Abstract
Infantile and childhood-onset cataracts form a heterogeneous group of disorders; among the many genetic causes, numerous pathogenic variants in additional genes associated with autosomal-recessive infantile cataracts remain to be discovered. We identified three consanguineous families affected by bilateral infantile cataracts. Using exome sequencing, we found homozygous loss-of-function variants in DNMBP: nonsense variant c.811C>T (p.Arg271∗) in large family F385 (nine affected individuals; LOD score = 5.18 at θ = 0), frameshift deletion c.2947_2948del (p.Asp983∗) in family F372 (two affected individuals), and frameshift variant c.2852_2855del (p.Thr951Metfs∗41) in family F3 (one affected individual). The phenotypes of all affected individuals include infantile-onset cataracts. RNAi-mediated knockdown of the Drosophila ortholog still life (sif), enriched in lens-secreting cells, affects the development of these cells as well as the localization of E-cadherin, alters the distribution of septate junctions in adjacent cone cells, and leads to a ∼50% reduction in electroretinography amplitudes in young flies. DNMBP regulates the shape of tight junctions, which correspond to the septate junctions in invertebrates, as well as the assembly pattern of E-cadherin in human epithelial cells. E-cadherin has an important role in lens vesicle separation and lens epithelial cell survival in humans. We therefore conclude that DNMBP loss-of-function variants cause infantile-onset cataracts in humans.
Keywords: cataract, DNMBP, Drosophila, still life, pigment cells, cornea, photoreceptors, bristles, ERG, eye development
Introduction
The development of the eye lens involves a complex morphogenetic and regulatory program of cellular signaling and differentiation.1 The lens, a transparent tissue that focuses light and images on the retina,2 is composed of epithelial and fiber cells that originate from the surface ectoderm during early embryogenesis. Epithelial cells elongate and differentiate into fiber cells upon migration, eventually filling the entire lens and becoming concentrated at the center. Terminal differentiation of lens fiber cells involves widespread remodeling of the cell membrane, elongation, compaction, and ultimately degradation of all cellular organelles, including the nuclei, to make it transparent.1, 3 Thus, in the absence of cell turnover, each lens fiber cell and its protein contents have a lifelong role in lens transparency. Normal growth of the lens and cornea plays a vital role in the development of the anterior segment of the eye. Abnormal development of the lens can cause ocular defects that include cataracts, glaucoma, microphthalmia, and anterior segment dysgenesis.4, 5 Infantile or early-childhood-onset cataracts—opacity that develops in the crystalline lens of the eye within the first year of life or before 5 years of age, respectively—is a major cause of visual impairment in children and is responsible for 5%–20% of visual impairment worldwide.6 A recent systematic analysis has estimated the frequency of infantile cataracts as 4.2 cases per 10,000 children.7 Visual impairment (VI) due to cataracts in children causes a substantial lifelong burden on quality of life and also contributes to a considerable socioeconomic burden.
Although surgical removal of cataracts and early interventions are required for the preservation of vision, the knowledge of the precise molecular etiology is also important for unraveling the underlying mechanism and proper management of the disease complications.8 Discovery of the underlying molecular basis of Mendelian disorders has been a powerful tool for improving genetic diagnosis in clinical settings, allowing accurate genetic counseling, and providing a better understanding of the underlying mechanism of disease phenotypes to lead to possible therapeutic interventions.8, 9, 10, 11 It is estimated that 34% of infantile cataracts are genetic.7 Pathogenic variants in more than 100 genes are known to cause syndromic and isolated infantile or early-childhood cataracts (see Cat-Map in Web Resources).12
Autosomal-dominant, autosomal-recessive, and X-linked forms have been described, but a substantial fraction of genetic causes remain unknown.5, 13, 14 Delineation of the precise underlying cause is important for clinical management8 and is now possible with advanced sequencing technologies.15 Recessive genetic disorders are more prevalent in consanguineous populations.16 The genetic cause of lenticular abnormalities in children of consanguineous couples is mostly autosomal recessive; therefore, such families are of considerable importance for identifying candidate genes, understanding the biology of lens development, and elucidating the mechanisms of disease pathogencity.5 By combining exome sequencing of one affected proband with homozygosity mapping and segregation analysis of family members, we have discovered candidate genes in families affected by suspected autosomal-recessive disorders.17, 18, 19, 20, 21, 22 Using this approach, we report three unrelated consanguineous Pakistani families (F372, F385, and F3) and 12 individuals who are affected by infantile-onset cataracts and harbor three different homozygous loss-of-function (LoF) variants in dynamin-binding protein (DNMBP [OMIM: 611282]). Knockdown of a DNMBP ortholog, still life (sif) in Drosophila, results in developmental defects of the cells that secrete the lens material as well as diminished electroretinography (ERG) amplitudes, suggesting a defect in the phototransduction cascade. These data provide compelling evidence for an evolutionarily conserved role of DNMBP (formerly known as Tuba) and its homologs in the visual system.
Subjects and Methods
Families
Family F372 was enrolled in the University of Karachi Department of Genetics, and family F385 was registered in the Khyber Medical University Institute of Basic Medical Sciences. These families were studied at the University of Geneva Department of Genetic Medicine and Development. The study was approved by the bioethics committee of the University Hospitals of Geneva (protocol CER 11-036) and by the ethical committees of the University of Karachi and Khyber Medical University. Family F3 is originally from Pakistan and was studied at the Faculty of Biology, Medicine, and Health of the Manchester Centre for Genomic Medicine. This research followed the tenets of the Declaration of Helsinki. Approval was obtained from the local research ethics committee (11/NW/0421), and informed consent was obtained as an essential prerequisite for study inclusion.
Genetic Analysis
In the samples from families F372 and F385, exome capture was performed with the SureSelect Human All Exon V6 reagent (Agilent Technologies), and sequencing was carried out on an Illumina HiSeq 4000 platform. The sequencing data were analyzed with a customized pipeline that comprised published algorithms including the Burrows-Wheeler Aligner (BWA),23 SAMtools,23 PICARD, and the Genome Analysis Toolkit (GATK).24 The reference human genome (UCSC Genome Browser hg19, assembly GRCh37)25 was used for the alignment of sequenced reads. Genotyping was performed with an Illumina 720K SNP array (HumanOmniExpress Bead Chip) on all members of families F372 and F385. Homozygosity mapping was performed with PLINK26 with a window of 50 consecutive homozygous SNPs, and a maximum of one mismatch was allowed in a homozygous region. Runs of homozygosity (ROHs) were defined by the first heterozygous SNP bordering each homozygous region. We used the program CATCH27 to identify the segregating pathogenic variants by using the family pedigree information, ROHs, and exome sequencing data. This program identifies and filters variants in the ROHs segregating with the disease phenotype of the family. After the initial filtering, all the remaining variants were evaluated as mentioned in previous studies.19, 22, 28 Sanger sequencing was performed to validate all candidate variants.
