Abstract
Using multipoint linkage analysis in 20 families segregating for X-linked retinitis pigmentosa (XLRP), the lod scores on a map of eight RFLP loci were obtained. Our results indicate that under the hypothesis of homogeneity the maximal multipoint lod score supports one disease locus located slightly distal to OTC at Xp21.1. Heterogeneity testing for two XLRP loci suggested that a second XLRP locus may be located 8.5 cM proximal to DXS28 at Xp21.3. Further heterogeneity testing for three disease loci failed to detect a third XLRP locus proximal to DXS7 in any of our 20 XLRP families.
INTRODUCTION
Prior linkage analyses of X-linked retinitis pigmentosa (XLRP) support the evidence for at least two loci on the short arm of the X chromosome. Initial localization of X LRP was near X p ll.3 (RP2) with close linkage to the locus DXS7, detected by DNA probe L1.28 (Bhattacharya et al., 1984). Since this first report on the assignment of the X LRP gene, other investigators have confirmed this mapping to X p ll.3 (Mukai et al., 1985; Friedrich et al, 1985; Wright et al., 1987). However, further linkage studies on numerous other families generally support an additional site for X LRP at Xp21 (RP3) (Nussbaum et al, 1985; Francke et al., 1985; Denton et al, 1988; Wirth et al., 1988; Musarella et al., 1988). Further support for an Xp21 locus for XLR P comes from descriptions of two rare male patients with multiple phenotypes including X LRP and deletions in the Xp21 region (Francke et al, 1985; de Saint-Basile et al., 1988). A third location of X LRP near the Duchenne muscular dystrophy gene (RP4) has been suggested by Musarella et al. (1988) and by Ott et al (1990).
In an attempt to further localize the XLR P loci on the short arm of the X chromosome, we report in the present study the results of a multipoint linkage analysis using polymorphic probes that detect RFLPs at eight loci from Xcen-Xp21 and heterogeneity testing in 20 X LR P families of various ethnic backgrounds. The clinical characteristics of each of the families were also considered in an attempt to correlate the gene location(s) and the phenotypes of carriers and affected males.
MATERIALS AND METHODS
Family Analysis
A total of 20 multigeneration families in which X LR P is segregating were typed for genetic linkage to loci defined by DNA markers between Xcen and Xp21. The kindreds were of the following ethnic groups: 1 Black, 1 Hispanic, 1 Italian, 1 Scottish, 2 English, 2 Mexican, 4 German, 2 Polish, and 6 unknown. Six of these families were previously reported (Musarella et al, 1988,1989). Available members of each family underwent careful ocular examination by G.A.F. or M.A.M. Affected males were diagnosed as having X LR P by pedigree analysis, which demonstrated absence of male-to-male transmission; history of early onset of night blindness; pigmentary degeneration of the retina; constricted visual fields; and appreciable amplitude reductions on ERG testing. The criteria used to identify female carriers were previously reported by Fishman et al (1986). In 14 of the 20 families, at least one of the heterozygotes had the tapetal-like reflex (metallic sheen). This reflex was absent in 3 families (F20, GHF-4, and GHF-14). In another 3 families (FI, F ll, and F38) not enough carriers were examined to determine the status of the tapetal reflex in the heterozygotes. The other carriers had degrees of atrophy of the retinal pigment epithelium and pigmentary migration to the inner retina. Males above the age of 13 years with no symptoms and normal ophthalmological findings were classified as unaffected.
DNA Isolation and RFLP Analysis
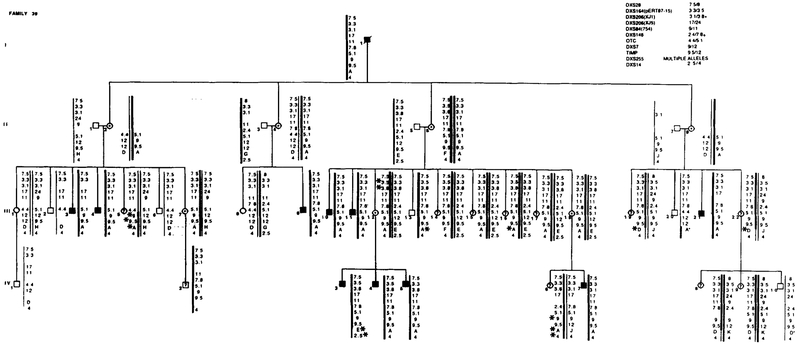
DNA was extracted from whole blood drawn in EDTA, and Southern blot analysis was carried out as described elsewhere (Musarella et ai, 1988). The 12 DNA probes used to detect the RFLPs and their locations on the short arm of the X chromosome are listed in Table 1. In families F20, GAF-17, and F38, an additional probe, p20, at locus DXS269 (Wapenaar et ai, 1987) was used and in F20, probe J66 (Ginjaar et al., 1987) was also used to determine the site of recombination in individual II-6.
TABLE 1.
X-Chromosome Probes Used for Analysis
| Locus | Probe | Location | Enzymes | Alleles (kb) | Frequencies |
|---|---|---|---|---|---|
| DXS28 | C7 | Xp21.3 | EcoRV | 7.5,8 | 0.85, 0.15 |
| DXS269 | p20 | Xp21.1−21.2 | EcoRV | 7,7.5 | 0.40, 0.60 |
| DXS164 | pERT87–15 | Xp21.2 | TaqI | 3.3, 3.5 | 0.67, 0.33 |
| DXS206 | XJ1.1 | Xp21.1−21.2 | TaqI | 3.1, 3.8 | 0.72, 0.28 |
| DXS206 | XJ5.1 | Xp21.1−21.2 | SphI | 24, 17 | 0.20, 0.80 |
| DXS84 | 754 | Xp21.1 | Pst | 11,9 | 0.55, 0.45 |
| DXS148 | cX5.7 | Xp21 | MspI | 7.8, 2.4 | 0.31, 0.69 |
| DXS141 | pERT145–12 | Xp21.1 | TaqI | 4.8, 3.7 | 0.25, 0.75 |
| Msp | 18, 4.8 | 0.15, 0.85 | |||
| OTC | HO731 | Xp21.1 | BamHI | 18, 5.4 | 0.80, 0.20 |
| Xp21.1 | MspI | 6.6, 6.2 | 0.61, 0.39 | ||
| MspI | 5.1, 4.4 | 0.73, 0.27 | |||
| DXS7 | L1.28 | Xp11.3 | TaqI | 12,9 | 0.77, 0.23 |
| TIMP | TIMP-3.9X | Xp11 | BglII | 12, 9.5 | 0.66, 0.34 |
| DXS255 | M27B | Xp11.3−cen | EcoRI | Multiallelic hypervariable systemsa | |
| PstI | |||||
| DXS14 | 58–1 | Xp11−cen | MspI | 4, 2.5 | 0.65, 0.35 |
Hypervariable fragments of M27B were recorded as three alleles in each family. The allelic frequencies used are 0.2286,0.3714, and 0.4000.
