Abstract
While a variety of human papillomavirus (HPV) tests and surrogate markers are available, currently there is no consensus on the best detection method(s) that should be used to identify HPV‐related oropharyngeal squamous cell carcinomas and serve as a standard test (or tests) for routine diagnostic use. As we begin to consider using the results of HPV testing for clinical purposes beyond simple prognostication, such as making decisions on treatment dose or duration or for targeted therapies that may be highly dependent on viral‐mediated pathways, we need to be more rigorous in assessing and ensuring the performance of the test (or tests) used. Here we provide an overview of the platforms and technologies, including the strengths and limitations of each test, and discuss what steps are needed to generate confidence in their performance for use in clinical practice.
Keywords: oropharyngeal cancer, oropharyngeal squamous cell carcinoma, OPSCC, head and neck cancer, human papillomavirus, HPV, p16, molecular diagnostics, biomarkers
Introduction
In addition to carcinomas of the uterine cervix, anus, vagina, vulva, and penis, squamous cell carcinoma of the oropharynx (OPSCC) is another tumor type that is recognized to be associated with human papillomavirus (HPV) infection 1, 2, 3. The oropharynx, which is the midportion of the pharynx, includes the soft palate, the lateral and posterior walls of the pharynx, the palatine tonsils, and the base of the tongue (see Figure 1). Approximately 60–70% of newly diagnosed OPSCCs are associated with HPV in the United States and some parts of Europe 4, 5, 6. It has been proposed that this preferential targeting of the oropharynx may reflect the biological interaction between HPV and the lymphoepithelium (i.e. reticulated epithelium) lining the tonsillar crypts, where strong expression of PD‐L1 results in a diminished cytotoxic T‐cell response to viral antigens and potentially creates an ‘immune‐privileged’ site for viral infection and persistence 7, 8.
Figure 1.
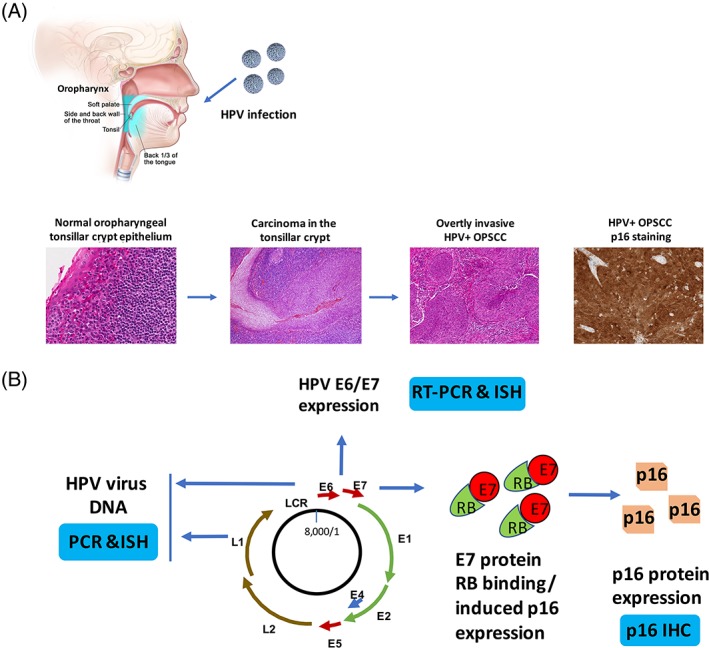
HPV‐associated carcinogenesis of head and neck cancer and molecular diagnostic tests. (A) The process of HPV‐related head and neck cancer development. HPV is contracted and introduced to the oral cavity and oropharynx. The susceptibility of the reticulated tonsillar crypt epithelium leads to the development of persistent viral infection and carcinogenesis, with neoplastic transformation of squamous cells in the epithelium that then expand the surface and eventually invade into the surrounding stroma and lymphatics, metastasizing to LNs early in the disease course. The tumor cells are strongly and diffusely positive for p16 by IHC with both nuclear and cytoplasmic staining. The top figure depicts the anatomy of oropharynx, including soft palate, lateral and posterior walls of the oropharynx (side and back wall of the throat), palatine tonsil (tonsil), and base of tongue (back 1/3 of the tongue). The histological images represent normal oropharyngeal tonsillar crypt epithelium (×300, left), carcinoma in the tonsillar crypt (×200, second left), overtly invasive HPV+ OPSCC (×100, second right), and HPV + OPSCC p16 IHC staining (×200, right). (B) Illustration of molecular diagnostic tests based on the HPV genome, E6/E7 gene expression and overexpression of p16 protein. The HPV viral genome is presented that the viral DNA can be detected by PCR or ISH (left). When HPV virus in its transcriptionally active form, the E6 or E7 gene can be detected by RT‐PCR or RNA ISH (top). HPV viral gene expression leads to abundant E7 protein overexpression with binding to Rb and mediating to its degradation. This allows for massive overexpression of p16 protein which is then detectable by IHC (right bottom). E: early genes; L: late genes; LCR: Long control region.
It is also well acknowledged that patients with HPV‐positive OPSCCs have better treatment responses and prognosis than patients with HPV‐negative tumors 9, 10, 11, 12, 13. Ragin et al performed a meta‐analysis of retrospective studies and reported that patients with HPV‐positive OPSCC had an estimated 50% reduction in risk of death when compared to patients with HPV‐negative tumors 13. A seeming exception is a variant form of oropharyngeal carcinoma with a high grade neuroendocrine phenotype (small cell carcinoma) which is indistinguishable from small cell carcinoma of the lung. These tumors seem to have aggressive clinical behavior regardless of HPV status 14, 15.
The National Comprehensive Cancer Network (NCCN) Clinical Practice Guidelines in Oncology for Head and Neck Cancers (NCCN Guidelines), Version 2.2017, state that the results of HPV testing should not change management decisions except in the context of a clinical trial. The NCCN Guidelines also state that, although HPV testing is not to be used to guide treatment, it is valuable in predicting oropharyngeal cancer prognosis, and tumor HPV testing is recommended as part of workup 16. The NCCN Guidelines note that p16 protein expression is highly correlated with HPV status and recommend assessment of p16 expression by immunohistochemistry (IHC) as another test option besides detection of HPV DNA in tumor cell nuclei by in situ hybridization (ISH). Prognosis is so dependent on HPV status that a College of American Pathologists (CAP) expert panel also recently recommended routine HPV testing (via p16 IHC) for all patients with newly diagnosed OPSCC 17. The American Joint Committee on Cancer (AJCC) and the Union for International Cancer Control (UICC) staging systems, which are based on equal grouping prognostication, were released in 2017 and now stage patients with oropharyngeal carcinoma differently by HPV status 18, 19.
