic variants in these genes were extracted from dbSNP, ExAC, 1000 Genomes, ESP, ClinVar, HGMD, and our internal manual curation database.8, 23, 24 All variants were annotated for MAF (from large-scale population databases), variant effect (intronic, UTR, splice-site, missense, nonsense, synonymous, inframe indels, frameshift indels, start loss, stop loss), deleteriousness predictions (dbNSFP), and classification (ClinVar, HGMD, our internal manual curation) (Figure 1).
All available data were used to classify variants computationally, with supplemental expert manual curation as detailed in the Material and Methods (Figure 1). We integrated predictions from six algorithms—two assessing conservation (PhyloP and GERP++) and four evaluating deleteriousness (SIFT, PolyPhen-2, MutationTaster, and LRT)—to calculate a composite pathogenicity score (PS) and annotate variants with MAF < 0.5%. Variants with MAFs above this threshold were automatically classified as benign with the exception of known common founder mutations (Figure 1C, Table S2).16 To validate the PS, we plotted all variants classified as pathogenic by MAF and PS (Figure S1) and found that of 3,591 pathogenic variants with predictions from at least five pathogenicity prediction tools, 95.4% have a composite PS ≥ 60%. The calculated sensitivity, specificity, PPV, and NPV were 0.95, 0.51, 0.74, and 0.88, respectively. We used this threshold for variant classification, labeling variants with a MAF < 0.5 and a PS ≤ 40%, based on at least five pathogenicity predictions, as LB.
In aggregate, DVD v.8.1 reports 876,139 variants from 152 genes and microRNAs. Of these variants, 7,502 (0.85%) are classified as P, 671 (0.077%) are LP, 15,287 (1.74%) are LB, 156,970 (17.9%) are B, and 695,709 (79.4%) are VUSs (Figure 2A). To assess only medically relevant variants for deafness, we considered only the 97,007 variants within coding and splice-site regions (exons as defined by RefSeq and Ensembl coding transcripts, ±20 bp from exon boundaries, 3′ and 5′ UTRs, and any deep intronic variant classified as P or LP) as these regions are routinely screened in clinical diagnostics settings. We also considered any variant that is P or LP for a phenotype other than deafness as a VUS for the purpose of this analysis. For example, 20 P/LP variants have been reported in MET (MIM: 164860), but only one has been linked to hearing loss. Of 97,007 variants we considered, 6.2% were P (6,045), 0.5% were LP (445), 14.2% were LB (13,823), 4.8% were B (4,628), and 74.3% were VUSs (72,066) (Figure 2B).
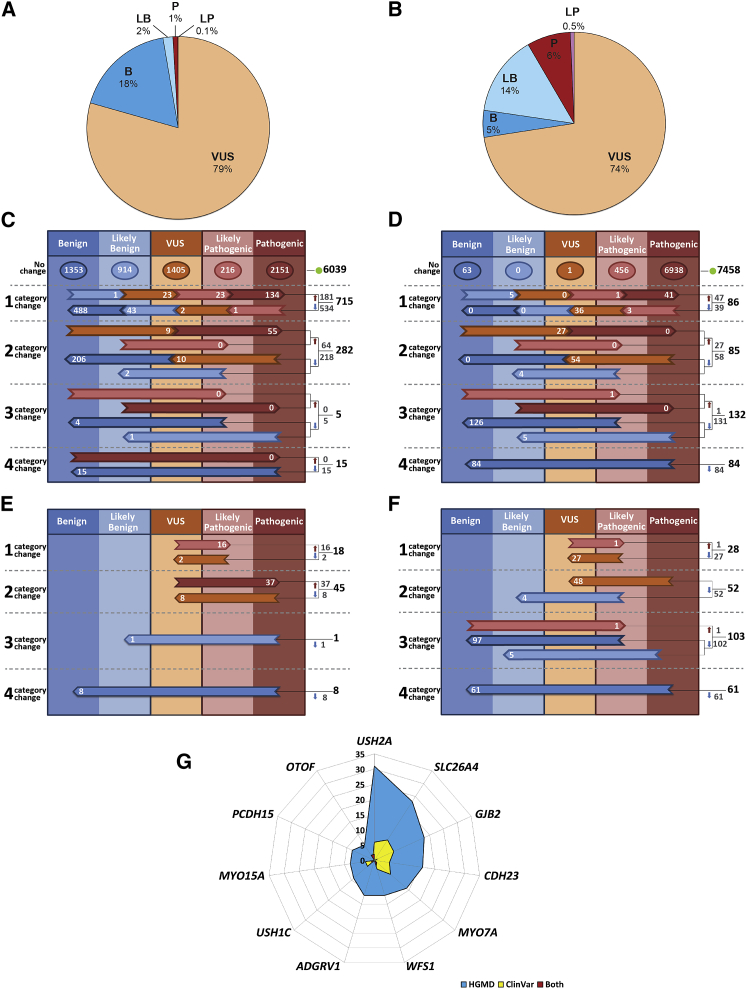
Figure 2.
Variant Classification by the DVD
(A) Fractions of different classification categories for variants in the whole DVD.
(B) A slightly different picture emerges when only clinically relevant regions and deafness-associated variants (variants that were associated with other non-related deafness phenotypes are excluded) are considered.
(C) Comparative overview of DVD versus ClinVar. 7,056 classifications from ClinVar were identified within our specified gene regions (each variant in ClinVar with multiple submissions for pathogenicity has been represented by its most pathogenic submission). Of this number, 6,039 ClinVar classifications agreed with the corresponding DVD classification whereas there was disagreement for 1,017 variants.
(D) Comparative overview of DVD versus HGMD. 7,845 classifications from HGMD were identified within our specified gene regions. Of this number, 7,458 classifications agreed with the corresponding DVD classification and discrepancies were found for 387 variants.
(E) There were 72 major categorical changes between ClinVar and DVD that resulted in medically significant differences (53 up-classifications and 19 down-classifications).
(F) 244 medically significant reclassifications were found when DVD was compared to HGMD (2 up-classifications and 242 down-classifications).
(G) Of the 20% of genes carrying the greatest numbers of medically significant changes, 6 are implicated in Usher syndrome.
For (C) through (F), the horizontal arrows show discordant calls, with the number of discordant classifications shown within each arrow; totals are listed to the right of the colored columns.
Computational and Expert Manual Curation Led to Medically Significant Changes in Pathogenicity
To assess differences in variant interpretation between the DVD and ClinVar and HGMD, we compared the number of downgraded (from more severe to more benign) and upgraded (from more benign to more severe) classifications (Figures 2C–2F). Of the variants listed in the DVD, 7,056 are found in ClinVar (filtered to represent each variant by only its most pathogenic classification). Of these variants, 175 are unique to ClinVar (Figure 1B). There was classification agreement for 6,039 (85.6%) variants. Of the 1,017 (14.4%) discordant calls, classification discrepancies of one degree were most common (715 of 1,017 changes), with the DVD being more likely to downgrade a ClinVar classification (772 downgrades versus 245 upgrades) (Figure 2C). Major classification changes for deafness-related variants that resulted in medically significant differences (variants that were upgraded to or downgraded from P/LP) were identified for 72 variants. Of these variants, there were 53 up-classifications of a variant by the DVD to P/LP and 19 down-classifications of a variant from P/LP (Figure 2E).