Multipoint Linkage Analysis
Multipoint linkage analysis was performed using the LINKAGE 5.03 (LINKM AP) computer program (Lathrop and Lalouel, 1988). Map distances between markers were obtained from several previously reported genetic distances (Drayna and White, 1985; Goodfellow et al., 1985; Mahtani and Willard, 1988). The estimated map locations of the marker loci are shown in Table 2. The values, x, designate map distances proximal to DXS28 in the region of Xp21.3 on the short arm of the X chromosome, where DXS28 is taken to be at map location x = 0. If there was more than one marker at a locus, the data from the most informative marker were used for calculations. Markers DXS84, DXS148, and DXS141 were regarded as being at one locus, but for most families calculations were performed using DXS84 instead of DXS148 and DXS141 because it was the most informative marker. The allelic frequencies used in the analysis are given in Table 1. Multipoint lod scores were derived by moving the disease locus between the markers and calculating the lod score for a given map distance. Each map consisted of overlapping maps of three or four marker loci and the disease locus. This was done to include all other markers. The recombination fraction estimates were converted to map distances using the Kosambi mapping function.
TABLE 2.
Locations of Marker Loci Used and Disease Loci under Hypothesis of Heterogeneity (H2) or Homogeneity (HI)
| Marker | x(cM) |
|---|---|
| DXS28(C7) | 0 |
| H2 | 8.48 |
| DXS164(pERT8715) | 14.38 |
| DXS2O6(XJ1, XJ5) | 15.38 |
| DXS84(754), DXS148(cx5.7), DXS141(145) | 19.19 |
| H1 | 22.80 |
| H2 | 24.01 |
| OTC | 25.22 |
| DXS7 | 36.40 |
| TIMP | 46.54 |
| DXS255(M27B) | 49.54 |
| DXS14(58–1) | 55.57 |
Note, x = Kosambi map locations in cM; H1 = location of XLRP locus under hypothesis of homogeneity; H2 = locations of two XLRPs loci under hypothesis of heterogeneity.
Heterogeneity Testing
To test for genetic heterogeneity, the admixture test was applied to the 20 XLR P families. This test was performed by the computer program HOMOG2, an adaptation of the HOMOG program (Ott, 1985). This program tests for two family types that are linked but are at different genetic locations. Type 1 has a disease location at x1 and type 2 has a disease location at x2. The H 0M 0G 2 program also calculates α1, the proportion of families belonging to type 1 and the conditional probability of each family belonging to type 1.
A test of homogeneity with one family type, hypothesis H i (α1 = 1), was carried out against the hypothesis of heterogeneity, hypothesis H2 (α1 < 1), for two linked family types.
Calculations under a model allowing for three disease loci (x1, x2, x3) were also performed using the HOMOG3 program. Type 3 has a disease location at x3.
RESULTS
Except for Family 39 and Family 22, only certain branches of each family were available for blood samples. In GAF-2 the alleles of four affected males in generation II are not shown but were included in the calculations. The segregation patterns of X LRP and the alleles of the RFLPs in 14 unpublished families are shown in Appendix I. Since the pedigrees in Appendix I depict only a portion of the family, Appendix II shows the full pedigree of each family and the X- linked inheritance is fully appreciated. Table 2 shows the marker loci used and their estimated map locations. In this table, H1 represents the location of the XLRP locus under the hypothesis of homogeneity, and H2 represents the two locations of the disease loci under the hypothesis that heterogeneity is present. Table 3 shows the posterior probability of being a type 1 family and the map distance where the maximum multipoint lod score occurs for each of the 20 families. The results of the heterogeneity analysis and χ2 testing are presented in Tables 4 and 5.
TABLE 3.
Admixture Test: Maximum Multipoint Lod Score (Z) at Map Distance (cM) and the Posterior Probabillity (w) of Being Type 1 Family
| Family | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F20a | F13a | F1a | F11a | F28a | F29a | GAF-1 | GAF-7 | GAF-10 | GAF-12 | GAF-17 | F38 | GAF-2 | GAF-4 | GAF-6 | GAF-11 | GAF-5 | GAF-14 | F22 | F39 | |
| z | 0.58 | 2.40 | 0.90 | 5.04 | 0.30 | 2.46 | 1.20 | 1.82 | 0.38 | 1.48 | 0.20 | 1.20 | 0.90 | 0.60 | 0.60 | 0.30 | 2.25 | 0.43 | 1.11 | 3.98 |
| cM | 8.50 | 19.19 | 25.20 | 25.22 | 25.22b | 5.62 | 19.19 | 20.39 | 22.79 | 20.39 | 36.40 | 46.54 | 19.19 | 49.54 | 25.22c | 25.22d | 19.19 | 49.54 | 15.38 | 36.40 |
| w | 0.98 | 0.06 | 0.08 | 0.00 | 0.09 | 0.39 | 0.01 | 0.02 | 0.02 | 0.00 | 0.10 | 0.06 | 0.08 | 0.04 | 0.01 | 0.09 | 0.05 | 0.12 | 0.09 | 0.00 |
These families were previously published.
Also peacked at 49.55 cM.
Also peacked at 49.55 and 55.57 cM.
Also peacked at 19.19 cM
TABLE 4.
Results of Heterogeneity Testing
| Hypothesis | Estimates of | |||
|---|---|---|---|---|
| Max In L | α1 | X1 (cM) | X2 (cM) | |
| H2: Linkage, heterogeneity | 55.985 | 0.1000 | 8.25 | 24.00 |
| H1: Linkage, homogeneity | 53.290 | (1) | 22.80 | 22.80 |
| H0: No linkage | (0) | (0) | (0.5) | (0.5) |
TABLE 5.