HPV testing is also critical in the workup of metastatic squamous cell carcinoma to cervical lymph nodes (LNs) of unknown primary. A positive HPV test result strongly suggests that an occult primary tumor is present in the palatine tonsil or base of tongue 20. Selected HPV testing in these situations is recommended by the CAP guidelines, and the AJCC guidelines actually recommend staging such patients as T0 OPSCC. Treatment‐wise, however, the NCCN Guidelines note that whether or not HPV positive status should be used to define radiation fields for occult primary tumors is still under investigation.
HPV detection technologies
Currently, the HPV tests that have been approved by the United States Food and Drug Administration (FDA) are those intended for use in cervical carcinoma. There are no HPV diagnostic tests with regulatory approval for oropharyngeal cancer. Rather, the HPV testing options available for use in the clinical setting for head and neck cancer are laboratory developed tests (LDTs), which can involve various techniques and platforms for the analysis of HPV DNA, HPV messenger RNA (mRNA), or the p16 protein. HPV E6 and E7 mRNA detection by reverse transcriptase polymerase chain reaction (RT‐PCR) is highly correlated with improved patient survival in OPSCC and is considered by many to be the ‘gold standard’ test for classifying an OPSCC as being positive for transcriptionally active high‐risk HPV 21, 22. This involves reverse transcribing the mRNA into complementary DNA (cDNA) and then subsequently amplifying the target DNA sequences of interest using PCR. The other tests are judged for sensitivity and specificity in the literature mostly in comparison to RT‐PCR mRNA detection from fresh frozen tissue. However, this requires technical expertise that is not routinely available in pathology laboratories and its interpretation is somewhat subjective based on amplification curve analysis. Use of fresh frozen tissues would be optimal. Recent advances have enabled the consistent and accurate measurement of E6/E7 mRNA in formalin‐fixed paraffin‐embedded (FFPE) tissue blocks going back 10–20 years or more, but this is not an established technique and is still technically challenging to perform reproducibly outside of the research laboratory setting 21.
Another test that is widely used is the detection of HPV DNA using PCR. The most commonly used primer sets used to amplify specific HPV DNA sequences are PGMY09/11, GP5+/GP6+, and SPF10 LiPA, which all target consensus sequences of varying lengths (different primer sets target sequences of different lengths) within the HPV L1 gene 22. These primer sets are designed to bind to a conserved region in multiple HPV types so that a large range of HPV types can be simultaneously identified. While PCR‐based methods are known for high sensitivity in the detection of HPV, different PCR assays will vary in their performance based on the choice of primer sets, PCR protocols, and the type of tissue analyzed (e.g. fresh frozen versus FFPE) 22. For example, because the fixation process often leads to DNA fragmentation, with strands that are shorter than 200bp, it is preferable to target short DNA sequences in FFPE samples to increase sensitivity 22. HPV genotyping may then be performed separately, using restriction fragment length polymorphism (RFLP), linear probe assays, direct sequencing, or genotype‐specific primers 23, in order identify specific oncogenic HPV types for risk stratification.
One limitation of PCR assays is the possibility of obtaining false positive test results due to contamination of negative specimens from previously amplified specimens in the laboratory or from surrounding nonneoplastic tissue that is HPV infected 22. Another is that there is no tissue context for the result (i.e. positivity cannot be identified directly in tumor cells in the tissue context). Perhaps the greatest limitation, though, is that PCR assays cannot distinguish ‘clinically significant’ HPV infections (i.e. HPV infections that results in transcriptional activity in tumor cells with expression of E6 and E7 genes) from those which are not 21. Transcriptional activity is the key differentiator between clinically significant and insignificant HPV infection in tumors. This is the most established in OPSCC, where there are abundant data showing that only the tumors that harbor transcriptionally active high‐risk HPV have significantly better survival 24.
Still another test is HPV DNA detection by ISH. The ISH technique is based on labeled probes that hybridize to target HPV DNA sequences in the nuclei of tumor cells, and the probes can be either HPV‐type specific or a cocktail to detect many different high‐risk HPV types concurrently. The signals are visualized directly in the tumor, which provides tumor cell specific, direct histologic context to the result. This assay is known for its high specificity, but a weakness is limited sensitivity compared to other detection methods such as PCR, RT‐PCR, and p16 immunostaining. False negative results are particularly likely when the viral load is low 25. In addition, signal interpretation can be subjective due to lack of clean, clear staining signals. Studies have shown that up to 10% of cases have interobserver discrepancies 26. Most cases are clearly positive or negative, but pathologists must evaluate patchy, focal signals and decide if they represent true results or false positives due to nonspecific signal.
A relatively newer technique is RNA ISH, which is highly sensitive and highly specific because it can identify the presence of transcriptionally active HPV. One such test that has been used on OPSCC is the RNAscope HPV test that detects E6/E7 mRNA from 18 high‐risk HPVs (Advanced Cell Diagnostics, Hayward, CA, USA). Ukpo et al have shown that RNA ISH is actually more sensitive than DNA ISH 27. Many other studies have also shown its excellent performance in comparison to RT‐PCR and in patient survival stratification. However, the research community's experience with the RNA ISH method is still somewhat limited relative to DNA‐based techniques and p16 IHC. Furthermore, this test has only recently become available as a test on clinical platforms so most laboratories around the United States and the rest of the world do not yet perform it.
Liquid‐phase assays for cytopathology specimens are another type of assay that may be an option, particularly for evaluation of cervical LN metastases. These are the most commonly used tests for HPV screening in cervical cancer, with several commercially available HPV test kits approved by the FDA for the detection of high‐risk HPV in cervical cytology: Hybrid Capture II (HC‐2) (Qiagen, Gaithersburg, MD, USA); Cervista HPV HR assay and Cervista HPV16/18 assay (Hologic Inc., Bedford, MA, USA); Roche Cobas HPV test (Roche Molecular Systems, Pleasanton, CA, USA); and APTIMA HPV assay (Gen‐Probe Inc., San Diego, CA, USA). The technical aspects of these assays are summarized in Table 1. These tests can also have technical and interpretation issues, however. For example, the primary issue with HC‐2 analysis is the lack of an internal control to verify the presence of adequate tumor DNA in the sample, which may lead to false negative results 28. Furthermore, the Roche Cobas test had lower specificity when compared to DNA ISH and p16 IHC 29.
Table 1.