DNA from the proband of family F3 was subject to initial screening of a panel of 115 known pediatric-cataract-related genes as previously described.15 No putative pathogenic variants were identified, and hence DNA from the proband was subjected to whole-exome sequencing (WES). Libraries were enriched with the SureSelect Human All Exon V5 Kit according to the manufacturer’s instructions (Agilent). Clonal amplification of enriched genomic regions was conducted on a c-Bot System (Illumina), and sequencing was performed on the HiSeq 2000 platform (Illumina). The BWA (v.0.6.2)23 was used to align sequencing reads to human reference sequence hg19. Annotation was performed with GATK v.2.0.39, and variants were called by the GATK UnifiedGenotyper. Variants with ≥5× coverage were annotated with Ensembl v.68, and functional consequences were defined against all RefSeq transcripts (release 61). SNPs were filtered at a novel allele depth of 50× and mapping quality variance (MQV) score of 45 (this in-house quality score is based on the probability that a read is misaligned). We used in silico scores from SIFT29 and PolyPhen-230 to predict the pathogenicity of variants. All putative pathogenic mutations were confirmed by Sanger sequencing with the BigDye Terminator v.3.1 Cycle Sequencing Kit (Life Technologies). Candidate rare homozygous variants (minor allele frequency [MAF] ≤ 0.01) were analyzed with an in-house hierarchy of functional consequence. To create gene expression profiles of putative disease-related genes, we consulted iSyTE31 (for hg19 microarray expression data from the embryonic mouse lens at embryonic days 10.5, 11.5, and 12.5) and NEIBank (for expressed-sequence-tag data from different tissues within the embryonic and adult eye).
ERG Recording of Fly Eye
ERG recordings were performed as described by Verstreken et al.32 In brief, flies were glued to a slide with Elmer’s Glue. A recording electrode filled with 100 mM NaCl was placed on the eye, and a reference electrode was placed on the fly head. During the recording, a 1 s pulse of light stimulation was given, and the ERG traces of ten flies for each genotype were recorded and analyzed with WinWCP v.5.3.3 software.
Drosophila Genetics
The following stocks were obtained from the Bloomington Drosophila Stock Center (BDSC) at Indiana University: y1w∗; Mi{PT-GFSTF.2}sifMI02376-GFSTF.2 (RRID: BDSC_59792), GMR-gal4 (on II), y1v1; P{TRiP.JF01795}attP2 (RRID: BDSC_25789), y1v1; P{TRiP.HMJ23517}attP40 (RRID: BDSC_61934), and y1v1;P{y[+t7.7]v[+t1.8] = UAS-LUC.VALIUM10}attP2 (RRID: BDSC_35788).
Immunohistochemistry
Antibody staining of larval eye discs, pupal eyes, and adult eyes was performed as described by Hsiao et al.33 with minor modifications. Tissues were dissected and fixed in 4% paraformaldehyde and 1× PBS fixative for 15 min at room temperature and incubated with primary antibodies overnight at 4°C. Primary antibodies were as follows: α-Elav (1:200, 7E8A10, DSHB),34 α-GFP (1:200, A-11122, Invitrogen), α-E-cad (1:50, DCAD2, DSHB),35 α-ATPase (1:200, a5, DSHB),36 α-Cut (1:200, 2B10, DSHB),37 and α-Nrx-IV (1:200).38 Secondary antibodies conjugated with Cy3, Cy5, or Alexa 488 were from Jackson ImmunoResearch and were incubated for imaging in Vectashield (Vector Laboratories) with 4’,6-diamidino-2-phenylindole. Fluorescent images were acquired on a Zeiss LSM880 confocal microscope.
Results
Clinical Evaluation
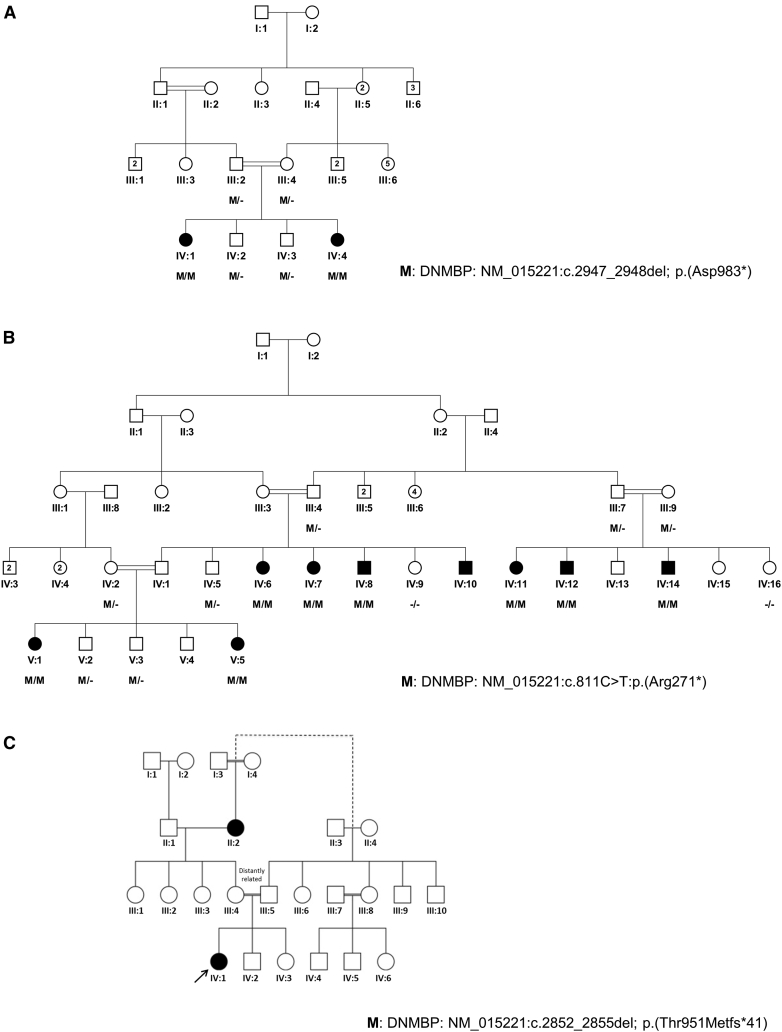
Family F372 (Figure 1A) originates from the Rahim Yar Khan region in Punjab, Pakistan. Among four siblings, individuals IV:1 (12 years old at last evaluation) and IV:4 (6 years old age at last evaluation) displayed visual impairment and bilateral cataracts in early childhood. Both affected individuals underwent lensectomy during early childhood. ERG after surgery in individual IV:1 suggested diminished scotopic (rod) and photopic (cones) responses and showed that scotopic responses were more affected.
Figure 1.
Family Pedigrees and Segregation of Variants
(A) Pedigree diagram of family F372 shows the segregation of DNMBP c.2947_2948del (p.Asp983∗).
(B) Pedigree diagram of family F385 shows the segregation of DNMBP nonsense mutation c.811C>T (p.Arg271∗).
(C) Pedigree diagram of family F3 shows the proband with the homozygous DNMBP mutation c.2852_2855del (p.Thr951Metfs∗41). The consanguineous parents indicated that they are distantly related but are uncertain as to exactly how the branches of their families are linked (indicated by the dashed line).
