Abstract
Excretion of albumin in urine, or albuminuria, is associated with the development of multiple cardiovascular and metabolic diseases. However, whether pathways leading to albuminuria are causal for cardiometabolic diseases is unclear. We addressed this question using a Mendelian randomization framework in the UK Biobank, a large population-based cohort. We first performed a genome-wide association study for albuminuria in 382,500 individuals and identified 32 new albuminuria loci. We constructed albuminuria genetic risk scores and tested for association with cardiometabolic diseases. Genetically elevated albuminuria was strongly associated with increased risk of hypertension (1.38 OR; 95% CI, 1.27–1.50 per 1 SD predicted increase in albuminuria, p = 7.01 × 10−14). We then examined bidirectional associations of albuminuria with blood pressure which suggested that genetically elevated albuminuria led to higher blood pressure (2.16 mmHg systolic blood pressure; 95% CI, 1.51–2.82 per 1 SD predicted increase in albuminuria, p = 1.22 × 10−10) and that genetically elevated blood pressure led to more albuminuria (0.005 SD; 95% CI 0.004–0.006 per 1 mmHg predicted increase in systolic blood pressure, p = 2.45 × 10−13). These results support the existence of a feed-forward loop between albuminuria and blood pressure and imply that albuminuria could increase risk of cardiovascular disease through blood pressure. Moreover, they suggest therapies that target albuminuria-increasing processes could have antihypertensive effects that are amplified through inhibition of this feed-forward loop.
Keywords: albuminuria, urine albumin excretion, urine albumin:creatinine ratio, cardiovascular disease, cardiometabolic disease, blood pressure, hypertension, genome-wide association study, Mendelian randomization, genetic risk score, polygenic risk score
Introduction
In observational epidemiologic studies, albuminuria, or the concentration of albumin excreted in urine, is associated with risk for multiple cardiometabolic diseases: elevations in albuminuria predict development of coronary artery disease, stroke, heart failure, type 2 diabetes, hypertension, and all-cause mortality.1, 2, 3, 4, 5, 6, 7, 8, 9 However, whether pathways leading to albuminuria are causally associated with cardiometabolic disease is unclear. Therapies lowering albuminuria are generally associated with reduced cardiovascular disease, for example. However, whether such effects are independent of concomitant reductions in blood pressure is ambiguous.10, 11, 12, 13, 70 Understanding whether associations of albuminuria pathways with disease reflect a causal relationship or mere correlation may inform whether targeting albuminuria-increasing processes could reduce risk for cardiometabolic diseases.
“Mendelian randomization” can provide evidence regarding the hypothesis that a given biomarker-disease relationship is causal.14 The strengths and limitations of Mendelian randomization can be considered via analogy with a randomized clinical trial. Individuals are assigned to lifelong increase or decrease in a disease risk factor due to the random segregation and independent assortment of genetic polymorphisms at conception, thus minimizing two key limitations of observational epidemiology, reverse causation and confounding. The effect of genetically modifying an exposure (here, albuminuria) can then be tested against increasing or decreasing risk of an outcome (here, cardiometabolic disease). Three assumptions must be met in order for a genetic variant to be a potentially valid instrumental variable in Mendelian randomization: (1) the variant must be strongly associated with the exposure, (2) the variant must not be associated with confounders, and (3) the variant must not be horizontally pleiotropic, i.e., cannot be associated with the outcome independent of the exposure pathway.15 While the second and third assumptions are hard to prove, many sensitivity analyses have been developed to improve the reliability of Mendelian randomization estimates.16
Here, we first identify genetic variants associated with albuminuria by conducting a genome-wide association study of albuminuria in 382,500 individuals in the UK Biobank. We subsequently utilized the identified genetic variants as instruments in a Mendelian randomization analysis to test the hypothesis that pathways increasing albuminuria are causal for cardiometabolic diseases.
Subjects and Methods
Study Design
This study had three main components. First, we examined epidemiological associations of baseline albuminuria with incident cardiometabolic disease in UK Biobank. Second, we conducted a genome-wide association study of baseline albuminuria and constructed a polygenic risk score. Finally, we performed a Mendelian randomization study to test the hypothesis that the associations between processes leading to albuminuria and cardiometabolic diseases are causal.
UK Biobank
Study Participants
Data from 382,500 unrelated individuals of European ancestry with albuminuria measurement in the UK Biobank were used. Samples were excluded for the following reasons: inferred sex did not match reported sex, kinship was not inferred, putative sex chromosome aneuploidy, consent withdrawn, or excessive heterozygosity or missingness, based on centralized sample quality control performed by UK Biobank.17 Excluded related individuals were defined as one individual in each pair with KING coefficient > 0.0884, indicating 2nd degree or closer relatedness. European ancestry was determined by self-reported ancestry of British, Irish, or other white, followed by outlier detection using the R package aberrant with lambda = 40 on genetic principal component (PC)1 and PC2, PC3 and PC4, and PC5 and PC6. Individuals who were outliers for any of the three pairs of PCs were removed from the European ancestry group. Kinship inference and genetic PCs were centrally calculated by UK Biobank.17
Albuminuria and Blood Pressure
Albuminuria was measured at the initial assessment visit (2006–2010); a Beckman Coulter AU5400 clinical chemistry analyzer was used to quantify urine albumin (df-30500, Randox Bioscience; immunoturbidimetric assay, detection range 6.7–200 mg/L) and urine creatinine (df-30510, Beckman Coulter; enzymatic assay, detection range 88–4,4200 μmol/L) concentrations. Urine albumin concentrations below the lower limit of detection (df-30505, n = 263654) were set to the lower limit of detection (6.7 mg/L). The resulting urine albumin:creatinine ratio (ACR, mg/g) was natural log-transformed to adjust for right skewedness. Microalbuminuria was defined as urine ACR of 25–355 mg/g in females and 17–250 mg/g in males; macroalbuminuria > 355 mg/g in females and > 250 mg/g in males.18 Baseline blood pressure was averaged from two measurements taken a few moments apart using an Omron 705 IT electronic blood pressure monitor (df-4079 and df-4080). A sphygmomanometer (df-93 and df-94) was used if a measurement could not be obtained with the electronic monitor. 381,833 individuals had both blood pressure and albuminuria measurements. Albuminuria and blood pressure can both be decreased by hypertensive medication, but there is no consensus about the magnitude of such effects on albuminuria. Therefore, neither variable was corrected for hypertensive medication use so as not to selectively skew one variable but not the other.
Disease Definitions
Prevalent cardiometabolic diseases were defined at study entry through the electronic health record and/or self-report with confirmation via verbal interview by a trained nurse. Detailed definitions for all disease classifications can be found in Table S2. Incident cardiometabolic diseases were ascertained among those not meeting disease criteria at baseline by applying phenotype definitions to longitudinal, in-patient hospital and death registry data linked to the UK Biobank. Participants were censored at the time of disease diagnosis, date of death, or date of last follow-up (i.e., February 9, 2016 for participants enrolled in Wales, February 16, 2016 for participants enrolled in England, and October 31, 2015 for participants enrolled in Scotland), whichever occurred first. Participants were presumed alive at last follow-up if there was no preceding report of death in the death register. UK Biobank was approved by the Research Ethics Committee (reference 16/NW/0274) and informed consent was obtained from all participants. Analysis of UK Biobank data was approved by the Partners HealthCare institutional review board (protocol 2013P001840).
Atherosclerosis Risk in Communities (ARIC)
10,235 unrelated individuals in the Atherosclerosis Risk in Communities study, genotyped using the Affymetrix Genome-wide Human SNP Array 5.0, were imputed to the Haplotype Reference Consortium using the Michigan Imputation Server. Phasing was performed using the Eagle2 algorithm. 4,954 variants were removed prior to imputation due to duplication, monomorphism, or allele mismatch. Imputation was then performed on 799,246 variants using the minimac3 algorithm. 39,235,157 variants in the Haplotype Reference Consortium were imputed. 6,398 individuals were of European ancestry as confirmed by centrally calculated, European-specific PC analysis and had albuminuria data.
An untimed urine sample was collected during the visit 4 clinical examination. Aliquots were frozen within 12 hr and stored at −70°C. Albumin and creatinine levels were measured in the University of Minnesota Physicians Outreach Laboratories, Minneapolis, Minnesota, with albumin by a nephelometric method either on the Dade Behring BN100 (assay sensitivity, 2.0 mg/L) or on the Beckman Image Nephelometer, and creatinine using the Jaffe method in order to determine the albumin-to-creatinine ratio (ACR; μg/mg) for participants. Blinded samples (n = 516) analyzed for quality assurance showed a correlation coefficient (r) of the loge-transformed ACR as r = 0.95. ACR was natural log-transformed for association analysis. Genotype and phenotype data were retrieved for analysis from NCBI dbGAP (phs000280.v3.p1) under procedures approved by the Partners HealthCare institutional review board (protocol 2016P002395).
Framingham Heart Study
8,825 individuals from the Offspring and Third Generation cohorts of the Framingham Heart Study, genotyped using the Affymetrix GeneChip Human Mapping 500K Array, were imputed to the Haplotype Reference Consortium using the Michigan Imputation Server. Genetic PCs were calculated on directly genotyped data using EIGENSOFT v7.2.1 after removing variants with MAF < 0.01 or genotype call rate < 0.99 and samples with sample call rate < 0.97 using PLINK-1.9. PCs were calculated in unrelated individuals only based on self-reported pedigree and projected onto related individuals. 6,534 individuals were of self-reported white ancestry confirmed by PC analysis. PCs used as covariates for association tests were recalculated in the white subgroup, and 21 individuals were removed as outliers on the basis of this analysis. 6,387 of the remaining individuals had albuminuria data available.