Commercially available assays and reagents for HPV detection
| Assay types* | Method/commercial kits | Technical details/interpretation/evaluation† | Web information |
|---|---|---|---|
| p16 IHC | CINtec® Histology‡ , § (Roche mtm Lab, Heidelberg, Germany) | Qualitative IHC test using mouse monoclonal anti‐p16 antibody clone E6H4. Used in light microscopic assessment of p16INK4a protein in FFPE tissues | https://usdiagnostics.roche.com/en/document/CINtec-Histology-Interpretation-Guide.pdf |
| CINtec® Histology § (Ventana Medical Systems, Tucson, AZ, USA) | http://www.ventana.com/product/1743?type=2202 | ||
| mAb clone JC8¶ (Santa Cruz Biotechnology Inc., Dallas, TX, USA); mAb clone G175‐405 (BD Biosciences, San Jose, CA, USA) | Other anti‐p16 antibodies for IHC (other than E6H4 clone) | http://www.nordiqc.org/downloads/assessments/26_59.pdf | |
| In situ hybridization for HPV DNA | GenPoint™ HPV DNA Probes Cocktail‡ (Y1443, Dako, CA, USA) | Probes contain HPV genomic clones in the form of double‐stranded fragments of 500 bp or less (biotinylated and unlabeled) and multiple biotinylated oligonucleotides from 25 to 40 bases in length. Staining procedure uses Dako GenPoint™ Detection System (code K0620), a nonfluorescent chromogen, and observed under light microscope. React with HPV types 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 68 on FFPE tissues and/or cells by ISH. Indicate for HPV genomic integration |
https://www.agilent.com/cs/library/packageinsert%20/public/128074001.PDF |
| GenPoint™ HPV type 16/18 biotinylated DNA probes (Y1412, Dako, CA, USA) | Use labeled DNA probes that specifically bind to a consensual or type‐specific HR‐HPV DNA sequences and detect amplified signals. React with HPV types 16, 18, on FFPE tissues and/or cells by ISH | http://www.finels.com/product/up_files/Y1412.pdf | |
| Bond™ Ready‐to‐Use ISH HPV Probe (subtypes 16, 18, 31, 33, 51)‡ (Leica Microsystems, IL, USA) | Qualitative identification of the HPV DNA in FFPE tissue by ISH using the automated BOND system (includes Leica BOND‐MAX system and Leica BOND‐III system). This probe binds to the five high‐risk HPV subtypes, 16, 18, 31, 33 and 51 | https://www.leicabiosystems.com/fileadmin/img_uploads/novocastra_reagents/Novocastra_datasheets/pb0829.pdf | |
| INFORM HPV III Family 16 Probe (B)§ (Ventana Medical Systems, AZ, USA) | Contains a cocktail of HPV genomic probes in a formamide‐based diluent. The intended targets are the common high‐risk HPV genotypes found to be associated with neoplasia. The probe cocktail has demonstrated affinity to the following genotypes: 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, and 66 | http://www.uclad.com/newsletters/HPV_ISH_Tissue-Probe-Interpretation_Guide.pdf | |
| In situ hybridization for HPV RNA | RNAscope® HPV Test (Advanced Cell Diagnostics, CA, USA) | RNAscope® HPV Biomarker Detection Reagents and its proprietary ‘double Z’ oligonucleotide probes specific for each subtype HPV E6/E7 mRNA enable high specificity detection of viral transcripts in routine FFPE tumor biopsies. Labeled probes contain chromogenic enzyme or fluorophore signal generating one punctate dot per RNA target. Hybridization of only three Z probe pairs is sufficient to obtain a detectable chromogenic signal by a brightfield microscope. Detect HPV 16, 18, 31, 33, 35, 52, and 58 | https://acdbio.com/science/applications/disease-areas/hpv-related-cancer |
| HPV 16 mRNA Probe (Ventana Medical System, Tucson, AZ, USA) | DNP hapten labeled probe designed to bind to the HPV 16 E6/E7 mRNA transcript. The probe has affinities to the HPV 16 genotype with no known cross reactivity to other common HPV genotypes | http://www.ventana.com/product/1830?type=2340 | |
| PCR and liquid‐phase assays | Digene® Hybrid Capture 2 (HC2) High‐Risk HPV DNA Test§ (Qiagen, Gaithersburg, MD, USA) | In vitro nucleic acid hybridization assay with signal amplification using antibodies and chemiluminescence for qualitative detection of 13 high‐risk types of HPV DNA. Probes are based on GP5+/GP6 primer set and detect HPV L1 open reading frame. Detects high‐risk HPV types 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 68. Cannot be used to determine specific HPV type present | https://www.qiagen.com/us/shop/detection-solutions/human-pathogens/digene-hc2-high-risk-%20hpv-dna-test/#orderinginformation |
| Digene® HPV Genotyping PS Test‡ | For use with Digene HC2 High‐Risk HPV DNA test for qualitative genotyping of HPV 16, 18, and 45 | https://www.qiagen.com/gb/shop/detection-solutions/human-pathogens/digene-hpv-%20genotyping-ps-test/#orderinginformation | |
| Digene® HC2 HPV DNA Test‡ , § | In vitro microplate assay based on an RNA probe cocktail and signal‐amplified nucleic acid hybridization that uses chemiluminescence for the qualitative detection of 13 high‐risk (16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 68) and 5 low‐risk types of HPV DNA (6, 11, 42, 43, 44). Can differentiate between two HPV DNA groups, low‐risk and high‐risk types | https://www.qiagen.com/us/shop/detection-solutions/human-pathogens/digene-hc2-hpv-dna-%20test/#productdetails | |
| LINEAR ARRAY® HPV Genotyping Test‡ (Roche Diagnostics, Indianapolis, IN, USA) | Based on PGMY09/11 primers set, detect HPV L1 open reading frame. A qualitative test that detects 37 high‐and low‐risk HPV genotypes | https://molecular.roche.com/assays/linear-array-hpv-genotyping-test | |
| INNO‐LiPA HPV Genotyping extra II(20T)‡ (Fujirebio Europe, Ghent, Belgium) | SPF10 Plus primers set provide high test sensitivity due to the precision of the short 65‐base pair PCR product. Permits simultaneous detection of multiple genotypes in a single sample. One probe line for one genotype, ready‐to‐use master mix for RT‐PCR application, and automation. Individually detect 32 HPV genotypes | http://search.cosmobio.co.jp/cosmo_search_p/search_gate2/docs/IGT_/81534.20160810.pdf | |
| Cobas® HPV Test‡ , § (Roche Molecular Diagnostics, Pleasanton, CA, USA) | Uses automated specimen preparation to extract DNA. Amplifies HPV DNA by PCR followed by nucleic acid hybridization for automated real‐time detection of 14 high‐risk HPV types in a single analysis. Specimens are limited to cells collected in PreservCyt® Solution, cobas® PCR Cell Collection Media and SurePath® Preservative Fluid. Test concurrently identifies 14 high‐risk HPV types (HPV types 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, and 68) and can also differentiate between HPV 16 and HPV 18 | https://diagnostics.roche.com/global/en/products/params/cobas-hpv.html | |