A total of 7,845 DVD variants are found in HGMD. DVD and HGMD classifications were concordant in 7,458 (95%) cases. Of the 387 (5%) discordant calls, classification discrepancies of three degrees were most common (132 of the 387 changes), with DVD downgrades of HGMD calls more common than upgrades (312 downgrades versus 75 upgrades) (Figure 2D). There were 244 major classification changes that resulted in medically significant differences, with all except two representing downgrades by the DVD from an HGMD call of P/LP (Figure 2F).
Following computational and manual curation, variants in 101 genes were reclassified in the DVD. These reclassifications included major categorical changes representing medically significant changes (P/LP versus VUS/LB/B) for 300 variants in 52 genes (Table S3). Of the 20% of genes carrying the greatest number of medically significant differences, six are associated with the diagnosis of Usher syndrome (Figure 2G, Table S4). For both ClinVar and HGMD, the same five genes carry the greatest number of major categorical changes (USH2A [MIM: 608400], SLC26A4 [MIM: 605646], GJB2, MYO7A [MIM: 276903], CDH23 [MIM: 605516]) (Figure 2G, Table S4). The remaining frequently impacted genes are WFS1 (MIM: 606201) (DFNA6/14/38 [MIM: 600965] and Wolfram syndrome), USH1C (MIM: 605242) (DFNB18A [MIM: 602092] and USH1C [MIM: 276904]), ADGRV1 (MIM: 602851) (USH2C [MIM: 605472]), and MYO15A (MIM: 602666) (DFNB3 [MIM: 600316]).
Most Genetic Variants in Deafness-Associated Genes Are Missense and Rare
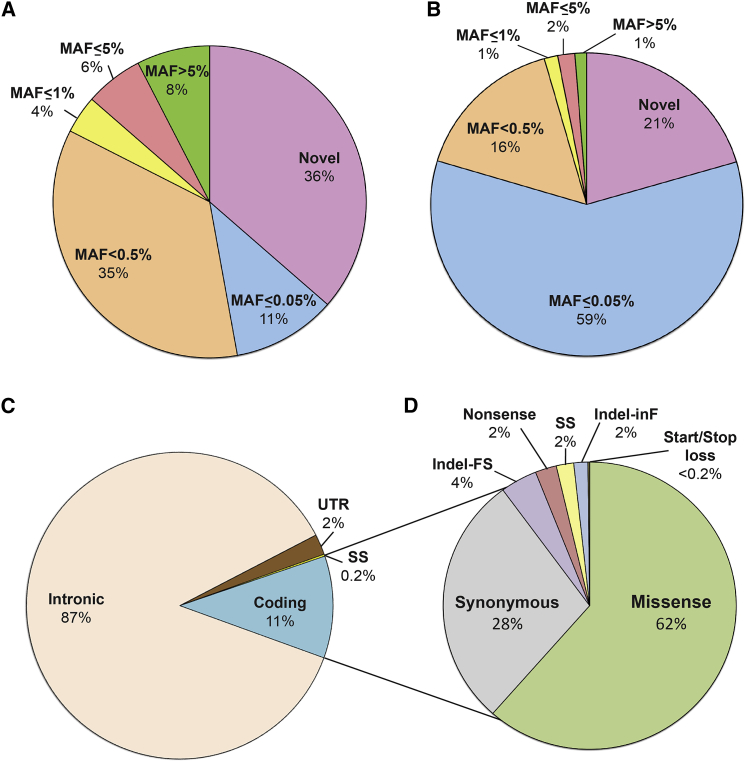
Having built a comprehensive resource that collates and annotates all variants in hearing loss genes and provides a clinical interpretation, we sought to explore the genomic and mutational landscape of deafness-associated genes. Our first objective was to evaluate the distribution of variants with respect to their MAF and type. Of all variants in the DVD, novel, ultra-rare (0% < MAF ≤ 0.05%), and rare (0.05% < MAF < 0.5%) variants represented 36%, 11%, and 35%, respectively (Figure 3A). When only clinically relevant variants within coding and splice-site regions were considered, the general tendency did not change. Variants with MAF < 0.5% remained the most prevalent (96%) although the distribution within this set changed, with ultra-rare variants (0% < MAF ≤ 0.05%) now representing the major category (59%) (Figure 3B). The finding that variants with a MAF < 0.5% (the threshold above which a variant is too common to be deafness causing16) account for 96% of all the variants falling within coding and splice-site regions implies that only 4% of variants can be excluded as deafness causing on the basis of MAF filtering.
Figure 3.
Distribution of Variants by Location, MAF, and Type
(A and B) MAF (all variants in DVD including intronic) (A) and only variants in gene coding regions (B). Most coding variants (96%) in deafness-associated genes are novel or rare (MAF < 0.5%).
(C) Distribution of variant by their gene location.
(D) Coding variant breakdown by type showing that missense variants constitute the major set of all coding variants.
Abbreviations: FS, frameshift; SS, splice-site; inF, in-frame.
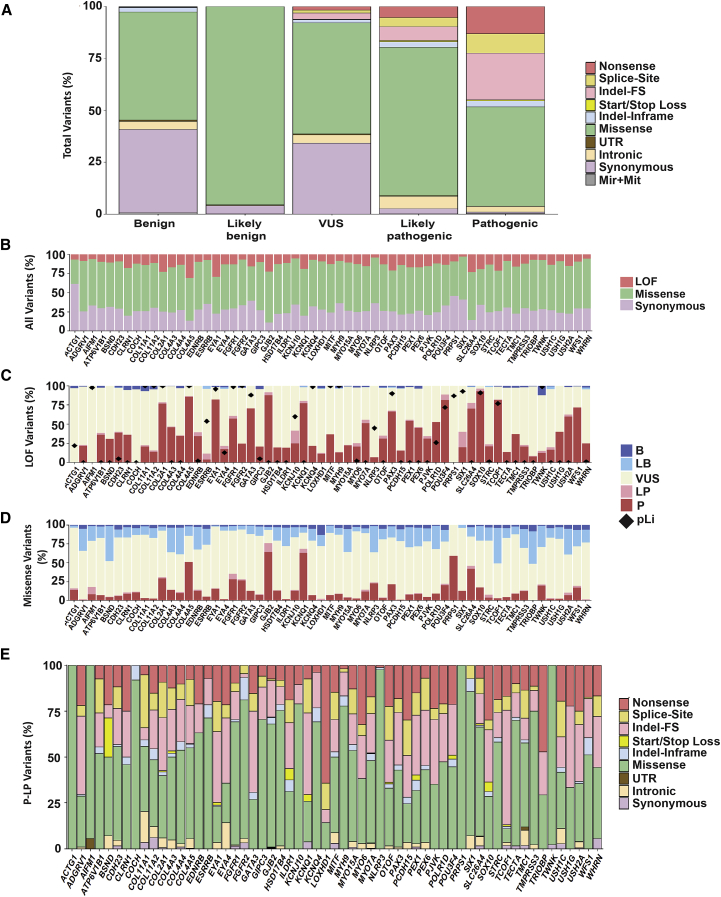
Of all variants within deafness-associated genes, ∼12% were located in the coding regions and canonical splice sites (Figure 3C). Missense variants represent the major set of all coding variants at 62%. The second most common type are synonymous variants (28%) followed by indels (4% frameshift and 2% inframe), nonsense (2%), canonical splice-site (2%), and start/stop loss (<1%) (Figure 3D).
