Abstract
Although recent studies provide evidence for a common genetic basis between complex traits and Mendelian disorders, a thorough quantification of their overlap in a phenotype-specific manner remains elusive. Here, we have quantified the overlap of genes identified through large-scale genome-wide association studies (GWASs) for 62 complex traits and diseases with genes containing mutations known to cause 20 broad categories of Mendelian disorders. We identified a significant enrichment of genes linked to phenotypically matched Mendelian disorders in GWAS gene sets; of the total 1,240 comparisons, a higher proportion of phenotypically matched or related pairs (n = 50 of 92 [54%]) than phenotypically unmatched pairs (n = 27 of 1,148 [2%]) demonstrated significant overlap, confirming a phenotype-specific enrichment pattern. Further, we observed elevated GWAS effect sizes near genes linked to phenotypically matched Mendelian disorders. Finally, we report examples of GWAS variants localized at the transcription start site or physically interacting with the promoters of genes linked to phenotypically matched Mendelian disorders. Our results are consistent with the hypothesis that genes that are disrupted in Mendelian disorders are dysregulated by non-coding variants in complex traits and demonstrate how leveraging findings from related Mendelian disorders and functional genomic datasets can prioritize genes that are putatively dysregulated by local and distal non-coding GWAS variants.
Keywords: polygenic, Mendelian, monogenic, complex traits, common disease, statistical genetics, GWAS, body mass index, Hi-C
Introduction
Genetic architectures of human traits have traditionally been classified into two major categories. Typically, complex traits demonstrate polygenic architectures arising from many low-effect common variants, whereas rare traits tend to have high-effect monogenic determinants.1 The underlying and practical distinction between these classes has historically been based on the presence of highly penetrant, rare, single-gene disruptive mutations causing recognizable clinical syndromes and monogenic diseases (e.g., cystic fibrosis [MIM: 219700]2), and the relative absence of such mutations in complex diseases, such as diabetes and schizophrenia.3 Accumulating evidence suggests that these two classes of phenotypes might not be as biologically distinct as previously thought.4 Multiple exceptions to the “common disease, common variant” hypothesis1 have been identified for complex traits5, 6, 7 and their molecular phenotypes,8, 9, 10, 11 and Mendelian disorders have also been found to be affected by multiple or common genetic variants.12, 13, 14, 15 This suggests that there exists a spectrum of genetic architectures rather than a dichotomous classification. Accordingly, the monogenic forms of complex traits (i.e., phenotypically matched Mendelian disorders) are increasingly used as a starting point for identifying genes relevant to complex traits for further study.16, 17, 18 Furthermore, overlap has been identified between genes, common variants, and copy-number variants linked to Mendelian disorders and genetic determinants of complex diseases and traits such as Parkinson disease (MIM: 68600),19 obesity,20 height,21 ototoxicity,22 and others.23 However, the overlap between each of these complex traits and Mendelian disorders has been examined individually with different metrics of overlap. In a large study of patient medical records, Blair et al. identified systematic, significant comorbidities between Mendelian disorders and complex diseases and found that association signals from genome-wide association studies (GWASs) for complex diseases were enriched in genomic regions with known roles in comorbid Mendelian disorders, suggesting a shared genetic basis.24 However, the study focused on Mendelian disorders comorbid with complex diseases in the same individual rather than examining Mendelian disorders demonstrating phenotypes similar to complex traits. Furthermore, advances in sequencing technology have greatly expanded the phenotypic spectrum in known Mendelian syndromes, allowing for deconstruction of syndromic diseases into component medical phenotypes. As such, it is now possible to identify all the component-phenotype consequences of genes linked to Mendelian disorders, allowing for greater resolution in identifying gene-phenotype relationships. However, to the best of our knowledge, no study has taken advantage of this to identify genes linked to any related component-phenotype regardless of the Mendelian disorder’s best-known or primary phenotype. Thus, a thorough quantification of the overlap between genes associated with complex traits and genes linked to Mendelian disorders in a phenotype-specific manner remains elusive.
Given that the majority of GWASs for complex traits and diseases have identified significant associations in non-coding genomic regions,25 we hypothesize that genes individually involved in Mendelian disease belong to the biological pathway(s) shared by both complex and Mendelian disease. Specifically, we hypothesize that large-effect coding variants disrupt individual genes and thus result in severe phenotypes (i.e., Mendelian disorders), whereas non-coding variants produce complex traits by collectively dysregulating expression of these same genes, allowing for nuanced or tissue-specific phenotypes. On the basis of this hypothesis, we expect to identify an enrichment of GWAS signal for a given complex trait near genes linked to Mendelian disorders demonstrating similar phenotypes but no enrichment near genes linked to Mendelian disorders with phenotypes unrelated to the complex trait of interest. To test this hypothesis, we defined “Mendelian disorder genes” as any genes linked to Mendelian disorders in the Online Mendelian Inheritance in Man (OMIM) database. We used the well-curated phenotypic breakdown of Mendelian disorders in OMIM to identify subsets of these genes linked to particular phenotypes (e.g., growth defects or immune dysregulation) expressed as part of any Mendelian disorder. We then examined publicly available GWASs across 62 complex traits (listed in Table 1 and detailed in Table S1) to identify risk genes (here called GWAS gene sets) for each complex trait and quantified the overlap between each GWAS gene set and 20 other sets of Mendelian disorder genes for particular phenotypes (detailed in Tables 1 and S1). We found a consistent, significant, and specific enrichment between GWAS gene sets for complex traits and Mendelian disorder genes for matched and related phenotypes (50 of 1,240 pairs; e.g., rheumatoid arthritis and immune dysregulation), supporting our hypothesis of a shared genetic basis between complex and Mendelian forms of disease. In addition, we observed instances of enrichment between GWAS gene sets for certain complex traits and Mendelian disorder genes for unrelated phenotypes (27 of 1,240 pairs; e.g., systemic lupus erythematosus [SLE (MIM: 152700)] and mature-onset diabetes of the young), suggestive of shared biological mechanisms yet to be examined. Furthermore, we found an increase in average effect size of GWAS variants near Mendelian disorder genes for matched phenotypes, and we identified examples of associated SNPs found directly at the transcription start sites (TSSs) of these phenotypically matched Mendelian disorder genes as candidates for functional follow-up. Finally, we report examples of significant body mass index (BMI)-associated variants directly interacting with phenotypically related Mendelian disorder genes CREBBP (MIM: 180849) and CYP19A1 (MIM: 139300 and 613546) by using human primary white-adipocyte-specific Hi-C data.66 Leveraging the growing body of well-curated phenotypic data from studies of Mendelian disorders, we provide a phenotype-driven approach to identifying genetic pathways shared by Mendelian diseases and complex traits.
Table 1.
Complex Traits and Corresponding Mendelian Disorders
| Complex Trait | Abbreviation | No. of GWAS Genes | Matched Mendelian Disorder(s) |
|---|---|---|---|
| Celiac disease26 | CEL | 34 | immune dysregulation |
| Crohn disease27 | CD | 239 | |
| Inflammatory bowel disease27 | IBD | 368 | |
| Ulcerative colitis28 | UC | 202 | |
| Primary biliary cirrhosis29 | PBC | 149 | |
| Rheumatoid arthritis (European)30 | RA | 297 | |
| Multiple sclerosis31 | MS | 160 | |
| Autism32 | AUT | 2 | monogenic autism |
| Hemoglobin33 | HB | 89 | hematologic disorders |
| Mean cell hemoglobin33 | MCH | 164 | |
| Mean cell hemoglobin concentration33 | MCHC | 12 | |
| Mean corpuscular volume33 | MCV | 180 | |
| Mean platelet volume28 | MPV | 102 | |
| Red blood cell count33 | RBC | 107 | |
| Systemic lupus erythematosus34 | SLE | 286 | |
| Birthweight35 | BW | 179 | growth defects |
| Height36 | HGT | 2361 | |
| Femoral neck bone mineral density37 | FN | 58 | bone and uric acid disorders |
| Forearm bone mineral density37 | FA | 8 | |
| Lumbar spine bone mineral density37 | LS | 67 | |
| Serum urate concentration38 | URT | 161 | |
| Packed cell volume33 | PCV | 53 | disorders of platelet function |
| Platelet count28 | PLT | 134 | |
| Coronary artery disease39 | CAD | 132 | cardiovascular disease |
| High-density lipoprotein40 | HDL | 464 | |
| Low-density lipoprotein40 | LDL | 370 | |
| Total cholesterol40 | TC | 500 | |
| Triglycerides40 | TG | 354 | |
| Hemoglobin A1C41 | HBA | 33 | monogenic diabetes |
| Type 2 diabetes42 | T2D | 28 | |
| Age-related macular degeneration43 | AMD | 215 | monogenic AMD |
| Age at menarche44 | MNR | 207 | female reproductive disorders |
| Age at menopause45 | MNP | 316 | |
| Fasting glucose46 | FG | 39 | insulin disorders |
| HOMA-B46 | HMB | 12 | |
| HOMA-IR46 | HMIR | 0 | |
| Micro-albuminuria47 | MA | 2 | microalbumin disorders |
| Fasting insulin46 | FI | 23 | mature-onset diabetes of the young |
| 2 hr glucose48 | 2HG | 2 | |
| Type 1 diabetes49 | T1D | 144 | |
| Alzheimer disease50 | ALZ | 58 | neurologic disease |
| Anxiety disorders (case-control)51 | ANXC | 2 | |
| Anxiety disorders (factor score)51 | ANXF | 3 | |
| Major depressive disorder52 | MDD | 4 | |
| Depressive symptoms53 | DS | 10 | |
| Neuroticism53 | NRT | 82 | |
| Bipolar disorder54 | BIP | 8 | psychiatric disease |
| Schizophrenia55 | SCZ | 479 | |
| Chronic kidney disease56 | CKD | 16 | renal disorders |
| Glomerular filtration rate (creatinine)56 | EGFR | 162 | |
| Urine albumin-to-creatinine ratio47 | UACR | 2 | |
| Resting heart rate57 | RHR | 304 | arrhythmias |
| Age at first birth58 | AFB | 45 | education and development disorders |
| College59 | COL | 12 | |
| Education years60 | EY | 554 | |
| Subjective well-being53 | SWB | 9 | positive mood disorders |
| Body fat percentage61 | BFP | 22 | body mass disorders |
| Body mass index62 | BMI | 231 | |
| Childhood BMI63 | CBMI | 49 | |
| Leptin (adjusted for BMI)64 | LEPB | 5 | |
| Leptin (not adjusted for BMI)64 | LEP | 0 | |
| Waist-to-hip ratio65 | WHR | 74 |
This table lists the phenotypically matched pairs of complex traits (n = 62) and groups of Mendelian disorders (n = 20) examined in our study. More details on these traits, including mean GWAS sample size, number of significant GWAS loci reported from original GWAS publications, and number of significant GWAS SNPs, are included in Table S1. GWAS genes for each complex trait were identified by the mapping approach described in the Material and Methods.
Material and Methods
Gene Coordinates and Symbols
We downloaded gene-body coordinates (GRCh37/hg19, UCSC Genes track) from the UCSC Table Browser67 (see Web Resources) and specifically selected the gene symbol from the “knownGene” table, transcription start and end sites for each gene from the “knownCanonical” table, and the longest transcript from the “knownGene” table for genes where no entry or multiple entries were listed in the “knownCanonical” table. We used these coordinates for all analyses in our study. Because many genes have been renamed over time, we standardized gene symbols across all analyses in our study by downloading a table of approved symbols, previous symbols, and locus group for each gene from the HUGO Gene Nomenclature Committee at the European Bioinformatics Institute (HGNC) (see Web Resources) and renaming any genes identified by previous symbols with approved gene symbols. We restricted all analyses in our study to genes classified as protein-coding according to the HGNC locus group from chromosomes 1–22. These processing steps resulted in a final single set of coordinates for 17,695 autosomal protein-coding genes (for data access, see Web Resources).