Albuminuria was measured at Offspring Exam 8 and Third Generation Exam 1 visits at the Framingham Heart Study Laboratory using a Roche Hitachi 911 Chemistry Analyzer. Urine albumin was quantified by the immunoturbidometric Tina-quant Albumin test (assay sensitivity, 3.0 mg/L); urine creatinine by colorimetric, modified Jaffe (rate blanked) creatinine test (assay sensitivity, 0.2 mg/100 mL). Urine albumin concentrations below the lower limit of detection (n = 1,682) were set to the lower limit of detection (3.0 mg/L). The resulting urine albumin:creatinine ratio (ACR, mg/g) was natural log-transformed for association analysis. Genotype and phenotype data were retrieved for analysis from NCBI dbGAP (phs000007.v26.p10) under procedures approved by the Partners HealthCare institutional review board (protocol 2016P002395).
International Consortium for Blood Pressure
The International Consortium for Blood Pressure is a large meta-analysis of study-specific results associating blood pressure with genotypes from the Cardio-MetaboChip SNP Array (nmax = 201,529)19 or imputed to 1000 Genomes Project Phase 1 haplotypes (nmax = 150,134).20 Summary statistics for SNP effects on systolic and diastolic blood pressure were corrected for anti-hypertensive medication use (+15 mmHg and +10 mmHg for systolic and diastolic blood pressure, respectively) and included body mass index, sex, age, and age2 as covariates, as previously described.19, 20 One limitation of this dataset is that adjustment for body mass index and anti-hypertensive medication may lead to associations between genetic variants and adjusted blood pressure being confounded with other factors that influence the adjustment variables (“collider effects”). This could bias Mendelian randomization analyses with albuminuria, which is not adjusted for these factors.21 SNPs from the Cardio-MetaboChip study19 were used to construct blood pressure genetic risk scores, whereas association of albuminuria variants with blood pressure was examined using blood pressure effects measured in the 1000G-based study.20
Statistical Analyses
Observational Epidemiology
In UK Biobank, Cox proportional hazards regression was used to determine the association of baseline albuminuria with incident cardiometabolic disease (average median follow-up time 7.0 years across diseases). Potential confounding variables were selected per prior epidemiological analyses of albuminuria and cardiometabolic disease,2, 5, 6, 9, 22, 23, 24 and a subsequent univariate screen of the selected traits in the UK Biobank was performed; these criteria yielded age at baseline, sex, current smoking status, body mass index, systolic blood pressure, diastolic blood pressure, baseline diabetes, and baseline hyperlipidemia as covariates for inclusion in most Cox proportional hazard models.
Genome-Wide Association Study
UK Biobank samples were genotyped by Affymetrix using either the UK BiLEVE or UK Biobank Axiom arrays. Genotyped variants were then imputed by the UK Biobank central analysis team onto the Haplotype Reference Consortium reference panel.17 Variant exclusion criteria were Hardy-Weinberg equilibrium p ≤ 1 × 10−20, QCTOOL INFOscore < 0.3, variant call rate ≤ 0.95, and MAF ≤ 0.001 yielding 11,709,857 variants in the analysis. Sex-specific residuals of natural log-transformed urine ACR were analyzed as a continuous trait with age, genotyping array, and the first ten genetic PCs as covariates via least-squares linear regression under an additive effects model using Hail v0.1 statistical software (Web Resources).25 The threshold for statistical significance was empirically determined using permutation testing according to a previous approach.26 Association of chromosome 21 variants with 1,000 random simulated continuous phenotypes were determined using Hail v0.1. The necessary significance threshold for a 5% family-wise error rate (FWER) was empirically estimated as the 5th percentile of the collection of the minimum variant p value from each simulated phenotype. The corresponding number of independent tests on chromosome 21 was calculated as p = 0.05/threshold5%FWER and was scaled to genome-wide using the proportion of the 11,709,857 genome-wide variants located on chromosome 21. This resulted in a genome-wide significance threshold of p < 9 × 10−9. Genomic inflation was calculated using the median estimator in the GenABEL package in R; LD score regression and common (MAF > 5%) SNP genetic correlation and heritability were calculated via LDSC v1.0.0 using standard variant filtering (MAF > 0.01 & INFOscore > 0.9), HapMap3 SNPs, and LD scores precomputed from European 1000 Genomes data.27 Variants were clumped into independent loci using PLINK-1.9 with R2 > 0.01 and < 1 MB from the index variant (smallest p value).
Albuminuria Genetic Risk Score
Up to 46 SNPs independently associated with albuminuria at a conventional p < 5 × 10−8 threshold in the genome-wide association study (Table S4) were used to construct weighted polygenic risk scores using PLINK 2.00a2LM. Each imputed genotype dosage was multiplied by the effect of the SNP on natural log-transformed urine ACR normalized to 1-SD albuminuria in UK Biobank (0.755 log(mg/g) urine ACR). The resulting weighted dosages were summed to create genetic risk scores. Association of the 46-SNP albuminuria genetic risk score with albuminuria in ARIC and Framingham Heart Study was determined using linear regression with age, sex, and the first ten genetic PCs as covariates. Sensitivity analysis excluding 1 and 10 poorly imputed variants in ARIC and Framingham Heart Study, respectively, did not substantially affect association results. Variance explained by each score was calculated as the adjusted R2 from the association of albuminuria with the albuminuria genetic risk score, age, sex, and ten genetic PCs minus the adjusted R2 from the association of albuminuria with age, sex, and ten genetic PCs.
Blood Pressure Genetic Risk Scores
Lead variants of genome-wide significant loci from Cardio-MetaboChip-based ICBP stage 4 meta-analysis19 were used to construct systolic blood pressure and diastolic blood pressure genetic risk scores. These results did not include UK Biobank. Only variants significantly (p < 5 × 10−8) associated with a specific blood pressure trait were included in that trait’s score. rs10164833 was excluded from the systolic blood pressure risk score as it did not replicate in further ICBP meta-analysis. In UK Biobank, each imputed genotype dosage was multiplied by the effect of the SNP on mmHg systolic or diastolic blood pressures from ICBP stage 4 meta-analysis, which were corrected for hypertensive medication use and body mass index,19 and the resulting weighted dosages were summed. Variance explained by each score in UK Biobank was calculated as the adjusted R2 from the association of blood pressure corrected for hypertensive medication use with the blood pressure genetic risk score, age, sex, and ten genetic PCs minus the adjusted R2 from the association of blood pressure corrected for hypertensive medication use with age, sex, and ten genetic PCs.
Mendelian Randomization
For individual-level data, association of the albuminuria or blood pressure genetic risk scores with outcomes were assessed using logistic (combined prevalent plus incident disease) or linear (continuous outcomes) two-stage least-squares regression in Stata v15. Age at baseline, sex, genotyping array, and the first ten genetic PCs to control for population structure were included as covariates. For summary-level data, the analogous approach is an inverse-variance-weighted (IVW) fixed-effects meta-analysis of the effect of each SNP on the outcome divided by the effect of this SNP on albuminuria.28, 29 Meta-analysis was conducted using the MendelianRandomization package30 in R. Effect estimates were normalized to 1 SD albuminuria in UK Biobank (0.755 log(mg/g) urine ACR). Power to detect associations with cardiometabolic disease in UK Biobank were calculated using an online tool (Web Resources, Table S13).
Sensitivity Analyses
We performed the following sensitivity analyses to address several limitations of Mendelian randomization: MR Steiger filtering to remove variants potentially acting through reverse causation,31, 32 calculation of heterogeneity and random-effects IVW meta-analysis to allow for variant effect size heterogeneity (Tables S6–S10 and S12),33, 34 median regressions which allow up to 50% of information from variants to violate Mendelian randomization assumptions,35 MR-Egger regression to detect directional pleiotropy,36 Cook’s distance to detect extreme outliers,37 and unweighted allele scores to minimize bias from internally derived weights in individual-level analyses.38
MR Steiger Filtering. The third assumption of Mendelian randomization (“no association independent of the exposure”) requires that a variant acts first through the exposure and not the outcome. Observational studies suggest that diseases such as diabetes and hypertension can increase albuminuria.39, 40 Some variants may therefore be associated with albuminuria via first increasing risk of such diseases and secondarily increasing albuminuria. These variants should not be included as instruments for testing the influence of albuminuria on those disease outcomes.
The MR Steiger method strengthens evidence regarding whether a variant acts first through the exposure or outcome under a model of vertical pleiotropy, where the SNP associates with two traits because one trait influences the other. The correlation of a variant with an outcome is a product of both the variant-exposure correlation and the exposure-outcome correlation. The variant-exposure correlation should therefore be greater than the variant-outcome correlation.32 The MR Steiger method determines the variant-exposure and variant-outcome correlations and removes variants where the variant-outcome correlation is greater than the variant-exposure correlation. The aim of this approach is to reduce the proportion of variants erroneously included in a Mendelian randomization analysis due to confounding or acting first through the outcome.31, 32 We note that MR Steiger is not designed to distinguish between vertical pleiotropy and horizontal pleiotropy, wherein a SNP influences both traits through independent pathways.