The large second family, F385 (Figure 1B), originates from the Khyber Pakhtunkhwa region of Pakistan. In total, nine affected individuals belong to three pedigree loops (IV:6, IV:7, IV:8, and IV:10 in the first; IV:11, IV:12, and IV:14 in the second; and V:1 and V:5 in the third); all affected individuals presented with infantile or early-childhood cataracts and visual impairment. The cataract lenses of all affected individuals were removed at different time points. ERG was performed in five individuals (IV:6, IV:7, IV:10, IV:11, and IV:14), and although photopic responses were decreased in individual IV:10, they were grossly normal in the other four affected individuals (IV:6, IV:7, IV:11, and IV:14). Individual clinical information is described in Table 1.
Table 1.
Clinical Evaluations of Affected Individuals
|
Family F372 |
Family F385 |
Family F3 |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IV:1 | IV:4 | IV:6 | IV:7 | IV:8 | IV:10 | IV:11 | IV:12 | IV:14 | V:1 | V:5 | IV-1 | |
| Age at last evaluation (years) | 12 | 6 | 26 | 22 | 32 | 34 | 45 | 46 | 34 | 5 | 6 | 4 |
| Gender | female | female | female | female | male | male | female | male | male | female | female | female |
| Consanguinity | yes | yes | yes | yes | yes | yes | yes | yes | yes | yes | yes | yes |
| DNMBP variation (GenBank: NM_015221.2) | c.2947_2948del (p.Asp983∗) | c.2947_2948del (p.Asp983∗) | c.811C>T (p.Arg271∗) | c.811C>T (p.Arg271∗) | c.811C>T (p.Arg271∗) | c.811C>T (p.Arg271∗) | c.811C>T (p.Arg271∗) | c.811C>T (p.Arg271∗) | c.811C>T (p.Arg271∗) | c.811C>T (p.Arg271∗) | c.811C>T (p.Arg271∗) | c.2852_2855del (p.Thr951Metfs∗41) |
| Vision at birth | reduced | reduced | reduced | partial blindness | reduced | reduced | partial blindness | partial blindness | partial blindness | partial blindness | partial blindness | reduced |
| Congenital cataracts | bilateral | bilateral | bilateral | bilateral | bilateral | bilateral | bilateral | bilateral | bilateral | bilateral, dense | bilateral | bilateral |
| Cataract surgery | yes | yes | yes (bilateral: 5 years of age) | yes (L: during childhood; R: 12 years of age) | yes | yes (L: 15 years of age; R: 16 years of age) | yes (bilateral: 8 years of age) | yes (9 years of age) | yes (L: 15 years of age; R: 18 years of age) | no | yes (15 months of age) | yes |
| Visual acuity | L: 1.06; R: 1.06 (LogMAR chart) | L: 6/18; R: 6/24 | L: 6/120; R: 6/75 | L: 6/240; R: 6/150 | no vision | L: 6/75; R: 6/90 | L: 6/60; R: 6/120 | bilateral reduced vision | L: 6/18; R: 6/60 | partial blindness | L: CF at 0.5 m; R: 6/38 | L: 6/18; R: 6/18 |
| ERG | diminished scotopic and photopic responses | NA | grossly normal | grossly normal | NA | decreased cone response | grossly normal | NA | not possible because of nystagmus | no fundus vision | could not be performed | NA |
| FFA | grossly normal | grossly normal | normal | could not be performed | NA | grossly normal | grossly normal | NA | not possible because of nystagmus | no fundus vision | R: normal; L: not visible because of dense PCO | NA |
| Other symptoms | NR | NR | L: exotropia, bilateral amblyopia, distorted pupils | L: exotropia, bilateral amblyopia, distorted pupils | left divergent squint | pendular nystagmus, constricted pupils |
L: exotropia, bilateral amblyopia, right divergent squint, distorted pupils | NR | bilateral amblyopia; L: exotropia left eye squint, pendular nystagmus, distorted pupils |
no view of fundus | constricted pupils | NR (developmentally normal) |
Detailed phenotypes of all affected individuals from three unrelated families F372, F385, and F3 are described. Abbreviations are as follows: PCO, posterior capsule opacification; FFA, fundus fluorescein angiography; ERG, electroretinography; L, left eye; R, right eye; CF, counting fingers; OCT, optical coherence tomography; NA, not available; NR, none reported.
The third family, F3, also originates from Pakistan. Individual IV:1 has been under ophthalmic care from a young age since bilateral cataracts were observed at 34 months of age (visual acuities 6/60 OD and 6/76 OS with Cardiff cards). Lensectomies were performed at 3 years of age. Examination showed a good foveal reflex and that her anterior chambers were formed and her optic nerves were healthy. Post-operative visual acuities were measured at 6/18 OD and 3/18 OS with K pictures. At 4 years of age, she was developmentally normal, and her general health was unremarkable. Family history revealed that her maternal grandmother (II:2), also born to consanguineous parents, had bilateral infantile-onset cataracts.
Genetic Analysis
Exome sequencing performed in one affected individual (IV:1) of family F372 revealed 22,406 high-quality exonic variants, and on average 96% of the exons were covered by at least ten reads. Similarly, in family F385, exome sequencing of an affected individual (IV:11) yielded 24,192 exonic sequences with a 10× average coverage of 99.4% of the target exomes. For individual IV-1 from family F3, WES followed by a filtering strategy identified 35,703 variants. After filters were applied, 689 homozygous variants were identified and predicted to have damaging effects on the encoded proteins. In the initial analysis, we did not detect any likely pathogenic variant in the panel of 115 cataract-related genes15 or in the 261 known genes related to various types of visual impairment documented in the Retinal Information Network.39 We subsequently performed SNP-array genotyping of all individuals from both families F372 and F385 and defined ROHs in every individual. By analyzing both the exome sequencing and homozygosity mapping data through CATCH,27 we identified a homozygous frameshift variant (c.2947_2948del [p.Asp983∗]) in family F372 and a homozygous stop-gain variant (c.811C>T [p.Arg271∗]) in DNMBP (OMIM: 611282; GenBank: NM_015221.2) in family 385. We performed Sanger sequencing to validate both variants and confirm the segregation of the variants in all members of the two families with the disease phenotype (Figure 1 and Figure S1). The homozygous frameshift variant in DNMBP (c.2947_2948del [p.Asp983∗]) segregated with the disease phenotype in family F372 with a LOD score of 0.85 (Figure 1A and Figure S2). However, the homozygous stop-gain variant in DNMBP (c.811C>T [p.Arg271∗]) segregated with a LOD score of 5.18 in all available members of the pedigree (Figure 1B). Neither variant was found in a local cohort of 240 control individuals of the same ethnicity or in gnomAD.40
Screening of the proband from family F3 for a panel of 115 known cataract-related genes15 did not identify any putative pathogenic variants. Subsequent WES of the proband in F3 yielded 18× read-depth coverage of 95.7% of targets. Enrichment of the data for homozygous variants identified 13,598 results. Further filtering of the data for relatively rare (MAF ≤ 0.01), protein-altering variants not present in a local database of 610 exomes identified 18 variants. Of these, two were predicted to damage protein function: c.2852_2855delCCAA (p.Thr951Metfs∗41) in DNMBP and c.388C>T (p.Leu130Phe) (GenBank: NM_194277) in FRMD7. Consultation of the ISyTE database found that of these two genes, DNMBP showed lens-enriched expression. Samples from parents or other family members were not available for study.