Mendelian Disorder Genes and LoF-Intolerant Genes
To identify Mendelian disorder genes, we downloaded the OMIM catalog and identified all genes linked to Mendelian disorders satisfying the following criteria: (1) disorder is Mendelian and fully penetrant, therefore excluding susceptibility phenotypes, and (2) molecular basis of the Mendelian disorder is known (i.e., phenotype mapping key = 3). We defined loss-of-function (LoF)-intolerant genes as all genes with greater than 90% probability of being LoF intolerant according to the pLI score (>0.9) from the Exome Aggregation Consortium (ExAC) Browser;68 this score is derived from the number of observed versus expected LoF variants in a given gene across approximately 60,000 healthy exomes. Following the same restriction and gene symbol standardization criteria described above resulted in a final set of 3,446 Mendelian disorder genes and 2,978 LoF-intolerant genes.
Phenotype-Specific Mendelian Disorder Gene Sets
To identify subsets of Mendelian disorder genes linked to particular phenotypes, for each complex trait we curated a set of standardized clinical phenotype terms to describe the full range of relevant Mendelian phenotypes. We used these terms to search OMIM for all Mendelian disorders demonstrating these phenotypes and then extracted the gene(s) linked to each Mendelian disorder. We restricted gene-phenotype associations to those satisfying criteria (1) and (2) described above and with the following additional criteria: (3) gene-phenotype association description does not contain “genome-wide association study” or other GWAS synonyms unless the description also contains any of the terms “missense,” “nonsense,” “nonsynonymous,” or “frameshift” or the gene contains at least one pathogenic or likely pathogenic allele in ClinVar. We include a full list of phenotype-specific Mendelian disorder gene sets and clinical phenotype terms used in Table S2.
A comparison of all phenotype-specific Mendelian disorder gene sets revealed a high degree of overlap among the gene sets for clinically related Mendelian phenotypes (Figure S1). Accordingly, we clustered gene sets on the basis of pairwise overlap and intersected gene sets clustering together by visual inspection at a hierarchical clustering threshold to create a single gene set for the representative group of Mendelian disorders. Each complex trait was thus matched with the single Mendelian disorder category in which the original specific Mendelian disorder gene set clustered, which ultimately best exemplified the phenotype.
Because of the systemic and pleiotropic nature of complex traits, some complex traits could conceivably be phenotypically related to more than one Mendelian disorder gene set. For example, we generated the Mendelian disorder gene set for SLE by using clinical keywords for both the driving immunological event and the clinical manifestations associated with SLE autoimmunity across a large number of organ systems (kidney, brain, skin, pleura, joints, etc.), such as “anemia.” Although the substantial contribution of Mendelian disorder genes related to anemia resulted in SLE pairing with the “hematological disorders” group, the immunological component of SLE is central to the disease. Thus, we identified immune dysregulation as a “relevant phenotype” for SLE and denoted it as such in Figure 2 and Table 2 (the same occurred with other traits).
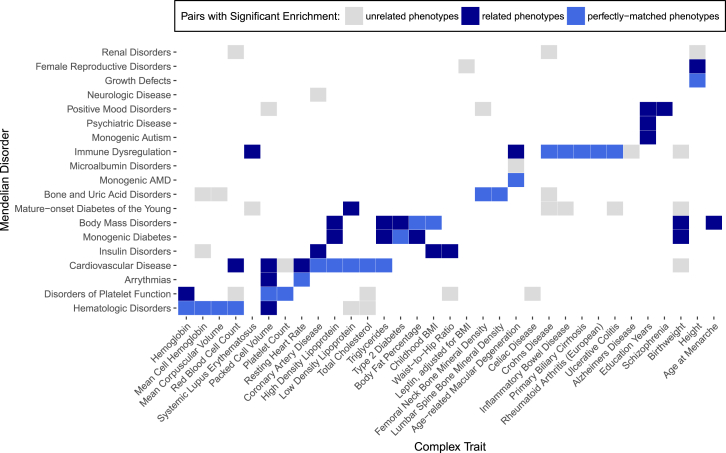
Figure 2.
Overlap between GWAS Genes and Mendelian Disorder Genes Demonstrates Trait Specificity
Significant overlaps from phenotypically matched pairs of complex traits and Mendelian disorders (blue) and pairs with unrelated phenotypes (gray) are shown. Phenotypically matched pairs are subdivided into pairs with perfectly matched phenotypes (light blue) and pairs with related phenotypes (dark blue). Complex traits and Mendelian disorders with no significant overlaps are excluded here; results from all traits are presented in Figure S2. We assessed significance by controlling for FDR < 5% at p < 0.00310.
Table 2.
Overlap between GWAS Genes and Phenotypically Matched Mendelian Disorder Genes
| Complex Trait (No. of Genes) | Matched or Related Mendelian Disorder (No. of Genes) | Shared Genes | OR for Matched Pair (CI) | Raw p Value | Average OR for Unmatched Pairs (CI) |
|---|---|---|---|---|---|
| AMD (215) | monogenic AMD (104) | 9 | 7.99 (3.50, 16.11) | 4.94 × 10−6 | 1.69 (1.21, 2.16) |
| immune dysregulation (550) | 17 | 2.73 (1.55, 4.52) | 4.13 × 10−4 | ||
| BFP (22) | body mass disorders (128) | 3 | 22.14 (4.14, 76.70) | 5.15 × 10−4 | 2.63 (0.75, 4.51) |
| monogenic diabetes (182) | 3 | 15.42 (2.90, 53.04) | 1.43 × 10−3 | ||
| BW (179) | body mass disorders (128) | 6 | 4.94 (1.76, 11.29) | 1.93 × 10−3 | 1.93 (1.48, 2.37) |
| monogenic diabetes (182) | 7 | 4.03 (1.57, 8.66) | 2.56 × 10−3 | ||
| CAD (132) | cardiovascular disease (598) | 13 | 3.17 (1.63, 5.67) | 5.36 × 10−4 | 2.13 (1.65, 2.61) |
| insulin disorders (623) | 13 | 3.04 (1.56, 5.43) | 7.83 × 10−4 | ||
| CBMI (49) | body mass disorders (128) | 5 | 16.18 (4.92, 41.63) | 2.71 × 10−5 | 1.55 (1.13, 1.97) |
| insulin disorders (623) | 7 | 4.61 (1.74, 10.41) | 1.54 × 10−3 | ||
| CD (239) | immune dysregulation (550) | 23 | 3.42 (2.10, 5.32) | 1.70 × 10−6 | 2.09 (−0.39, 4.58) |
| EY (554) | positive mood disorders (69) | 9 | 4.70 (2.04, 9.59) | 2.88 × 10−4 | 0.94 (0.81, 1.07) |
| monogenic autism (111) | 10 | 3.10 (1.44, 5.98) | 2.54 × 10−3 | ||
| psychiatric disease (264) | 19 | 2.45 (1.44, 3.95) | 8.96 × 10−4 | ||
| FN (58) | bone and uric acid disorders (220) | 5 | 7.64 (2.36, 19.24) | 7.61 × 10−4 | 3.79 (2.36, 5.22) |
| HB (89) | disorders of platelet function (443) | 12 | 6.21 (3.05, 11.59) | 2.17 × 10−6 | 2.38 (1.68, 3.09) |
| hematologic disorders (551) | 10 | 3.99 (1.83, 7.79) | 4.42 × 10−4 | ||
| HDL (464) | body mass disorders (128) | 12 | 3.92 (1.95, 7.17) | 1.39 × 10−4 | 1.18 (1.00, 1.37) |
| monogenic diabetes (182) | 15 | 3.41 (1.85, 5.85) | 9.36 × 10−5 | ||
| cardiovascular disease (598) | 31 | 2.10 (1.40, 3.06) | 3.45 × 10−4 | ||
| HGT (2361) | female reproductive disorders (288) | 61 | 1.76 (1.30, 2.36) | 2.18 × 10−4 | 1.31 (1.19, 1.42) |
| growth defects (723) | 126 | 1.39 (1.13, 1.70) | 1.43 × 10−3 | ||
| IBD (368) | immune dysregulation (550) | 34 | 3.32 (2.23, 4.79) | 1.58 × 10−8 | 1.38 (1.10, 1.66) |
| LDL (370) | cardiovascular disease (598) | 31 | 2.70 (1.79, 3.95) | 3.45 × 10−6 | 1.66 (1.24, 2.08) |
| mature-onset diabetes of the young (561) | 24 | 2.17 (1.36, 3.32) | 8.54 × 10−4 | ||
| LS (67) | bone and uric acid disorders (220) | 6 | 8.00 (2.80, 18.72) | 1.83 × 10−4 | 2.53 (1.98, 3.08) |
| MCH (164) | hematologic disorders (551) | 15 | 3.19 (1.73, 5.48) | 1.92 × 10−4 | 1.64 (1.16, 2.11) |
| MCV (180) | hematologic disorders (551) | 20 | 4.00 (2.36, 6.44) | 8.90 × 10−7 | 1.61 (1.18, 2.04) |
| MNR (207) | body mass disorders (128) | 7 | 5.02 (1.95, 10.86) | 7.76 × 10−4 | 1.03 (0.78, 1.27) |
| PBC (149) | immune dysregulation (550) | 13 | 3.03 (1.56, 5.40) | 7.77 × 10−4 | 1.07 (0.76, 1.38) |
| PCV (53) | disorders of platelet function (443) | 10 | 9.24 (4.11, 18.83) | 6.50 × 10−7 | 4.04 (2.22, 5.87) |
| arrhythmias (275) | 5 | 6.70 (2.07, 16.94) | 1.36 × 10−3 | ||
| hematologic disorders (551) | 8 | 5.60 (2.27, 12.08) | 2.17 × 10−4 | ||
| cardiovascular disease (598) | 7 | 4.39 (1.66, 9.84) | 1.94 × 10−3 | ||
| PLT (134) | disorders of platelet function (443) | 12 | 3.91 (1.95, 7.15) | 1.40 × 10−4 | 1.42 (0.98, 1.87) |
| RA (297) | immune dysregulation (550) | 25 | 2.95 (1.86, 4.50) | 6.80 × 10−6 | 0.83 (0.65, 1.01) |
| RBC (107) | hematologic disorders (551) | 14 | 4.78 (2.50, 8.50) | 5.82 × 10−6 | 2.76 (2.20, 3.33) |
| cardiovascular disease (598) | 13 | 4.02 (2.05, 7.26) | 6.49 × 10−5 | ||
| RHR (304) | arrhythmias (275) | 17 | 3.93 (2.23, 6.53) | 5.87 × 10−6 | 1.29 (0.95, 1.64) |
| cardiovascular disease (598) | 26 | 2.75 (1.75, 4.16) | 1.47 × 10−5 | ||
| SCZ (479) | positive mood disorders (69) | 9 | 5.47 (2.37, 11.19) | 9.71 × 10−5 | 1.11 (0.94, 1.28) |
| SLE (286) | immune dysregulation (550) | 24 | 2.94 (1.83, 4.52) | 1.09 × 10−5 | 1.41 (1.10, 1.72) |
| T2D (28) | body mass disorders (128) | 4 | 23.55 (5.85, 69.99) | 4.67 × 10−5 | 2.43 (1.57, 3.30) |
| monogenic diabetes (182) | 3 | 11.72 (2.24, 38.93) | 2.90 × 10−3 | ||
| TC (500) | cardiovascular disease (598) | 38 | 2.44 (1.69, 3.45) | 3.46 × 10−6 | 1.40 (1.10, 1.70) |
| TG (354) | body mass disorders (128) | 9 | 3.77 (1.67, 7.49) | 1.10 × 10−3 | 1.26 (1.01, 1.51) |
| monogenic diabetes (182) | 12 | 3.54 (1.78, 6.43) | 3.10 × 10−4 | ||
| cardiovascular disease (598) | 25 | 2.22 (1.41, 3.38) | 5.06 × 10−4 | ||
| UC (202) | immune dysregulation (550) | 21 | 3.72 (2.23, 5.92) | 1.39 × 10−6 | 1.45 (1.12, 1.78) |
| WHR (74) | insulin disorders (623) | 9 | 3.83 (1.67, 7.78) | 1.13 × 10−3 | 2.37 (1.71, 3.04) |
For each pair of complex trait and Mendelian disorder, Fisher’s exact test was used to quantify the enrichment of shared genes with an OR and p value (see Material and Methods). Pairs with significant enrichment passed the cutoff of FDR < 5% at p < 0.00310. This table lists pairs of complex traits and phenotypically matched or related Mendelian disorders with significant overlap. For comparison, the average OR and 95% CI for pairings of each complex trait with all unrelated Mendelian disorder gene sets are included.