To perform MR Steiger filtering, the correlation of a variant with each exposure and outcome was first determined. For continuous traits from studies with individual-level data, the squared correlation of each variant with a continuous exposure or outcome was calculated as the R2 from association of the trait with the variant and covariates minus the R2 from association of the trait with covariates. For continuous traits from studies with summary statistics, the correlation R was estimated using get_r_from_pn in the TwoSampleMR package41 in R. Correlation R of each variant with binary traits was estimated on the logit liability scale32, 42 using get_r_from_lor in TwoSampleMR modified to use allele frequency measured in UK Biobank. To apply directional MR Steiger filtering, variants with R2exposure < R2outcome were removed.
As an additional sensitivity analysis for one-sample Mendelian randomizations with individual-level data, the Steiger test of correlated correlations was used to calculate the probability that the variant-exposure and variant-outcome correlations were different. Variants whose correlation with the exposure was not significantly different from correlation with the outcome, defined as Steiger p value > 0.05, were removed. Steiger tests were calculated using the r.test in the psych package in R. This analysis was not performed for two-sample Mendelian randomizations, as correlations of a variant with an exposure or outcome from summary statistics are estimated only in separate cohorts and therefore may be less appropriate for detecting significant differences between the two measurements.
Other Sensitivity Analyses. IVW random-effects, simple median, weighted median, and MR-Egger random-effects regression were calculated with normal distributions using the MendelianRandomization v0.2.0 package in R. For these analyses of individual-level data, associations of score SNPs with each outcome were determined via linear or logistic (Wald) regression using age at baseline, sex, genotyping array, and the first ten genetic PCs using Hail v0.1 statistical software. While MR-Egger can be particularly biased by weak instruments in individual-level or one-sample Mendelian randomization analyses,43 the fact that MR-Egger regression results were roughly similar between one-sample and two-sample analyses (Tables S8 and S10) suggests that weak instrument bias is not disproportionately affecting these results. Graphs of each variant’s effect on exposure versus outcome and IVW-based leave-one-out analyses (Figures S2–S4, S6, and S7) were created using the TwoSampleMR package in R. Variant effect heterogeneity was assessed via Cochran’s Q and MR-PRESSO residual sum of squares (RSS), which shows improved false-positive rates.33 These were calculated using the MendelianRandomization and MRPRESSO33 v1.0 packages, respectively, in R. For outlier detection, Cook’s distance was calculated on IVW meta-analysis; SNPs with a Cook’s distance greater than twice the nominal outlier cutoff 4/nSNPs were considered for outlier exclusion. Unweighted allele scores were constructed by summing the number of albuminuria- or blood pressure-increasing alleles per individual. Two-stage least-squares regression was used to determine the association of the unweighted allele score with cardiometabolic outcomes as described above. Linkage disequilibrium between variants in albuminuria genetic risk scores and blood pressure genetic risk scores was defined as R2 > 0.2 and < 1 MB using linkage disequilibrium calculated in the UK Biobank study population via PLINK-1.9.
Anti-hypertensive medications reduce albuminuria in addition to lowering blood pressure.44 We wanted to determine whether not correcting blood pressure and albuminuria for hypertensive medication use confounded albuminuria-blood pressure association results. As a sensitivity analysis, we therefore excluded individuals on hypertensive medication and re-tested association of an albuminuria genetic risk score with blood pressure. Hypertensive medication use was defined by self-report with confirmation via verbal interview by a trained nurse (df-6177 and df-6153). A genome-wide association study for albuminuria was performed in 302,687 individuals in UK Biobank not on hypertensive medication and who had blood pressure and albuminuria measurements. This yielded 23 independent loci (p < 5 × 10−8, R2 > 0.01 and < 1 MB from the index variant, Table S11). Effects of the 23 SNPs were used to construct an albuminuria risk score normalized to 1 SD albuminuria in this population (0.713 log (mg/g)). Associations of this albuminuria risk score with blood pressure were determined as above. Directional MR Steiger filtering removed one variant from the risk score for association with both systolic and diastolic blood pressure.
Results
382,500 unrelated individuals of European ancestry in the UK Biobank, a population-based cohort, were used in this study. 54% of participants were female, and the mean age was 56.9 (SD 7.9) years at baseline. Mean baseline systolic blood pressure and diastolic blood pressures were 138.3 (SD 18.6) and 82.3 (SD 10.1) mmHg, respectively; 18,940 (5.0%) individuals had diabetes at baseline and 53,004 (13.9%) had hyperlipidemia at baseline. The median baseline urine albumin:creatinine ratio (ACR) was 9.8 mg/g (IQR 6.1–16.5). 14.3% had microalbuminuria and 0.4% macroalbuminuria (Tables S1 and S2). Baseline urine ACR was natural log-transformed and is referred to as albuminuria (mean 2.3, SD 0.755 log(mg/g)) in subsequent analyses.
Association of Albuminuria with Development of Cardiometabolic Diseases
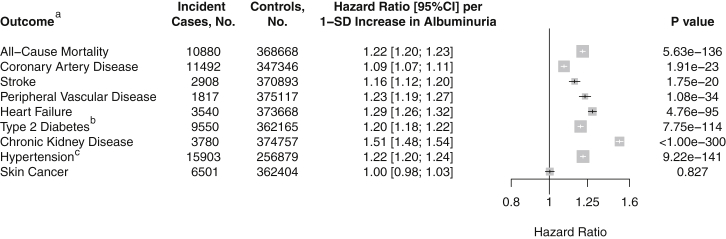
In UK Biobank, we first examined the association of baseline albuminuria with risk of incident cardiometabolic diseases using Cox proportional hazard regression (average median follow-up time across all diseases, 7.0 years). Baseline albuminuria was strongly associated with subsequent development of cardiometabolic disease (Figure 1): a 1 SD increase in albuminuria was associated with higher hazard of all-cause mortality (1.22 HR; 95%CI 1.20–1.23), coronary artery disease (1.09 HR; 95%CI 1.07–1.11), stroke (1.16 HR; 95%CI 1.12–1.20), peripheral vascular disease (1.23 HR; 95%CI 1.19–1.27), heart failure (1.29 HR; 95%CI 1.26–1.32), type 2 diabetes (1.20 HR; 95%CI 1.18–1.22), chronic kidney disease (1.51 HR; 95%CI 1.48–1.54), and hypertension (1.22 HR; 95%CI 1.20–1.24) but not with other diseases such as skin cancer (1.00 HR; 95%CI 0.98–1.03), even after adjustment for standard metabolic risk factors. Thus, albuminuria measured in UK Biobank is associated with cardiometabolic disease in a manner consistent with previous observational studies.1, 6, 7, 9, 22
Figure 1.
Association of Albuminuria with Incident Disease Endpoints in UK Biobank
aIncident disease adjusted for age, sex, current smoking status, body mass index, systolic blood pressure, diastolic blood pressure, baseline diabetes, and baseline hyperlipidemia unless otherwise specified.
bAdjusted for age, sex, current smoking status, body mass index, systolic blood pressure, diastolic blood pressure, waist-to-hip ratio, and baseline hyperlipidemia.
cAdjusted for age, age2, current smoking status, body mass index, baseline diabetes, and baseline hyperlipidemia.
Bars indicate 95% confidence interval for hazard ratio.
Genome-wide Association Study for Albuminuria
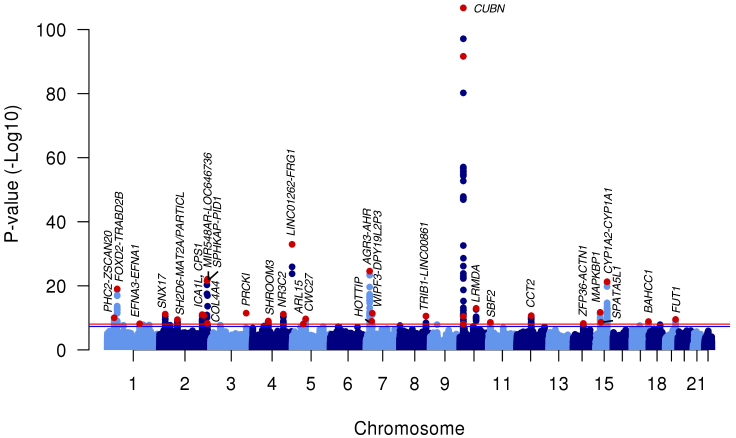
To identify variants to be used as genetic instruments for albuminuria, we conducted a discovery genome-wide association study of albuminuria in the 382,500 UK Biobank participants. Minimal genomic inflation was observed (lambdaGC = 1.17, LD score regression intercept27 = 1.02, Figure S1). The common SNP heritability of albuminuria was 0.045 (SE 0.002). In addition to replicating the previous association45 at the CUBN locus (rs10795433: beta = 0.024 log(mg/g) for C allele; p = 1.37 × 10−24, Table S3), we discovered an additional 1,246 genome-wide significant (p < 9 × 10−9) associations representing 32 novel (i.e., not previously published) independent loci, for a total of 33 genome-wide significant loci (Figure 2, Table 1). Novel associations of potential clinical interest include the NR3C2 and COL4A4 loci. NR3C2 encodes the mineralocorticoid receptor, and mineralocorticoid receptor antagonists such as spironolactone and eplerenone reduce albuminuria when added to other anti-hypertensive medications.46, 47 Mutations in COL4A4 and neighboring gene COL4A3 can cause autosomal Alport syndrome, which is characterized by kidney disease that can include proteinuria.48 22 of the 33 loci (or their proxies R2 > 0.8) were available in a smaller previously published genome-wide association study;45 of these, 20 had a consistent direction of effect and 7 were nominally significant (p < 0.05, Table S3).