| The Abbott RealTime High Risk (HR)‡ HPV assay (Abbott Park, IL, USA) | Qualitative in vitro PCR assay that utilizes homogeneous target amplification and detection technology for detection of high‐risk HPV DNA in cells collected in liquid cytology media. Detect 14 high‐risk HPV types: 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, 68 and to partially genotype 16, 18 from other 12 high‐risk genotypes | https://www.molecular.abbott/int/en/products/%20infectious-disease/realtime-high-risk-hpv | |
| Cervista® HPV HR Test‡ , § (Hologic Inc., Bedford, MA, USA) | Uses Invader™ chemistry, a signal amplification method for detection of specific nucleic acid sequences. This method uses two types of isothermal reactions: a primary reaction that occurs on the targeted DNA sequence and a secondary reaction that produces a fluorescent signal. Qualitative detection of DNA from 14 high‐risk HPV types 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, and 68. Cannot determine the specific HPV type | https://www.hologic.com/sites/default/files/package-insert/15-3100_105_01.pdf | |
| Cervista™ HPV 16/18 Test‡ , § | Detects HPV DNA 16, 18 | https://www.hologic.com/sites/default/files/package-insert/15-3101_102_01.pdf | |
| Aptima® HPV assay‡ , § (Hologic Inc., Bedford, CA, USA) | Nucleic acid amplification involving 3 steps that take place in a single tube: target capture; target amplification of RNA by transcription‐mediated amplification (TMA); and detection of amplification products (amplicons) by hybridization protection assay (HPA). Assay incorporates an internal control to monitor nucleic acid capture, amplification, and detection, as well as operator or instrument error. Detects E6/E7 viral mRNA from 14 high‐risk HPV types: 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, and 68 | https://www.hologic.com/sites/default/files/2018-02/AW-12821_002_01.pdf | |
| Onclarity™ HPV assay‡ , § (BD Diagnostics Systems, Baltimore, MD, USA) | Amplified DNA test for the qualitative detection of high risk types of human HPV. Using BD Onclarity HPV Cervical Brush Collection Kit, BD SurePathT™ Preservative Fluid, andPreservCyt® Solution. Performed with the BD Viper™ LT System. Detects all high‐risk HPV types (16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, and 68), and provides the capability for genotyping of six high risk types (HPV 16, 18, 31, 45, 51, and 52) | http://www.puls-norge.no/media/285557/pakningsvedlegg-hpv.pdf |
Most assays are tested and validated in cervical cancer. The assays are listed from the most commonly use pathological tests to the more technical challenged or advanced methodologies.
Technical details only describe the main technical principles of the methods or commercial kits. Each methods or kits may have different protocol details and reagents.
CE marked for HPV detection for cervical cancer in Europe. CE marking is a certification mark that indicates conformity with health, safety, and environmental protection standards for products sold within the European Economic Area (EEA). CE Marking indicates a product's compliance with the applicable EU regulations and enables the commercialization of products in 32 European countries.
FDA approved for HPV detection for cervical cancer in United States.
Additional antibodies available for p16 IHC were tested and used in clinical pathology laboratory. The information are summerized in the weblink to NordiQC website.
While investigators have begun exploring these tests for primary head and neck squamous cell carcinoma (HNSCC) specimens and for nodal metastases 28, 29, 30, these tests are used sparingly in routine clinical practice for head and neck cancers and none of them are FDA approved for this purpose. Since a large fraction of HNSCC patients (and in particular OPSCC) are initially diagnosed based on cytology specimens from nodal metastases, these may be the only specimens available to establish a diagnosis. Thus, establishing an accurate and reliable method of HPV detection in cytologic samples is critical 31. Table 2 summarizes the many of the studies conducted in recent years comparing commercially available tests for their performance in HNSCC specimens 28, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44.
Table 2.
Studies comparing assays for HPV in head and neck squamous cell carcinoma specimens
| Assays* | Method/commercial kits | Samples | Comparison† | Conclusion/concordance | References |
|---|---|---|---|---|---|
| p16 IHC | E6H4 clone, CINtec® p16 Histology§ (Ventana, Tucson, AZ, USA) | 199 FFPE OPSCC | Other p16 antibodies, JC8, G175‐405 | E6H4 clone performs best, strongest staining intensity, greatest differential in outcomes, lowest interobserver variability, and lowest background, nonspecific staining. A 75% cutoff is very functional, 50% and any staining cutoffs may be more effective, particularly for the non‐E6H4 clones | Shelton et al [43] |
| G175‐405 clone (BD Biosciences, San Jose, CA, USA) | 81 archival OPSCC | Anti‐p16 E6H4 clone CINtec® p16 Histology (Ventana Medical Systems) | Developed criteria of >75% p16 positivity or 25–75% positivity with >75% confluence, can effectively risk stratify a cohort of 81 OPSCC cases into two prognostically relevant p16(+) or (−) groups | Barasch et al [33] | |
| RNA in situ hybridization | RNAscope® HPV Test HPV‐HR18 Probe (Advanced Cell Diagnostics, CA, USA) | 82 HNSCC specimens | p16 IHC‡ | RISH(+) in 100% p16(+)/DISH(+), and in 88% p16(+)/DISH(−) samples | Rooper et al [42] |
| RNAscope® HPV Test HPV‐HR18 Probe | 105 OPC specimens | p16 IHC‡ alone, p16 IHC‡ + PCR (INNO‐LiPA HPV Genotyping Extra II kit), p16 IHC‡ + ISH (Ventana HPV III Family 16, Probe B) | RNAscope HPV‐test and p16 combined tests are better than p16 alone. The RNAscope HPV‐test has the advantage of being a single test | Mirghani et al [39] | |
| RNAscope® HPV Test HPV‐HR18 Probe | 41 FFPE OPSCC samples | p16 IHC‡, DNA ISH (INFORM‐HPV III family 16 probe), HPV DNA PCR (INNO‐LiPA HPV genotyping assay version Extra), HPV E6 DNA amplification | Concordance between mRNA ISH versus other assays: sensitivity and specificity were 100 and 90% for p16; 90 and 100% for DNA ISH; 70 and 76% for SPF10 LiPA; 90 and 76% for E6 amplification | Morbini et al [40] | |
| RNAscope® HPV Test HPV‐HR18 Probe | 70 OSCC cases | p16 IHC‡, HR‐HPV DNA ISH (Inform HPV III family probe), HPV 16 E6/E7 mRNA real time RT‐PCR | RNAscope gives the most accurate approach to decipher the presence of integrated and transcriptionally active virus in FFPE samples of OSCC | Volpi et al [44] | |