Disparity in Gene Variation Rates
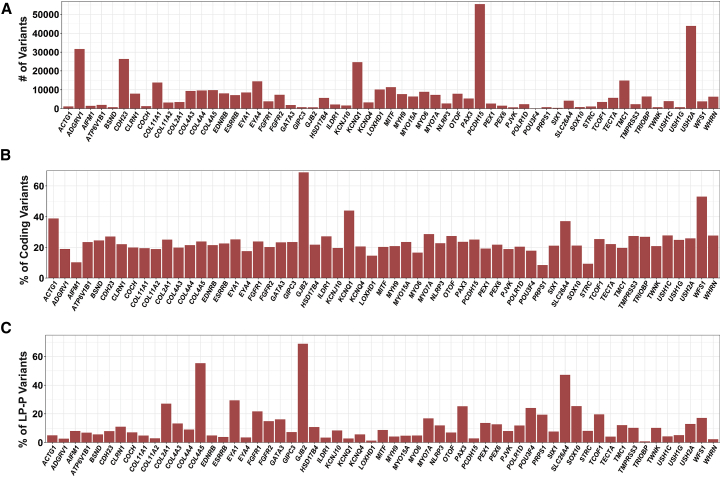
As expected, the number of variants per gene correlated with gene size, with larger genes carrying higher numbers of variants (Figures 4A and S2A, Table S5). The greatest variant load was found in PCDH15 (MIM: 605514), USH2A, ADGRV1, and CDH23, but when the number of variants was normalized for gene size, different trends emerged (Figures S2B and S2C, Table S5). ACTG1 (MIM: 102560) had the highest variant rate at 41% (4 of 10 bases carry reported variants), with most genes (85%) having a variation rate below 10%. If we restricted the analysis to coding and splice-site regions, again there was a correlation between the number of variants and the size of the coding regions, with USH2A, ADGRV1, and CDH23 carrying the highest number of variants (Table S5). Normalizing to the size of the coding region, however, gave strikingly different results: GJB2 carried the greatest variation at ∼69% (nearly 7 of 10 bases carry reported variants) and six other genes had variation rates higher than 30%: WFS1 (53%), KCNQ1 (MIM: 607542) (44%), ACTG1 (39%), SLC26A4 (37%), and KCNE1 (MIM: 176261) (36%). The average variation rate was ∼22% (Figures 4B and S3A).
Figure 4.
Variation Rate for Deafness-Associated Genes
(A) Total number of variants per gene.
(B) Normalized number of coding variants based on the size of the coding and splice regions.
(C) Normalized number of deafness-associated variants (P+LP) based on the total number of coding variants.
Only genes with ≥14 reported deafness-associated variants are included in this figure; the remaining genes are shown in Figures S2 and S3.
To determine whether gene-specific variation rates correlated with tolerance or intolerance to variation, we focused on the 6,490 variants classified as P and LP for deafness and normalized to the total number of coding variants. We found that ∼69% of coding variants in GJB2 are disease causing (P and LP variants), meaning that for any new variant identified in the coding sequence of GJB2, there is a 70% chance that it is pathogenic. Both COL4A5 (MIM: 303630) (55.3%) and SLC26A4 (47.2%) also had high (P+LP)/(Coding Variant) ratios (Figures 4C and S3B, Table S5).
Variants Are Differentially Distributed across Classifications
To characterize the molecular profile of variants within different classification categories, we focused on variants in coding and splice regions and grouped them by type (nonsense, splice-site, frameshift indels, start loss, stop loss, in-frame indels, missense, UTRs, intronic, synonymous) across variant classifications (P, LP, VUS, LB, B). Overall, missense variants were most prevalent in all categories (Figure 5A). For P variants, loss-of-function (LoF) variants and non-LoF were equally represented (∼50%). Of LoF variants, frameshift indels were most common (47.8%), followed by nonsense and splice-site at 27.65% and 22.27%, respectively. LP variants showed a slightly different profile with mostly missense variants at ∼70%. As expected, P variants are enriched in LoF and B variants are depleted. VUSs are enriched for missense (53.5%) and synonymous (34.1%) variants, with LoF variants representing only ∼7%. We found an enrichment of LoF variants in the P (50%) and LP (16.9%) categories, whereas they represent only 7% of VUSs and a negligible proportion of LB (0.03%) and B (0.47%) variants (Figure 5A). Missense variants are most common in the LB classification at ∼95% and represent ∼70% of the LP variants and ∼50% of P variants.
Figure 5.
Genomic Landscape of Deafness-Associated Genes
(A) Variant architecture by each classification category shows a strikingly distinct distribution of variant types across the five classifications.
(B) Distribution of LoF, missense, and synonymous variants is different across genes.
(C) Most LoF variants are P/LP and some genes are highly enriched in this type of variant.
(D) The contribution of missense variants to the mutational pool of hearing loss is variable across genes. However, in most genes, the majority of missense variants are VUSs.
(E) The mutational spectrum is gene specific. Splice-site indicates variants in canonical splice sites.
Only genes with ≥14 reported deafness-associated variants are included in this figure; the remaining genes are shown in Figures S4 and S5.
Diverse Mutational Spectrum across Deafness-Associated Genes
We next examined the distribution of LoFs and missense and synonymous variants by gene and observed disparity among genes as some are depleted of LoF (such as SIX1 [MIM: 601205]) whereas others are enriched in synonymous (such as ACTG1) or missense (such as ADGRV1) variants (Figures 5B and S4A). Of all LoF variants, the fraction contributing to the P/LP pool differs across genes, showing that for some genes a LoF variant is most likely to be pathogenic (SOX10 [MIM: 602229], TCOF1 [MIM: 606847], COL2A1 [MIM: 120140], COL4A5 [MIM: 303630], EYA1 [MIM: 601653], GATA3 [MIM: 131320], POU3F4 [MIM: 300039]), whereas for others it is not (e.g., ACTG1, AIFM1 [MIM: 300169]) (Figures 5C and S4B). A similar disparity is also observed for missense variants, where for some genes more than half of all missense variants are P/LP (GJB2, KCNQ1, PRPS1 [MIM: 311850]) and for others this contribution is marginal (e.g., TRIOBP [MIM: 609761], ADGRV1) (Figures 5D and S4C). Interestingly, for some genes such as BSND (MIM: 606412), TCOF1, and TRIOBP, approximately half of all missense variants are classified as B/LB, implying that a missense variant in those genes is more likely to be non-disease causing.