Components of χ2
| Component | df | X2 | P |
|---|---|---|---|
| H2 vs H1 heterogeneity | 2 | 5.391 | 0.0338 |
| H1 vs H0 linkage | 1 | 106.580 | 0.0000 |
| H2 vs H0 total | 3 | 111.971 | 0.0000 |
Under the hypothesis of homogeneity (hypothesis Hj), the location of the disease locus in our 20 XLRP families was found at a map distance of 22.8 cM (Table 2), which is 2.5 cM distal to OTC. The maximum lod score at this map distance is 53.29 (Table 4). Testing for heterogeneity under the alternative hypothesis of heterogeneity (H2) provides evidence for two XLRP loci that are linked: one 6 cM distal to DXS164 at a map distance of 8.5 cM (between markers DXS28 and DXS164) and the other 1.2 cM distal to OTC at a map distance of 24.0 cM. An α1 of 0.10 was found, denoting the estimated proportion of families of type 1. Testing H2 against Hi gives an odds ratio of 15:1 in favor of heterogeneity.
Extending the analysis to a three-locus mixture of families did not alter the results obtained by the mixture of two loci. Hence, no evidence is furnished by these families for a locus at or proximal to DXS7.
DISCUSSION
In our multipoint linkage analysis of 20 families with XLRP, we found that the disease locus is located close to OTC in almost all families. Although the results for heterogeneity under HOMOG2 are suggestive of the second disease locus with odds of 15 to 1, the most likely location of this locus is between DXS164 and DXS28. Although the evidence for this locus near DXS28 in our families is only suggestive, a recent multipoint linkage analysis of 40 families derived from several worldwide centers found a likelihood ratio of 293:1 for this locus midway between DXS28 and DXS164 (Ott et al., 1990). To determine whether this locus is indeed an XLRP locus, it would be useful to test all XLR P families with markers extending into the distal Xp21 region.
In contrast to several previously published reports (Freidrich et al., 1985; Wright et al., 1987), we could not detect any statistical evidence for a more proximally located XLRP disease locus between DXS7 and the centromere. Recently, Chen et al. (1989) performed multipoint linkage studies on nine families with XLRP and found evidence for an X LRP locus centromeric to DXS7 in two families.
An important objective has also been to correlate the clinical findings found in carriers and in affected males with the location of the XLR P disease locus. Previous reports have failed to distinguish any phe notypes with the gene location. In our families at least one heterozygote in 14 of the 20 families had the ta-petal-like reflex. Because of the peculiarities of the ta-petal-like reflex, it is extremely difficult to draw any conclusion on its significance to a particular genotype. For instance, the tapetal-like reflex may not be present in all heterozygotes in the same family; hence, it is necessary to examine as many women as possible to determine its presence in an XLRP family. In addition, it can be age dependent, generally being most visible between the second and the fourth decade of life and least detectable in older women and young girls. Although several prior linkage studies describe the existence of the tapetal-like reflex in their families, others have not addressed this issue. This is particularly true of families with the more proximal XLR P location. To date, only one XLRP family with this X p ll localization has been reported to have the tapetal-like reflex in the carrier (Friedrich et al., 1985). Since none of our families mapped to this region, we cannot draw a firm conclusion about the relationship of genotype to the retinal phenotypes of carriers. It is interesting to note that most of our families with the XLRP locus located distal to OTC have the tapetal-like reflex, and F20 in which the X LRP gene maps near DXS28, does not demonstrate this reflex. Unfortunately, therefore, determining the phenotype of the carrier state does not appear to aid further in determining the genotype of an XLRP family. Moreover, within pedigrees there are variations in clinical severity of affected males, and this also does not provide any further clues to the location of the disease locus.
We studied 20 X LRP families for genetic evidence of heterogeneity and for correlation of the retinal phenotype of carriers and affected males. Our results support convincing evidence for an X LRP gene distal to OTC, known as RP3, and perhaps a more distally placed second locus between DXS28 and DXS164, suggested as RP4. We did not find any statistical evidence for a more proximal X LRP locus, RP2, near DXS7 and the centromere.
ACKNOWLEDGMENTS
This research was supported by a grant from the National Retinitis Pigmentosa Foundation Fighting Blindness (U.S.A.). We thank Drs. J. L. Mandel, P. Pearson, L. Kunkel, G. Bruns, Ian Craig, and M. W. Hofker for the gifts of their probes. W e are particularly grateful to the X L R P families for their dedication to this research.
APPENDIX I
Segregation Patterns of XLRP and the Alleles of RFLPs in 14 Families
FIG. 1.