The pLI (probability of LoF intolerance) score for DNMBP in the ExAC Browser40 is 0.00. This suggests that heterozygous LoF mutations in DNMBP are tolerated and that a single functional copy of a gene is sufficient to maintain normal function of protein; thus, bi-allelic pathogenic variants in this gene are likely to cause a recessive disorder.
The Fly Homolog of DNMBP, sif, Is Required for Proper Differentiation of Lens-Secreting Cells and Phototransduction
The sole homolog of DNMBP in the fly is sif. Although sif is unique in the fly, the human genome carries at least three homologs of sif (TIAM1, TIAM2, and DNMBP). Overall, Sif and DNMBP share 33% similarity and 19% identity.41 DNMBP and Sif share a well-conserved RhoGEF domain with 48% homology and 30% amino acid identity. The RhoGEF domain present in the middle of DNMBP is known to activate Cdc42 and cooperate with the C terminus of the protein to control actin assembly.42
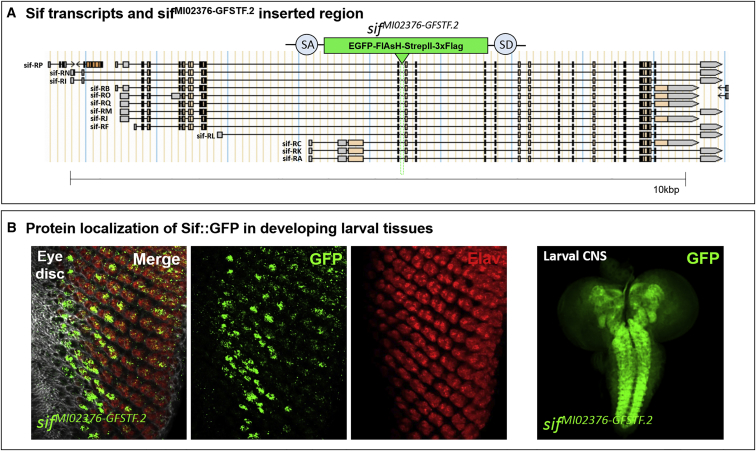
To assess the endogenous subcellular localization of Sif, we utilized a fly strain in which we inserted an artificial exon encoding a GFP in a coding intron (an intron between two coding exons). We used recombination-mediated cassette exchange of a Minos-mediated integration cassette (MiMIC; MI02376) to integrate the GFP-encoding exon SA-EGFP-FlAsH-StrepII-TEV-3xFlag tag-SD flanked by two inverted attB sites43, 44 (Figure 2A). This stock is healthy and viable in the homozygous state, indicating that this internal GFP tag is not toxic because loss of the gene shows very severe motor deficits.45 As shown in Figure 2B, Sif-GFP is enriched in a single photoreceptor in third-instar larval eye discs but is widely localized in the neuropil of the larval CNS.
Figure 2.
sifMI02376-GFSTF.2 Is a Functional GFP Trap
(A) sifMI02376-GFSTF.2 was inserted in a coding intron tagging most or all sif transcripts through the integration of a SA-GFP-SD artificial exon.
(B) Developing larval eye discs stained with antibodies against GFP (green) and Elav, a pan-neuronal nuclear marker (red). Sif::GFP is enriched in a single photoreceptor. Right: larval CNS stained with GFP; Sif::GFP is present in most neuropils of third-instar larvae.
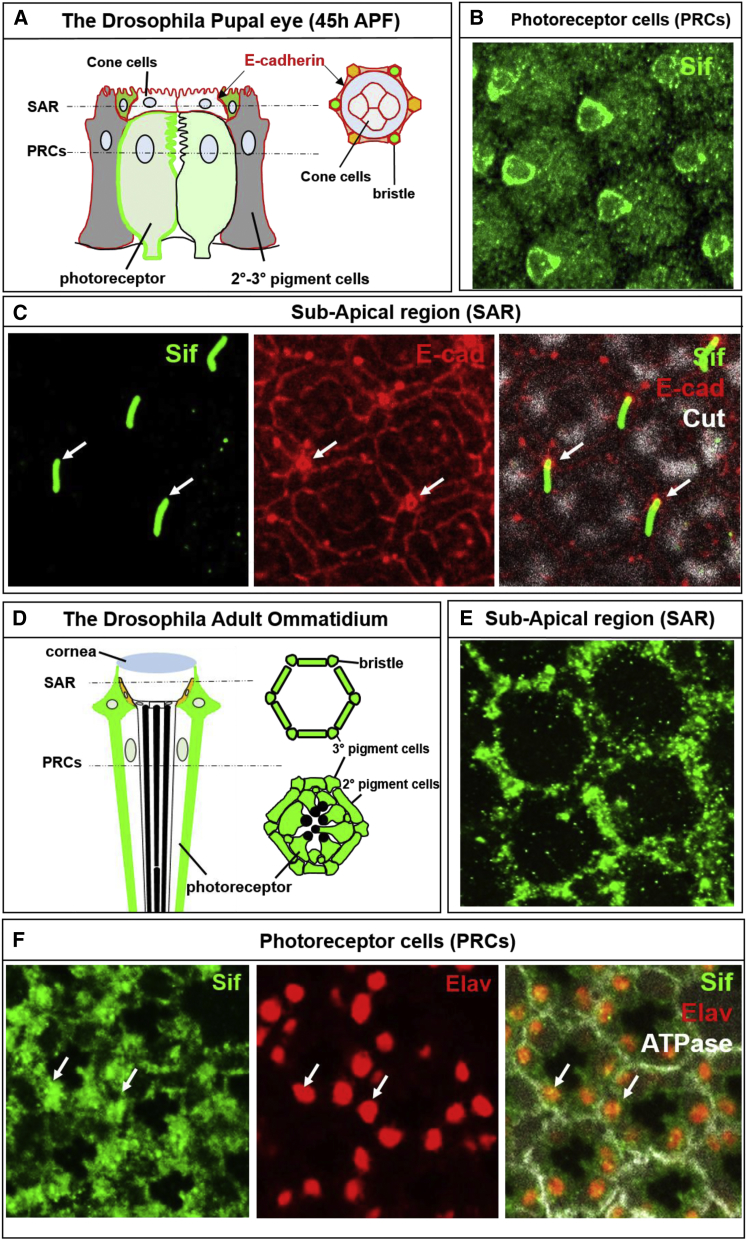
During pupal development, the cells of the third instar eye disc differentiate, the photoreceptors elongate, pigment cells at the apical area of the photoreceptors secrete a lens, and bristle cells at the apical end start forming hair-like protrusions that depend on extensive actin polymerization (Figure 3). Cell-adhesion molecules N- and E-cadherin are broadly expressed in the developing eye. E-cadherin is present in all accessory cells (pigment cells, cone cells, and bristle cells), whereas N-cadherin is restricted to cone cells and the underlying photoreceptors.46 Vertebrate lens development shares many features with fly pupal eye development,47 and we therefore investigated the localization of Sif-GFP in the pupal eye (45 hr after pupae formation [APF]). During pupal eye development, cone cells grow over the photoreceptors (R1–R8), and E-cadherin is expressed in the subcellular membrane, where the cone cells touch the photoreceptors, pigment cells, and bristle cells. Cut, an apical determinant, is specifically expressed in cone cells48 (Figure 3A). Note that Sif-GFP is also localized in photoreceptors but is clearly more abundant in one of them (Figure 3B). Sif-GFP is also enriched in the shaft of the bristle cells during bristle formation (Figure 3C).