After combining similar gene sets, we were left with 20 non-disjoint phenotype-specific Mendelian disorder gene sets with an average of 375 genes per set; we include a description of each cluster in Table S3.
Complex-Trait Gene Sets
We downloaded publicly available summary statistics (per-allele SNP effect sizes, or log odds ratios [OR] for case-control traits, with standard errors69) for large-scale GWASs of 62 traits66 (Tables 1 and S1; average n = 83,170, minimum n = 10,610, maximum n = 298,420; some GWASs were imputed with the 1000 Genomes Project as a reference panel by their respective consortia, whereas others were not). For each trait, we identified a gene set by mapping each autosomal genome-wide-significant SNP (p < 5 × 10−8) to the closest up- and downstream protein-coding genes as defined above, resulting in a total of 62 non-disjoint GWAS gene sets. Because GWAS regions often contain multiple genome-wide-significant SNPs and the relevant gene might not lie adjacent to the lead SNP in a region,70, 71 we defined GWAS gene sets by mapping genes with respect to every genome-wide-significant SNP rather than only the index GWAS SNPs at each genomic risk region.
Quantifying Overlap between Complex Traits and Mendelian Disorders
For each pair of complex trait and Mendelian disorder, we compared the GWAS gene set and phenotype-specific Mendelian disorder gene set with a 2 × 2 contingency table (indicating the counts of gene membership in the GWAS gene set only, in the Mendelian disorder gene set only, in both, or in neither) whereby the set of autosomal protein-coding genes (n = 17,695) represented the total sample. We used Fisher’s exact test72 to determine significance. We assessed phenotype specificity of overlap significance by comparing the GWAS gene sets for each complex trait (n = 62) with all phenotype-specific Mendelian disorder gene sets (n = 20), a total of 1,240 pairs. Significance was assessed at a false-discovery rate (FDR) threshold < 5% (p < 0.00310).
To assess the robustness and stability of our SNP-gene mapping approach for complex traits, we performed an overlap quantification between phenotype-specific Mendelian disorder genes and GWAS gene sets derived from three additional SNP-gene mapping methods: mapping each genome-wide-significant SNP to all genes within a 50 Mb window, mapping each genome-wide-significant SNP to all genes within a 500 Mb window, and mapping all SNPs in the credible set to the closest two genes. Comparison of the ORs produced by Fisher’s exact test demonstrated no major difference in outcomes from different mapping methods (Table S4); thus, we found that even more conservative gene sets, such as the GWAS gene sets derived from the credible set for each complex trait, still demonstrate the pattern of trait-specific enrichment.
Estimating Enrichment of GWAS SNP Association Signal
We created genomic annotations to capture the regions spanning 50 kb upstream through 50 kb downstream of gene bodies for four categories of genes: all protein-coding genes (n = 17,695), all Mendelian disorder genes (n = 3,446), all LoF-intolerant genes (n = 2,978), and the phenotype-specific Mendelian disorder gene sets (average n = 609). For each pair of complex trait and gene category, we computed enrichment of GWAS signal within category c with respect to the set of all protein-coding genes according to Equation 1:
| Equation 1 |
Here, Nc is the number of genes in category c, Mj is the number of SNPs within 50 kb of gene j, Zi is the GWAS effect size of SNP i divided by the standard error, and Np is the total number of protein-coding genes. Thus, ac is the enrichment of the average SNP effect size (Z2) per gene in category c (compared with the average Z2 for any protein-coding gene). The percent increase in average SNP effect size per gene for category c, or (ac − 1) × 100, is shown in Figure 3. We performed similar comparisons for median SNP effect size per gene for category c and maximum SNP effect size per gene for category c (Table S5).
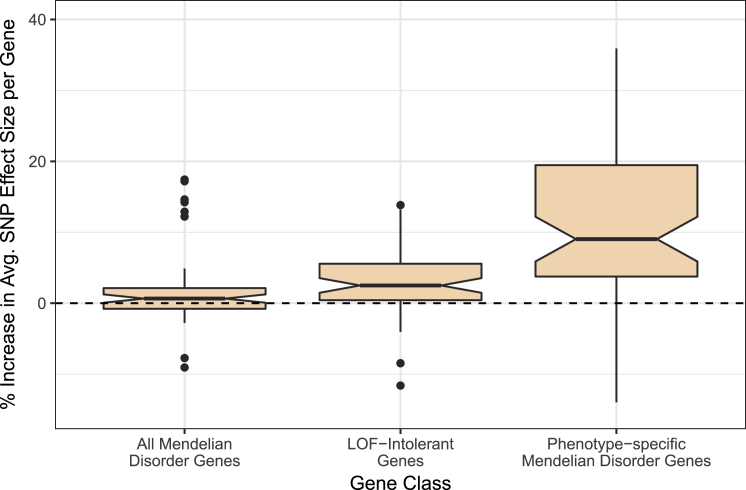
Figure 3.
Effect Sizes for SNPs on Complex Traits from GWASs Are Higher for LoF-Intolerant Genes and for Phenotypically Relevant Mendelian Disorder Genes
The increase in average SNP effect size per gene across gene categories, as compared with all protein-coding genes (dashed line), is shown. We averaged effect size (Z2) across all SNPs falling within 50 kb of a gene to obtain an average SNP effect size per gene and averaged across all genes in each category (all protein-coding genes, all Mendelian disorder genes, all LoF-intolerant genes, and all phenotypically relevant Mendelian disorder genes for each trait). We normalized these averages to the average SNP effect per gene for any protein-coding genes. The boxplots represent the distribution of increases in average effect size per gene across all traits, and notches designate the confidence intervals (CIs). From left to right, CIs read (0.07, 1.24), (1.47, 3.54), and (5.88, 12.19).
To ensure that this signal was not driven by linkage disequilibrium (LD), minor allele frequency (MAF), or average gene length per category, we compared these three properties across the gene categories for each complex trait. We calculated LD scores73 reflecting the amount of LD tagged by each SNP in the HapMap 3 reference panel; then, for each gene category, we averaged the LD scores of SNPs falling within 50 kb of each gene. We performed similar analyses to examine average MAF per gene and average gene length per category across each complex trait (Table S6). For comparison, we additionally performed a permutation test by drawing 100 sets of random genes for each Mendelian disorder gene set (matched for number and length of genes) and computing the average effect size per gene for each phenotypically matched complex trait across all 100 random sets.
Putative Causal Mechanisms at GWAS Risk Regions
We performed statistical fine-mapping of the genome-wide-significant regions (p < 5 × 10−8) for each GWAS by using fgwas74 with no functional annotations and default parameter settings. For each GWAS, we constructed a 95% credible set (defined as the minimum set of SNPs where 95% of the probability of causation at a region is accumulated) for each region of 500 SNPs containing a significant GWAS association. We achieved this by adding SNPs one at a time with a decreasing posterior probability of causation (posterior probability of association for the SNP, conditioned on there being an association in the region) until a cumulative 95% probability of causation was reached.
Identification of Candidate Regulatory Variants
We intersected credible sets for each complex trait with genomic regions 1 kb upstream of each phenotypically relevant Mendelian disorder gene to identify SNPs localizing at the TSS. To identify genes whose expression the GWAS SNPs might regulate, we queried the UCSC GTEx Combined eQTL Track (version 2017-10-25) and filtered by SNP rsID. This table describes all gene-tissue pairs where a SNP has evidence of regulatory function. We restricted results to phenotype-matched Mendelian genes whose promoter contained a genome-wide-significant SNP in our GWAS fine-mapped results. To identify candidate regulatory variants interacting with promoters of phenotype-matched Mendelian disorder genes, we used interactions from promoter capture Hi-C in human primary white adipocytes66 for each complex trait and filtered interactions to pairs of interacting regions where at least one region contained a promoter of a phenotype-specific Mendelian disorder gene. We then intersected interaction pairs for each of these regions with credible sets for each complex trait to identify credible SNPs interacting with regions containing promoters of phenotype-specific Mendelian disorder genes.
Estimating the Enrichment of SNP Heritability of Complex Traits within Mendelian Disorder Gene-Set Annotations
We used stratified LD score regression (s-LDSC)75 to estimate the enrichment of SNP heritability of 47 complex traits and diseases within each of 20 Mendelian disorder gene-set annotations corresponding to the regions spanning 50 kb upstream through 50 kb downstream of gene bodies for each Mendelian disorder gene set. Of the 62 total GWAS traits analyzed in this study, the 47 complex traits and diseases are a subset that meet the criteria for running s-LDSC (i.e., the GWAS did not use custom genotyping arrays). The annotation value for SNP i and gene set k is defined as aik = 1 if SNP i is within 50 kb upstream or 50 kb downstream of any of the gene bodies in gene set k and aik = 0 otherwise. For each of the 20 annotations, we computed LD scores within 1 cM blocks with default parameters and LD estimated from the European individuals in the 1000 Genomes Phase 3 reference panel. For each GWAS-annotation pair, we ran s-LDSC by using the recommended “baseline model”75 as covariates in the regression for a total of 53 annotations per run (52 “baseline” annotations plus the gene-set annotation of interest).
Results
GWAS Risk Genes Show Specific, Significant Overlap with Phenotypically Matched Mendelian Disorder Genes
We first sought to examine the degree of overlap between phenotype-matched Mendelian disorder genes with risk genes for complex traits as identified through GWASs. For each complex trait, we identified corresponding Mendelian forms, often as familial forms or rare phenotypic extremes, and curated Mendelian disorder gene sets composed of Mendelian disorder genes linked to those specific phenotypes from OMIM (see Material and Methods and Figure 1). We combined similar Mendelian disorder gene sets to create one gene set for the representative Mendelian disorder(s) (for a total of 20 Mendelian disorder gene sets). We separately ascertained GWAS gene sets for each complex trait by identifying the closest up- and downstream genes to each GWAS SNP meeting genome-wide significance (see Material and Methods and Figure 1). Overlap between each phenotype-specific Mendelian disorder gene set (n = 20) and each GWAS gene set (n = 62) was assessed by Fisher’s exact test for a total of 1,240 comparisons (Tables 1 and S7). We hypothesized that GWAS gene sets would have a specific significant enrichment of Mendelian disorder genes for perfectly matched Mendelian disorders (as identified in Table 1; 62 of the 1,240 comparisons) or related Mendelian disorders (an additional 30 of the 1,240 comparisons; 92 of 1,240 total) but no enrichment for unrelated Mendelian disorders (the remaining 1,148 of 1,240 comparisons). Among all 1,240 pairs of complex and Mendelian disorder gene sets assessed, we identified 77 pairs with significant overlap crossing an FDR < 5% cutoff at p < 0.00310 (Figure 2). An examination of the log ORs for each overlap comparison revealed more extreme enrichments among phenotypically matched pairs than among phenotypically unmatched pairs (Table 2), which is consistent with our hypothesis. 50 out of the 77 significantly overlapping pairs showed perfectly matching phenotypes (as defined in Table 1; see Material and Methods) or reflected known shared biology (identified in dark blue within Figure 2). Specifically, in many of these pairs, monogenic forms of the complex trait have been well established in the genetics literature; examples include age-related macular degeneration and cholesterol traits (high-density lipoprotein [HDL], low-density lipoprotein [LDL], total cholesterol [TC], and triglycerides [TG]).7, 76, 77, 78, 79 We confirmed significant enrichment between many of these previously reported pairs; for example, we found significant enrichment between the complex and monogenic forms of height80 (OR = 1.39, p = 1.43 × 10−3) and between HDL and Mendelian forms of cardiovascular disease81 (OR = 2.10, p = 3.45 × 10−4). We also identified previously unreported enrichments; for example, we found a strong enrichment between inflammatory bowel disease and Mendelian forms of immune dysregulation (OR = 3.32, p = 1.58 × 10−8) and between hemoglobin and Mendelian hematologic disorders (OR = 3.99, p = 4.42 × 10− 4). The remaining 27 pairs with significant overlap suggested shared biological mechanisms yet to be established between complex traits and Mendelian disorders (Table 3). For example, we observed an enrichment between height and renal disorders (OR = 1.48, p = 3.75 × 10−5) and enrichment between Crohn disease and mature-onset diabetes of the young (OR = 2.69, p = 2.32 × 10−4). Strikingly, a higher proportion of phenotypically matched or related pairs (n = 50 of 92 [54%]) than phenotypically unmatched pairs (n = 27 of 1,148 [2%]) demonstrated significant overlap, consistent with our hypothesis of a phenotype-specific enrichment pattern (Table S7 and Figure S2).