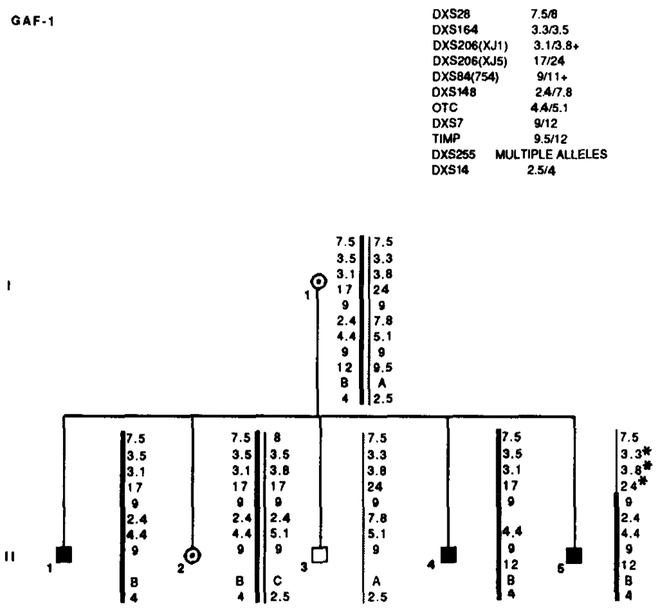
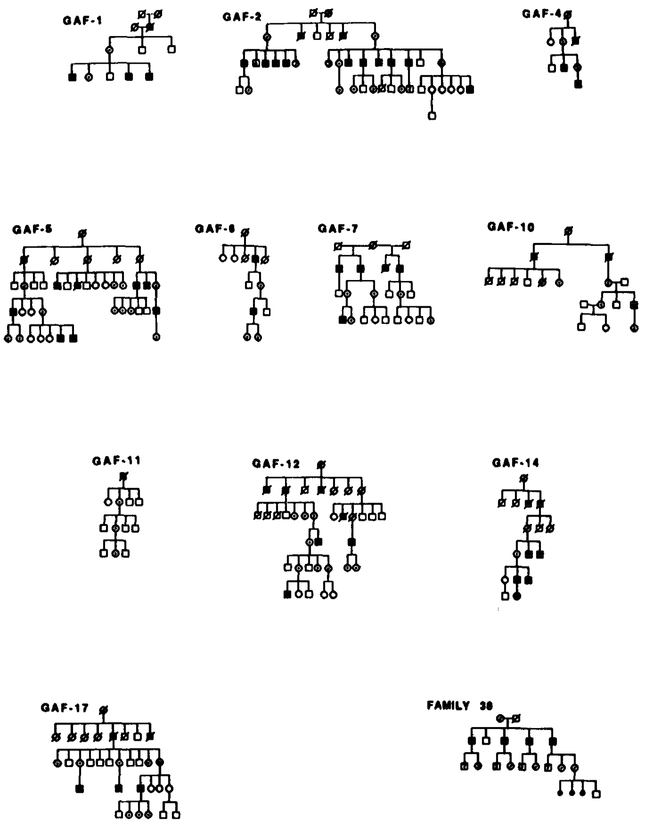
Results of RFLP analysis in GAF-1. Summary of the probes and alleles is given in the upper right table in the same order as the data presented along the sides of the chromosome stick figures. Asterisks (*) denote recombinations. Individual II-5 is recombined for markers distal to DXS84. Data from the most informative marker (+) were used in the calculations when more than one marker at a locus.
FIG. 2.
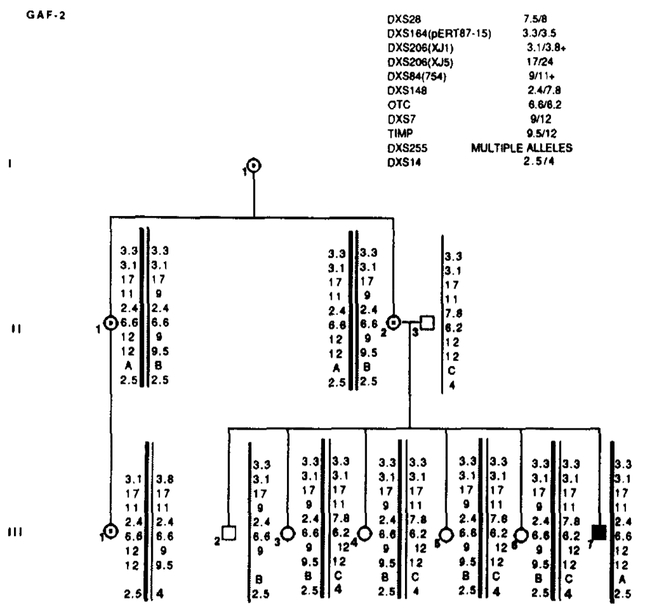
Results of RFLP analysis in GAF-2. Marker loci with their alleles are listed in the upper right table. No recombinations were detected. Data from the most informative marker (+) were used in the calculations when more than one marker at a locus.
FIG. 3.
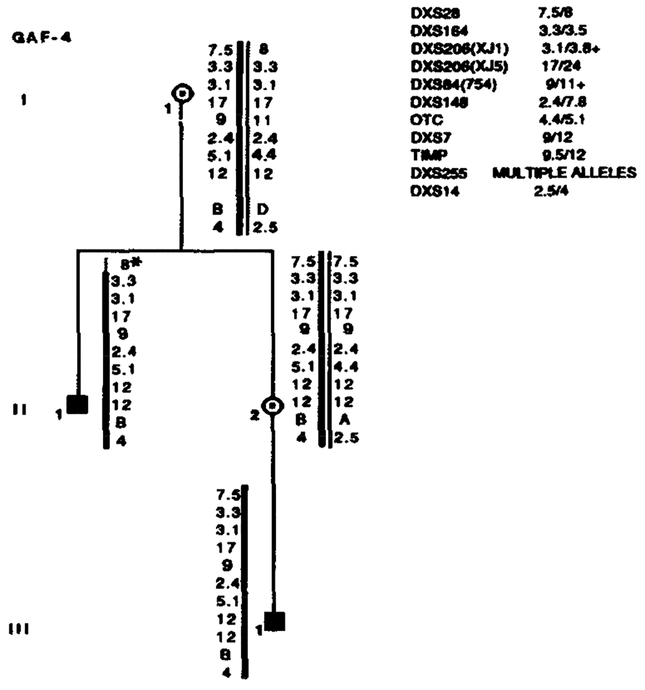
Segregation of RFLP alleles in GAF-4. Marker loci with their alleles are listed in the upper right table. Asterisks (*) denote recombinations. Data from the most informative marker (+) were used in the calculations when more than one marker at a locus.
FIG. 4.
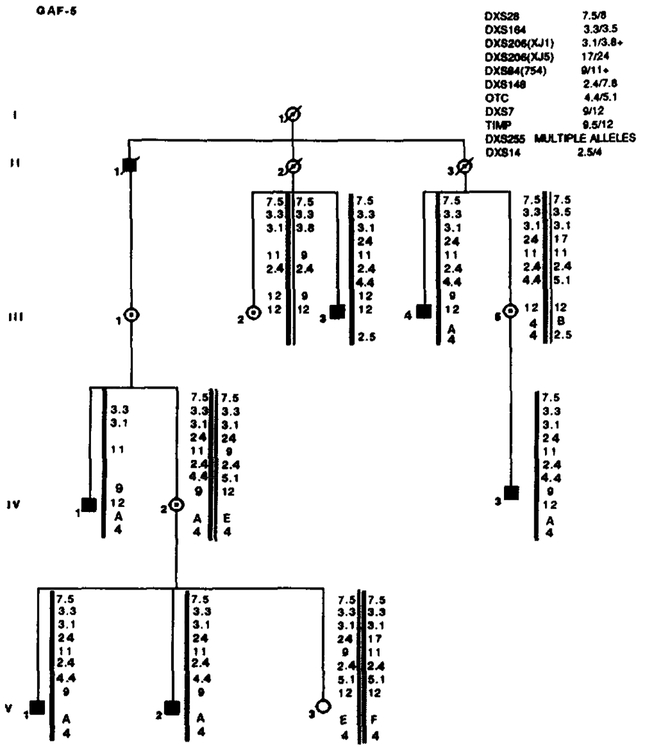
Results of RFLP analysis in GAF-5. Marker loci with their alleles are listed in the upper right table. No recombinations were detected. Data from the most informative marker (+) were used in the calculations when more than one marker at a locus.