Figure 2.
Genome-wide Association Study of Albuminuria in UK Biobank Identifies 32 Loci
33 genome-wide significant loci (including one previously published) are indicated by red points. Red line indicates genome-wide significance threshold (p = 9 × 10−9); blue line indicates conventional significance threshold (p = 5 × 10−8).
Table 1.
Albuminuria Loci from GWAS of 382,500 Individuals in UK Biobank
| Lead Variant | Nearest Gene(s) | Description | Chr | Position (hg19) | Effect Allele | Noneffect Allele | EAF | Beta (log(mg/g)) | SE (log (mg/g)) | p Value |
|---|---|---|---|---|---|---|---|---|---|---|
| rs12032996 | PHC2-ZSCAN20 | intergenic | 1 | 33920586 | G | A | 0.838 | 0.01463 | 0.00226 | 9.33E−11 |
| rs10157710 | FOXD2-TRABD2B | intergenic | 1 | 47961691 | T | C | 0.802 | 0.01900 | 0.00209 | 9.69E−20 |
| rs11264327 | EFNA3-EFNA1 | intergenic | 1 | 155095107 | A | G | 0.399 | 0.00987 | 0.00171 | 7.03E−09 |
| rs4665972 | SNX17 | intronic | 2 | 27598097 | T | C | 0.393 | 0.01176 | 0.00172 | 6.96E−12 |
| rs13394343 | SH2D6-MAT2A/PARTICL | intergenic | 2 | 85754342 | C | A | 0.570 | 0.01053 | 0.00168 | 3.86E−10 |
| rs10207567 | ICA1L | intronic | 2 | 203714973 | C | G | 0.813 | 0.01455 | 0.00214 | 1.00E−11 |
| rs1047891 | CPS1 | missense | 2 | 211540507 | C | A | 0.684 | 0.01205 | 0.00179 | 1.71E−11 |
| rs183131780 | MIR548AR-LOC646736 | intergenic | 2 | 226684886 | T | C | 0.002 | 0.19055 | 0.01959 | 2.33E−22 |
| rs35483183 | COL4A4 | intronic | 2 | 227876687 | A | G | 0.123 | 0.01490 | 0.00255 | 5.19E−09 |
| rs35924503 | SPHKAP-PID1 | intergenic | 2 | 229131286 | C | T | 0.001 | 0.24742 | 0.02518 | 8.68E−23 |
| rs112607182 | PRKCI | downstream variant | 3 | 170027407 | T | C | 0.077 | 0.02279 | 0.00327 | 3.39E−12 |
| rs7654754 | SHROOM3 | intronic | 4 | 77409795 | G | A | 0.462 | 0.01020 | 0.00167 | 9.96E−10 |
| rs6535594 | NR3C2 | intronic | 4 | 149132756 | A | G | 0.496 | 0.01146 | 0.00167 | 7.12E−12 |
| rs189107782 | LINC01262-FRG1 | intergenic | 4 | 190729009 | T | C | 0.002 | 0.24502 | 0.02026 | 1.12E−33 |
| rs702634 | ARL15 | intronic | 5 | 53271420 | A | G | 0.692 | 0.01042 | 0.00181 | 8.03E−09 |
| rs7731168 | CWC27 | intronic | 5 | 64296471 | C | G | 0.233 | 0.01253 | 0.00197 | 2.19E−10 |
| rs4410790 | AGR3-AHR | intergenic | 7 | 17284577 | C | T | 0.634 | 0.01798 | 0.00173 | 2.63E−25 |
| rs2023844 | HOTTIP | intronic | 7 | 27243238 | A | G | 0.926 | 0.01934 | 0.00318 | 1.18E−09 |
| rs17158386 | WIPF3-DPY19L2P3 | intergenic | 7 | 29805361 | A | G | 0.262 | 0.01330 | 0.00191 | 3.65E−12 |
| rs28601761 | TRIB1-LINC00861 | intergenic | 8 | 126500031 | C | G | 0.579 | 0.01136 | 0.00171 | 2.81E−11 |
| rs45551835 | CUBN | missense | 10 | 16932384 | A | G | 0.014 | 0.14237 | 0.00698 | 2.28E−92 |
| rs144360241 | CUBN | missense | 10 | 16967417 | C | T | 0.005 | 0.08186 | 0.01234 | 3.31E−11 |
| rs1276720 | CUBN | intronic | 10 | 16971426 | T | C | 0.745 | 0.01109 | 0.00193 | 8.98E−09 |
| rs141640975 | CUBN | missense | 10 | 16992011 | A | G | 0.003 | 0.35876 | 0.01629 | 1.75E−107 |
| rs67339103 | LRMDA | intronic | 10 | 77893686 | A | G | 0.212 | 0.01522 | 0.00205 | 1.07E−13 |
| rs17368443 | SBF2 | intronic | 11 | 10296836 | C | G | 0.061 | 0.02071 | 0.00348 | 2.58E−09 |
| rs2601006 | CCT2 | 5′ UTR variant | 12 | 69979517 | C | T | 0.657 | 0.01176 | 0.00176 | 2.13E−11 |
| rs4288924 | ZFP36L1-ACTN1 | intergenic | 14 | 69302399 | G | A | 0.480 | 0.00980 | 0.00168 | 5.66E−09 |
| rs8035855 | MAPKBP1 | intronic | 15 | 42077961 | A | G | 0.644 | 0.01227 | 0.00174 | 1.91E−12 |
| rs1145074 | SPATA5L1 | intronic | 15 | 45703824 | T | A | 0.745 | 0.01140 | 0.00191 | 2.41E−09 |
| rs2472297 | CYP1A2-CYP1A1 | intergenic | 15 | 75027880 | T | C | 0.267 | 0.01812 | 0.00188 | 5.31E−22 |
| rs35572189 | BAHCC1 | missense | 17 | 79419025 | G | A | 0.638 | 0.01051 | 0.00174 | 1.44E−09 |
| rs838142 | FUT1 | 3′ UTR variant | 19 | 49252151 | A | G | 0.723 | 0.01174 | 0.00187 | 3.13E−10 |
Abbreviations: Chr, chromosome; EAF, effect allele frequency. For intergenic loci, nearest upstream and downstream RefSeq genes are indicated. Nearest gene should not be taken as evidence of causal gene. Description, most-severe consequence of nearest RefSeq gene.
Albuminuria Genetic Instrument Strength
A 46-SNP genetic risk score constructed from the 33 genome-wide significant loci plus an additional 13 loci meeting a conventional significance level of p < 5 × 10−8 (Table S4) explained 0.7% of the variance in albuminuria in UK Biobank (F-statistic, 2,928). The genetic risk score was validated in two additional North American cohorts of European ancestry and non-inflated estimates were obtained. The 46-SNP score was associated with albuminuria in both the Atherosclerosis Risk in Communities study (n = 6,398, p = 6.7 × 10−5, 0.2% variance in albuminuria explained) and the Framingham Heart Study (n = 6,387, p = 4.4 × 10−4, 0.2% variance in albuminuria explained; Table S5).
Association of Albuminuria Genetic Risk Score with Cardiometabolic Disease
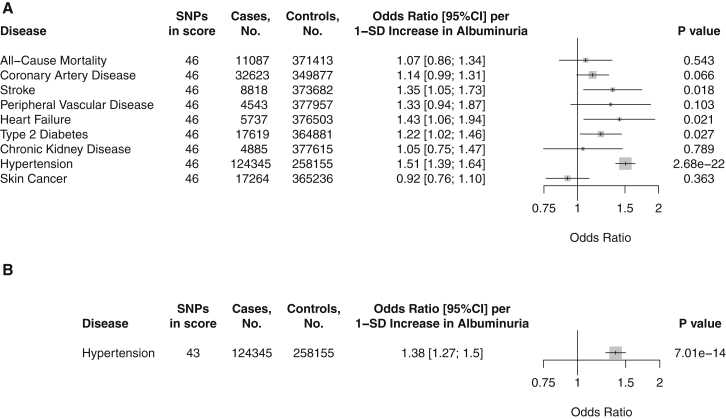
We examined whether genetically elevated albuminuria due to the 46-SNP risk score associated with increased risk of cardiometabolic disease in UK Biobank. Genetic predisposition to elevated albuminuria was associated with increased risk of hypertension (1.51 OR; 95%CI 1.39–1.64 per 1-SD predicted increase in albuminuria due to the 46-SNP score, p = 2.68 × 10−22). However, no significant associations were observed between the albuminuria genetic risk score and risk of all-cause mortality, coronary artery disease, stroke, heart failure, type 2 diabetes, chronic kidney disease, or skin cancer (Figure 3).
Figure 3.
Association of Genetic Predisposition to Increased Albuminuria with Risk of Cardiometabolic Disease in UK Biobank
Two-stage least-squares regression using albuminuria genetic risk score as instrumental variable; age, sex, genotyping array, and first ten genetic PCs as covariates. Results are standardized to 1-SD increase in albuminuria due to the genetic risk score.
(A) Genetic risk score composed of all 46 albuminuria variants.
(B) Genetic risk score composed of 43 albuminuria variants after applying directional MR Steiger filtering to remove variants potentially acting in the incorrect direction.
Bars indicate 95% confidence interval for odds ratio.