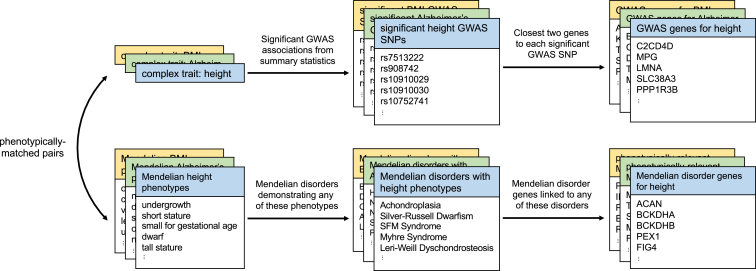
Figure 1.
GWAS Gene Sets and Phenotype-Specific Mendelian Disorder Gene Sets
For each complex trait (e.g., height), we first identified matched Mendelian phenotypes (e.g., undergrowth and short stature; Table S2). Using publicly available GWAS data, we defined the “GWAS genes” for a given complex trait to be the closest upstream and closest downstream protein-coding genes for every genome-wide-significant variant in the GWAS. We selected phenotype-matched Mendelian disorder genes by first identifying Mendelian disorders expressing any of the matched Mendelian phenotypes and then identifying all genes linked to any of those disorders.
Table 3.
Instances of Significant Overlap between GWAS Genes and Unrelated Mendelian Disorder Genes
| Complex Trait (No. of Genes) | Matched or Related Mendelian Disorder (No. of Genes) | Shared Genes | OR (CI) | Raw p Value | Average OR for Remaining Unrelated Pairs (CI) |
|---|---|---|---|---|---|
| ALZ (58) | immune dysregulation (550) | 7 | 4.32 (1.65, 9.62) | 2.06 × 10−3 | 1.95 (1.36, 2.53) |
| AMD (215) | microalbumin disorders (159) | 8 | 4.43 (1.86, 9.13) | 7.37 × 10−4 | 1.59 (1.03, 2.15) |
| BW (179) | mature-onset diabetes of the young (561) | 16 | 3.06 (1.69, 5.16) | 1.91 × 10−4 | 2.35 (1.94, 2.75) |
| immune dysregulation (550) | 14 | 2.69 (1.43, 4.68) | 1.44 × 10−3 | ||
| cardiovascular disease (598) | 14 | 2.46 (1.31, 4.28) | 3.10 × 10−3 | ||
| CAD (132) | neurologic disease (222) | 7 | 4.52 (1.76, 9.75) | 1.39 × 10−3 | 2.34 (1.83, 2.86) |
| CD (239) | bone and uric acid disorders (220) | 9 | 3.20 (1.42, 6.29) | 3.10 × 10−3 | 2.09 (−0.39, 4.58) |
| mature-onset diabetes of the young (561) | 19 | 2.69 (1.58, 4.35) | 2.32 × 10−4 | ||
| renal disorders (838) | 23 | 2.17 (1.34, 3.37) | 1.13 × 10−3 | ||
| CEL (34) | disorders of platelet function (443) | 5 | 6.78 (2.04, 17.83) | 1.47 × 10−3 | 1.90 (1.28, 2.52) |
| FN (58) | positive mood disorders (69) | 3 | 14.51 (2.83, 46.53) | 1.50 × 10−3 | 3.79 (2.36, 5.22) |
| HGT (2361) | renal disorders (838) | 153 | 1.48 (1.23, 1.78) | 3.75 × 10−5 | 1.31 (1.19, 1.42) |
| IBD (368) | mature-onset diabetes of the young (561) | 30 | 2.81 (1.85, 4.13) | 2.40 × 10−6 | 1.38 (1.10, 1.66) |
| LDL (370) | hematologic disorders (551) | 25 | 2.31 (1.46, 3.51) | 3.50 × 10−4 | 1.66 (1.24, 2.08) |
| LEPB (5) | female reproductive disorders (288) | 2 | 40.52 (3.37, 353.67) | 2.56 × 10−3 | 10.83 (3.75, 17.91) |
| MCH (164) | bone and uric acid disorders (220) | 8 | 4.19 (1.75, 8.61) | 1.05 × 10−3 | 1.65 (1.15, 2.16) |
| insulin disorders (623) | 15 | 2.80 (1.52, 4.81) | 6.99 × 10−4 | ||
| MCV (180) | bone and uric acid disorders (220) | 8 | 3.80 (1.59, 7.79) | 1.89 × 10−3 | 1.74 (1.25, 2.22) |
| PCV (53) | positive mood disorders (69) | 3 | 15.97 (3.11, 51.37) | 1.16 × 10−3 | 4.04 (2.22, 5.87) |
| PLT (134) | cardiovascular disease (598) | 12 | 2.85 (1.42, 5.20) | 1.97 × 10−3 | 1.42 (0.98, 1.87) |
| RBC (107) | disorders of platelet function (443) | 12 | 5.03 (2.49, 9.29) | 1.51 × 10−5 | 2.90 (2.13, 3.67) |
| renal disorders (838) | 18 | 4.14 (2.33, 6.96) | 2.60 × 10−6 | ||
| SLE (286) | mature-onset diabetes of the young (561) | 22 | 2.61 (1.59, 4.07) | 1.24 × 10−4 | 1.59 (1.24, 1.94) |
| TC (500) | hematologic disorders (551) | 32 | 2.20 (1.47, 3.18) | 1.18 × 10−4 | 1.35 (1.05, 1.66) |
| disorders of platelet function (443) | 25 | 2.11 (1.34, 3.20) | 1.13 × 10−3 | ||
| UC (202) | mature-onset diabetes of the young (561) | 19 | 3.25 (1.90, 5.27) | 2.44 × 10−5 | 1.41 (1.05, 1.77) |
| WHR (74) | disorders of platelet function (443) | 7 | 4.12 (1.59, 9.03) | 2.50 × 10−3 | 2.38 (1.58, 3.18) |
As in Table 2, Fisher’s exact test was used to quantify the enrichment of shared genes between complex traits and Mendelian disorders with an OR and p value (see Material and Methods). Pairs with significant enrichment passed the cutoff of FDR < 5% at p < 0.00310. This table lists pairs of complex traits and phenotypically unrelated Mendelian disorders that demonstrated significant overlap. For comparison, the average OR and 95% CI for pairings of each complex trait with all remaining unrelated Mendelian disorder gene sets are included.
To investigate whether intra-chromosomal proximal clustering of genes with similar functionality was confounding our results, we pruned our dataset of all protein-coding genes to include only one gene per 33.2 kb window across the genome (determined by the average distance to the next closest gene in our dataset) and re-computed overlap ORs. The results were highly similar to those of our original approach (Pearson r = 0.96). Moreover, we found the average distance to the next closest gene among the sets of genes shared by a phenotypically matched pair of complex trait and Mendelian disorder to be 21.1 Mb (Table S8).
SNPs near Phenotypically Matched Mendelian Disorder Genes Show Increased Effect Size on Complex Traits
Because Mendelian disorder genes are linked with severe biological effects when either one or both alleles are disrupted, dysregulation of the gene through changes in expression or other mechanisms might have a more significant effect than dysregulation of another protein-coding gene not linked to any Mendelian disorders. We hypothesized that SNPs near these phenotype-specific Mendelian disorder genes have further increased effects on complex traits as a result of the increased biological relevance of these gene categories. From the publicly available GWAS summary statistics for each complex trait, we computed the average GWAS effect sizes of SNPs falling within each protein-coding gene and compared the average effect sizes per gene across all Mendelian disorder genes and across phenotypically relevant Mendelian disorder genes (see Material and Methods). Across complex traits, we found an increased average effect size per gene for all Mendelian disorder genes and a further increased average effect size per gene for phenotypically relevant Mendelian disorder genes (Figure 3 and Table S5). This suggests that the genomic regions containing the most biologically relevant genes for each trait contribute most significantly to the biology of complex traits. We also confirmed that LoF-intolerant genes (as defined by ExAC’s pLI score > 0.9; see Material and Methods) demonstrate a higher average effect size across most complex traits examined.68 Given the extreme intolerance of deleterious mutations in these genes, it is possible that LoF-intolerant genes are linked with embryonically lethal mutant phenotypes and are thus undiscovered as Mendelian disorder genes at this time.
We found no significant difference in LD or average MAF between the SNPs within each category and the SNPs within all protein-coding genes (Table S6), suggesting that the observed signal is not driven by any of these potential confounders. In particular, we found the average LD tagged for all protein-coding genes to be 24.38 (95% confidence interval [CI] = (24.35, 24.42)); for none of the other three gene classes did the CIs fall above the upper bound, including the average across all Mendelian disorder gene sets (24.10; 95% CI = (23.47, 24.72)). Similarly, we found the average MAF for all protein-coding genes to be 0.238 (95% CI = (0.237, 0.239)); for none of the other gene classes did the CIs fall below the lower bound, including the average across all Mendelian disorder gene sets (0.238; 95% CI = (0.237, 0.240)). Details for each gene set are included in Table S6. Of note, we did observe an increase in average gene length across gene categories, particularly between all protein-coding genes (159.94 kb; 95% CI = (158.16 kb, 161.71 kb)) and the average across all phenotype-specific Mendelian disorder gene sets (177.87 kb; 95% CI = (168.49 kb, 187.24 kb)) (Table S6). To ensure that our findings of enriched GWAS signal in these gene categories were not due to longer genes’ being more likely to tag causal variation, we performed a permutation test comparing the average effect size per gene for phenotype-matched Mendelian disorder genes with the same metric across 100 sets of random genes (matched for number of genes and gene length) (Table S6). We found no evidence that gene length confounded our results given that across 58 of 62 complex traits, the average effect size per gene was higher for phenotype-matched Mendelian disorder genes than for random genes of the same length. Also of note, we did not find evidence of a pervasive phenotype-specific enrichment of SNP heritability within 50 kb of Mendelian disorder genes (Table S9; see Material and Methods). Thus, we can conclude that the average GWAS effect size per gene for phenotypically relevant Mendelian disorder genes is higher than that of all protein-coding genes, all Mendelian disorder genes, and LoF-intolerant genes.