FIG. 5.
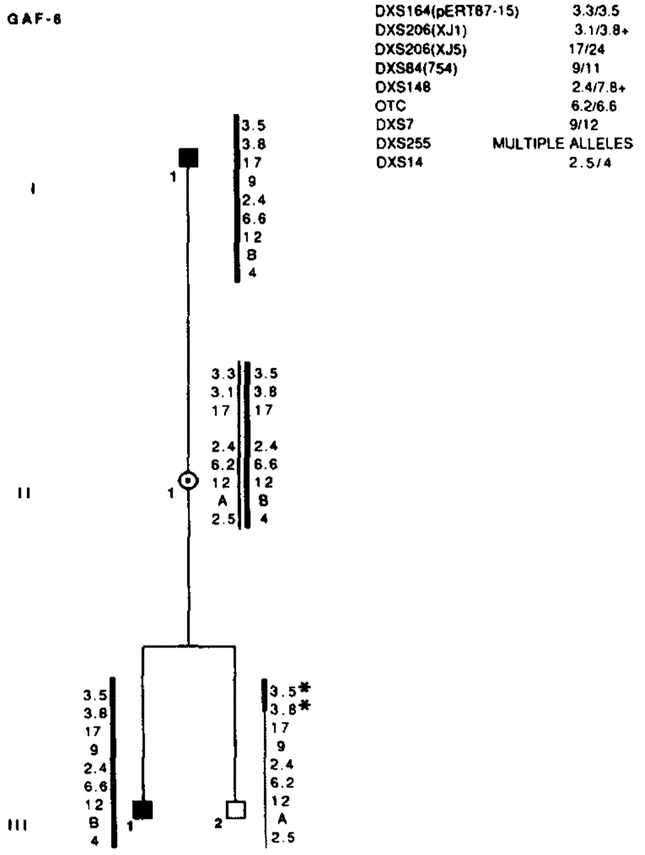
Results of RFLP analysis in GAF-6. Marker loci with their alleles are listed in the upper right table. Asterisks (*) denote recombinations. Data from the most informative marker (+) were used in the calculations when more than one marker at a locus.
FIG. 6.
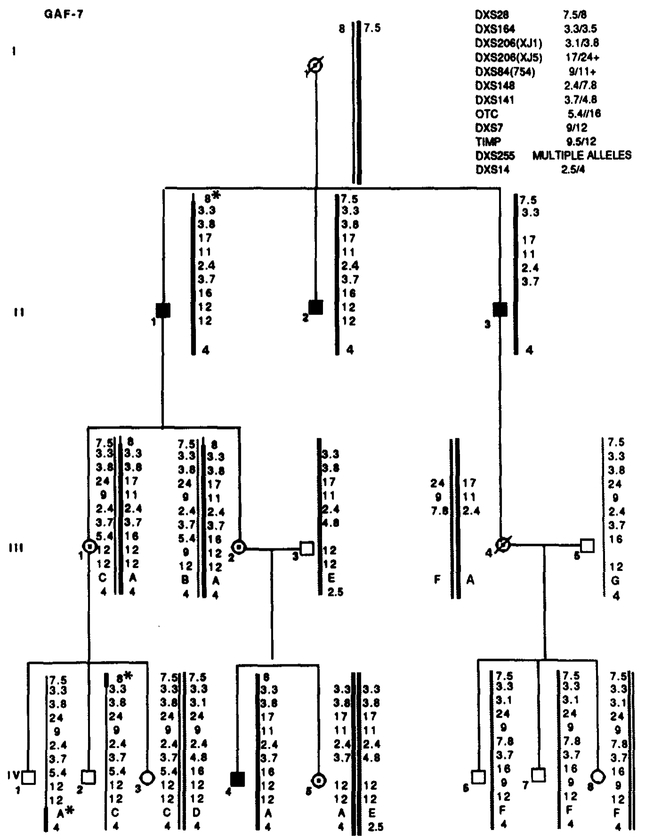
Segregation of RFLP alleles in GAF-7. Marker loci with their alleles are listed in the upper right table. Asterisks (*) denote recombinations. Data from the most informative marker (+) were used in the calculations when more than one marker at a locus.
FIG. 7.
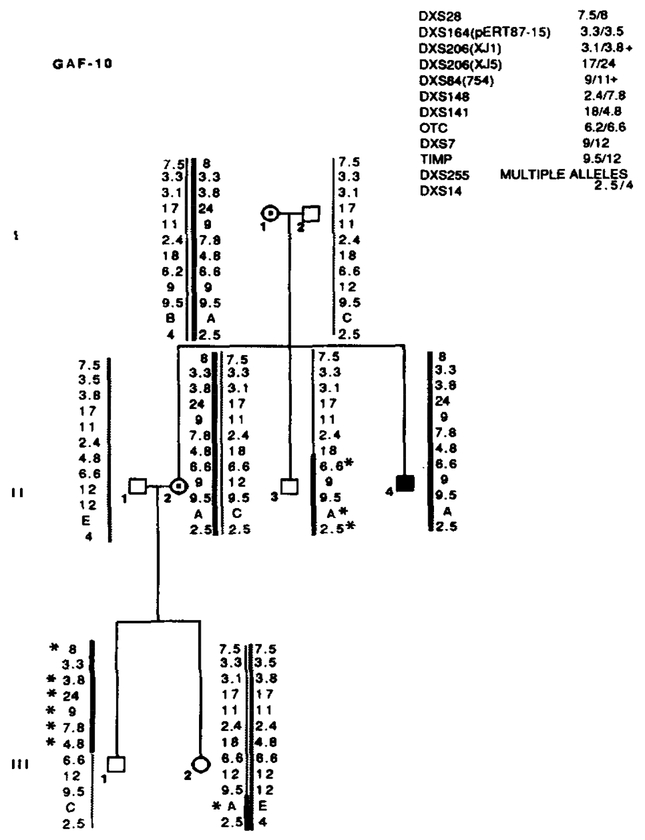
Segregation of RFLP alleles in GAF-10. Marker loci with their alleles are listed in the upper right table. Asterisks (*) denote recombinations. Individual II-3 is recombined for OTC and more proximal markers. His nephew, III-l, is recombined for all markers distal to OTC. Neither male is affected, suggesting that the X L R P mutation is between OTC and DXS141. Data from the most informative marker (+) were used in the calculations when more than one marker at a locus.