To remove variants that may act through reverse causation—that is, influence albuminuria through hypertension—from the albuminuria score, we applied MR Steiger filtering.31, 32 This approach removed three variants more directly associated with hypertension. After filtering, the 43-SNP score was still associated with increased risk of hypertension (1.38 OR; 95%CI 1.27–1.50 per 1-SD predicted increase in albuminuria, p = 7.01 × 10−14, Figure 3).
Bidirectional Mendelian Randomization of Albuminuria and Blood Pressure
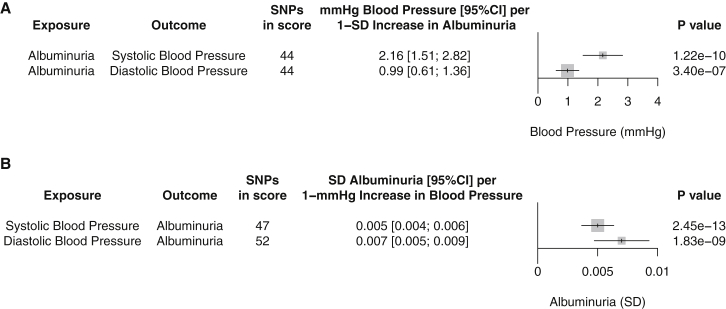
To further examine the association of albuminuria with hypertension, we investigated the genetic correlations between albuminuria, blood pressure, and hypertension. We found significant common SNP genetic correlations between albuminuria and hypertension (rg = 0.16; SE 0.03, p = 2.06 × 10−8), systolic blood pressure (rg = 0.20; SE 0.03, p = 3.6 × 10−15), or diastolic blood pressure (rg = 0.10; SE 0.03, p = 3.2 × 10−4). We performed bidirectional Mendelian randomization between albuminuria and blood pressure to understand the determinants of these correlations. First, we examined association of the albuminuria genetic risk score with blood pressure outcomes. After MR Steiger filtering, albuminuria genetic risk scores remained associated with increased systolic blood pressure (2.16 mmHg; 95%CI 1.51–2.82 per 1-SD predicted increase in albuminuria, p = 1.22 × 10−10) and diastolic blood pressure (0.99 mmHg; 0.61–1.36 per 1-SD predicted increase in albuminuria, p = 3.40 × 10−7, Figure 4). Next, we investigated the reverse association—blood pressure affecting albuminuria—using a blood pressure risk score as the exposure and albuminuria as the outcome. 47 and 52 variants significantly associated with blood pressure in ICBP,19 which did not include UK Biobank, explained 1.2% and 1.4% of the variance in systolic and diastolic blood pressure, respectively, in UK Biobank. Both blood pressure risk scores were associated with elevated albuminuria (0.005 SD albuminuria; 95%CI 0.004–0.006 per 1 mmHg predicted increase in systolic blood pressure, p = 2.45 × 10−13 and 0.007 SD albuminuria; 95%CI 0.005–0.009 per 1 mmHg predicted increase in diastolic blood pressure, p = 1.83 × 10−9, Figure 4), validating a previous suggestive report.19
Figure 4.
Bidirectional Mendelian Randomization Identifies Suggestive Causal Effects of Albuminuria on Blood Pressure and of Blood Pressure on Albuminuria
(A) Mendelian randomization of albuminuria genetic risk scores on blood pressure in UK Biobank (n = 381,833). Two-stage least-squares regression using albuminuria genetic risk score as instrumental variable on blood pressure outcome; age, sex, genotyping array, and first ten genetic PCs as covariates. Results are standardized to 1-SD increase in albuminuria due to the genetic risk score. Genetic risk scores were composed of 44 albuminuria variants after applying directional MR Steiger filtering to remove variants potentially acting in the incorrect direction.
(B) Mendelian randomization of blood pressure genetic risk scores on albuminuria. Effects of variants on systolic or diastolic blood pressure were determined in ICBP19 (nmax = 201,529) and thus corrected for hypertensive medication use and adjusted for body mass index. Two-stage least-squares regression using blood pressure genetic risk score as instrumental variable on albuminuria outcome in UK Biobank (n = 381,833); age, sex, genotyping array, and first ten genetic PCs as covariates. Results are standardized to 1-mmHg increase in blood pressure due to the genetic risk score. 47 or 52 variants were used to construct scores specific for systolic or diastolic blood pressure, respectively. Directional MR Steiger filtering removed no variants.
SNPs in score, number of SNPs remaining after directional MR Steiger filtering applied. Bars indicate 95% confidence interval for effect on blood pressure (top) or albuminuria (bottom).
Sensitivity Analyses
Seven sets of sensitivity analyses were used to verify the robustness of the associations between albuminuria and hypertension or blood pressure. First, Mendelian randomization results were consistent for a restricted score at the genome-wide significance level of p < 9 × 10−9 (Table S6). Second, we used several methods to detect and mitigate the effects of pleiotropic variants: (1) MR Egger regression to detect the presence of directional pleiotropy;36 (2) Cook’s distance to detect outlier variants, which can also indicate pleiotropy;37, 49 (3) leave-one-out analyses to determine whether associations are biased by a single, potentially pleiotropic SNP;41 and (4) median-based regressions, which are robust when up to 50% of information comes from invalid variant instruments, including due to pleiotropy.35 Some directional pleiotropy was observed in the associations between albuminuria and blood pressure but not other associations (Tables S7–S9). MR-Egger regression is especially sensitive to influential points,50, 51 so the observed directional pleiotropy could be due in part to a potential outlier, rs141640975 in the CUBN locus (Cook’s distance = 0.6–0.7, Figures S2 and S3). This variant was the top SNP in the albuminuria GWAS, raising the possibility that it could derive its large effect via aggregating potentially pleiotropic effects of multiple pathways. Excluding this variant reduced directional pleiotropy while maintaining associations between the albuminuria risk score and blood pressure or hypertension (Tables S7 and S8). Leave-one-out analyses suggested that the observed associations were not biased by other single variants (Figures S2–S4, Tables S7–S9). Notably, albuminuria risk scores also remained associated with hypertension and blood pressure, and blood pressure risk scores with albuminuria, using one or more forms of median regression that allow for many pleiotropic variants (Tables S7–S9).
Third, to be more confident variants were not acting through reverse causation, we used a more stringent MR Steiger filter. This removed variants that were not significantly more associated with albuminuria than outcomes. While effect estimates were slightly attenuated, the associations of albuminuria with blood pressure and hypertension persisted even after this additional filtering (Tables S7 and S8).
Fourth, for bidirectional Mendelian randomization it is important that variants in the albuminuria score are not in linkage disequilibrium with variants in the blood pressure score.21 One pair of variants at the HOTTIP locus was in linkage disequilibrium (rs2023844-rs3735533 R2 = 0.99). MR Steiger analysis suggests that this variant is more directly associated with blood pressure. It was therefore removed from all albuminuria risk scores by directional MR Steiger filtering. Additionally, sensitivity analyses excluding this variant from blood pressure risk scores did not affect association of blood pressure risk scores with albuminuria (Table S9).
Fifth, using the same samples for both discovery of a genetic risk score and analysis of score effects can bias association results toward the observational estimate.52 Unweighted genetic risk scores can reduce this bias.38 Unweighted albuminuria risk scores were also associated with increased risk of hypertension and elevated systolic and diastolic blood pressure in UK Biobank (Tables S7 and S8).
Sixth, to further mitigate bias from score discovery-analysis overlap in UK Biobank, we examined the effects of albuminuria-associated variants on blood pressure measured in a separate cohort. Blood pressure effects in ICBP were corrected for hypertensive medication use.20 In this cohort, albuminuria variants were associated with increased systolic blood pressure (2.69 mmHg; 95%CI 1.18–4.19 per 1-SD predicted increase in albuminuria, p = 4.64 × 10−4) and nominally with increased diastolic blood pressure (1.03 mmHg; 0.10–1.97 per 1-SD predicted increase in albuminuria, p = 0.030, Figures S5 and S6, Table S10).
Finally, hypertensive medications lower both albuminuria and blood pressure. To investigate this source of potential bias, we excluded any individuals in UK Biobank taking hypertensive medication. The effects of the resulting albuminuria genetic risk score on increased blood pressure were largely consistent (Figures S5 and S7, Tables S11 and S12), albeit with reduced power in this n = 302,687 subset.
Discussion
We used Mendelian randomization to examine whether processes leading to elevated albuminuria lead to increased risk of cardiometabolic disease. A genome-wide association study of albuminuria in UK Biobank identified 33 albuminuria loci, including one previously published. A genetic risk score of up to 46 albuminuria variants was strongly associated with increased risk of hypertension and elevated blood pressure but showed only weak associations with other cardiometabolic diseases.
These results permit several conclusions. First, processes that increase albuminuria appear to increase risk of hypertension and blood pressure. Although hypertension is commonly thought to increase albuminuria, previous epidemiological studies also suggest that albuminuria predicts development of hypertension.7, 8 Our data add genetic and observational evidence supporting this association. Multiple pathways leading to albuminuria may contribute to hypertension. Albuminuria may arise as a result of generalized endothelial dysfunction,53, 54, 55 which can contribute to development of hypertension.56, 57 Albuminuria can also result from kidney damage. In damaged kidneys, increased blood pressure is thought to help the subfunctional kidney excrete sufficient sodium to maintain sodium homeostasis.58, 59 Consistent with this, severe kidney injury leads to experimental hypertension60 and mild kidney damage precedes the development of hypertension in multiple experimental models.58, 61 The intrinsic role of the kidney in blood pressure regulation is also supported by the observation that kidney transplantation from hypertensive donors can cause hypertension in previously normotensive recipients.62, 63 Further work is needed to determine the mechanisms by which risk score variants contribute to elevated albuminuria.