| PCR and liquid‐phase assays | Digene® HC2 High‐Risk HPV DNA Test§ (Qiagen, MD, USA) | 22 HNSCC LN metastasis (Met), cytology needle rinse material | p16 IHC (BD Pharmingen mouse antihuman p16 antibody) | 55% were p16 positive, of which 58% tested positive for HPV by HC2. All cases negative for p16 were negative for HPV by HC2 | Hakima et al [35] |
| Digene® HC2 High‐Risk HPV DNA Test§ | 25 HNSCC fine needle aspiration (FNA) LN Met | p16 IHC‡, HPV DNA ISH (HPV III Family16 probe set) | Accuracy was improved to 100% when cytologic evaluation confirmed the presence of cancer cells in the test samples | Smith et al [28] | |
| INNO‐LIPA‡ HPV Genotyping Extra Assay (Fujirebio Europe, Ghent, Belgium) | 71 HNSCC patients, tumor biopsies FFPE, RNAlater | p16 IHC‡ | p16 IHC or HPV‐PCR used alone appear to be insufficient | Fonmarty et al [34] | |
| Cobas® 4800 HPV Test‡ , § (Roche Molecular Diagnostics, Pleasanton, CA, USA) | 42 FNA specimens from 37 patients | p16 IHC‡ and HPV 16 ISH INFORM HPV III Family 16 probe (B) | HR‐HPV detection and genotyping can be performed on LN FNAs with metastatic SCC using the Roche Cobas 4800 system | Baldassarri et al [32] | |
| Cobas® 4800 HPV Test‡ , § | 123 FFPE HNSCC (44 excisions, 63 biopsies, and 16 FNAs) | p16 IHC‡ | High concordance with p16 IHC (96%), positive agreement (91.5%), negative agreement (100%) | Huho et al [37] | |
| Cobas® 4800 HPV Test‡ , § | 62 FFPE HNSCC | p16 IHC‡ and DNA ISH (Inform HPV III Family 16 Probe) | Concordance between Cobas and ISH was >90%; Detection of HR‐HPV, Cobas' sensitivity: 100%; specificity: 91% | Kerr et al [38] | |
| Cobas® 4800 HPV Test‡ , § | 28 FFPE from 25 OPSCC patients | p16 IHC‡, Roche Linear Array® HPV genotyping test kit | Interassay concordance (96.2%), 100% concordance for HPV‐16/18 positive samples | Pettus et al [41] | |
| Aptima® HPV assay‡, § (Hologic Inc., Bedford, MA, USA) | 50 OPSCC surgical biopsy/resection and FNA of the nodal metastasis | p16 IHC‡ | 87.5% p16(+) are Aptima(+) 100% p16(−) are Apitima(−) | Han et al [36] |
Most assays are tested and validated in cervical cancer. The assays are listed from the most commonly use pathological tests to the more technical challenged or advanced methodologies. Using the terminology and designs described by authors.
The comparisons were made with p16 IHC using anti‐p16 antibody (E6H4 clone from CINtec® Histology, Roche mtm Lab, or from Ventana Medical Systems, Tucson, AZ, USA). The comparison with other assays is described specifically.
CE marked for use for detection of HPV in cervical cancer in Europe. CE marking is a certification mark that indicates conformity with health, safety, and environmental protection standards for products sold within the European Economic Area (EEA). CE Marking indicates a product's compliance with the applicable EU regulations and enables the commercialization of products in 32 European countries.
FDA approved for HPV detection for cervical cancer in USA.
The test that is most widely available in clinical laboratories and most widely used in clinical practice is p16 (INK4A) detection by IHC. This protein is a surrogate marker of transcriptionally active high‐risk HPV infection. It functions as a tumor suppressor by binding to and inhibiting cyclin dependent kinase 4 and 6 (CDK4/6) so that it cannot phosphorylate the retinoblastoma protein (pRB). Hypophosphorylated pRB remains bound to E2F. Phosphorylation of pRB releases it from binding to E2F transcription factor to allow cell cycle progression 22. Expression of p16 normally leads to the accumulation of hypophosphorylated pRB and thus to sequestration of E2F, with resulting G1 arrest 45. In HPV‐related carcinomas, viral E7 protein binds to pRB and leads to its degradation, which then allows E2F to bypass pRB‐dependent cell cycle arrest 21, 46. A secondary effect of pRB degradation is that E2F then stimulates the expression of p16, which accumulates in large amounts in the nuclei and cytoplasm of tumor cells (Figure 1).
p16 IHC is inexpensive, is performed on FFPE samples which are routinely generated in pathology laboratories as part of standard clinical surgical pathology practice, and has a sensitivity for transcriptionally active high‐risk HPV that is almost 100% 47. Different clones of p16 monoclonal antibodies are commercially available from different vendors. The most widely used is the E6H4 clone, originally linked with the CINtec p16 Histology kit (Roche, Heidelberg, Germany) which is specifically designed for evaluation of cervical dysplasia. This kit has CE‐marking (i.e. the product can be legally placed on the market in the European Union), but the FDA approval for marketing in the United States is limited to intended uses in cervical cancer.
p16 IHC has demonstrated good agreement with HPV E6/E7 mRNA expression detected by RT‐PCR 22 and RNA ISH. Extensive literature shows that p16 expression in OPSCC is associated with improved overall and disease specific survival independent of all other known prognostic factors with two to five times lower risk of adverse outcomes. A significant issue with p16 IHC is that, until recently, there has been no consensus on the definition of a positive p16 IHC result. It has been recognized for decades that there should be extensive overexpression of p16 for it to suitably be a surrogate of high‐risk HPV, but how extensive has been debatable. Publications have varied and different laboratories have used different threshold levels (i.e. percentage of tumor cells that were stained) 22. Cutoffs for the percentage of positive tumor cells needed have ranged from 1 to 5, 10, 50, 60, 70, or 75% of tumor cells 9, 20, 26, 27, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58. Since very few tumors have borderline p16 expression (10–70% range), this has not been a formidable issue. However, newer guidelines are helping to clarify this, such as the CAP Guidelines recommending 70% nuclear and cytoplasmic expression of at least moderate or strong intensity for a definitive positive result. Borderline nuclear and cytoplasmic p16 overexpression (50–70%) and diffuse, but weak, overexpression are uncommon but potentially significant problems. Since the small studies examining this issue have shown that most of these patients have transcriptionally active high‐risk HPV in their tumors, the CAP guidelines suggest that patients with these borderline p16 results can undergo HPV specific testing to arbitrate whether or not to be classified as HPV‐positive/HPV‐related 17.