Figure 3.
Sif::GFP Is Localized to Subsets of Cells in the Pupal Eye and Adult Eye
(A) Schematic representation of the Drosophila pupal eye (45 hr APF).
(B) Sif::GFP is present in all photoreceptors but was clearly more abundant in one of them.
(C) Sif::GFP is enriched in the shaft of bristle cells in the sub-apical region (SAR) during their development. E-cadherin marks the membranes of pigment cells, cone cells, and bristle cells (red labels in A and C), whereas Cut (white) marks the apical area of cone cells. White arrows point to bristles.
(D) Schematic representation of a longitudinal section of an adult ommatidium.
(E and F) Sif::GFP is localized in secondary and tertiary pigment cells in the SAR (E) but is also localized in the cytoplasm and nuclei of photoreceptors (F). Elav (red) marks the nuclei of photoreceptors. White arrows point to the area where Sif::GFP and Elav colocalize.
The adult eye consists of ∼750 ommatidia each with 8 photoreceptors and 11 accessory cells (Figure 3D). Two of the primary pigment cells and the four underlying cone cells secrete the corneal lenses.49 Interestingly, Sif-GFP is enriched in two primary pigment cells in the subapical region of the eye (Figure 3E). In addition, Sif-GFP is also present in photoreceptors (Figure 3F).
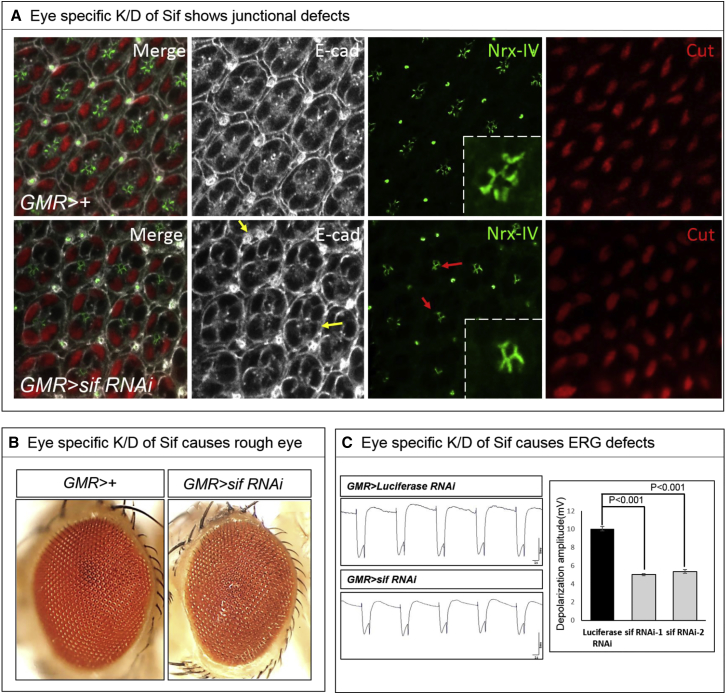
To analyze the function of Sif in the developing eye, we expressed two different UAS-sif RNAi constructs (TRiP JF01795 and TRiP HMJ23517) by using the GMR Gal4 driver, which expresses the yeast transcription factor GAL4 under the control of the glass multiple reporter (GMR) promoter, during eye development in all cells of the visual system. To determine whether knockdown of sif causes defects in pupal eye development, we analyzed the pupal eye of GMR>+ and GMR>Sif RNAi pupae at 45 hr of pupal development. We used E-cadherin35 to mark the membrane of the accessory cells, Nrx-IV to mark septate junctions that functionally correspond to tight junctions,38 and Cut to mark the apical region of the cone cells.37 Knockdown of Sif causes an abnormal distribution of Nrx-IV (red arrows in Figure 4A) and affects the distribution pattern of E-cadherin (yellow arrows in Figure 3A). The E-cadherin distribution indicates that some of the pigment cells that secrete the lenses are misshapen or lost. In addition, the position of some bristle cells is aberrant or lacking. Finally, the distribution of Nrx-IV is aberrant, showing that the junction between photoreceptors and cone cells is aberrant. These defects are also associated with a subtle but obvious roughness of the external eye, as shown in Figure 4B.
Figure 4.
Knockdown (K/D) of sif Causes Morphological and Functional Defects in Developing Eyes
(A) Pupal eye stained at 45 hr APF for E-cad (white), Nrx-IV (green), and Cut (red). Yellow arrows highlight abnormal bristle and pigment cells. Red arrows point to the region of adjacent photoreceptors.
(B) Adult eyes from GMR>+ (control) and GMR>sif RNAi. Expression of sif RNAi causes rough eyes.
(C) Reduced ERG amplitude of GMR>sif RNAi flies and quantification of the amplitude of ERG traces (n = 10; error bars represent ± SEM; ∗∗∗p < 0.001, t test).
We performed ERG in young flies to assess whether GMR>sif RNAi leads to a defect of the function of the photoreceptors. As shown in Figure 3C, expression of two independent RNAi constructs reduces ERG amplitude to about half of that of the control (GMR>Luciferase RNAi) (Figure 4C). These data implicate that the phototransduction cascade in the fly is impaired. However, the on and off transients, which are a measure of synaptic transmission between the photoreceptor cells and their postsynaptic partners, are unaffected (data not shown). These data argue that flies are still able to see but that they have less visual input. In summary, these data show that Sif is required for proper eye development and most likely affects corneal lens-forming cells and that its loss leads to some vision loss.
Discussion
We report 12 individuals (from three families) affected by three different homozygous LoF variants at the DNMBP locus and presenting with bilateral infantile- or early-childhood-onset cataracts. The segregation LOD score for DNMBP in family F385 is 5.18, suggesting that the gene is associated with the observed cataracts. In addition, the identification of two additional families affected by infantile- or early-childhood-onset cataracts, as well as homozygous truncating variants in DNMBP, provides further evidence that these variants are indeed associated with infantile- or early-childhood-onset cataracts. The data are also in agreement with an ExAC40 pLI score of 0.00, suggesting that loss of DNMBP is not haploinsufficient, and thus heterozygosity of the parents for the LoF variants is compatible with an absence of the phenotype and recessive inheritance. Finally, the claim that pathogenic variants in DNMBP are causative is bolstered by protein localization and phenotypic data associated with the loss of the homolog of DNMBP, sif, in flies. Indeed, when sif expression is reduced during eye development in flies, defects in the corneal lens-forming cells are evident. Moreover, we have shown that reduced ocular expression of sif in Drosophila results in reduced ERG amplitude, potentially indicating a defect of the visual transduction pathway in young flies. Clinical data revealed that full-field ERG was subnormal in 2 of 12 individuals. The significance of this will require further clinical and functional validation for assessing whether this represents a conserved process supporting photoreceptor function. DNMBP and Sif contain a RhoGEF domain that activates Cdc42 to facilitate actin assembly in conjunction with actin regulatory proteins,42 and DNMBP has been reported to be abundantly expressed during eye development in mice and zebrafish.50, 51 The results from Eurexpress,50 a project of gene expression patterns in mouse embryos by in situ hybridization at embryonic day 14.5, showed that DNMBP is exclusively expressed in the lens during eye development at this stage (Figure S1). We found that Sif is obviously enriched in the corneal lens-forming cells during fly eye development (Figure 3E), suggesting that the proteins might have an evolutionarily conserved function in lens development.