We also noted wide variation across genes in the fractional contribution of missense versus LoF variants to the P/LP category (Figures 5E and S4D). Some genes have exclusively missense mutations (ACTG1, PRPS1, COCH [MIM: 603196], and AIFM1) while other genes were enriched in LoF mutations (TCOF1, LOXHD1 [MIM: 613072], ADGRV1, EYA1, and PCDH15). A more detailed analysis of the different types of mutations within the LoF group revealed greater variability in the fractions of nonsense, splice-site, and frameshift indels across genes (Figures 5E and S4D). For example, the majority of LoF mutations in LOXHD1 are nonsense, whereas for COL11A1 (MIM: 120280) they are splice sites.
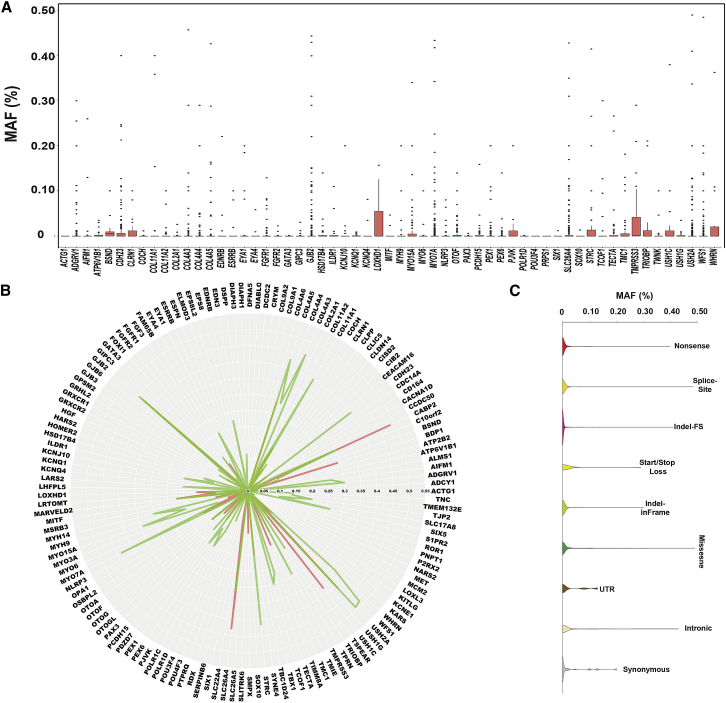
MAF Thresholds for Disease-Causing Variants Are Gene Specific
Gene-specific MAF thresholds for P+LP variants ranged from 0% to 7.34%. GJB2, MYO15A, OTOF (MIM: 603681), PEX6 (MIM: 601498), and CLRN1 (MIM: 601498) had the highest MAFs at 7.34%, 2.45%, 0.79%, 0.71%, and 0.69%, respectively (Table S6). However, these maximum MAFs are misleading and do not provide an accurate MAF for the majority of disease-causing variants associated with these genes. For example, while the maximum MAF for any pathogenic variant reported in GJB2 (GenBank: NM_004004.5) is 7.3% for c.109G>A (p.Val37Ile), the median MAF for all mutations in GJB2 is surprisingly 0, reflecting the huge number of ultra-rare P+LP variants in this gene (Figures 6A, 6B, and S5, Table S6). Similar results were found for SLC26A4, USH2A, and WFS1. These discrepancies also reflect founder effects as some mutations occur solely in a single population or ethnicity and account for a large portion of that population’s hearing loss (Table S2). These critical exceptions emphasize the importance of expert curation and review of variants that exceed the 0.5% MAF cut-off.
Figure 6.
MAFs Thresholds for Deafness-Associated Variants Are Gene and Type Specific
(A) Plot of MAFs of all P/LP variants in each deafness-associated gene.
(B) Maximum MAF is gene specific and there is a clear distinction between LoF versus missense variants.
(C) Overall, missense variants exhibit the highest MAFs when compared to all other variants.
Only genes with ≥14 reported deafness-associated variants are included in this figure; the remaining genes are shown in Figure S5.
MAF Thresholds for Disease-Causing Variants Are Type Specific
To determine whether MAFs for P+LP variants were mutation-type dependent, we subdivided all variants by effect and plotted against their MAF. Although the median MAF is 0 for all variant types, synonymous and UTR variants had the highest mean MAF (0.023% and 0.027%, respectively), followed by missense (0.017%), nonsense (0.009%), splice-site (0.0047%), in-frame indels (0.0036%), and frameshift indels (0.0028%) (Figure 6C, Table S7). These results compare closely to the gene-level results and demonstrate that regardless of type and gene, disease-causing mutations are ultra-rare and are heavily comprised of novel/private variants.
Kafeen and the DVD Are Configurable, Customizable, and Open-Access Resources
The DVD is freely available. It is widely used by the scientific and clinical communities worldwide with ∼3,000 users and 13,000 sessions over the past 12 months (Figure S6). The Kafeen bioinformatic pipeline, upon which the DVD was built, is configurable, adaptable, and extensible, allowing incorporation of additional variant and annotation sources as well as deleteriousness prediction tools. It also allows for customizable thresholds of MAF to classify variants. Consequently, its use is not limited to deafness and could be implemented for a variety of other genetic disorders.
Discussion
Genetic variant classification is crucial to accurate genetic diagnoses and represents a major challenge in the post-genome era, particularly for a disorder with genetic and phenotypic heterogeneity like deafness. The DVD was designed as a deafness-specific, comprehensive, open-access database that collates and summarizes all available data in addition to providing expert curation of genetic variants implicated in deafness (Figure 1). We integrate its use into a weekly multidisciplinary conference where a person’s genotypic data are reviewed in the context of available phenotypic data to provide expert contextual interpretation of the genetic results. As a first step, DVD annotations are used for prioritization of a person’s variant list, automatically flagging variants known to be reported as pathogenic in the DVD, as well as retaining DVD-classified LP/P variants that may have been filtered out of our NGS processing pipeline due to poor quality or ambiguous mapping. This type of curation reduces false negative rates and highlights the importance of disease-specific knowledge and disease-specific databases. CNVs are not integrated in the current release of the DVD. We have shown previously that they are major contributor to hearing loss and are implicated in ∼18% of all positive diagnoses.25 The challenge to their incorporation in the DVD resides in the lack of data regarding their exact breakpoint junctions. As more data become available, integration of CNVs should be an integral part of any variant database.
While it is difficult to provide “universal” MAF thresholds, the ACMG does recommend using MAF data as a key filter in their guidelines for variant interpretation. We deem a MAF ≥ 0.5% to be incompatible with a classification of P/LP for hereditary hearing loss aside from specific cases such as variants in GJB2 and SLC26A4, and we use this threshold to automatically classify any variant as B (Table S2).16 It is important to note that with the availability of new datasets from large-population sequencing projects such as gnomAD, MAF for some variants will change, which in turn may affect their clinical significance. While using a universal MAF cutoff is beneficial, for a common disease such as deafness this filter aided in classifying only 4% of coding variants as benign, illustrating that MAF cutoffs and rarity alone are not sufficient to determine deleteriousness.