Examples of Credible SNPs for GWAS Regions near Phenotypically Matched Mendelian Disorder Genes
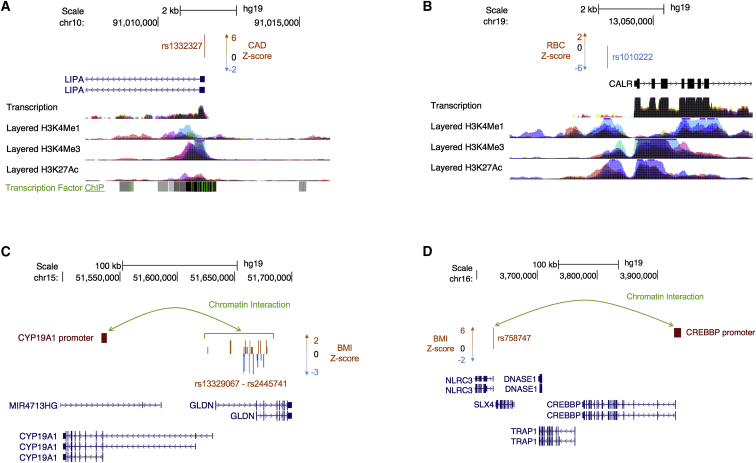
We next sought to identify common non-coding variants that might causally affect complex-trait phenotypes by dysregulating phenotypically relevant Mendelian disorder genes. For each complex trait, we performed statistical fine-mapping of significant GWAS regions to construct 95% credible sets for each region (see Material and Methods) and identified SNPs from the credible set located at the TSS of a gene from the phenotypically relevant Mendelian disorder gene set. We found a total of 786 credible-set SNPs (out of approximately 3.5 million) localizing at the TSS of a phenotypically relevant Mendelian disorder gene (an average of 20 SNPs per trait for 38 traits where at least one such SNP was found; Tables S10 and S11). We identified 25 promising candidate SNPs (attaining genome-wide significance in a GWAS) at TSSs that could be regulating the proximal Mendelian disorder gene (Table 4). We further examined the GTEx Portal to determine whether any of these SNPs were also expression quantitative trail loci (eQTLs) for the corresponding gene; we found 12 variants to be significant eQTLs for the corresponding gene in at least one tissue (Table 4). We highlight two examples: first, we found a significantly associated SNP from the credible set for coronary artery disease (rs1332327, Z = 6.80) at the promoter of LIPA (MIM: 278000), a Mendelian disorder gene linked to Wolman disease and cholesteryl ester storage disease (both lysosomal acid lipase deficiencies [MIM: 278000]), which involve hypercholesterolemia and hypertriglyceridemia as part of cholesteryl ester- and triglyceride-filled macrophage infiltration syndromes (Figure 4A). Additional analyses identified rs1332327 (along with other SNPs in LD with the variant) as a cis-eQTL for LIPA in the METSIM adipose RNA-sequencing dataset (Table S13); this finding is consistent with eQTL results reported in the GTEx Portal for these SNPs and LIPA. Second, from the credible set for red blood cell count, we found a significantly associated SNP (rs1010222, Z = −5.97) at the promoter of CALR (MIM: 109091), a Mendelian disorder gene linked to myelofibrosis (MIM: 254450), which involves generalized bone marrow fibrosis, reduced hemopoiesis, a lack of hemophagocytosis, and myeloproliferative disease (Figure 4B). In both cases, the putative causal SNP for the complex trait lies immediately upstream of the TSS of the phenotypically relevant Mendelian disorder gene, in addition to falling within regions containing regulatory epigenetic marks.
Table 4.
Genome-wide-Significant SNPs Localizing at the TSS of Phenotypically Relevant Mendelian Disorder Genes
| Complex Trait | SNP ID | Chromosomal Position | Z Score | Gene (MIM) | Maximum eQTL Effect | Maximum −log10 p Value | Tissues |
|---|---|---|---|---|---|---|---|
| PBC | rs13239597 | chr7: 128,695,982 | 9.85 | TNPO3 (610032) | – | – | – |
| HGT | rs8028537 | chr15: 89,345,946 | −9.33 | ACAN (155760) | – | – | – |
| HGT | rs10853751 | chr19: 41,903,219 | 8.71 | BCKDHA (608348) | – | – | – |
| CD | rs59283234 | chr5: 150,225,586 | −8.45 | IRGM (608212) | −0.042 | 6.733 | whole blood |
| CD | rs751627 | chr5: 150,225,112 | −8.45 | IRGM (608212) | −0.042 | 6.733 | whole blood |
| CD | rs35707106 | chr5: 150,225,376 | −8.33 | IRGM (608212) | −0.041 | 6.412 | whole blood |
| HGT | rs2298307 | chr6: 80,816,295 | 8.28 | BCKDHB (248611) | – | – | – |
| HGT | rs12386601 | chr7: 92,157,885 | 8.20 | PEX1 (602136) | – | – | – |
| BMI | rs17066842 | chr18: 58,040,623 | −7.54 | MC4R (155541) | – | – | – |
| HGT | rs12192268 | chr6: 110,011,457 | −7.00 | FIG4 (609390) | – | – | – |
| CAD | rs1332327 | chr10: 91,011,680 | 6.80 | LIPA (613497) | 12.541 | 5.132 | subcutaneous adipose, visceral adipose, adrenal gland, transverse colon, lung, spleen, thyroid, whole blood |
| RA | rs13239597 | chr7: 128,695,982 | 6.66 | TNPO3 (610032) | – | – | – |
| IBD | rs59283234 | chr5: 150,225,586 | 6.51 | IRGM (608212) | −0.042 | 6.733 | whole blood |
| IBD | rs751627 | chr5: 150,225,112 | 6.51 | IRGM (608212) | −0.042 | 6.733 | whole blood |
| HGT | rs7592246 | chr2: 219,926,220 | 6.45 | IHH (600726) | 0.056 | 6.199 | brain cerebellum |
| IBD | rs34005003 | chr5: 150,225,198 | 6.43 | IRGM (608212) | −0.043 | 6.637 | whole blood |
| IBD | rs35707106 | chr5: 150,225,376 | 6.33 | IRGM (608212) | −0.041 | 6.412 | whole blood |
| MNR | rs3775971 | chr4: 104,641,919 | 6.20 | TACR3 (162332) | −0.019 | 8.588 | lung |
| IBD | rs27741 | chr16: 28,504,180 | 6.11 | CLN3 (607042) | – | – | – |
| HGT | rs4244808 | chr11: 2,163,109 | 6.06 | IGF2 (147470) | – | – | – |
| RBC | rs1010222 | chr19: 13,048,607 | −5.96 | CALR (109091) | 17.142 | 6.851 | lung |
| CD | rs27741 | chr16: 28,504,180 | −5.87 | CLN3 (607042) | – | – | – |
| HGT | rs613924 | chr11: 65,769,294 | −5.86 | BANF1 (603811) | – | – | – |
| AFB | rs4845357 | chr1: 153,896,211 | −5.78 | GATAD2B (614998) | −0.108 | 4.443 | skin not exposed |
| HGT | rs6591226 | chr11: 66,675,989 | 5.52 | PC (608786) | – | – | – |
GWAS SNPs from the credible set for each complex trait were intersected with TSS regions 1 kb upstream of phenotypically matched Mendelian disorder genes. This table lists all genome-wide-significant SNPs (p < 5 × 10−8 from GWASs) with chromosomal location from all complex traits localizing at the TSS of a phenotypically matched Mendelian disorder gene.
Figure 4.
Candidate Regulatory SNPs Fall at TSSs and Long-Range Promoters of Phenotypically Relevant Mendelian Disorder Genes
(A and B) Shown here are two examples of putative causal SNPs localizing at a TSS of a phenotypically relevant Mendelian disorder gene. (A) Putative causal SNP rs1332327, associated with coronary artery disease (Z = 6.80), lies at the TSS of LIPA. (B) Putative causal SNP rs1010222, associated with red blood cell count (Z = −5.97), lies at the TSS of CALR.
(C and D) Shown here are two representations of chromatin interactions in white adipose tissue. (C) A cluster of SNPs from the credible set of variants associated with BMI (Z score plotted in orange and blue) physically interacts with the promoter of a particular isoform of CYP19A1. (D) A single SNP (rs758747) from the credible set, associated with BMI (Z = 6.08), physically interacts with the promoter of a distant gene, CREBBP.
Putative Causal SNPs for GWAS Regions Interacting with Promoters of Phenotypically Relevant Mendelian Disorder Genes
Functional genomic datasets, such as chromatin interactions identified through Hi-C, can give us insight into the functional interpretation of GWAS variants and how they might regulate Mendelian disorder genes. Examination of chromatin interactions in human primary white adipocytes66 revealed further candidate credible-set, metabolic-trait SNPs physically interacting with promoters of phenotypically relevant Mendelian disorder genes (Table S12). Specifically, we report that a genome-wide-significant SNP for BMI (rs758747, Z = 6.08) physically interacts with the promoter of CREBBP, a gene linked to Rubinstein-Taybi syndrome 1 (MIM: 180849), in which obesity is one of the syndromic features (Figure 4C). These interactions can also identify the relevant isoforms of genes in disease. From the credible set of variants associated with BMI, we identified a cluster of SNPs that physically interact with the promoter of a specific isoform of CYP19A1, a gene linked to aromatase excess syndrome (MIM: 139300), which involves short stature and excess fat storage in the chest (gynecomastia) (Figure 4D). Although longer isoforms of CYP19A1 are by default chosen to represent the gene, our data suggest that the shorter isoform is likely to be more relevant in obesity. Together, these results demonstrate examples of GWAS variants localizing in regulatory regions for phenotypically relevant Mendelian disorder genes, consistent with the hypothesis that low-effect common variants contribute to complex traits by regulating genes known to cause Mendelian disorders.
Discussion
In this work, we used GWAS summary statistics from 62 complex traits and genes linked to specific phenotypes within 20 Mendelian broad disorders to quantify the shared genetic basis of complex traits and Mendelian disorders. We identified a pervasive, specific enrichment of phenotypically matched and related Mendelian disorder genes in GWAS regions for complex traits; we also identified some pairs of complex traits and phenotypically unmatched Mendelian disorders with significant enrichment. We further found that phenotypically relevant Mendelian disorder genes are enriched with GWAS signal across complex traits in comparison with all Mendelian disorder genes and other protein-coding genes. Finally, we report examples of putative causal SNPs for GWAS regions in potentially regulating phenotypically relevant Mendelian disorder genes. We conclude with four considerations about how our results contribute to the understanding of genetic architectures and biological mechanisms across complex traits and Mendelian disorders.
First, our finding of a specific enrichment of phenotypically matched and related Mendelian disorder genes in GWAS regions for complex traits suggests that, across complex-trait architectures, many complex traits share a genetic basis (and by extension, biological mechanisms) with their Mendelian forms. This supports our hypothesis that the shared genes contribute to both extreme and common genetic phenotypes and suggests an important role of gene regulation by non-coding variants in complex traits. However, we note that our findings are limited by the power of each GWAS to detect significant associations. As GWASs become better powered, we anticipate being able to identify phenotype-specific enrichments of Mendelian disorder genes in GWAS regions for more complex traits.
Second, we identified a subset of pairs of complex traits and Mendelian disorders that, despite having no known shared biology, still demonstrated significant enrichment of Mendelian disorder genes in GWAS regions. This subset of pairs can offer us insight into the biological mechanisms of complex traits and Mendelian disorders. A high degree of co-morbidity between complex traits and Mendelian disorders has been previously observed regardless of phenotype similarity;24 these findings together suggest that many complex traits and Mendelian disorders might also be linked by the pleiotropic properties of the underlying genes, in addition to regulatory differences. These observations are also consistent with a multigenic or oligogenic architecture of human disease; the pervasive pleiotropic effects that are seen across complex traits are consistent with the widespread prevalence of multi-system, syndromic phenotypes observed across a majority of Mendelian disorders. We also confirm that LoF-intolerant genes harbor an enrichment of GWAS signal;68 because genes with pLI > 0.9 exhibit extreme intolerance of deleterious mutation, it is possible that these genes demonstrate embryonically lethal mutant phenotypes and are thus undiscovered as Mendelian disorder genes at this time. Our findings provide further motivation to explore phenotypic consequences of mutations in LoF-intolerant genes (particularly those enriched with GWAS signal for a particular complex trait) for phenotypically relevant Mendelian disorders.