Drosophila bristles have been utilized as a model for studying the function of the F-actin cytoskeleton.52 In flies, bristle shafts are long extensions of bristle cells and are heavily dependent on proper actin dynamics during their development.52 Loss of actin-assembly proteins leads to abnormal bristle morphology as well as reductions in bristle number.53 We observed that fly Sif is highly enriched in the shaft of developing bristle cells (Figure 3C) and that loss of sif results in polarity defects and a decreased number of bristles cells (data not shown), consistent with defects in actin dynamics. As shown in Figure 4B, knockdown of Sif in the developing eye (GMR>sif RNAi) also causes a mild rough-eye phenotype (Figure 4B). These results again support the notion that DNMBP and Sif have evolutionary conserved function in F-actin assembly and eye development.
In human epithelial cells, RNAi-mediated knockdown of DNMBP results in a disorganization of the junctional domains of cells in association with an aberrant distribution of F-actin and E-cadherin. DNMPB is recruited to the apical margin of cell-cell junctions54 and directly binds to TJP1 (tight junction protein 1) to regulate the shape of tight junctions. Interestingly, knockdown of Sif in the developing fly eye leads to the abnormal distribution of Nrx-IV, an established septate junction (SJ) marker.38 Note that tight junctions do not exist in invertebrates. However, SJs have been shown to play an analogous role to that of tight junctions in vertebrates,55 and disruption of SJs affects cellular junction in flies,56 consistent with our observations reported in Figure 4A. These results support the notion that DNMBP and Sif might play very similar roles in maintaining proper tight-junction functions and SJs. Interestingly, a cohort of 52 individuals with cataracts has been reported to have remarkably reduced amounts of TJP1 in lens epithelial cells, suggesting that these junctions might play an important role in lens formation.57 Moreover, DNMBP is a scaffold protein that interacts with dynamin to regulate the actin cytoskeleton.42 It is well known that a sophisticated cytoskeleton network within lens fiber cells is crucial for their proper migration, elongation, and differentiation, as well as lens fiber maintenance.58, 59 Mutations in lens cytoskeleton components beaded filament protein 1 (BFSP1 [OMIM: 603307]) and 2 (BFSP2 [OMIM: 603212]) are known to cause cataracts in humans.60, 61 Therefore, loss of DNMBP could affect cytoskeleton dynamics in lens fiber cells, which in itself could be a mechanism contributing to cataract formation. Future work in this area would provide further insight into the role of DNMBP and the lenticular cytoskeleton in lens development and transparency.
In summary, we have identified three unrelated consanguineous families with a total of 12 individuals who are affected by bilateral infantile- or early-childhood-onset cataracts and harbor homozygous LoF mutations in DNMBP. Flies lacking the ortholog Sif present with an ocular phenotype that is strikingly similar to that observed in humans, thereby providing strong supporting evidence that DNMBP LoF variants cause infantile- or early-childhood-onset cataracts in humans.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We thank the Swiss Government Excellence Scholarships program, which provided M.A. the opportunity to work at the University of Geneva Medical School in Switzerland. R.L.T is supported by the Medical Research Council through a UK Research and Innovation Fellowship (MR/R024952/1). We thank the Pro Visu Foundation for the grant support to S.E.A. This project was partially supported by European Research Council grant 219968 to S.E.A. H.J.B. is supported by the NIH (R24OD022005) and is an investigator of the Howard Hughes Medical Institute. The work conducted at Manchester Centre for Genomic Medicine (G.C.B and R.L.T) was supported by Fight for Sight UK (grant 1831). Confocal microscopy was performed in the neurovisualization core of the Baylor College of Medicine Intellectual and Developmental Disabilities Research Center (supported by National Institute of Child Health and Human Development [NICHD] grant U54HD083092). Drosophila stocks were obtained from the Bloomington Stock Center (NIH P40OD018537) at Indiana University. Monoclonal antibodies were obtained from the Developmental Studies Hybridoma Bank, created by the NICHD and maintained at the University of Iowa. We are thankful to all the members of the families reported in this study.
Published: October 4, 2018
Footnotes
Supplemental Data include two figures and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.09.004.
Contributor Information
Hugo J. Bellen, Email: hbellen@bcm.edu.
Stylianos E. Antonarakis, Email: stylianos.antonarakis@unige.ch.
Web Resources
Cat-Map, http://cat-map.wustl.edu/
Eurexpress, http://www.eurexpress.org
ExAC Browser, http://exac.broadinstitute.org/
MARRVEL, http://marrvel.org/
OMIM, http://www.omim.org/
UCSC Genome Browser, https://genome.ucsc.edu/
Supplemental Data
References
- 1.Srivastava R., Budak G., Dash S., Lachke S.A., Janga S.C. Transcriptome analysis of developing lens reveals abundance of novel transcripts and extensive splicing alterations. Sci. Rep. 2017;7:11572. doi: 10.1038/s41598-017-10615-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharma K.K., Santhoshkumar P. Lens aging: effects of crystallins. Biochim. Biophys. Acta. 2009;1790:1095–1108. doi: 10.1016/j.bbagen.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cvekl A., Ashery-Padan R. The cellular and molecular mechanisms of vertebrate lens development. Development. 2014;141:4432–4447. doi: 10.1242/dev.107953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Idrees F., Vaideanu D., Fraser S.G., Sowden J.C., Khaw P.T. A review of anterior segment dysgeneses. Surv. Ophthalmol. 2006;51:213–231. doi: 10.1016/j.survophthal.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Gillespie R.L., Lloyd I.C., Black G.C. The use of autozygosity mapping and next-generation sequencing in understanding anterior segment defects caused by an abnormal development of the lens. Hum. Hered. 2014;77:118–137. doi: 10.1159/000362599. [DOI] [PubMed] [Google Scholar]
- 6.Gilbert C., Foster A. Childhood blindness in the context of VISION 2020--the right to sight. Bull. World Health Organ. 2001;79:227–232. [PMC free article] [PubMed] [Google Scholar]