As an additional aid, the DVD integrates predictions from six algorithms—two assessing conservation (PhyloP and GERP++) and four evaluating deleteriousness (SIFT, PolyPhen-2, MutationTaster, and LRT)—from which to calculate a composite PS. As more than 95% of known P variants have a pathogenicity score > 40%, the PPV of this approach reaches 0.995 (Figure S1). Using this threshold, we classify variants as either VUS (PS ≥ 60%) or LB (PS ≤ 40%). ACMG guidelines also endorse predictions from in silico algorithms as one of the eight evidence criteria recommended for variant clinical interpretation, and although outcomes and results from several studies vary depending on the algorithms used, these studies all agree on the utility of such tools for improving accuracy and reducing VUS burden in clinical diagnosis.3, 26
Questions remain regarding the strength and amount of evidence needed to sway a classification from a VUS to P/LP or B/LB. Since in clinical settings substantial evidence is needed to reach a P/LP classification, we have opted to use in silico algorithms exclusively to shift a VUS classification to LB.27 We require additional evidence (genotype, phenotype, family history, segregation, and functional studies) to upgrade a VUS to a P or LP variant.
Discrepancies in variant classification between the DVD versus ClinVar and HGMD were observed at 14.5% and 5%, respectively (Figure 2). Differences were due in part to the misclassification of B and LB variants as P or LP and have been reported in other studies highlighting the limitations of ClinVar and HGMD.28, 29, 30 ClinVar is based on submissions from researchers and clinical diagnostic laboratories. It is an invaluable resource that creates an open platform for sharing genetic data and variant interpretation, but it has some disadvantages. Most obvious are the differences in the methods used to detect, validate, curate, and derive variant interpretation, which understandably vary between groups and thus can lead to conflicting classifications.27, 31, 32, 33 Unlike ClinVar, HGMD relies on published literature and is primarily a disease-causing focused variant database. Although the variants reported in HGMD have been published and therefore have undergone peer review, the HGMD curation process is error prone due to the potential for subjective misinterpretation of the literature and a lack of disease-specific experts reviewing the material.
We implemented major categorical reclassifications that led to medically significant changes in 52 genes. In 33 genes, the change affected three or fewer variants (20 genes, 1 variant change; 11 genes, 2 variant changes; 2 genes, 3 variant changes); however, of the top 20% of genes carrying the greatest number of reclassifications, six cause Usher syndrome (ADGRV1 [USH2C], CDH23 [USH1D and DFNB12], MYO7A [USH1B, DFNB2, and DFNA11], PCDH15 [USH1F and DFNB23], USH2A [USH2A], and USH1C [USH1C and DFNB18A]) (Figure 2G, Table S4). Differentiating USH1 from NSHL is possible if a directed developmental history is obtained, because sitting and walking milestones are significantly delayed in USH1 due to the associated vestibular dysfunction, emphasizing the need to correlate clinical history with the interpretation of genetic data. We also consider audioprofiles, noting any progression of hearing loss, age at diagnosis and symmetry; imaging studies if available; and family history, as it is often possible to refine a diagnosis when more clinical information is provided. For example, a genetic diagnosis consistent with either USH1C or DNFB18A would be changed to USH1C if the child had delayed developmental milestones.
Recognizing the importance of more stringent filtering strategies to improve variant classification prompted us to use the DVD to define the molecular landscape of deafness-associated genes. When normalized to genomic size, some genes show remarkably high variation rates, such as ACTG1, although for the majority of genes the variation rate is below 10% (Figures 4B and S2B). This trend changes dramatically when only clinically relevant regions (coding and splice regions) are considered, implying that most variation is intronic. The coding/splice-site variation rate is highest for GJB2 (∼69%) and ranges from 8.5% to 53% for all other genes (Figures 4B and S2B, Table S5). Other studies, notably by Petrovski et al.,34 Lek et al.,12 and Samocha et al.,35 have used population-scale databases of variant numbers and allele frequencies to infer gene constraint or tolerance to genetic variation. Their assumption is that genes carrying more variants than expected have low constraint, while those with lesser variants have higher constraint and are intolerant to genetic variation. Our data showed that GJB2 does not fit into this model. Although it has the highest variation rate, it also carries the highest fraction of pathogenic variants (Figure 4C). This observation contrasts with its z-score of −1.07 (ExAC), which implies tolerance to variation and decreased constraint (Table S5). Similar findings are seen for SLC26A4, where every other variant is disease causing although its Z score is −3.23. These findings highlight the need to integrate real variant clinical interpretation data for each gene-phenotype association as large-scale population data can be misleading.
Several studies have also emphasized the importance of moving from gene-wide constraint calculations to protein domain-specific constraints as a method of identifying regions of functional importance.35, 36, 37, 38 This refinement is particularly important for proteins involved in hearing loss, as most have various structurally different domains with distinct functions. Furthermore, some show an extraordinary pleiotropy and cause both autosomal-dominant NSHL (ADNSHL) and autosomal-recessive NSHL (ARNSHL) (TECTA [MIM: 602574] and TMC1 [MIM: 606706]) or both syndromic hearing loss and NSHL (Usher type 1-associated genes, WFS1, TBC1D24 [MIM: 613577], and COL11A1 [MIM: 120290]).39, 40, 41, 42, 43, 44 Classifying variants by domain or regional constraint can minimize both false-positive and false-negative pathogenicity predictions and facilitate proper diagnosis, especially for genes associated with NSHL mimics.
Our assessment of variant distribution by mutation type, classification, and gene-specific MAFs across all 152 genes and microRNAs uncovered gene-specific variant architecture (Figures 5, 6, S4, and S5). For example, some genes (GJB2, SLC26A4, and COL4A5) are relatively depleted of synonymous changes when compared to other genes (Figure 5B). Interestingly, these same genes possess the highest intolerance to variation with 70%, 55%, and 47% of all coding variants being P/LP for GJB2, COL4A5, and SLC26A4, respectively (Figure 4C, Table S5). The involvement of synonymous variants in disease is secondary to splice alteration by changing exonic splice enhancers or silencers, or through codon usage bias that impacts gene expression by affecting mRNA folding and stability, messenger ribonucleoprotein (mRNP) complex formation, translation rate, and protein folding and function.45, 46 Synonymous variants may be under selective pressure in GJB2, COL4A5, and SLC26A4, implying a potential unrecognized disease mechanism that would affect their proper expression in inner ear. This highlights the need to carefully review these variants when interpreting sequence data from persons with hearing loss. Conversely, for other genes like ACTG1, synonymous variants predominate, while the only reported deafness-associated pathogenic variants are missense, suggesting intolerance to changes at the protein level, which is in line with the reported gain-of-function mechanism for these variants.