Another issue is that while p16 IHC is highly sensitive, its specificity for transcriptionally active HPV, even in the indicated clinical settings of oropharyngeal primary tumor and/or upper jugular nodal metastasis of unknown primary, can range from 79 to 95% because HNSCCs that are not HPV‐related as well as benign squamous cells can overexpress p16 31, 47, 49. For example, almost one‐half of benign squamous cells from branchial cleft cysts of the lateral neck overexpress p16 59, although it is rarely strong and diffuse. It should be kept in mind that the positive predictive value of any test depends highly on the disease prevalence. In the United States, where HPV‐positivity amongst OPSCC patients is very high (70–90% in recent studies), the positive predictive value/correlation with HPV is higher. However, in areas where HPV‐positive rates are lower, such as certain European populations (as low as 20%) and other geographic regions outside of the United States, the positive predictive value is lower, and p16 alone may not be the best approach for classifying patients 60. Also, up to 26% of nonsquamous, basaloid carcinomas such as solid type adenoid cystic carcinoma, which are not associated with HPV, can be diffusely positive for p16 61. While some of these sinonasal carcinomas have been more recently shown to be the unique tumor ‘HPV‐related multiphenotypic carcinoma’, the rest across other anatomic sites are not HPV‐related 62, 63. It has also been reported that p16 IHC is positive (using the high expression cutoff) in as many as 20–30% of head and neck cutaneous and primary lung SCCs that are unrelated to HPV 64. These findings, although they do not obviate it, underscore the need for caution with a p16 IHC only approach. In primary OPSCC patients and in the tightly defined locations and scenarios for patients with neck nodal metastatic SCC of unknown or uninvestigated primary site, p16 IHC alone is appropriate as recommended by the CAP, AJCC, and the World Health Organization 17, 18, 19, 65.
Finally, investigators are in the early stages of development in testing new specimen types for HPV, such as saliva and plasma samples. For example, quantitative PCR‐based HPV‐16 DNA (E6 and E7 genes) in the saliva and plasma of OPSCC samples before and after treatment has been explored as a biomarker of recurrence 66.
HPV assays in clinical trials
Standard therapies for OPSCC can involve high doses of toxic radiation and chemotherapy, which may be unnecessary for HPV‐positive OPSCC, so clinicians are actively investigating whether reduced doses of radiation and chemotherapy (i.e. deintensification) may be appropriate for HPV‐positive oropharyngeal cancers. There are substantial data supporting this idea in other HPV‐related SCCs, such as cervical and anal SCC, as well as indirect data from head and neck cancer trials designed for other reasons. However, direct (and in particular prospective controlled clinical trial) data for OPSCC patients are limited. Recently, Chera et al published the results of their prospective clinical trial from which they concluded, ‘for patients with favorable‐risk human papillomavirus‐associated oropharyngeal squamous cell carcinoma, a substantially decreased intensity of therapy with 60 gray of intensity‐modulated radiotherapy and weekly low‐dose cisplatin produced better preservation of quality of life compared with standard therapies while maintaining excellent 3‐year tumor control and survival’ 67. It should be noted that this was a small, single‐arm phase II trial (1) without a control group receiving standard radiation and chemotherapy doses and (2) without a biomarker negative group. In these patients receiving reduced intensity chemoradiotherapy, the pathologic complete response rate was 86% and 3‐year overall survival was 95%. The 44 patients enrolled in this trial were all p16‐positive (>70% of carcinoma cells showing nuclear reactivity) but 36% were negative for HPV DNA (using fluorescence ISH), and it would have been interesting to see whether there was a difference in the outcome between the HPV+/p16+ and HPV−/p16+ groups. We should soon hear about the results from other ongoing clinical trials involving deintensification treatment protocols for HPV‐associated locally advanced oropharyngeal cancer.
The National Clinical Trials Network (NCTN)/cooperative group clinical trials focused on HPV‐positive OPSCC require documented p16 or HPV positivity as eligibility criteria. In the RTOG 1016 Phase III Trial of Radiotherapy Plus Cetuximab versus Chemoradiotherapy in HPV‐Associated Oropharynx Cancer (http://clinicaltrials.gov Identifier: NCT01302834), the overall goal was to identify a less toxic approach for HPV‐related OPSCC patients, in order to reduce the burden of acute toxicity and to facilitate quicker recovery of patients. For this trial, HPV‐positive tumor status was determined by p16 expression, and all patients were required to submit their tissues for mandatory p16 analysis by a central laboratory. The reason for this mandatory testing was that p16 testing had not been standardized at a national level, resulting in up to 30% discordance between local and central testing. In this setting, tumor p16 expression was evaluated by means of IHC with a mouse monoclonal antibody (MTM Laboratories, Westborough, MA, USA) visualized with use of an autostainer (Discovery XT, Ventana, Tucson, AZ, USA) and a secondary detection kit (iVIEW DAB Detection Kit, Ventana) by standard protocol. Positive p16 expression was defined for these clinical trials as a strong and diffuse nuclear and cytoplasmic staining in 70% or more of the tumor cells.
The ECOG‐ACRIN E1308 trial (A Phase II Trial of Induction Chemotherapy Followed by Cetuximab [Erbitux] with Low Dose Versus Standard Dose IMRT in Patients with HPV‐Associated Resectable Squamous Cell Carcinoma of the Oropharynx) required central testing of HPV positive status by HPV‐16 ISH and p16 IHC for determination of patient eligibility. If p16 IHC was negative, the HPV status was called negative. In their recent publication reporting on the trial's promising findings, which showed that radiotherapy deintensification may result in equivalent or similar response in selected patients compared to full‐dose radiotherapy 68, the investigators did not provide assay details other than citing the Jordan et al paper 49. In the Jordan et al paper, p16 IHC was performed using the monoclonal anti‐p16INK4a (MTM Laboratories, Heidelberg, Germany) and HPV16 DNA was detected in paraffin‐embedded tumor samples using the GenPoint catalyzed ISH signal amplification system for biotinylated probes (Dako, Carpinteria, CA, USA). In this paper, the optimal cutpoint was determined to be the H score cutoff point of 60 indicating ‘that a tumor with diffuse low‐intensity nuclear and cytoplasmic p16 staining in the majority of the tumor is a true positive’ and that ‘the optimal cutoff point for percentage staining was lower at ≥35%’ instead of the recommended cutoff point of ≥70% 49. Because p16 IHC can lead to false positive results, the authors recommended ‘For clinical trials evaluating ‘deintensification’ strategies for patients with HPV‐associated OSCC, combined p16 IHC and HPV16 ISH is necessary to provide the high specificity required to avoid possible undertreatment of patients without a diagnosis of HPV‐associated OSCC’ 49.
Need for harmonization of HPV tests
As described in previous sections, clinical HPV testing practices for OPSCC are variable, and there are many methods for HPV detection/patient prognostication, which vary in their platform, target, and performance with respect to sensitivity, specificity, and ability to stratify patient outcomes for positive versus negative results. While there are commonly used assays, there is no clear consensus among the pathologists and head and neck oncologists on which HPV test or tests should be required to reliably identify patients with transcriptionally active high‐risk HPV‐positive OPSCC. Although p16 IHC is easy to perform and every clinical histopathology lab should already have the capability to perform this test, we cannot take analytical validity for granted. As we move toward using HPV tests to help us make clinical treatment decisions regarding the type of therapy or intensity of treatment, we need to ensure the accuracy and reproducibility of the HPV test result through further studies.