- 7.Wu X., Long E., Lin H., Liu Y. Prevalence and epidemiological characteristics of congenital cataract: a systematic review and meta-analysis. Sci. Rep. 2016;6:28564. doi: 10.1038/srep28564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gillespie R.L., Urquhart J., Anderson B., Williams S., Waller S., Ashworth J., Biswas S., Jones S., Stewart F., Lloyd I.C. Next-generation Sequencing in the Diagnosis of Metabolic Disease Marked by Pediatric Cataract. Ophthalmology. 2016;123:217–220. doi: 10.1016/j.ophtha.2015.06.035. [DOI] [PubMed] [Google Scholar]
- 9.Graw J. The genetic and molecular basis of congenital eye defects. Nat. Rev. Genet. 2003;4:876–888. doi: 10.1038/nrg1202. [DOI] [PubMed] [Google Scholar]
- 10.Pierce E.A., Bennett J. The Status of RPE65 Gene Therapy Trials: Safety and Efficacy. Cold Spring Harb. Perspect. Med. 2015;5:a017285. doi: 10.1101/cshperspect.a017285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Antonarakis S.E., Beckmann J.S. Mendelian disorders deserve more attention. Nat. Rev. Genet. 2006;7:277–282. doi: 10.1038/nrg1826. [DOI] [PubMed] [Google Scholar]
- 12.Shiels A., Bennett T.M., Hejtmancik J.F. Cat-Map: putting cataract on the map. Mol. Vis. 2010;16:2007–2015. [PMC free article] [PubMed] [Google Scholar]
- 13.Hejtmancik J.F. Congenital cataracts and their molecular genetics. Semin. Cell Dev. Biol. 2008;19:134–149. doi: 10.1016/j.semcdb.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J., Xia C.H., Wang E., Yao K., Gong X. Screening, genetics, risk factors, and treatment of neonatal cataracts. Birth Defects Res. 2017;109:734–743. doi: 10.1002/bdr2.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gillespie R.L., O’Sullivan J., Ashworth J., Bhaskar S., Williams S., Biswas S., Kehdi E., Ramsden S.C., Clayton-Smith J., Black G.C., Lloyd I.C. Personalized diagnosis and management of congenital cataract by next-generation sequencing. Ophthalmology. 2014;121:2124–2137. doi: 10.1016/j.ophtha.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 16.Basel-Vanagaite L., Taub E., Halpern G.J., Drasinover V., Magal N., Davidov B., Zlotogora J., Shohat M. Genetic screening for autosomal recessive nonsyndromic mental retardation in an isolated population in Israel. Eur. J. Hum. Genet. 2007;15:250–253. doi: 10.1038/sj.ejhg.5201750. [DOI] [PubMed] [Google Scholar]
- 17.Iqbal Z., Willemsen M.H., Papon M.A., Musante L., Benevento M., Hu H., Venselaar H., Wissink-Lindhout W.M., Vulto-van Silfhout A.T., Vissers L.E. Homozygous SLC6A17 mutations cause autosomal-recessive intellectual disability with progressive tremor, speech impairment, and behavioral problems. Am. J. Hum. Genet. 2015;96:386–396. doi: 10.1016/j.ajhg.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Makrythanasis P., Kato M., Zaki M.S., Saitsu H., Nakamura K., Santoni F.A., Miyatake S., Nakashima M., Issa M.Y., Guipponi M. Pathogenic Variants in PIGG Cause Intellectual Disability with Seizures and Hypotonia. Am. J. Hum. Genet. 2016;98:615–626. doi: 10.1016/j.ajhg.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Makrythanasis P., Nelis M., Santoni F.A., Guipponi M., Vannier A., Béna F., Gimelli S., Stathaki E., Temtamy S., Mégarbané A. Diagnostic exome sequencing to elucidate the genetic basis of likely recessive disorders in consanguineous families. Hum. Mutat. 2014;35:1203–1210. doi: 10.1002/humu.22617. [DOI] [PubMed] [Google Scholar]
- 20.Riazuddin S., Hussain M., Razzaq A., Iqbal Z., Shahzad M., Polla D.L., Song Y., van Beusekom E., Khan A.A., Tomas-Roca L., UK10K Exome sequencing of Pakistani consanguineous families identifies 30 novel candidate genes for recessive intellectual disability. Mol. Psychiatry. 2017;22:1604–1614. doi: 10.1038/mp.2016.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Najmabadi H., Hu H., Garshasbi M., Zemojtel T., Abedini S.S., Chen W., Hosseini M., Behjati F., Haas S., Jamali P. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature. 2011;478:57–63. doi: 10.1038/nature10423. [DOI] [PubMed] [Google Scholar]
- 22.Ansar M., Riazuddin S., Sarwar M.T., Makrythanasis P., Paracha S.A., Iqbal Z., Khan J., Assir M.Z., Hussain M., Razzaq A. Biallelic variants in LINGO1 are associated with autosomal recessive intellectual disability, microcephaly, speech and motor delay. Genet. Med. 2018;20:778–784. doi: 10.1038/gim.2017.113. [DOI] [PubMed] [Google Scholar]
- 23.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pruitt K.D., Tatusova T., Maglott D.R. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Santoni F.A., Makrythanasis P., Antonarakis S.E. CATCHing putative causative variants in consanguineous families. BMC Bioinformatics. 2015;16:310. doi: 10.1186/s12859-015-0727-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ansar M., Chung H., Waryah Y.M., Makrythanasis P., Falconnet E., Rao A.R., Guipponi M., Narsani A.K., Fingerhut R., Santoni F.A. Visual impairment and progressive phthisis bulbi caused by recessive pathogenic variant in MARK3. Hum. Mol. Genet. 2018 doi: 10.1093/hmg/ddy180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar P., Henikoff S., Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 30.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lachke S.A., Ho J.W., Kryukov G.V., O’Connell D.J., Aboukhalil A., Bulyk M.L., Park P.J., Maas R.L. iSyTE: Integrated Systems Tool for Eye gene discovery. Invest. Ophthalmol. Vis. Sci. 2012;53:1617–1627. doi: 10.1167/iovs.11-8839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Verstreken P., Koh T.W., Schulze K.L., Zhai R.G., Hiesinger P.R., Zhou Y., Mehta S.Q., Cao Y., Roos J., Bellen H.J. Synaptojanin is recruited by endophilin to promote synaptic vesicle uncoating. Neuron. 2003;40:733–748. doi: 10.1016/s0896-6273(03)00644-5. [DOI] [PubMed] [Google Scholar]
- 33.Hsiao H.Y., Johnston R.J., Jr., Jukam D., Vasiliauskas D., Desplan C., Rister J. Dissection and immunohistochemistry of larval, pupal and adult Drosophila retinas. J. Vis. Exp. 2012;69:4347. doi: 10.3791/4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinow S., White K. Characterization and spatial distribution of the ELAV protein during Drosophila melanogaster development. J. Neurobiol. 1991;22:443–461. doi: 10.1002/neu.480220503. [DOI] [PubMed] [Google Scholar]
- 35.Oda H., Uemura T., Harada Y., Iwai Y., Takeichi M. A Drosophila homolog of cadherin associated with armadillo and essential for embryonic cell-cell adhesion. Dev. Biol. 1994;165:716–726. doi: 10.1006/dbio.1994.1287. [DOI] [PubMed] [Google Scholar]