Third, linking Mendelian disorder genes with complex traits can help with characterization of the genetic architecture of complex traits—specifically with genes and pathways that can be functionally characterized for the identification of molecular mechanisms.7 Identifying causal variants from large-scale GWASs is particularly challenging given that most GWAS loci lie in non-coding regions of the genome; although thousands of genomic loci have been significantly associated with specific diseases, few casual SNPs have been functionally verified.82, 83 Many approaches have been used to tie a particular variant to a relevant gene or genes,84, 85, 86 including newer methods that directly link gene expression to a trait (e.g., transcriptome-wide association studies70 and PrediXcan87); we have found that leveraging GWAS findings with functional data to identify candidate regulatory variants for Mendelian disorder genes can potentially lead to better interpretation of relevant genes and isoforms. Here, we demonstrate the heterogeneity of mechanisms potentially underlying causal variation by showing roles for TSS promoter regions of Mendelian disorder genes and long-range interactions involving significant GWAS regions. We expand on recent work showing that BMI-associated variants interact with genes in GWAS regions to demonstrate similar findings for Mendelian disorder genes.66 With the appropriate functional data from relevant tissues and cell types, this phenotype-driven approach can identify relevant candidate regulatory variants and their targets. Further, from the perspective of monogenic diseases, identifying common variants that might modify the expressivity of phenotypes can provide insights into gene function in addition to putative drug targets. Many drugs approved by the FDA and developed by pharmaceutical companies are targeted toward the treatment of complex traits and diseases; by identifying underlying links between Mendelian disorders and complex traits through their effects on the same biological genes and pathways, we can systematically and rationally target existing drugs for complex traits and diseases toward those with rare Mendelian disorders that largely do not have any rationally targeted treatments.88, 89, 90
Fourth and finally, we note that our approach of examining traits and disorders at the component-phenotype level offers us valuable resolution into the specific pathways involved in the overall trait or disorder. In clinical genetics, genome-wide sequencing has expanded the clinical phenotypic spectrum associated with a gene91, 92 through identification of pleiotropic effects due to mutations in specific protein domains.93, 94 These advances have detected a genetic predisposition for diseases previously considered to be due to environment13 and uncovered variable penetrance for genetic mutations previously thought to be sufficient to cause disease. Furthermore, the findings from clinical genome-wide sequencing have suggested that genetic background influences the phenotypic variability of monogenic diseases.95, 96 The phenotypic characterizations of Mendelian syndromes are deconstructed by expert clinical geneticists into component phenotypes, labeled by standardized clinical terms that identify both the primary phenotypes and phenotypes that have variable penetrance and expressivity.97 Recent work has demonstrated that incorporating such dense phenotype information into ranking putative disease-causing genetic mutations improves diagnostic rates in clinical exome sequencing tests;98, 99 using component Mendelian phenotypes to identify Mendelian disorders that might be phenotypically relevant to a variety of complex traits can be similarly impactful in identifying biological pathways for complex traits. Ultimately, identification of GWAS-significant regions with biologically relevant genes and pathways will enable effective utilization of GWAS data in medical settings.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We would like to thank Ruth Johnson, Megan Major, Megan Roytman, Claudia Giambartolomei, Arunabha Majumdar, Jazlyn Mooney, Brendan Freund, Robert Brown, Tommer Schwarz, and Robert Smith for helpful discussions. This work was funded by National Institutes of Health (NIH) Training Grant in Genomic Analysis and Interpretation T32HG002536 to M.K.F.; NIH Early Independence Award DP5OD024579 to V.A.A.; NIH awards R01HG009120, R01MH115676, R01HG006399, and U01CA194393 to B.P.; NIH National Institute of Mental Health grant T32MH073526 to K.S.B.; NIH grant F31HL142180 to K.M.G.; NIH National Cancer Institute grant T32LM012424 to D.Z.P.; and NIH grants HL-095056 and HL-28481 to P.P. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Published: October 4, 2018
Footnotes
Supplemental Data include 2 figures and 14 tables and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.08.017.
Contributor Information
Malika Kumar Freund, Email: kumarm@ucla.edu.
Valerie A. Arboleda, Email: varboleda@mednet.ucla.edu.
Web Resources
1000 Genomes Phase 3, http://www.internationalgenome.org/category/phase-3/
ClinVar, ftp://ftp.ncbi.nlm.nih.gov/pub/clinvar/tab_delimited/gene_specific_summary.txt
ExAC Browser, http://exac.broadinstitute.org/downloads
Gene sets, https://github.com/bogdanlab/gene_sets
GTEx Portal, https://www.gtexportal.org/home/
HapMap 3, https://www.sanger.ac.uk/resources/downloads/human/hapmap3.html
OMIM, https://omim.org/
UCSC GTEx Combined eQTL Track, http://genome.ucsc.edu/cgi-bin/hgGtexTrackSettings?db=hg19&g=gtexEqtlCluster
UCSC Table Browser, https://genome.ucsc.edu/cgi-bin/hgTables
Supplemental Data
References
- 1.Schork N.J., Murray S.S., Frazer K.A., Topol E.J. Common vs. rare allele hypotheses for complex diseases. Curr. Opin. Genet. Dev. 2009;19:212–219. doi: 10.1016/j.gde.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cutting G.R., Kasch L.M., Rosenstein B.J., Zielenski J., Tsui L.C., Antonarakis S.E., Kazazian H.H., Jr. A cluster of cystic fibrosis mutations in the first nucleotide-binding fold of the cystic fibrosis conductance regulator protein. Nature. 1990;346:366–369. doi: 10.1038/346366a0. [DOI] [PubMed] [Google Scholar]
- 3.Henriksen M.G., Nordgaard J., Jansson L.B. Genetics of schizophrenia: Overview of methods, findings and limitations. Front. Hum. Neurosci. 2017;11:322. doi: 10.3389/fnhum.2017.00322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Katsanis N. The continuum of causality in human genetic disorders. Genome Biol. 2016;17:233. doi: 10.1186/s13059-016-1107-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Auer P.L., Teumer A., Schick U., O’Shaughnessy A., Lo K.S., Chami N., Carlson C., de Denus S., Dubé M.P., Haessler J. Rare and low-frequency coding variants in CXCR2 and other genes are associated with hematological traits. Nat. Genet. 2014;46:629–634. doi: 10.1038/ng.2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen J., Pertsemlidis A., Kotowski I.K., Graham R., Garcia C.K., Hobbs H.H. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat. Genet. 2005;37:161–165. doi: 10.1038/ng1509. [DOI] [PubMed] [Google Scholar]
- 7.Tsai C.W., North K.E., Tin A., Haack K., Franceschini N., Saroja Voruganti V., Laston S., Zhang Y., Best L.G., MacCluer J.W. Both rare and common variants in PCSK9 influence plasma low-density lipoprotein cholesterol level in American Indians. J. Clin. Endocrinol. Metab. 2015;100:E345–E349. doi: 10.1210/jc.2014-3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han K., Holder J.L., Jr., Schaaf C.P., Lu H., Chen H., Kang H., Tang J., Wu Z., Hao S., Cheung S.W. SHANK3 overexpression causes manic-like behaviour with unique pharmacogenetic properties. Nature. 2013;503:72–77. doi: 10.1038/nature12630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sztainberg Y., Zoghbi H.Y. Lessons learned from studying syndromic autism spectrum disorders. Nat. Neurosci. 2016;19:1408–1417. doi: 10.1038/nn.4420. [DOI] [PubMed] [Google Scholar]
- 10.Lin A., Ching C.R.K., Vajdi A., Sun D., Jonas R.K., Jalbrzikowski M., Kushan-Wells L., Pacheco Hansen L., Krikorian E., Gutman B. Mapping 22q11.2 gene dosage effects on brain morphometry. J. Neurosci. 2017;37:6183–6199. doi: 10.1523/JNEUROSCI.3759-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toro R., Konyukh M., Delorme R., Leblond C., Chaste P., Fauchereau F., Coleman M., Leboyer M., Gillberg C., Bourgeron T. Key role for gene dosage and synaptic homeostasis in autism spectrum disorders. Trends Genet. 2010;26:363–372. doi: 10.1016/j.tig.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 12.Posey J.E., Harel T., Liu P., Rosenfeld J.A., James R.A., Coban Akdemir Z.H., Walkiewicz M., Bi W., Xiao R., Ding Y. Resolution of disease phenotypes resulting from multilocus genomic variation. N. Engl. J. Med. 2017;376:21–31. doi: 10.1056/NEJMoa1516767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corvol H., Blackman S.M., Boëlle P.Y., Gallins P.J., Pace R.G., Stonebraker J.R., Accurso F.J., Clement A., Collaco J.M., Dang H. Genome-wide association meta-analysis identifies five modifier loci of lung disease severity in cystic fibrosis. Nat. Commun. 2015;6:8382. doi: 10.1038/ncomms9382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emond M.J., Louie T., Emerson J., Zhao W., Mathias R.A., Knowles M.R., Wright F.A., Rieder M.J., Tabor H.K., Nickerson D.A., National Heart, Lung, and Blood Institute (NHLBI) GO Exome Sequencing Project. Lung GO Exome sequencing of extreme phenotypes identifies DCTN4 as a modifier of chronic Pseudomonas aeruginosa infection in cystic fibrosis. Nat. Genet. 2012;44:886–889. doi: 10.1038/ng.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dorfman R., Sandford A., Taylor C., Huang B., Frangolias D., Wang Y., Sang R., Pereira L., Sun L., Berthiaume Y. Complex two-gene modulation of lung disease severity in children with cystic fibrosis. J. Clin. Invest. 2008;118:1040–1049. doi: 10.1172/JCI33754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dron J.S., Hegele R.A. Genetics of triglycerides and the risk of atherosclerosis. Curr. Atheroscler. Rep. 2017;19:31. doi: 10.1007/s11883-017-0667-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hegele R.A. Learning from patients with ultrarare conditions: Cholesterol hoof beats. J. Am. Coll. Cardiol. 2018;71:289–291. doi: 10.1016/j.jacc.2017.09.1158. [DOI] [PubMed] [Google Scholar]
- 18.Peltonen L., Perola M., Naukkarinen J., Palotie A. Lessons from studying monogenic disease for common disease. Hum. Mol. Genet. 2006;15:R67–R74. doi: 10.1093/hmg/ddl060. [DOI] [PubMed] [Google Scholar]
- 19.Hernandez D.G., Reed X., Singleton A.B. Genetics in Parkinson disease: Mendelian versus non-Mendelian inheritance. J. Neurochem. 2016;139(Suppl 1):59–74. doi: 10.1111/jnc.13593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lim E.T., Liu Y.P., Chan Y., Tiinamaija T., Käräjämäki A., Madsen E., Altshuler D.M., Raychaudhuri S., Groop L., Flannick J., Go-T2D Consortium A novel test for recessive contributions to complex diseases implicates Bardet-Biedl syndrome gene BBS10 in idiopathic type 2 diabetes and obesity. Am. J. Hum. Genet. 2014;95:509–520. doi: 10.1016/j.ajhg.2014.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chan Y., Salem R.M., Hsu Y.H., McMahon G., Pers T.H., Vedantam S., Esko T., Guo M.H., Lim E.T., Franke L., GIANT Consortium Genome-wide analysis of body proportion classifies height-associated variants by mechanism of action and implicates genes important for skeletal development. Am. J. Hum. Genet. 2015;96:695–708. doi: 10.1016/j.ajhg.2015.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wheeler H.E., Gamazon E.R., Frisina R.D., Perez-Cervantes C., El Charif O., Mapes B., Fossa S.D., Feldman D.R., Hamilton R.J., Vaughn D.J. Variants in WFS1 and other Mendelian deafness genes are associated with cisplatin-associated ototoxicity. Clin. Cancer Res. 2017;23:3325–3333. doi: 10.1158/1078-0432.CCR-16-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amininejad L., Charloteaux B., Theatre E., Liefferinckx C., Dmitrieva J., Hayard P., Muls V., Maisin J.M., Schapira M., Ghislain J.M., International IBD Genetics Consortium Analysis of genes associated with monogenic primary immunodeficiency identifies rare variants in XIAP in patients with Crohn’s disease. Gastroenterology. 2018;154:2165–2177. doi: 10.1053/j.gastro.2018.02.028. [DOI] [PubMed] [Google Scholar]
- 24.Blair D.R., Lyttle C.S., Mortensen J.M., Bearden C.F., Jensen A.B., Khiabanian H., Melamed R., Rabadan R., Bernstam E.V., Brunak S. A nondegenerate code of deleterious variants in Mendelian loci contributes to complex disease risk. Cell. 2013;155:70–80. doi: 10.1016/j.cell.2013.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Welter D., MacArthur J., Morales J., Burdett T., Hall P., Junkins H., Klemm A., Flicek P., Manolio T., Hindorff L., Parkinson H. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dubois P.C., Trynka G., Franke L., Hunt K.A., Romanos J., Curtotti A., Zhernakova A., Heap G.A., Adány R., Aromaa A. Multiple common variants for celiac disease influencing immune gene expression. Nat. Genet. 2010;42:295–302. doi: 10.1038/ng.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu J.Z., van Sommeren S., Huang H., Ng S.C., Alberts R., Takahashi A., Ripke S., Lee J.C., Jostins L., Shah T., International Multiple Sclerosis Genetics Consortium. International IBD Genetics Consortium Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet. 2015;47:979–986. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gieger C., Radhakrishnan A., Cvejic A., Tang W., Porcu E., Pistis G., Serbanovic-Canic J., Elling U., Goodall A.H., Labrune Y. New gene functions in megakaryopoiesis and platelet formation. Nature. 2011;480:201–208. doi: 10.1038/nature10659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cordell H.J., Han Y., Mells G.F., Li Y., Hirschfield G.M., Greene C.S., Xie G., Juran B.D., Zhu D., Qian D.C., Canadian-US PBC Consortium. Italian PBC Genetics Study Group. UK-PBC Consortium International genome-wide meta-analysis identifies new primary biliary cirrhosis risk loci and targetable pathogenic pathways. Nat. Commun. 2015;6:8019. doi: 10.1038/ncomms9019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okada Y., Wu D., Trynka G., Raj T., Terao C., Ikari K., Kochi Y., Ohmura K., Suzuki A., Yoshida S., RACI consortium. GARNET consortium Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506:376–381. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sawcer S., Hellenthal G., Pirinen M., Spencer C.C., Patsopoulos N.A., Moutsianas L., Dilthey A., Su Z., Freeman C., Hunt S.E., International Multiple Sclerosis Genetics Consortium. Wellcome Trust Case Control Consortium 2 Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–219. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Autism Spectrum Disorder Working Group of the Psychiatry Genomics Consortium (2015). Dataset: PGC-ASD summary statistics from a meta-analysis of 5,305 ASD-diagnosed cases and 5,305 pseudocontrols of European descent (based on similarity to CEPH reference genotypes). March 2015. http://www.med.unc.edu/pgc/results-and-downloads.