In the newly published CAP guidelines for HPV testing in head and neck carcinomas in routine clinical practice, the recommendation for workup of patient specimens from oropharyngeal primary tumors is to perform p16 IHC testing, with lack of staining or <70% nuclear and cytoplasmic staining classified as HPV‐negative and ≥70% nuclear and cytoplasmic staining classified as HPV‐positive 17. In addition, it is specified that the intensity should be moderate or strong. However, the CAP Guideline does not recommend one antibody, platform, or set of test conditions over another. The guideline is specifically focused on creating consistent, data‐driven, HPV testing practices across practice settings. Admittedly, however, not having a standard test or tests (nor single platform for such tests or strong interassay concordance data) limits the ability to assure results for individual patients. And for p16 and HPV specific tests, needing a visual assessment of staining distribution and/or intensity brings risk for subjectivity into play.
In order to help support the needed harmonization studies, the National Cancer Institute (NCI) has active funding opportunity announcements (FOAs) for both cooperative grants and supplement grants. PAR‐17‐003 is the FOA for supplement grants to support projects that will clinically validate molecular assays and prepare for their use in clinical studies or trials for the treatment, prevention or control of cancer. Projects that improve standardization or harmonization of assay performance among laboratories are eligible for funding, and these types of project may involve continued efforts in assay optimization and ensuring concordance of assay results from different laboratories. Because these grants are meant to ‘supplement’ existing grants, the investigators need to be current recipients of an active R01 research project grant. PAR‐18‐317 and PAR‐18‐310 are FOAs for grants to support assay validation through the UH2 and UH3 cooperative agreement mechanisms. Efforts to harmonize clinical laboratory tests, including investigation into the performance and reproducibility of assays across multiple clinical laboratories, are appropriate for these funding opportunities as well.
In conclusion, now that there is real clinical utility of HPV testing in the management OPSCC patients, more test validation and method comparison studies are needed to decide what specific tests to perform and in what settings, and to assess the concordance the tests between different clinical laboratories and establish procedures for harmonization. There is a great opportunity for standardizing practices.
Acknowledgements
This study was supported by the Intramural Research Program of The National Institute on Deafness and Other Communication Disorders (ZC, ZIA‐DC‐000016), Extramural Cancer Diagnosis Program of National Cancer Institute (KYK), National Institute of Health, and NCI grant 1R21CA179327‐01A1 (JSL). We thank Dr. Carter Van Waes (Head and Neck Surgery Branch, NIDCD, NIH) and Dr. Cheng‐Ming Chiang (UT Southwestern Medical Center) for their helpful comments and support for the submission of the manuscript.
Author contributions statement
KK, ZC and JSL conceived and designed the manuscript. KK, ZC and JSL acquired and interpreted literature data. KK drafted the manuscript, and ZC and JSL reviewed and revised the manuscript. ZC designed and made the figure and tables, and KK and JSL reviewed and revised the figure and tables. JSL provided pathological images of OPSCC tissues. All authors read and approved the submitted and published versions of the manuscript.
No conflicts of interest were declared.
References
- 1. Gillison ML, Koch WM, Capone RB, et al Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J Natl Cancer Inst 2000; 92: 709–720. [DOI] [PubMed] [Google Scholar]
- 2. van Houten VM, Snijders PJ, van den Brekel MW, et al Biological evidence that human papillomaviruses are etiologically involved in a subgroup of head and neck squamous cell carcinomas. Int J Cancer 2001; 93: 232–235. [DOI] [PubMed] [Google Scholar]
- 3. Marur S, D'Souza G, Westra WH, et al HPV‐associated head and neck cancer: a virus‐related cancer epidemic. Lancet Oncol 2010; 11: 781–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chaturvedi AK, Engels EA, Pfeiffer RM, et al Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol 2011; 29: 4294–4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mehanna H, Beech T, Nicholson T, et al Prevalence of human papillomavirus in oropharyngeal and nonoropharyngeal head and neck cancer – systematic review and meta‐analysis of trends by time and region. Head Neck 2013; 35: 747–755. [DOI] [PubMed] [Google Scholar]
- 6. Nasman A, Attner P, Hammarstedt L, et al Incidence of human papillomavirus (HPV) positive tonsillar carcinoma in Stockholm, Sweden: an epidemic of viral‐induced carcinoma? Int J Cancer 2009; 125: 362–366. [DOI] [PubMed] [Google Scholar]
- 7. Lyford‐Pike S, Peng S, Young GD, et al Evidence for a role of the PD‐1:PD‐L1 pathway in immune resistance of HPV‐associated head and neck squamous cell carcinoma. Cancer Res 2013; 73: 1733–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Westra WH. The pathology of HPV‐related head and neck cancer: implications for the diagnostic pathologist. Semin Diagn Pathol 2015; 32: 42–53. [DOI] [PubMed] [Google Scholar]
- 9. Ang KK, Harris J, Wheeler R, et al Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med 2010; 363: 24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dayyani F, Etzel CJ, Liu M, et al Meta‐analysis of the impact of human papillomavirus (HPV) on cancer risk and overall survival in head and neck squamous cell carcinomas (HNSCC). Head Neck Oncol 2010; 2: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fakhry C, Zhang Q, Nguyen‐Tan PF, et al Human papillomavirus and overall survival after progression of oropharyngeal squamous cell carcinoma. J Clin Oncol 2014; 32: 3365–3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rischin D, Young RJ, Fisher R, et al Prognostic significance of p16INK4A and human papillomavirus in patients with oropharyngeal cancer treated on TROG 02.02 phase III trial. J Clin Oncol 2010; 28: 4142–4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ragin CC, Taioli E. Survival of squamous cell carcinoma of the head and neck in relation to human papillomavirus infection: review and meta‐analysis. Int J Cancer 2007; 121: 1813–1820. [DOI] [PubMed] [Google Scholar]