- 36.Lebovitz R.M., Takeyasu K., Fambrough D.M. Molecular characterization and expression of the (Na+ + K+)-ATPase alpha-subunit in Drosophila melanogaster. EMBO J. 1989;8:193–202. doi: 10.1002/j.1460-2075.1989.tb03364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blochlinger K., Bodmer R., Jan L.Y., Jan Y.N. Patterns of expression of cut, a protein required for external sensory organ development in wild-type and cut mutant Drosophila embryos. Genes Dev. 1990;4:1322–1331. doi: 10.1101/gad.4.8.1322. [DOI] [PubMed] [Google Scholar]
- 38.Baumgartner S., Littleton J.T., Broadie K., Bhat M.A., Harbecke R., Lengyel J.A., Chiquet-Ehrismann R., Prokop A., Bellen H.J. A Drosophila neurexin is required for septate junction and blood-nerve barrier formation and function. Cell. 1996;87:1059–1068. doi: 10.1016/s0092-8674(00)81800-0. [DOI] [PubMed] [Google Scholar]
- 39.Daiger S.P., Rossiter B.J.F., Greenberg J., Christoffels A., Hide W. Data services and software for identifying genes and mutations causing retinal degeneration. Invest. Ophthalmol. Vis. Sci. 1998;39:S295. [Google Scholar]
- 40.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang J., Al-Ouran R., Hu Y., Kim S.Y., Wan Y.W., Wangler M.F., Yamamoto S., Chao H.T., Comjean A., Mohr S.E., UDN MARRVEL: Integration of Human and Model Organism Genetic Resources to Facilitate Functional Annotation of the Human Genome. Am. J. Hum. Genet. 2017;100:843–853. doi: 10.1016/j.ajhg.2017.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salazar M.A., Kwiatkowski A.V., Pellegrini L., Cestra G., Butler M.H., Rossman K.L., Serna D.M., Sondek J., Gertler F.B., De Camilli P. Tuba, a novel protein containing bin/amphiphysin/Rvs and Dbl homology domains, links dynamin to regulation of the actin cytoskeleton. J. Biol. Chem. 2003;278:49031–49043. doi: 10.1074/jbc.M308104200. [DOI] [PubMed] [Google Scholar]
- 43.Venken K.J., Schulze K.L., Haelterman N.A., Pan H., He Y., Evans-Holm M., Carlson J.W., Levis R.W., Spradling A.C., Hoskins R.A., Bellen H.J. MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat. Methods. 2011;8:737–743. doi: 10.1038/nmeth.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nagarkar-Jaiswal S., Lee P.T., Campbell M.E., Chen K., Anguiano-Zarate S., Gutierrez M.C., Busby T., Lin W.W., He Y., Schulze K.L. A library of MiMICs allows tagging of genes and reversible, spatial and temporal knockdown of proteins in Drosophila. eLife. 2015;4 doi: 10.7554/eLife.05338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sone M., Hoshino M., Suzuki E., Kuroda S., Kaibuchi K., Nakagoshi H., Saigo K., Nabeshima Y., Hama C. Still life, a protein in synaptic terminals of Drosophila homologous to GDP-GTP exchangers. Science. 1997;275:543–547. doi: 10.1126/science.275.5299.543. [DOI] [PubMed] [Google Scholar]
- 46.Hayashi T., Carthew R.W. Surface mechanics mediate pattern formation in the developing retina. Nature. 2004;431:647–652. doi: 10.1038/nature02952. [DOI] [PubMed] [Google Scholar]
- 47.Charlton-Perkins M., Brown N.L., Cook T.A. The lens in focus: a comparison of lens development in Drosophila and vertebrates. Mol. Genet. Genomics. 2011;286:189–213. doi: 10.1007/s00438-011-0643-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Izaddoost S., Nam S.C., Bhat M.A., Bellen H.J., Choi K.W. Drosophila Crumbs is a positional cue in photoreceptor adherens junctions and rhabdomeres. Nature. 2002;416:178–183. doi: 10.1038/nature720. [DOI] [PubMed] [Google Scholar]
- 49.Wolff T., Ready D.F. The beginning of pattern formation in the Drosophila compound eye: the morphogenetic furrow and the second mitotic wave. Development. 1991;113:841–850. doi: 10.1242/dev.113.3.841. [DOI] [PubMed] [Google Scholar]
- 50.Diez-Roux G., Banfi S., Sultan M., Geffers L., Anand S., Rozado D., Magen A., Canidio E., Pagani M., Peluso I. A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol. 2011;9:e1000582. doi: 10.1371/journal.pbio.1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baek J.I., Kwon S.H., Zuo X., Choi S.Y., Kim S.H., Lipschutz J.H. Dynamin Binding Protein (Tuba) Deficiency Inhibits Ciliogenesis and Nephrogenesis in Vitro and in Vivo. J. Biol. Chem. 2016;291:8632–8643. doi: 10.1074/jbc.M115.688663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wulfkuhle J.D., Petersen N.S., Otto J.J. Changes in the F-actin cytoskeleton during neurosensory bristle development in Drosophila: the role of singed and forked proteins. Cell Motil. Cytoskeleton. 1998;40:119–132. doi: 10.1002/(SICI)1097-0169(1998)40:2<119::AID-CM2>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 53.Wu J., Wang H., Guo X., Chen J. Cofilin-mediated actin dynamics promotes actin bundle formation during Drosophila bristle development. Mol. Biol. Cell. 2016;27:2554–2564. doi: 10.1091/mbc.E16-02-0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Otani T., Ichii T., Aono S., Takeichi M. Cdc42 GEF Tuba regulates the junctional configuration of simple epithelial cells. J. Cell Biol. 2006;175:135–146. doi: 10.1083/jcb.200605012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bellen H.J., Lu Y., Beckstead R., Bhat M.A. Neurexin IV, caspr and paranodin--novel members of the neurexin family: encounters of axons and glia. Trends Neurosci. 1998;21:444–449. doi: 10.1016/s0166-2236(98)01267-3. [DOI] [PubMed] [Google Scholar]
- 56.Littleton J.T., Bhat M.A., Bellen H.J. Deciphering the function of neurexins at cellular junctions. J. Cell Biol. 1997;137:793–796. doi: 10.1083/jcb.137.4.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arora A.I., Johar K., Gajjar D.U., Ganatra D.A., Kayastha F.B., Pal A.K., Patel A.R., Rajkumar S., Vasavada A.R. Cx43, ZO-1, alpha-catenin and beta-catenin in cataractous lens epithelial cells. J. Biosci. 2012;37:979–987. doi: 10.1007/s12038-012-9264-9. [DOI] [PubMed] [Google Scholar]
- 58.Cheng C., Nowak R.B., Fowler V.M. The lens actin filament cytoskeleton: Diverse structures for complex functions. Exp. Eye Res. 2017;156:58–71. doi: 10.1016/j.exer.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Quinlan R.A., Sandilands A., Procter J.E., Prescott A.R., Hutcheson A.M., Dahm R., Gribbon C., Wallace P., Carter J.M. The eye lens cytoskeleton. Eye (Lond.) 1999;13(Pt 3b):409–416. doi: 10.1038/eye.1999.115. [DOI] [PubMed] [Google Scholar]
- 60.Jakobs P.M., Hess J.F., FitzGerald P.G., Kramer P., Weleber R.G., Litt M. Autosomal-dominant congenital cataract associated with a deletion mutation in the human beaded filament protein gene BFSP2. Am. J. Hum. Genet. 2000;66:1432–1436. doi: 10.1086/302872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ramachandran R.D., Perumalsamy V., Hejtmancik J.F. Autosomal recessive juvenile onset cataract associated with mutation in BFSP1. Hum. Genet. 2007;121:475–482. doi: 10.1007/s00439-006-0319-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.