- 33.van der Harst P., Zhang W., Mateo Leach I., Rendon A., Verweij N., Sehmi J., Paul D.S., Elling U., Allayee H., Li X. Seventy-five genetic loci influencing the human red blood cell. Nature. 2012;492:369–375. doi: 10.1038/nature11677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bentham J., Morris D.L., Graham D.S.C., Pinder C.L., Tombleson P., Behrens T.W., Martín J., Fairfax B.P., Knight J.C., Chen L. Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat. Genet. 2015;47:1457–1464. doi: 10.1038/ng.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horikoshi M., Beaumont R.N., Day F.R., Warrington N.M., Kooijman M.N., Fernandez-Tajes J., Feenstra B., van Zuydam N.R., Gaulton K.J., Grarup N., CHARGE Consortium Hematology Working Group. Early Growth Genetics (EGG) Consortium Genome-wide associations for birth weight and correlations with adult disease. Nature. 2016;538:248–252. doi: 10.1038/nature19806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wood A.R., Esko T., Yang J., Vedantam S., Pers T.H., Gustafsson S., Chu A.Y., Estrada K., Luan J., Kutalik Z., Electronic Medical Records and Genomics (eMEMERGEGE) Consortium. MIGen Consortium. PAGEGE Consortium. LifeLines Cohort Study Defining the role of common variation in the genomic and biological architecture of adult human height. Nat. Genet. 2014;46:1173–1186. doi: 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng H.F., Forgetta V., Hsu Y.H., Estrada K., Rosello-Diez A., Leo P.J., Dahia C.L., Park-Min K.H., Tobias J.H., Kooperberg C., AOGC Consortium. UK10K Consortium Whole-genome sequencing identifies EN1 as a determinant of bone density and fracture. Nature. 2015;526:112–117. doi: 10.1038/nature14878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Köttgen A., Albrecht E., Teumer A., Vitart V., Krumsiek J., Hundertmark C., Pistis G., Ruggiero D., O’Seaghdha C.M., Haller T., LifeLines Cohort Study. CARDIoGRAM Consortium. DIAGRAM Consortium. ICBP Consortium. MAGIC Consortium Genome-wide association analyses identify 18 new loci associated with serum urate concentrations. Nat. Genet. 2013;45:145–154. doi: 10.1038/ng.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nikpay M., Goel A., Won H.H., Hall L.M., Willenborg C., Kanoni S., Saleheen D., Kyriakou T., Nelson C.P., Hopewell J.C. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat. Genet. 2015;47:1121–1130. doi: 10.1038/ng.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Willer C.J., Schmidt E.M., Sengupta S., Peloso G.M., Gustafsson S., Kanoni S., Ganna A., Chen J., Buchkovich M.L., Mora S., Global Lipids Genetics Consortium Discovery and refinement of loci associated with lipid levels. Nat. Genet. 2013;45:1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Soranzo N., Sanna S., Wheeler E., Gieger C., Radke D., Dupuis J., Bouatia-Naji N., Langenberg C., Prokopenko I., Stolerman E., WTCCC Common variants at 10 genomic loci influence hemoglobin A1(C) levels via glycemic and nonglycemic pathways. Diabetes. 2010;59:3229–3239. doi: 10.2337/db10-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morris A.P., Voight B.F., Teslovich T.M., Ferreira T., Segrè A.V., Steinthorsdottir V., Strawbridge R.J., Khan H., Grallert H., Mahajan A., Wellcome Trust Case Control Consortium. Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC) Investigators. Genetic Investigation of ANthropometric Traits (GIANT) Consortium. Asian Genetic Epidemiology Network–Type 2 Diabetes (AGEN-T2D) Consortium. South Asian Type 2 Diabetes (SAT2D) Consortium. DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fritsche L.G., Igl W., Bailey J.N., Grassmann F., Sengupta S., Bragg-Gresham J.L., Burdon K.P., Hebbring S.J., Wen C., Gorski M. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat. Genet. 2016;48:134–143. doi: 10.1038/ng.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perry J.R., Day F., Elks C.E., Sulem P., Thompson D.J., Ferreira T., He C., Chasman D.I., Esko T., Thorleifsson G., Australian Ovarian Cancer Study. GENICA Network. kConFab. LifeLines Cohort Study. InterAct Consortium. Early Growth Genetics (EGG) Consortium Parent-of-origin-specific allelic associations among 106 genomic loci for age at menarche. Nature. 2014;514:92–97. doi: 10.1038/nature13545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Day F.R., Ruth K.S., Thompson D.J., Lunetta K.L., Pervjakova N., Chasman D.I., Stolk L., Finucane H.K., Sulem P., Bulik-Sullivan B., PRACTICAL consortium. kConFab Investigators. AOCS Investigators. Generation Scotland. EPIC-InterAct Consortium. LifeLines Cohort Study Large-scale genomic analyses link reproductive aging to hypothalamic signaling, breast cancer susceptibility and BRCA1-mediated DNA repair. Nat. Genet. 2015;47:1294–1303. doi: 10.1038/ng.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dupuis J., Langenberg C., Prokopenko I., Saxena R., Soranzo N., Jackson A.U., Wheeler E., Glazer N.L., Bouatia-Naji N., Gloyn A.L., DIAGRAM Consortium. GIANT Consortium. Global BPgen Consortium. Anders Hamsten on behalf of Procardis Consortium. MAGIC investigators New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat. Genet. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Teumer A., Tin A., Sorice R., Gorski M., Yeo N.C., Chu A.Y., Li M., Li Y., Mijatovic V., Ko Y.A., DCCT/EDIC Genome-wide association studies identify genetic loci associated with albuminuria in diabetes. Diabetes. 2016;65:803–817. doi: 10.2337/db15-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saxena R., Hivert M.F., Langenberg C., Tanaka T., Pankow J.S., Vollenweider P., Lyssenko V., Bouatia-Naji N., Dupuis J., Jackson A.U., GIANT consortium. MAGIC investigators Genetic variation in GIPR influences the glucose and insulin responses to an oral glucose challenge. Nat. Genet. 2010;42:142–148. doi: 10.1038/ng.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Onengut-Gumuscu S., Chen W.M., Burren O., Cooper N.J., Quinlan A.R., Mychaleckyj J.C., Farber E., Bonnie J.K., Szpak M., Schofield E., Type 1 Diabetes Genetics Consortium Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat. Genet. 2015;47:381–386. doi: 10.1038/ng.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lambert J.C., Ibrahim-Verbaas C.A., Harold D., Naj A.C., Sims R., Bellenguez C., DeStafano A.L., Bis J.C., Beecham G.W., Grenier-Boley B., European Alzheimer’s Disease Initiative (EADI) Genetic and Environmental Risk in Alzheimer’s Disease. Alzheimer’s Disease Genetic Consortium. Cohorts for Heart and Aging Research in Genomic Epidemiology Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Otowa T., Hek K., Lee M., Byrne E.M., Mirza S.S., Nivard M.G., Bigdeli T., Aggen S.H., Adkins D., Wolen A. Meta-analysis of genome-wide association studies of anxiety disorders. Mol. Psychiatry. 2016;21:1391–1399. doi: 10.1038/mp.2015.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ripke S., Wray N.R., Lewis C.M., Hamilton S.P., Weissman M.M., Breen G., Byrne E.M., Blackwood D.H., Boomsma D.I., Cichon S., Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium A mega-analysis of genome-wide association studies for major depressive disorder. Mol. Psychiatry. 2013;18:497–511. doi: 10.1038/mp.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Okbay A., Baselmans B.M., De Neve J.E., Turley P., Nivard M.G., Fontana M.A., Meddens S.F., Linnér R.K., Rietveld C.A., Derringer J., LifeLines Cohort Study Genetic variants associated with subjective well-being, depressive symptoms, and neuroticism identified through genome-wide analyses. Nat. Genet. 2016;48:624–633. doi: 10.1038/ng.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Psychiatric G.C.B.D.W.G., Psychiatric GWAS Consortium Bipolar Disorder Working Group Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat. Genet. 2011;43:977–983. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schizophrenia Working Group of the Psychiatric Genomics Consortium Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pattaro C., Teumer A., Gorski M., Chu A.Y., Li M., Mijatovic V., Garnaas M., Tin A., Sorice R., Li Y., ICBP Consortium. AGEN Consortium. CARDIOGRAM. CHARGe-Heart Failure Group. ECHOGen Consortium Genetic associations at 53 loci highlight cell types and biological pathways relevant for kidney function. Nat. Commun. 2016;7:10023. doi: 10.1038/ncomms10023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eppinga R.N., Hagemeijer Y., Burgess S., Hinds D.A., Stefansson K., Gudbjartsson D.F., van Veldhuisen D.J., Munroe P.B., Verweij N., van der Harst P. Identification of genomic loci associated with resting heart rate and shared genetic predictors with all-cause mortality. Nat. Genet. 2016;48:1557–1563. doi: 10.1038/ng.3708. [DOI] [PubMed] [Google Scholar]
- 58.Barban N., Jansen R., de Vlaming R., Vaez A., Mandemakers J.J., Tropf F.C., Shen X., Wilson J.F., Chasman D.I., Nolte I.M., BIOS Consortium. LifeLines Cohort Study Genome-wide analysis identifies 12 loci influencing human reproductive behavior. Nat. Genet. 2016;48:1462–1472. doi: 10.1038/ng.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rietveld C.A., Medland S.E., Derringer J., Yang J., Esko T., Martin N.W., Westra H.J., Shakhbazov K., Abdellaoui A., Agrawal A., LifeLines Cohort Study GWAS of 126,559 individuals identifies genetic variants associated with educational attainment. Science. 2013;340:1467–1471. doi: 10.1126/science.1235488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Okbay A., Beauchamp J.P., Fontana M.A., Lee J.J., Pers T.H., Rietveld C.A., Turley P., Chen G.B., Emilsson V., Meddens S.F., LifeLines Cohort Study Genome-wide association study identifies 74 loci associated with educational attainment. Nature. 2016;533:539–542. doi: 10.1038/nature17671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lu Y., Day F.R., Gustafsson S., Buchkovich M.L., Na J., Bataille V., Cousminer D.L., Dastani Z., Drong A.W., Esko T. New loci for body fat percentage reveal link between adiposity and cardiometabolic disease risk. Nat. Commun. 