FIG. 8.
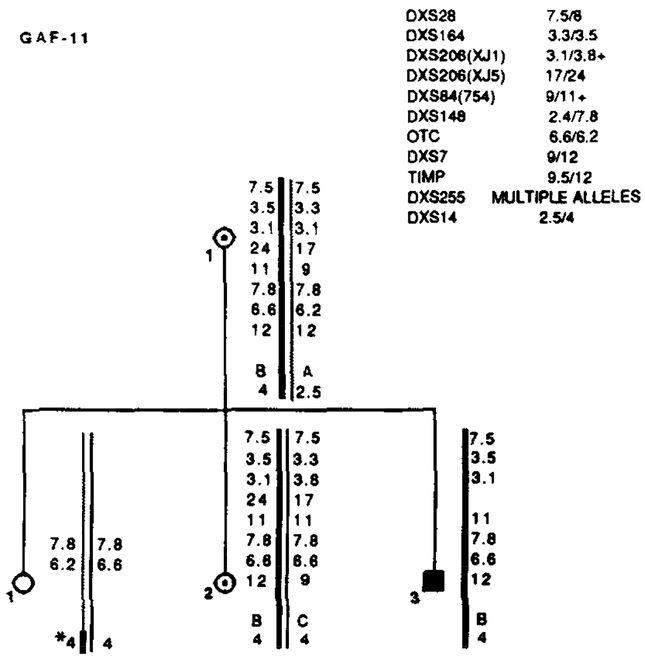
Segregation of RFLP alleles in GAF-11. Marker loci with their alleles are listed in the upper right table. Asterisks (*) denote recombinations. Data from the most informative marker (+) were used in the calculations when more than one marker at a locus.
FIG. 9.
Results of RFLP analysis in GAF-12. Marker loci with their alleles are listed in the upper right table. Asterisks (*) denote recombinations. Data from the most informative marker (+) were used in the calculations when more than one marker at a locus.
FIG. 10.
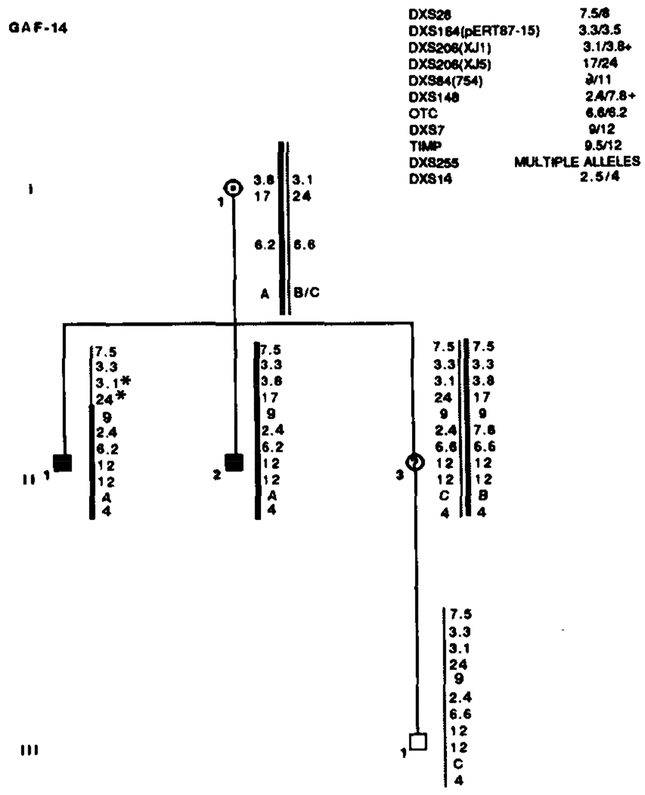
Results of RFLP analysis in GAF-14. Marker loci with their alleles are listed in the upper right table. Asterisks (*) denote recombinations. Data from the most informative marker (+) were used in the calculations when more than one marker at a locus.
FIG. 11.
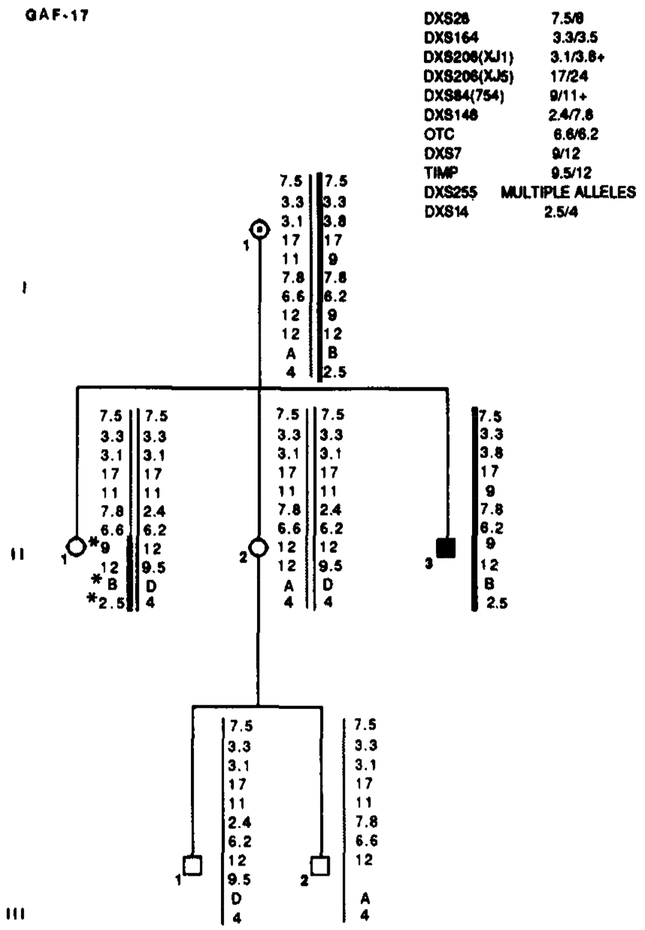
Segregation of RFLP alleles in GAF-17. Marker loci with their alleles are listed in the upper right table. Asterisks (*) denote recombinations. Data from the most informative marker (+) were used in the calculations when more than one marker at a locus.