Second, application of MR Steiger filtering31, 32 enabled the discovery of evidence for bidirectional effects between albuminuria and blood pressure. The associations of genetically elevated albuminuria with increased blood pressure and of genetically elevated blood pressure with increased albuminuria suggest that the relationship between albuminuria and blood pressure is bidirectional. This would imply the existence of a feed-forward loop, in which elevated blood pressure leads to increased albuminuria, which in turn would further increase blood pressure. It is important to note that because each Mendelian randomization analysis estimates the effects in one direction, this feedback loop is not formally modeled by such analyses.52 These results suggest that therapies targeting processes that lower albuminuria could have antihypertensive effects that are further amplified by inhibiting this feed-forward loop. Determining the specific genes and pathways affected by albuminuria variants could assist in rational design of such therapies.
Third, these results imply that processes leading to albuminuria can influence cardiovascular disease through blood pressure. Observational and genetic evidence establishes blood pressure as an important causal risk factor for multiple cardiovascular diseases.19, 64, 65, 66 By the principle of two-step Mendelian randomization,15, 67 significant associations between an albuminuria risk score and blood pressure and between blood pressure risk scores and diseases such as stroke or coronary artery disease19 imply that the albuminuria risk score is associated with these diseases at least via blood pressure. This raises the question of why we did not observe significant associations between albuminuria and such diseases. Stronger genetic risk scores may be necessary to detect downstream consequences of a causal relationship between albuminuria and blood pressure: since blood pressure explains only some of the variance in cardiovascular outcomes,68 albuminuria should have a smaller effect size on cardiovascular disease than on blood pressure. We therefore may have been underpowered to detect such downstream effects of albuminuria-induced hypertension on cardiovascular diseases (Table S13). Larger datasets that generate stronger albuminuria genetic risk scores should help clarify this issue.
A key strength of this study is that albuminuria and genotypes were measured in 382,500 individuals, seven times more than the next largest genome-wide association study,45 enabling construction of a polygenic risk score that explained 0.2% of the variance in albuminuria in two validation cohorts. We were also able to validate the associations between albuminuria and blood pressure in an outside cohort. Access to individual-level data in UK Biobank allowed us to interrogate whether these associations were confounded by hypertensive medication use. Finally, we used MR Steiger filtering to remove variants that potentially acted through reverse causation, and multiple sensitivity analyses to detect and mitigate pleiotropic variants.
Several limitations should be acknowledged. First, reliance on internally derived weights in our albuminuria genetic risk score may have biased our results toward the observational associations.43 To address this, we replicated significant associations using unweighted allele scores and/or in two-sample analyses. Second, an alternate explanation for the bidirectional associations observed is that a shared genetic basis underlies the two traits. If so, SNPs that influence both traits through a shared mechanism could violate the instrument strength independent of direct effect (InSIDE) assumption of standard Mendelian randomization and MR-Egger analyses.69 Although the associations were consistent using median-based regressions, which do not require the InSIDE assumption,35, 41 we cannot rule out the possibility that associations between albuminuria and blood pressure are due to a shared genetic basis of the two traits rather than causal effects. Third, there was substantial heterogeneity in the causal effect estimates from different variants (Tables S6–S10 and S12); i.e., for association of albuminuria variants with hypertension, Cochran’s Q = 160 (p = 9.5 × 10−16). This is perhaps not surprising considering the hypothesis under investigation was whether pathways that lead to albuminuria can increase blood pressure and hypertension risk. It is plausible—and quite likely—that multiple albuminuria-inducing pathways exist which could elevate blood pressure to different degrees (i.e., endothelial dysfunction and kidney damage) or not at all (i.e., pathways involved in albumin metabolism or post-renal urine regulation).50 However, other sources of heterogeneity could nevertheless be present, although these do not necessarily lead to bias.16, 50 Fourth, UK Biobank is a population-based longitudinal cohort. Our study was likely underpowered to detect associations in diseases less common than hypertension (Table S13); therefore, lack of association of albuminuria with other diseases should not be over-interpreted. Finally, it is important to note that UK Biobank is an older cohort of European ancestry; therefore, results may differ in younger populations or in other ethnic backgrounds.
In conclusion, an albuminuria genetic risk score of up to 46 SNPs was associated with increased risk of hypertension and elevated blood pressure. Application of recently developed Mendelian randomization methods identified evidence of bidirectional effects from albuminuria-increasing pathways to blood pressure and from blood pressure to albuminuria. These results provide genetic data to refine and highlight the complex interplay between albuminuria and hypertension.
Declaration of Interests
S.K. has received grants from Bayer Healthcare, Aegerion Pharmaceuticals, and Regeneron Pharmaceuticals; has received consulting fees from Merck, Novartis, Sano, AstraZeneca, Alnylam Pharmaceuticals, Leerink Partners, Noble Insights, Quest Diagnostics, Genomics PLC, and Eli Lilly and Company; and holds equity in San Therapeutics and Catabasis Pharmaceuticals.
Acknowledgments
This work was funded by the National Institutes of Health R01 HL127564 to S.K. and was conducted using the UK Biobank resource under application 7089. S.K. is supported by a research scholar award from Massachusetts General Hospital and the Donovan Family Foundation. K.G.A. is supported by the American Heart Association Institute for Precision Cardiovascular Medicine (17IFUNP33840012). G.D.S. and G.H. work within the Medical Research Council Integrative Epidemiology Unit, supported by the Medical Research Council (MC_UU_00011/1) and the University of Bristol. G.H. is supported by the Wellcome Trust (208806/Z/17/Z). The Atherosclerosis Risk in Communities Study is carried out as a collaborative study supported by the National Heart, Lung, and Blood Institute (NHLBI, contracts HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C); funding for GENEVA was provided by National Human Genome Research Institute grant U01HG004402. The authors thank the staff and participants of the ARIC study for their important contributions. The Framingham Heart Study is conducted and supported by the NHLBI in collaboration with Boston University (contracts N01-HC-25195, N02-HL64278, and HHSN268201500001I). This manuscript was not prepared in collaboration with investigators of the Framingham Heart Study and does not necessarily reflect the opinions or views of the Framingham Heart Study, Boston University, or NHLBI.
Published: September 13, 2018
Footnotes
Supplemental Data include 7 figure and 13 tables and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.08.004.
Web Resources
Hail, https://github.com/hail-is/hail
Mendelian randomization power calculator, https://sb452.shinyapps.io/power
Supplemental Data
References
- 1.Gerstein H.C., Mann J.F.E., Yi Q., Zinman B., Dinneen S.F., Hoogwerf B., Hallé J.P., Young J., Rashkow A., Joyce C., HOPE Study Investigators Albuminuria and risk of cardiovascular events, death, and heart failure in diabetic and nondiabetic individuals. JAMA. 2001;286:421–426. doi: 10.1001/jama.286.4.421. [DOI] [PubMed] [Google Scholar]
- 2.Klausen K., Borch-Johnsen K., Feldt-Rasmussen B., Jensen G., Clausen P., Scharling H., Appleyard M., Jensen J.S. Very low levels of microalbuminuria are associated with increased risk of coronary heart disease and death independently of renal function, hypertension, and diabetes. Circulation. 2004;110:32–35. doi: 10.1161/01.CIR.0000133312.96477.48. [DOI] [PubMed] [Google Scholar]
- 3.Hillege H.L., Fidler V., Diercks G.F.H., van Gilst W.H., de Zeeuw D., van Veldhuisen D.J., Gans R.O.B., Janssen W.M.T., Grobbee D.E., de Jong P.E., Prevention of Renal and Vascular End Stage Disease (PREVEND) Study Group Urinary albumin excretion predicts cardiovascular and noncardiovascular mortality in general population. Circulation. 2002;106:1777–1782. doi: 10.1161/01.cir.0000031732.78052.81. [DOI] [PubMed] [Google Scholar]
- 4.Hemmelgarn B.R., Manns B.J., Lloyd A., James M.T., Klarenbach S., Quinn R.R., Wiebe N., Tonelli M., Alberta Kidney Disease Network Relation between kidney function, proteinuria, and adverse outcomes. JAMA. 2010;303:423–429. doi: 10.1001/jama.2010.39. [DOI] [PubMed] [Google Scholar]