2016;7:10495. doi: 10.1038/ncomms10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Locke A.E., Kahali B., Berndt S.I., Justice A.E., Pers T.H., Day F.R., Powell C., Vedantam S., Buchkovich M.L., Yang J., LifeLines Cohort Study. ADIPOGen Consortium. AGEN-BMI Working Group. CARDIOGRAMplusC4D Consortium. CKDGen Consortium. GLGC. ICBP. MAGIC Investigators. MuTHER Consortium. MIGen Consortium. PAGE Consortium. ReproGen Consortium. GENIE Consortium. International Endogene Consortium Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Felix J.F., Bradfield J.P., Monnereau C., van der Valk R.J., Stergiakouli E., Chesi A., Gaillard R., Feenstra B., Thiering E., Kreiner-Møller E., Bone Mineral Density in Childhood Study (BMDCS) Early Genetics and Lifecourse Epidemiology (EAGLE) consortium. Early Growth Genetics (EGG) Consortium. Bone Mineral Density in Childhood Study BMDCS Genome-wide association analysis identifies three new susceptibility loci for childhood body mass index. Hum. Mol. Genet. 2016;25:389–403. doi: 10.1093/hmg/ddv472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kilpeläinen T.O., Carli J.F., Skowronski A.A., Sun Q., Kriebel J., Feitosa M.F., Hedman A.K., Drong A.W., Hayes J.E., Zhao J. Genome-wide meta-analysis uncovers novel loci influencing circulating leptin levels. Nat. Commun. 2016;7:10494. doi: 10.1038/ncomms10494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shungin D., Winkler T.W., Croteau-Chonka D.C., Ferreira T., Locke A.E., Mägi R., Strawbridge R.J., Pers T.H., Fischer K., Justice A.E., ADIPOGen Consortium. CARDIOGRAMplusC4D Consortium. CKDGen Consortium. GEFOS Consortium. GENIE Consortium. GLGC. ICBP. International Endogene Consortium. LifeLines Cohort Study. MAGIC Investigators. MuTHER Consortium. PAGE Consortium. ReproGen Consortium New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518:187–196. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pan D.Z., Garske K.M., Alvarez M., Bhagat Y.V., Boocock J., Nikkola E., Miao Z., Raulerson C.K., Cantor R.M., Civelek M. Integration of human adipocyte chromosomal interactions with adipose gene expression prioritizes obesity-related genes from GWAS. Nat. Commun. 2018;9:1512. doi: 10.1038/s41467-018-03554-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Karolchik D., Hinrichs A.S., Furey T.S., Roskin K.M., Sugnet C.W., Haussler D., Kent W.J. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pasaniuc B., Price A.L. Dissecting the genetics of complex traits using summary association statistics. Nat. Rev. Genet. 2017;18:117–127. doi: 10.1038/nrg.2016.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mancuso N., Shi H., Goddard P., Kichaev G., Gusev A., Pasaniuc B. Integrating gene expression with summary association statistics to identify genes associated with 30 complex traits. Am. J. Hum. Genet. 2017;100:473–487. doi: 10.1016/j.ajhg.2017.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gusev A., Ko A., Shi H., Bhatia G., Chung W., Penninx B.W., Jansen R., de Geus E.J., Boomsma D.I., Wright F.A. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 2016;48:245–252. doi: 10.1038/ng.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fisher R.A. On the interpretation of χ2 from contingency tables, and the calculation of P. J. R. Stat. Soc. 1922;85:87–94. [Google Scholar]
- 73.Bulik-Sullivan B.K., Loh P.R., Finucane H.K., Ripke S., Yang J., Patterson N., Daly M.J., Price A.L., Neale B.M., Schizophrenia Working Group of the Psychiatric Genomics Consortium LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pickrell J.K. Joint analysis of functional genomic data and genome-wide association studies of 18 human traits. Am. J. Hum. Genet. 2014;94:559–573. doi: 10.1016/j.ajhg.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Finucane H.K., Bulik-Sullivan B., Gusev A., Trynka G., Reshef Y., Loh P.R., Anttila V., Xu H., Zang C., Farh K., ReproGen Consortium. Schizophrenia Working Group of the Psychiatric Genomics Consortium. RACI Consortium Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 2015;47:1228–1235. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fritsche L.G., Chen W., Schu M., Yaspan B.L., Yu Y., Thorleifsson G., Zack D.J., Arakawa S., Cipriani V., Ripke S., AMD Gene Consortium Seven new loci associated with age-related macular degeneration. Nat. Genet. 2013;45:433–439. doi: 10.1038/ng.2578. e1–e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Helgason H., Sulem P., Duvvari M.R., Luo H., Thorleifsson G., Stefansson H., Jonsdottir I., Masson G., Gudbjartsson D.F., Walters G.B. A rare nonsynonymous sequence variant in C3 is associated with high risk of age-related macular degeneration. Nat. Genet. 2013;45:1371–1374. doi: 10.1038/ng.2740. [DOI] [PubMed] [Google Scholar]
- 78.Brunham L.R., Singaraja R.R., Hayden M.R. Variations on a gene: rare and common variants in ABCA1 and their impact on HDL cholesterol levels and atherosclerosis. Annu. Rev. Nutr. 2006;26:105–129. doi: 10.1146/annurev.nutr.26.061505.111214. [DOI] [PubMed] [Google Scholar]
- 79.Kanoni S., Masca N.G., Stirrups K.E., Varga T.V., Warren H.R., Scott R.A., Southam L., Zhang W., Yaghootkar H., Müller-Nurasyid M., Wellcome Trust Case Control Consortium Analysis with the exome array identifies multiple new independent variants in lipid loci. Hum. Mol. Genet. 2016;25:4094–4106. doi: 10.1093/hmg/ddw227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yang J., Benyamin B., McEvoy B.P., Gordon S., Henders A.K., Nyholt D.R., Madden P.A., Heath A.C., Martin N.G., Montgomery G.W. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rosenson R.S., Brewer H.B., Jr., Barter P.J., Björkegren J.L.M., Chapman M.J., Gaudet D., Kim D.S., Niesor E., Rye K.A., Sacks F.M. HDL and atherosclerotic cardiovascular disease: genetic insights into complex biology. Nat. Rev. Cardiol. 2018;15:9–19. doi: 10.1038/nrcardio.2017.115. [DOI] [PubMed] [Google Scholar]
- 82.Sekar A., Bialas A.R., de Rivera H., Davis A., Hammond T.R., Kamitaki N., Tooley K., Presumey J., Baum M., Van Doren V., Schizophrenia Working Group of the Psychiatric Genomics Consortium Schizophrenia risk from complex variation of complement component 4. Nature. 2016;530:177–183. doi: 10.1038/nature16549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Smemo S., Tena J.J., Kim K.H., Gamazon E.R., Sakabe N.J., Gómez-Marín C., Aneas I., Credidio F.L., Sobreira D.R., Wasserman N.F. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 2014;507:371–375. doi: 10.1038/nature13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lango Allen H., Estrada K., Lettre G., Berndt S.I., Weedon M.N., Rivadeneira F., Willer C.J., Jackson A.U., Vedantam S., Raychaudhuri S. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.de Leeuw C.A., Mooij J.M., Heskes T., Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 2015;11:e1004219. doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Watanabe K., Taskesen E., van Bochoven A., Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 2017;8:1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gamazon E.R., Wheeler H.E., Shah K.P., Mozaffari S.V., Aquino-Michaels K., Carroll R.J., Eyler A.E., Denny J.C., Nicolae D.L., Cox N.J., Im H.K., GTEx Consortium A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet. 2015;47:1091–1098. doi: 10.1038/ng.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pascual V., Allantaz F., Arce E., Punaro M., Banchereau J. Role of interleukin-1 (IL-1) in the pathogenesis of systemic onset juvenile idiopathic arthritis and clinical response to IL-1 blockade. J. Exp. Med. 2005;201:1479–1486. doi: 10.1084/jem.20050473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pardoll D.M. Immunology beats cancer: a blueprint for successful translation. Nat. Immunol. 2012;13:1129–1132. doi: 10.1038/ni.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gerich M.E., McGovern D.P. Towards personalized care in IBD. Nat. Rev. Gastroenterol. Hepatol. 2014;11:287–299. doi: 10.1038/nrgastro.2013.242. [DOI] [PubMed] [Google Scholar]
- 91.Chong J.X., Buckingham K.J., Jhangiani S.N., Boehm C., Sobreira N., Smith J.D., Harrell T.M., McMillin M.J., Wiszniewski W., Gambin T., Centers for Mendelian Genomics The genetic basis of Mendelian phenotypes: Discoveries, challenges, and opportunities. Am. J. Hum. Genet. 2015;97:199–215. doi: 10.1016/j.ajhg.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Reuter C.M., Brimble E., DeFilippo C., Dries A.M., Enns G.M., Ashley E.A., Bernstein J.A., Fisher P.G., Wheeler M.T., Undiagnosed Diseases Network A new approach to rare diseases of children: The Undiagnosed Diseases Network. J. Pediatr. 2018;196:291–297.e2. doi: 10.1016/j.jpeds.2017.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Goudie D.R., D’Alessandro M., Merriman B., Lee H., Szeverényi I., Avery S., O’Connor B.D., Nelson S.F., Coats S.E., Stewart A. Multiple self-healing squamous epithelioma is caused by a disease-specific spectrum of mutations in TGFBR1. Nat. Genet. 2011;43:365–369. doi: 10.1038/ng.780. [DOI] [PubMed] [Google Scholar]
- 94.Arboleda V.A., Lee H., Parnaik R., Fleming A., Banerjee A., Ferraz-de-Souza B., Délot E.C., Rodriguez-Fernandez I.A., Braslavsky D., Bergadá I. Mutations in the PCNA-binding domain of CDKN1C cause IMAGe syndrome. Nat. Genet. 2012;44:788–792. doi: 10.1038/ng.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Born H.A., Dao A.T., Levine A.T., Lee W.L., Mehta N.M., Mehra S., Weeber E.J., Anderson A.E. Strain-dependence of the Angelman Syndrome phenotypes in Ube3a maternal deficiency mice. Sci. Rep. 2017;7:8451. doi: 10.1038/s41598-017-08825-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hensman Moss D.J., Pardiñas A.F., Langbehn D., Lo K., Leavitt B.R., Roos R., Durr A., Mead S., Holmans P., Jones L., Tabrizi S.J., TRACK-HD investigators. REGISTRY investigators Identification of genetic variants associated with Huntington’s disease progression: a genome-wide association study. Lancet Neurol. 2017;16:701–711. doi: 10.1016/S1474-4422(17)30161-8. [DOI] [PubMed] [Google Scholar]
- 97.Köhler S., Doelken S.C., Mungall C.J., Bauer S., Firth H.V., Bailleul-Forestier I., Black G.C., Brown D.L., Brudno M., Campbell J. The Human Phenotype Ontology project: linking molecular biology and disease through phenotype data. Nucleic Acids Res. 2014;42:D966–D974. doi: 10.1093/nar/gkt1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yang H., Robinson P.N., Wang K. Phenolyzer: phenotype-based prioritization of candidate genes for human diseases. Nat. Methods. 2015;12:841–843. doi: 10.1038/nmeth.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zemojtel T., Köhler S., Mackenroth L., Jäger M., Hecht J., Krawitz P., Graul-Neumann L., Doelken S., Ehmke N., Spielmann M. Effective diagnosis of genetic disease by computational phenotype analysis of the disease-associated genome. Sci. Transl. Med. 2014;6:252ra123. doi: 10.1126/scitranslmed.3009262. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.