FIG. 12.
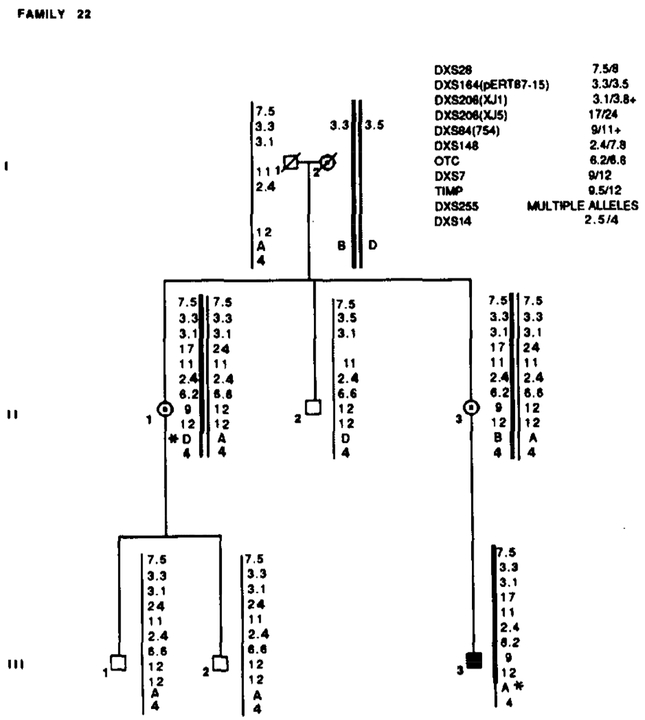
Family 22 segregated with X L R P and RFLP alleles. Marker loci with their alleles are listed in the upper right table. Asterisks (*) denote recombinations. Data from the most informative marker (+) were used in the calculations when more than one marker at a locus.
FIG. 13.
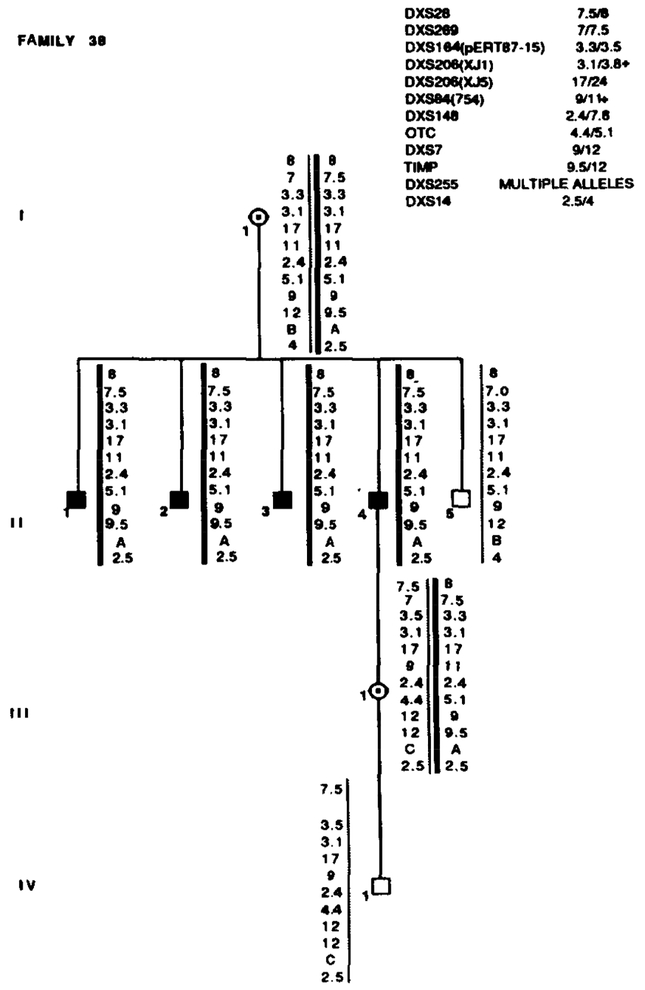
Family 38 segregated with X L R P and RFLP alleles. Marker loci with their alleles are listed in the upper right table. Data from the most informative marker (+) were used in the calculations when more than one marker at a locus.
FIG. 14.
Segregation of RFLP alleles in Family 39. Marker loci with their alleles are listed in the upper right table. Asterisks (*) denote recombinations. Data from the most informative marker (+) were used in the calculations when more than one marker at a locus.
APPENDIX II
Since in some families only certain members of each family were available for blood testing, the X-linked inhertance pattern in each pedigree may not be clearly demonstrated in Appendix I. The full pedigrees of each family are shown below.