Overall, there was a great diversity in the contribution of LoF and missense variants to the mutational load across genes (Figures 5C–5E and S4). Of all the LoF variants, the fraction contributing to the P/LP group was highest for genes for which haploinsufficiency is the mechanism of action (autosomal-dominant and X-linked genes) such as EYA1, TCOF1, SOX10, and GATA3 (Figure 5C). This trend was further accentuated when assessing the contribution of LoF variants to the mutational spectrum of these genes (Figure 5E). For example, LoF mutations in TCOF1 and EYA1 represent ∼90% and 80% of all reported pathogenic variants, respectively.
The variability in the fractional contribution of nonsense, splice-site, and frameshift indels to the mutational load across genes is intriguing. Although for autosomal-recessive genes this variability may not affect the outcome at the protein level (as most of these variants are expected to result in null alleles), the story is quite different for autosomal-dominant genes. The latter exert their effect via haploinsufficieny or a gain-of-function/dominant-negative mode of action, and the specific type of mutation might be crucial. LoF variants in genes known to have a dominant-negative/gain-of-function mechanism of action are not traditionally predicted to be pathogenic for a dominant disease. However, this caveat ignores the position-dependent effect of these variants on nonsense-mediated mRNA decay (NMD).47 For example, truncating pathogenic variants in DIAPH1 are linked to two different phenotypes: (1) autosomal-recessive seizures, cortical blindness, and microcephaly syndrome (MIM: 616632) due to null alleles through NMD and (2) autosomal-dominant DFNA1 hearing loss with thrombocytopenia due to gain-of-function truncating variants (that escape NMD) in the C-terminal DAD domain, which disrupt the autoinhibitory activity of the DAD and renders the protein constitutively active.48, 49 SOX10 and PTPRQ are other examples where the impact of a LoF variant is position dependent.50, 51
COL11A1 has the largest proportion of splice-site pathogenic variants when compared to all other deafness-associated genes. The majority of these variants are located in the triple-helical domain and cause inframe exon skipping rather than frameshifts. The mutant proteins exert their effect through a dominant-negative mechanism to cause Marshall syndrome (MRSHS) and Stickler syndrome type II (STL2).52, 53 However, biallelic null alleles cause fibrochondrogenesis (FBCG1), a severe recessive often neonatally lethal disease.54 This genotype-phenotype correlation explains the enrichement of splice-altering disease variants in COL11A1.
For genes where most missense variants are classified as B/LB, we estimate that a majority of the variants that are currently classified as VUS will be subsequently downgraded to B/LB. Similar to the diversity of variant distribution across classifications, we exposed clear distinctions in the maximum MAFs of P/LP variants depending on the gene and variant type (LoF versus missense) (Figures 6 and S5).
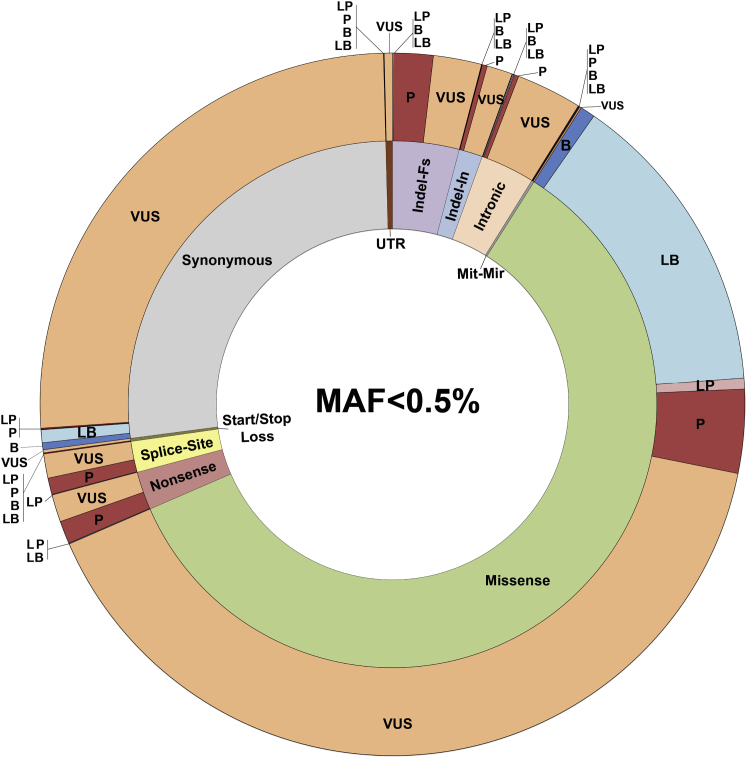
The emerging global picture from our findings is an intricate and complex portrait of the genomic landscape and mutational signature of deafness-associated genes. Although this work lays the foundation for improved variant interpretation, which greatly enhances clinical decision making, significant challenges remain. For example, of coding variants with a MAF < 0.5%, missense variants predominate. They constitute 70% of all VUSs and their accurate reclassification will require better computational tools (Figure 7). The non-coding pathogenic landscape also must be defined, warranting coordinated studies to integrate expression and genomic data.
Figure 7.
The Challenge of VUSs
Variant architecture correlating variant type (inner ring) and clinical significance (outer ring) for variants with MAF less than 0.5% and located within the clinically relevant regions. Of all coding variants with MAF < 0.5%, missense variants represent the majority at 61.5%; of these missense variants, 70% are classified as VUSs. Abbreviations: Indel-In, in-frame indel; Indel-Fs, frameshift indel; Mit-Mir, mitochondrial and microRNA.
In summary, using decision support tools and human expert curation, we have developed an integrated approach to facilitate the application of comprehensive genetic testing to the clinical care of persons with hearing loss. We believe that detailed disease-specific knowledge of the genomic landscape is requisite to establish a framework for variant interpretation and show that there are gene-specific mutational signatures, the knowledge of which will refine guidelines for variant interpretation for deafness and advance our understanding of disease biology. This resource is freely available to the public and configurable to allow its implementation for any Mendelian genetic disorder.
Declaration of Interests
R.J.H.S. directs the Molecular Otolaryngology and Renal Research Laboratories (MORL) which developed and offers comprehensive genetic testing for persons with hearing loss.
Acknowledgments
The authors thank Julie S. Wertz and Andrea Hallier for help with bioinformatic troubleshooting. The authors are also grateful to all DVD users who have submitted data and provided feedback. This work was supported by NIDCD RO1s DC003544, DC002842, and DC012049 to R.J.H.S.
Published: September 20, 2018
Footnotes
Supplemental Data include six figures and seven tables and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.08.006.