- 5.Matsushita K., Coresh J., Sang Y., Chalmers J., Fox C., Guallar E., Jafar T., Jassal S.K., Landman G.W.D., Muntner P., CKD Prognosis Consortium Estimated glomerular filtration rate and albuminuria for prediction of cardiovascular outcomes: a collaborative meta-analysis of individual participant data. Lancet Diabetes Endocrinol. 2015;3:514–525. doi: 10.1016/S2213-8587(15)00040-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brantsma A.H., Bakker S.J.L., Hillege H.L., de Zeeuw D., de Jong P.E., Gansevoort R.T., PREVEND Study Group Urinary albumin excretion and its relation with C-reactive protein and the metabolic syndrome in the prediction of type 2 diabetes. Diabetes Care. 2005;28:2525–2530. doi: 10.2337/diacare.28.10.2525. [DOI] [PubMed] [Google Scholar]
- 7.Wang T.J., Evans J.C., Meigs J.B., Rifai N., Fox C.S., D’Agostino R.B., Levy D., Vasan R.S. Low-grade albuminuria and the risks of hypertension and blood pressure progression. Circulation. 2005;111:1370–1376. doi: 10.1161/01.CIR.0000158434.69180.2D. [DOI] [PubMed] [Google Scholar]
- 8.Brantsma A.H., Bakker S.J.L., de Zeeuw D., de Jong P.E., Gansevoort R.T. Urinary albumin excretion as a predictor of the development of hypertension in the general population. J. Am. Soc. Nephrol. 2006;17:331–335. doi: 10.1681/ASN.2005111153. [DOI] [PubMed] [Google Scholar]
- 9.Scirica B.M., Mosenzon O., Bhatt D.L., Udell J.A., Steg P.G., McGuire D.K., Im K., Kanevsky E., Stahre C., Sjöstrand M. Cardiovascular outcomes according to urinary albumin and kidney disease in patients with type 2 diabetes at high cardiovascular risk: observations from the SAVOR-TIMI 53 trial. JAMA Cardiol. 2018;3:155–163. doi: 10.1001/jamacardio.2017.4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Asselbergs F.W., Diercks G.F.H., Hillege H.L., van Boven A.J., Janssen W.M.T., Voors A.A., de Zeeuw D., de Jong P.E., van Veldhuisen D.J., van Gilst W.H., Prevention of Renal and Vascular Endstage Disease Intervention Trial (PREVEND IT) Investigators Effects of fosinopril and pravastatin on cardiovascular events in subjects with microalbuminuria. Circulation. 2004;110:2809–2816. doi: 10.1161/01.CIR.0000146378.65439.7A. [DOI] [PubMed] [Google Scholar]
- 11.Ibsen H., Olsen M.H., Wachtell K., Borch-Johnsen K., Lindholm L.H., Mogensen C.E., Dahlöf B., Devereux R.B., de Faire U., Fyhrquist F. Reduction in albuminuria translates to reduction in cardiovascular events in hypertensive patients: losartan intervention for endpoint reduction in hypertension study. Hypertension. 2005;45:198–202. doi: 10.1161/01.HYP.0000154082.72286.2a. [DOI] [PubMed] [Google Scholar]
- 12.de Zeeuw D., Remuzzi G., Parving H.-H., Keane W.F., Zhang Z., Shahinfar S., Snapinn S., Cooper M.E., Mitch W.E., Brenner B.M. Albuminuria, a therapeutic target for cardiovascular protection in type 2 diabetic patients with nephropathy. Circulation. 2004;110:921–927. doi: 10.1161/01.CIR.0000139860.33974.28. [DOI] [PubMed] [Google Scholar]
- 13.Holtkamp F.A., de Zeeuw D., de Graeff P.A., Laverman G.D., Berl T., Remuzzi G., Packham D., Lewis J.B., Parving H.H., Lambers Heerspink H.J. Albuminuria and blood pressure, independent targets for cardioprotective therapy in patients with diabetes and nephropathy: a post hoc analysis of the combined RENAAL and IDNT trials. Eur. Heart J. 2011;32:1493–1499. doi: 10.1093/eurheartj/ehr017. [DOI] [PubMed] [Google Scholar]
- 14.Smith G.D., Ebrahim S. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int. J. Epidemiol. 2003;32:1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 15.Zheng J., Baird D., Borges M.-C., Bowden J., Hemani G., Haycock P., Evans D.M., Smith G.D. Recent developments in Mendelian randomization studies. Curr. Epidemiol. Rep. 2017;4:330–345. doi: 10.1007/s40471-017-0128-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hemani G., Bowden J., Davey Smith G. Evaluating the potential role of pleiotropy in Mendelian randomization studies. Hum. Mol. Genet. 2018;27(R2):R195–R208. doi: 10.1093/hmg/ddy163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bycroft C., Freeman C., Petkova D., Band G., Elliott L.T., Sharp K., Motyer A., Vukcevic D., Delaneau O., O’Connell J. Genome-wide genetic data on ∼500,000 UK Biobank participants. bioRxiv. 2017 [Google Scholar]
- 18.Mattix H.J., Hsu C.-Y., Shaykevich S., Curhan G. Use of the albumin/creatinine ratio to detect microalbuminuria: implications of sex and race. J. Am. Soc. Nephrol. 2002;13:1034–1039. doi: 10.1681/ASN.V1341034. [DOI] [PubMed] [Google Scholar]
- 19.Ehret G.B., Ferreira T., Chasman D.I., Jackson A.U., Schmidt E.M., Johnson T., Thorleifsson G., Luan J., Donnelly L.A., Kanoni S., CHARGE-EchoGen consortium. CHARGE-HF consortium. Wellcome Trust Case Control Consortium The genetics of blood pressure regulation and its target organs from association studies in 342,415 individuals. Nat. Genet. 2016;48:1171–1184. doi: 10.1038/ng.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wain L.V., Vaez A., Jansen R., Joehanes R., van der Most P.J., Erzurumluoglu A.M., O’Reilly P.F., Cabrera C.P., Warren H.R., Rose L.M. Novel blood pressure locus and gene discovery using genome-wide association study and expression data sets from blood and the kidney. Hypertension. 2017;70:e4–e19. doi: 10.1161/HYPERTENSIONAHA.117.09438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davey Smith G., Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum. Mol. Genet. 2014;23(R1):R89–R98. doi: 10.1093/hmg/ddu328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsushita K., Ballew S.H., Coresh J., Arima H., Ärnlöv J., Cirillo M., Ebert N., Hiramoto J.S., Kimm H., Shlipak M.G., Chronic Kidney Disease Prognosis Consortium Measures of chronic kidney disease and risk of incident peripheral artery disease: a collaborative meta-analysis of individual participant data. Lancet Diabetes Endocrinol. 2017;5:718–728. doi: 10.1016/S2213-8587(17)30183-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Halimi J.-M., Bonnet F., Lange C., Balkau B., Tichet J., Marre M., DESIR Study Group Urinary albumin excretion is a risk factor for diabetes mellitus in men, independently of initial metabolic profile and development of insulin resistance. J. Hypertens. 2008;26:2198–2206. doi: 10.1097/HJH.0b013e328310ddff. [DOI] [PubMed] [Google Scholar]
- 24.Ishani A., Grandits G.A., Grimm R.H., Svendsen K.H., Collins A.J., Prineas R.J., Neaton J.D. Association of single measurements of dipstick proteinuria, estimated glomerular filtration rate, and hematocrit with 25-year incidence of end-stage renal disease in the multiple risk factor intervention trial. J. Am. Soc. Nephrol. 2006;17:1444–1452. doi: 10.1681/ASN.2005091012. [DOI] [PubMed] [Google Scholar]
- 25.Ganna A., Genovese G., Howrigan D.P., Byrnes A., Kurki M., Zekavat S.M., Whelan C.W., Kals M., Nivard M.G., Bloemendal A. Ultra-rare disruptive and damaging mutations influence educational attainment in the general population. Nat. Neurosci. 2016;19:1563–1565. doi: 10.1038/nn.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu C., Tachmazidou I., Walter K., Ciampi A., Zeggini E., Greenwood C.M.T., UK10K Consortium Estimating genome-wide significance for whole-genome sequencing studies. Genet. Epidemiol. 2014;38:281–290. doi: 10.1002/gepi.21797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bulik-Sullivan B.K., Loh P.-R., Finucane H.K., Ripke S., Yang J., Patterson N., Daly M.J., Price A.L., Neale B.M., Schizophrenia Working Group of the Psychiatric Genomics Consortium LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emdin C.A., Khera A.V., Natarajan P., Klarin D., Zekavat S.M., Hsiao A.J., Kathiresan S. Genetic association of waist-to-hip ratio with cardiometabolic traits, type 2 diabetes, and coronary heart disease. JAMA. 2017;317:626–634. doi: 10.1001/jama.2016.21042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burgess S., Butterworth A., Thompson S.G. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 2013;37:658–665. doi: 10.1002/gepi.21758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yavorska O.O., Burgess S. MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data. Int. J. Epidemiol. 2017;46:1734–1739. doi: 10.1093/ije/dyx034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hemani G., Bowden J., Haycock P.C., Zheng J., Davis O., Flach P., Gaunt T.R., Davey Smith G. Automating Mendelian randomization through machine learning to construct a putative causal map of the human phenome. bioRxiv. 2017 [Google Scholar]
- 32.Hemani G., Tilling K., Davey Smith G. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet. 2017;13:e1007081. doi: 10.1371/journal.pgen.1007081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verbanck M., Chen C.-Y., Neale B., Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 2018;50:693–698. doi: 10.1038/s41588-018-0099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burgess, S., and Bowden, J. (2015). Integrating summarized data from multiple genetic variants in Mendelian randomization: bias and coverage properties of inverse-variance weighted methods. arXiv. https://arxiv.org/abs/1512.04486.