REFERENCES
- 1.Bhattacharya SS, Wright AF, Clayton JF, Price WH, Phillips CI, Mckeown CME, Jay M, Bird AC, Pearson PL, Southern EM, AND Evans HJ (1984). Close genetic linkage between X-linked retinitis pigmentosa and a recombinant DNA probe L1.28. Nature (London) 309:253–255. [DOI] [PubMed] [Google Scholar]
- 2.Chen JD, Halliday F, Keith G, Sheffield L, Dickinson P, Gray R, Constable I, AND Denton M (1989). Linkage heterogeneity between X-linked retinitis pigmentosa and a map of 10 RFLP loci. Amer. J. Hum. Genet 45: 401411. [PMC free article] [PubMed] [Google Scholar]
- 3.Denton MJ, Chen JD, Serravalle S, Colley P, Halliday FB, AND Donald J (1988). Analysis of linkage relationships of X-linked retinitis pigmentosa with the following Xp loci: L1.28, OTC, XJ1.1, pERT87, and C7. Hum. Genet 78:60–64. [DOI] [PubMed] [Google Scholar]
- 4.de Saint-Basile G, Bohlek MC, Fischer A, Cartron J, Dufier JL, Griselli C, AND Orkin SH (1988). X p21 D N A microdeletion in a patient with chronic granulomatous disease, retinitis pigmentosa, and the McLeod phenotype. Hum. Genet 80: 85–89. [DOI] [PubMed] [Google Scholar]
- 5.Drayna D, AND White R (1985). The genetic linkage map of the X chromosome. Science 230: 753–758. [DOI] [PubMed] [Google Scholar]
- 6.Fishman GA, Weinberg AW, AND Mcmahon TT (1986). X-linked recessive retinitis pigmentosa: Clinical characteristics of carriers. Arch. Ophthalmol 104: 1329–1335. [DOI] [PubMed] [Google Scholar]
- 7.Francke U, Ochs HD, De Martinville B, Giacalone J, Lindegren V, Disteche C, Pagon R, van Ommen GJB, Pearson RL, and Wedgewood RJ (1985). Minor X p21 chromosome deletion in a male associated with expression of Duchenne muscular dystrophy and McLeod syndrome. Amer. J. Hum. Genet 37: 250–267. [PMC free article] [PubMed] [Google Scholar]
- 8.Friedrich U, Warburg M, Wieacker P, Wienker TF, Gal A, AND Ropers H-H (1985). X-linked retinitis pigmentosa: Linkage with the centromere and a cloned DNA sequence from the proximal short arm of the X chromosome. Hum. Genet 71: 93–99. [DOI] [PubMed] [Google Scholar]
- 9.Glnjaar HB, den Dunnen JTH, van Passen HMB Bakker E, Bertelson C, Boyd Y, Pearson PL, AND van Ommen GJB (1987). Cosmid cloning, FIGE mapping, chromosome walking and RFLP study of a distal intragenic DMD-deletion endpoint, expanding the DM D gene to 1.8 million bp (Abstract). Cytogenet. Cell Genet 46: 620. [Google Scholar]
- 10.Goodfellow PN, Davies KE, AND Ropers H-H (1985). Report of the committee on genetic constitution of the X and Y chromosomes (8th International Workshop on Human Gene Mapping). Cytogenet. Cell Genet 40: 296–352. [DOI] [PubMed] [Google Scholar]
- 11.Lathrop GM, AND Lalouel J-M (1988). Efficient com putations in multipoint linkage analysis. Amer. J. Hum. Genet 42: 498–505. [PMC free article] [PubMed] [Google Scholar]
- 12.Mahtani ME, AND Willard HF (1988). A primary genetic map of the pericentric region of the human X chromosome. Genomics 2: 294–301. [DOI] [PubMed] [Google Scholar]
- 13.Mukai S, Dryja TP, Bruns GAP, Aldridge JF, AND Berson EL (1985). Linkage between the X-linked retinitis pigmentosa locus and the L1.28 locus. Amer. J. Ophthalmol 100: 225–229. [DOI] [PubMed] [Google Scholar]
- 14.Musarella MA, Burghes A, Anson-CARTWRIGHT L, Mahtani MM, Argonza R, Tsui L-C, AND Worton R (1988). Localization of the gene for X-linked recessive type of retinitis pigmentosa (XLRP) to Xp21 by linkage analysis. Amer. J. Hum. Genet 43: 484–494. [PMC free article] [PubMed] [Google Scholar]
- 15.Musarella MA, Anson-Cartwright L, Burghes A, Worton R, Lesko JG, AND Nussbaum RL (1989). Linkage analysis of a large Latin-American family with X -linked retinitis pigmentosa and metallic sheen in the heterozygote carrier. Genomics 4: 601–605. [DOI] [PubMed] [Google Scholar]
- 16.Nussbaum RL, Lewis RA, Lesko JG, AND Ferrel R (1985). Mapping ophthalmological disease. II. Linkage of relationship of X-linked retinitis pigmentosa to X chromosome short arm markers. Hum. Genet 70: 45–50. [DOI] [PubMed] [Google Scholar]
- 17.Ott J (1985). “Analysis of Human Genetic Linkage,” Johns Hopkins Univ. Press, Baltimore, MD. [Google Scholar]
- 18.Ott J, Bhattacharya S, Chen JD, Denton MJ, Donald J Dubay C, Farrar G, Felix JS, Fishman GA, Frey D, Gal A, Humphries P, Jay B, Jay M, Litt M, Musarella MA, Neugebauer M, Worton RG, Nussbaum RL, Terwilliger JD, Weleber RG, Wirth B, Wong F, AND Wright AF (1990). Localizing multiple X - linked retinitis pigmentosa loci using extended multi-locush omogeneity tests. Proc. Natl. Acad. Sci. USA 87: 701–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wapenaar M, Klevits CT, Blonden LAJ, Baker E, den Dunnen JT, van Ommen GJB, AND Pearson PL (1987). Hybrid cell-mediated cloning of a new intragenic sequence from the 3’ region of the DMD gene (Abstract). Cytogenet. Cell Genet 4 6: 711. [Google Scholar]
- 20.Wirth B, Denton MJ, Chen J, Neugebauer M, Halliday FB „ van Schooneveld M, Donald F, Bleeker-Wagemakers EM, Pearson PL, AND Gal A (1988). Two different genes for X-linked retinitis pigmentosa. Genomics 2: 263–266. [DOI] [PubMed] [Google Scholar]
- 21.Wright AF, Shom S, Bhattacharya JF, Clayton M, Dempster P, Tippett C, Mckeown ME, Jay M, AND Bird AC (1987). Linkage relationships between X-linked retinitis pigmentosa and nine short-arm markers: Exclusion of the disease locus from Xp21 and localization to between DXS7 and DXS14. Amer. J. Hum. Genet 41: 635–644. [PMC free article] [PubMed] [Google Scholar]