Web Resources
1000 Genomes, http://www.internationalgenome.org/
Deafness Variation Database, http://deafnessvariationdatabase.com/
ExAC Browser, http://exac.broadinstitute.org/
gnomAD Browser, http://gnomad.broadinstitute.org/
Hereditary Hearing Loss Homepage, http://hereditaryhearingloss.org
Human Gene Mutation Database (HGMD), https://www.qiagenbioinformatics.com/products/human-gene-mutation-database/
Kafeen, https://github.com/clcg/Kafeen
MutationTaster, http://www.mutationtaster.org/
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
Ruby programming language, https://www.ruby-lang.org/
Supplemental Data
References
- 1.Collins F.S., Varmus H. A new initiative on precision medicine. N. Engl. J. Med. 2015;372:793–795. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patel R.Y., Shah N., Jackson A.R., Ghosh R., Pawliczek P., Paithankar S., Baker A., Riehle K., Chen H., Milosavljevic S., ClinGen Resource ClinGen Pathogenicity Calculator: a configurable system for assessing pathogenicity of genetic variants. Genome Med. 2017;9:3. doi: 10.1186/s13073-016-0391-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Landrum M.J., Lee J.M., Benson M., Brown G., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Hoover J. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44(D1):D862–D868. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stenson P.D., Mort M., Ball E.V., Shaw K., Phillips A., Cooper D.N. The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum. Genet. 2014;133:1–9. doi: 10.1007/s00439-013-1358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stenson P.D., Mort M., Ball E.V., Evans K., Hayden M., Heywood S., Hussain M., Phillips A.D., Cooper D.N. The Human Gene Mutation Database: towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum. Genet. 2017;136:665–677. doi: 10.1007/s00439-017-1779-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith R.J., Bale J.F., Jr., White K.R. Sensorineural hearing loss in children. Lancet. 2005;365:879–890. doi: 10.1016/S0140-6736(05)71047-3. [DOI] [PubMed] [Google Scholar]
- 8.Sloan-Heggen C.M., Bierer A.O., Shearer A.E., Kolbe D.L., Nishimura C.J., Frees K.L., Ephraim S.S., Shibata S.B., Booth K.T., Campbell C.A. Comprehensive genetic testing in the clinical evaluation of 1119 patients with hearing loss. Hum. Genet. 2016;135:441–450. doi: 10.1007/s00439-016-1648-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sudmant P.H., Rausch T., Gardner E.J., Handsaker R.E., Abyzov A., Huddleston J., Zhang Y., Ye K., Jun G., Fritz M.H.-Y., 1000 Genomes Project Consortium An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526:75–81. doi: 10.1038/nature15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sherry S.T., Ward M.-H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu X., Jian X., Boerwinkle E. dbNSFP: a lightweight database of human nonsynonymous SNPs and their functional predictions. Hum. Mutat. 2011;32:894–899. doi: 10.1002/humu.21517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu X., Wu C., Li C., Boerwinkle E. dbNSFP v3.0: a one-stop database of functional predictions and annotations for human nonsynonymous and splice-site SNVs. Hum. Mutat. 2016;37:235–241. doi: 10.1002/humu.22932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shearer A.E., Eppsteiner R.W., Booth K.T., Ephraim S.S., Gurrola J., 2nd, Simpson A., Black-Ziegelbein E.A., Joshi S., Ravi H., Giuffre A.C. Utilizing ethnic-specific differences in minor allele frequency to recategorize reported pathogenic deafness variants. Am. J. Hum. Genet. 2014;95:445–453. doi: 10.1016/j.ajhg.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siepel A., Pollard K.S., Haussler D. New methods for detecting lineage-specific selection. Lect. Notes Comput. Sci. (Including Subser. Lect. Notes Artif. Intell. Lect. Notes Bioinformatics) 2006;3909 LNBI:190–205. [Google Scholar]
- 18.Davydov E.V., Goode D.L., Sirota M., Cooper G.M., Sidow A., Batzoglou S. Identifying a high fraction of the human genome to be under selective constraint using GERP++ PLoS Comput. Biol. 2010;6:e1001025. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar P., Henikoff S., Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 20.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwarz J.M., Rödelsperger C., Schuelke M., Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 22.Chun S., Fay J.C. Identification of deleterious mutations within three human genomes. Genome Res. 2009;19:1553–1561. doi: 10.1101/gr.092619.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sloan-Heggen C.M., Babanejad M., Beheshtian M., Simpson A.C., Booth K.T., Ardalani F., Frees K.L., Mohseni M., Mozafari R., Mehrjoo Z. Characterising the spectrum of autosomal recessive hereditary hearing loss in Iran. J. Med. Genet. 2015;52:823–829. doi: 10.1136/jmedgenet-2015-103389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moteki H., Azaiez H., Booth K.T.T., Shearer A.E.E., Sloan C.M.M., Kolbe D.L.L., Nishio S., Hattori M., Usami S., Smith R.J.H. Comprehensive genetic testing with ethnic-specific filtering by allele frequency in a Japanese hearing-loss population. Clin. Genet. 2016;89:466–472. doi: 10.1111/cge.12677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shearer A.E.E., Kolbe D.L., Azaiez H., Sloan C.M., Frees K.L., Weaver A.E., Clark E.T., Nishimura C.J., Black-Ziegelbein E.A.A., Smith R.J.H. Copy number variants are a common cause of non-syndromic hearing loss. Genome Med. 2014;6:37. doi: 10.1186/gm554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bean L.J.H., Hegde M.R. Clinical implications and considerations for evaluation of in silico algorithms for use with ACMG/AMP clinical variant interpretation guidelines. Genome Med. 2017;9:111. doi: 10.1186/s13073-017-0508-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garber K.B., Vincent L.M., Alexander J.J., Bean L.J.H., Bale S., Hegde M. Reassessment of genomic sequence variation to harmonize interpretation for personalized medicine. Am. J. Hum. Genet. 2016;99:1140–1149. doi: 10.1016/j.ajhg.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacArthur D.G., Manolio T.A., Dimmock D.P., Rehm H.L., Shendure J., Abecasis G.R., Adams D.R., Altman R.B., Antonarakis S.E., Ashley E.A. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508:469–476. doi: 10.1038/nature13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Q., Wang K. InterVar: Clinical Interpretation of Genetic Variants by the 2015 ACMG-AMP Guidelines. Am. J. Hum. Genet. 2017;100:267–280. doi: 10.1016/j.ajhg.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Editorial Improving databases for human variation. Nat. Methods. 2016;13:103. doi: 10.1038/nmeth.3762. [DOI] [PubMed] [Google Scholar]