- 35.Bowden J., Davey Smith G., Haycock P.C., Burgess S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet. Epidemiol. 2016;40:304–314. doi: 10.1002/gepi.21965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bowden J., Davey Smith G., Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 2015;44:512–525. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Richmond R., Wade K., Corbin L., Bowden J., Hemani G., Timpson N., Davey Smith G. Investigating the role of insulin in increased adiposity: Bi-directional Mendelian randomization study. bioRxiv. 2017 [Google Scholar]
- 38.Burgess S., Thompson S.G. Use of allele scores as instrumental variables for Mendelian randomization. Int. J. Epidemiol. 2013;42:1134–1144. doi: 10.1093/ije/dyt093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bianchi S., Bigazzi R., Campese V.M. Microalbuminuria in essential hypertension: significance, pathophysiology, and therapeutic implications. Am. J. Kidney Dis. 1999;34:973–995. doi: 10.1016/S0272-6386(99)70002-8. [DOI] [PubMed] [Google Scholar]
- 40.Williams M.E. Diabetic nephropathy: the proteinuria hypothesis. Am. J. Nephrol. 2005;25:77–94. doi: 10.1159/000084286. [DOI] [PubMed] [Google Scholar]
- 41.Hemani G., Zheng J., Elsworth B., Wade K.H., Haberland V., Baird D., Laurin C., Burgess S., Bowden J., Langdon R. The MR-Base platform supports systematic causal inference across the human phenome. eLife. 2018;7:68. doi: 10.7554/eLife.34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee S.H., Goddard M.E., Wray N.R., Visscher P.M. A better coefficient of determination for genetic profile analysis. Genet. Epidemiol. 2012;36:214–224. doi: 10.1002/gepi.21614. [DOI] [PubMed] [Google Scholar]
- 43.Hartwig F.P., Davies N.M. Why internal weights should be avoided (not only) in MR-Egger regression. Int. J. Epidemiol. 2016;45:1676–1678. doi: 10.1093/ije/dyw240. [DOI] [PubMed] [Google Scholar]
- 44.Sarafidis P.A., Khosla N., Bakris G.L. Antihypertensive therapy in the presence of proteinuria. Am. J. Kidney Dis. 2007;49:12–26. doi: 10.1053/j.ajkd.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 45.Teumer A., Tin A., Sorice R., Gorski M., Yeo N.C., Chu A.Y., Li M., Li Y., Mijatovic V., Ko Y.-A., DCCT/EDIC Genome-wide association studies identify genetic loci associated with albuminuria in diabetes. Diabetes. 2016;65:803–817. doi: 10.2337/db15-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma T.K.-W., Szeto C.-C. Mineralocorticoid receptor antagonist for renal protection. Ren. Fail. 2012;34:810–817. doi: 10.3109/0886022X.2012.672156. [DOI] [PubMed] [Google Scholar]
- 47.Currie G., Taylor A.H.M., Fujita T., Ohtsu H., Lindhardt M., Rossing P., Boesby L., Edwards N.C., Ferro C.J., Townend J.N. Effect of mineralocorticoid receptor antagonists on proteinuria and progression of chronic kidney disease: a systematic review and meta-analysis. BMC Nephrol. 2016;17:127. doi: 10.1186/s12882-016-0337-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kruegel J., Rubel D., Gross O. Alport syndrome--insights from basic and clinical research. Nat. Rev. Nephrol. 2013;9:170–178. doi: 10.1038/nrneph.2012.259. [DOI] [PubMed] [Google Scholar]
- 49.Corbin L.J., Richmond R.C., Wade K.H., Burgess S., Bowden J., Smith G.D., Timpson N.J. BMI as a modifiable risk factor for type 2 diabetes: refining and understanding causal estimates using Mendelian randomization. Diabetes. 2016;65:3002–3007. doi: 10.2337/db16-0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Burgess S., Foley C.N., Zuber V. Inferring causal relationships between risk factors and outcomes from genome-wide association study data. Annu. Rev. Genomics Hum. Genet. 2018 doi: 10.1146/annurev-genom-083117-021731. Published online April 25, 2018. 10.1146/annurev-genom-083117-021731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burgess S., Thompson S.G. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur. J. Epidemiol. 2017;32:377–389. doi: 10.1007/s10654-017-0255-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haycock P.C., Burgess S., Wade K.H., Bowden J., Relton C., Davey Smith G. Best (but oft-forgotten) practices: the design, analysis, and interpretation of Mendelian randomization studies. Am. J. Clin. Nutr. 2016;103:965–978. doi: 10.3945/ajcn.115.118216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deckert T., Feldt-Rasmussen B., Borch-Johnsen K., Jensen T., Kofoed-Enevoldsen A. Albuminuria reflects widespread vascular damage. The Steno hypothesis. Diabetologia. 1989;32:219–226. doi: 10.1007/BF00285287. [DOI] [PubMed] [Google Scholar]
- 54.Clausen P., Feldt-Rasmussen B., Jensen G., Jensen J.S. Endothelial haemostatic factors are associated with progression of urinary albumin excretion in clinically healthy subjects: a 4-year prospective study. Clin. Sci. 1999;97:37–43. [PubMed] [Google Scholar]
- 55.Stehouwer C.D.A., Smulders Y.M. Microalbuminuria and risk for cardiovascular disease: Analysis of potential mechanisms. J. Am. Soc. Nephrol. 2006;17:2106–2111. doi: 10.1681/ASN.2005121288. [DOI] [PubMed] [Google Scholar]
- 56.Rossi R., Chiurlia E., Nuzzo A., Cioni E., Origliani G., Modena M.G. Flow-mediated vasodilation and the risk of developing hypertension in healthy postmenopausal women. J. Am. Coll. Cardiol. 2004;44:1636–1640. doi: 10.1016/j.jacc.2004.07.027. [DOI] [PubMed] [Google Scholar]
- 57.Sander M., Chavoshan B., Victor R.G. A large blood pressure-raising effect of nitric oxide synthase inhibition in humans. Hypertension. 1999;33:937–942. doi: 10.1161/01.hyp.33.4.937. [DOI] [PubMed] [Google Scholar]
- 58.Cowley A.W., Jr., Roman R.J. The role of the kidney in hypertension. JAMA. 1996;275:1581–1589. [PubMed] [Google Scholar]
- 59.Coffman T.M. The inextricable role of the kidney in hypertension. J. Clin. Invest. 2014;124:2341–2347. doi: 10.1172/JCI72274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goldblatt H., Lynch J., Hanzal R.F., Summerville W.W. Studies on experimental hypertension: I. the production of persistent elevation of systolic blood pressure by means of renal ischemia. J. Exp. Med. 1934;59:347–379. doi: 10.1084/jem.59.3.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Johnson R.J., Herrera-Acosta J., Schreiner G.F., Rodriguez-Iturbe B. Subtle acquired renal injury as a mechanism of salt-sensitive hypertension. N. Engl. J. Med. 2002;346:913–923. doi: 10.1056/NEJMra011078. [DOI] [PubMed] [Google Scholar]
- 62.Strandgaard S., Hansen U. Hypertension in renal allograft recipients may be conveyed by cadaveric kidneys from donors with subarachnoid haemorrhage. Br. Med. J. (Clin. Res. Ed.) 1986;292:1041–1044. doi: 10.1136/bmj.292.6527.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guidi E., Menghetti D., Milani S., Montagnino G., Palazzi P., Bianchi G. Hypertension may be transplanted with the kidney in humans: a long-term historical prospective follow-up of recipients grafted with kidneys coming from donors with or without hypertension in their families. J. Am. Soc. Nephrol. 1996;7:1131–1138. doi: 10.1681/ASN.V781131. [DOI] [PubMed] [Google Scholar]
- 64.Rapsomaniki E., Timmis A., George J., Pujades-Rodriguez M., Shah A.D., Denaxas S., White I.R., Caulfield M.J., Deanfield J.E., Smeeth L. Blood pressure and incidence of twelve cardiovascular diseases: lifetime risks, healthy life-years lost, and age-specific associations in 1·25 million people. Lancet. 2014;383:1899–1911. doi: 10.1016/S0140-6736(14)60685-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lewington S., Clarke R., Qizilbash N., Peto R., Collins R., Prospective Studies Collaboration Age-specific relevance of usual blood pressure to vascular mortality: a meta-analysis of individual data for one million adults in 61 prospective studies. Lancet. 2002;360:1903–1913. doi: 10.1016/s0140-6736(02)11911-8. [DOI] [PubMed] [Google Scholar]
- 66.Levy D., Larson M.G., Vasan R.S., Kannel W.B., Ho K.K.L. The progression from hypertension to congestive heart failure. JAMA. 1996;275:1557–1562. [PubMed] [Google Scholar]
- 67.Relton C.L., Davey Smith G. Two-step epigenetic Mendelian randomization: a strategy for establishing the causal role of epigenetic processes in pathways to disease. Int. J. Epidemiol. 2012;41:161–176. doi: 10.1093/ije/dyr233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cheng S., Claggett B., Correia A.W., Shah A.M., Gupta D.K., Skali H., Ni H., Rosamond W.D., Heiss G., Folsom A.R. Temporal trends in the population attributable risk for cardiovascular disease: the Atherosclerosis Risk in Communities Study. Circulation. 2014;130:820–828. doi: 10.1161/CIRCULATIONAHA.113.008506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bowden J., Del Greco M F., Minelli C., Davey Smith G., Sheehan N., Thompson J. A framework for the investigation of pleiotropy in two-sample summary data Mendelian randomization. Stat. Med. 2017;36:1783–1802. doi: 10.1002/sim.7221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bakris G.L. Moderately increased albuminuria (microalbuminuria) and cardiovascular disease. UpToDate. 2016 https://www.uptodate.com/contents/moderately-increased-albuminuria-microalbuminuria-and-cardiovascular-disease#H220350. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.