- 31.Harrison S.M., Dolinsky J.S., Knight Johnson A.E., Pesaran T., Azzariti D.R., Bale S., Chao E.C., Das S., Vincent L., Rehm H.L. Clinical laboratories collaborate to resolve differences in variant interpretations submitted to ClinVar. Genet. Med. 2017;19:1096–1104. doi: 10.1038/gim.2017.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amendola L.M., Jarvik G.P., Leo M.C., McLaughlin H.M., Akkari Y., Amaral M.D., Berg J.S., Biswas S., Bowling K.M., Conlin L.K. Performance of ACMG-AMP variant-interpretation guidelines among nine laboratories in the clinical sequencing exploratory research consortium. Am. J. Hum. Genet. 2016;98:1067–1076. doi: 10.1016/j.ajhg.2016.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoskinson D.C., Dubuc A.M., Mason-Suares H. The current state of clinical interpretation of sequence variants. Curr. Opin. Genet. Dev. 2017;42:33–39. doi: 10.1016/j.gde.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petrovski S., Wang Q., Heinzen E.L., Allen A.S., Goldstein D.B. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 2013;9:e1003709. doi: 10.1371/journal.pgen.1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Samocha K.E., Robinson E.B., Sanders S.J., Stevens C., Sabo A., McGrath L.M., Kosmicki J.A., Rehnström K., Mallick S., Kirby A. A framework for the interpretation of de novo mutation in human disease. Nat. Genet. 2014;46:944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peterson T.A., Nehrt N.L., Park D., Kann M.G. Incorporating molecular and functional context into the analysis and prioritization of human variants associated with cancer. J. Am. Med. Inform. Assoc. 2012;19:275–283. doi: 10.1136/amiajnl-2011-000655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sivley R.M., Dou X., Meiler J., Bush W.S., Capra J.A. Comprehensive analysis of constraint on the spatial distribution of missense variants in human protein structures. Am. J. Hum. Genet. 2018;102:415–426. doi: 10.1016/j.ajhg.2018.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eilbeck K., Quinlan A., Yandell M. Settling the score: variant prioritization and Mendelian disease. Nat. Rev. Genet. 2017;18:599–612. doi: 10.1038/nrg.2017.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bork J.M., Peters L.M., Riazuddin S., Bernstein S.L., Ahmed Z.M., Ness S.L., Polomeno R., Ramesh A., Schloss M., Srisailpathy C.R.S. Usher syndrome 1D and nonsyndromic autosomal recessive deafness DFNB12 are caused by allelic mutations of the novel cadherin-like gene CDH23. Am. J. Hum. Genet. 2001;68:26–37. doi: 10.1086/316954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Azaiez H., Booth K.T., Bu F., Huygen P., Shibata S.B., Shearer A.E., Kolbe D., Meyer N., Black-Ziegelbein E.A., Smith R.J.H. TBC1D24 mutation causes autosomal-dominant nonsyndromic hearing loss. Hum. Mutat. 2014;35:819–823. doi: 10.1002/humu.22557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cryns K., Sivakumaran T.A., Van den Ouweland J.M.W., Pennings R.J.E., Cremers C.W.R.J., Flothmann K., Young T.L., Smith R.J.H., Lesperance M.M., Van Camp G. Mutational spectrum of the WFS1 gene in Wolfram syndrome, nonsyndromic hearing impairment, diabetes mellitus, and psychiatric disease. Hum. Mutat. 2003;22:275–287. doi: 10.1002/humu.10258. [DOI] [PubMed] [Google Scholar]
- 42.Rigoli L., Lombardo F., Di Bella C. Wolfram syndrome and WFS1 gene. Clin. Genet. 2011;79:103–117. doi: 10.1111/j.1399-0004.2010.01522.x. [DOI] [PubMed] [Google Scholar]
- 43.Richardson G.P., de Monvel J.B., Petit C. How the genetics of deafness illuminates auditory physiology. Annu. Rev. Physiol. 2011;73:311–334. doi: 10.1146/annurev-physiol-012110-142228. [DOI] [PubMed] [Google Scholar]
- 44.Acke F.R.E., Dhooge I.J.M., Malfait F., De Leenheer E.M.R. Hearing impairment in Stickler syndrome: a systematic review. Orphanet J. Rare Dis. 2012;7:84. doi: 10.1186/1750-1172-7-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hanson G., Coller J. Codon optimality, bias and usage in translation and mRNA decay. Nat. Rev. Mol. Cell Biol. 2018;19:20–30. doi: 10.1038/nrm.2017.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hunt R.C., Simhadri V.L., Iandoli M., Sauna Z.E., Kimchi-Sarfaty C. Exposing synonymous mutations. Trends Genet. 2014;30:308–321. doi: 10.1016/j.tig.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 47.Coban-Akdemir Z., White J.J., Song X., Jhangiani S.N., Fatih J.M., Gambin T., Bayram Y., Chinn I.K., Karaca E., Punetha J., Baylor-Hopkins Center for Mendelian Genomics Identifying genes whose mutant transcripts cause dominant disease traits by potential gain-of-function alleles. Am. J. Hum. Genet. 2018;103:171–187. doi: 10.1016/j.ajhg.2018.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ueyama T., Ninoyu Y., Nishio S.-Y., Miyoshi T., Torii H., Nishimura K., Sugahara K., Sakata H., Thumkeo D., Sakaguchi H. Constitutive activation of DIA1 (DIAPH1) via C-terminal truncation causes human sensorineural hearing loss. EMBO Mol. Med. 2016;8:1310–1324. doi: 10.15252/emmm.201606609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Neuhaus C., Lang-Roth R., Zimmermann U., Heller R., Eisenberger T., Weikert M., Markus S., Knipper M., Bolz H.J. Extension of the clinical and molecular phenotype of DIAPH1-associated autosomal dominant hearing loss (DFNA1) Clin. Genet. 2017;91:892–901. doi: 10.1111/cge.12915. [DOI] [PubMed] [Google Scholar]
- 50.Inoue K., Khajavi M., Ohyama T., Hirabayashi S., Wilson J., Reggin J.D., Mancias P., Butler I.J., Wilkinson M.F., Wegner M., Lupski J.R. Molecular mechanism for distinct neurological phenotypes conveyed by allelic truncating mutations. Nat. Genet. 2004;36:361–369. doi: 10.1038/ng1322. [DOI] [PubMed] [Google Scholar]
- 51.Eisenberger T., Di Donato N., Decker C., Delle Vedove A., Neuhaus C., Nürnberg G., Toliat M., Nürnberg P., Mürbe D., Bolz H.J. A C-terminal nonsense mutation links PTPRQ with autosomal-dominant hearing loss, DFNA73. Genet. Med. 2018;20:614–621. doi: 10.1038/gim.2017.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Annunen S., Körkkö J., Czarny M., Warman M.L., Brunner H.G., Kääriäinen H., Mulliken J.B., Tranebjaerg L., Brooks D.G., Cox G.F. Splicing mutations of 54-bp exons in the COL11A1 gene cause Marshall syndrome, but other mutations cause overlapping Marshall/Stickler phenotypes. Am. J. Hum. Genet. 1999;65:974–983. doi: 10.1086/302585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rose P.S., Levy H.P., Liberfarb R.M., Davis J., Szymko-Bennett Y., Rubin B.I., Tsilou E., Griffith A.J., Francomano C.A. Stickler syndrome: clinical characteristics and diagnostic criteria. Am. J. Med. Genet. A. 2005;138A:199–207. doi: 10.1002/ajmg.a.30955. [DOI] [PubMed] [Google Scholar]
- 54.Tompson S.W., Bacino C.A., Safina N.P., Bober M.B., Proud V.K., Funari T., Wangler M.F., Nevarez L., Ala-Kokko L., Wilcox W.R. Fibrochondrogenesis results from mutations in the COL11A1 type XI collagen gene. Am. J. Hum. Genet. 2010;87:708–712. doi: 10.1016/j.ajhg.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.