- 14. Bishop JA, Westra WH. Human papillomavirus‐related small cell carcinoma of the oropharynx. Am J Surg Pathol 2011; 35: 1679–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kraft S, Faquin WC, Krane JF. HPV‐associated neuroendocrine carcinoma of the oropharynx: a rare new entity with potentially aggressive clinical behavior. Am J Surg Pathol 2012; 36: 321–330. [DOI] [PubMed] [Google Scholar]
- 16. NCCN . NCCN Guidelines Version 2.2017 Head and Neck Cancers. National Comprehensive Cancer Network Inc.: Fort Washington, PA, 2017. [Accessed 12 August 2018]. Available from: https://www.nccn.org/professionals/physician_gls/pdf/head-and-neck.pdf. [Google Scholar]
- 17. Lewis JS Jr, Beadle B, Bishop JA, et al Human papillomavirus testing in head and neck carcinomas: guideline from the College of American Pathologists. Arch Pathol Lab Med 2018; 142: 559–597. [DOI] [PubMed] [Google Scholar]
- 18. Lydiatt WM, Patel SG, O'Sullivan B, et al Head and neck cancers‐major changes in the American joint committee on cancer eighth edition cancer staging manual. CA Cancer J Clin 2017; 67: 122–137. [DOI] [PubMed] [Google Scholar]
- 19. O'Sullivan B, Lydiatt WM, Haughey BH, et al HPV‐Mediated (p16+) oropharyngeal cancer In: AJCC Cancer Staging Manual (8th edn)Amin MB. (Ed). Springer Nature: Switzerland, 2016. [Google Scholar]
- 20. Begum S, Gillison ML, Nicol TL, et al Detection of human papillomavirus‐16 in fine‐needle aspirates to determine tumor origin in patients with metastatic squamous cell carcinoma of the head and neck. Clin Cancer Res 2007; 13: 1186–1191. [DOI] [PubMed] [Google Scholar]
- 21. Bishop JA, Lewis JS Jr, Rocco JW, et al HPV‐related squamous cell carcinoma of the head and neck: an update on testing in routine pathology practice. Semin Diagn Pathol 2015; 32: 344–351. [DOI] [PubMed] [Google Scholar]
- 22. Mirghani H, Amen F, Moreau F, et al Human papilloma virus testing in oropharyngeal squamous cell carcinoma: what the clinician should know. Oral Oncol 2014; 50: 1–9. [DOI] [PubMed] [Google Scholar]
- 23. Abreu AL, Souza RP, Gimenes F, et al A review of methods for detect human papillomavirus infection. Virol J 2012; 9: 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jung AC, Briolat J, Millon R, et al Biological and clinical relevance of transcriptionally active human papillomavirus (HPV) infection in oropharynx squamous cell carcinoma. Int J Cancer 2010; 126: 1882–1894. [DOI] [PubMed] [Google Scholar]
- 25. Stevens TM, Caughron SK, Dunn ST, et al Detection of high‐risk HPV in head and neck squamous cell carcinomas: comparison of chromogenic in situ hybridization and a reverse line blot method. Appl Immunohistochem Mol Morphol 2011; 19: 574–578. [DOI] [PubMed] [Google Scholar]
- 26. Thavaraj S, Stokes A, Guerra E, et al Evaluation of human papillomavirus testing for squamous cell carcinoma of the tonsil in clinical practice. J Clin Pathol 2011; 64: 308–312. [DOI] [PubMed] [Google Scholar]
- 27. Ukpo OC, Flanagan JJ, Ma XJ, et al High‐risk human papillomavirus E6/E7 mRNA detection by a novel in situ hybridization assay strongly correlates with p16 expression and patient outcomes in oropharyngeal squamous cell carcinoma. Am J Surg Pathol 2011; 35: 1343–1350. [DOI] [PubMed] [Google Scholar]
- 28. Smith DF, Maleki Z, Coughlan D, et al Human papillomavirus status of head and neck cancer as determined in cytologic specimens using the hybrid‐capture 2 assay. Oral Oncol 2014; 50: 600–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kerr DA, Pitman MB, Sweeney B, et al Performance of the Roche cobas 4800 high‐risk human papillomavirus test in cytologic preparations of squamous cell carcinoma of the head and neck. Cancer Cytopathol 2014; 122: 167–174. [DOI] [PubMed] [Google Scholar]
- 30. Guo M, Khanna A, Dhillon J, et al Cervista HPV assays for fine‐needle aspiration specimens are a valid option for human papillomavirus testing in patients with oropharyngeal carcinoma. Cancer Cytopathol 2014; 122: 96–103. [DOI] [PubMed] [Google Scholar]
- 31. Roy‐Chowdhuri S, Krishnamurthy S. The role of cytology in the era of HPV‐related head and neck carcinoma. Semin Diagn Pathol 2015; 32: 250–257. [DOI] [PubMed] [Google Scholar]
- 32. Baldassarri R, Aronberg R, Levi AW, et al Detection and genotype of high‐risk human papillomavirus in fine‐needle aspirates of patients with metastatic squamous cell carcinoma is helpful in determining tumor origin. Am J Clin Pathol 2015; 143: 694–700. [DOI] [PubMed] [Google Scholar]
- 33. Barasch S, Mohindra P, Hennrick K, et al Assessing p16 status of oropharyngeal squamous cell carcinoma by combined assessment of the number of cells stained and the confluence of p16 staining: a validation by clinical outcomes. Am J Surg Pathol 2016; 40: 1261–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fonmarty D, Cherriere S, Fleury H, et al Study of the concordance between p16 immunohistochemistry and HPV‐PCR genotyping for the viral diagnosis of oropharyngeal squamous cell carcinoma. Eur Ann Otorhinolaryngol Head Neck Dis 2015; 132: 135–139. [DOI] [PubMed] [Google Scholar]
- 35. Hakima L, Adler E, Prystowsky M, et al Hybrid capture 2 human papillomavirus testing of fine needle aspiration cytology of head and neck squamous cell carcinomas. Diagn Cytopathol 2015; 43: 683–687. [DOI] [PubMed] [Google Scholar]
- 36. Han M, Bernadt CT, Murray B, et al Aptima HR‐HPV testing from Diff‐Quick‐stained fine‐needle aspiration smears of oropharyngeal squamous cell carcinoma. J Am Soc Cytopathol 2016; 5: 221–226. [DOI] [PubMed] [Google Scholar]
- 37. Huho AN, Yadak N, Bocklage TJ, et al Evaluation of diagnostic utility of a high‐risk human papillomavirus PCR test on formalin‐fixed, paraffin‐embedded head and neck tumor tissues. J Mol Diagn 2018; 20: 232–239. [DOI] [PubMed] [Google Scholar]
- 38. Kerr DA, Sweeney B, Arpin RN III, et al Automated extraction of formalin‐fixed, paraffin‐embedded tissue for high‐risk human papillomavirus testing of head and neck squamous cell carcinomas using the Roche Cobas 4800 system. Arch Pathol Lab Med 2016; 140: 844–848. [DOI] [PubMed] [Google Scholar]
- 39. Mirghani H, Casiraghi O, Guerlain J, et al Diagnosis of HPV driven oropharyngeal cancers: comparing p16 based algorithms with the RNAscope HPV‐test. Oral Oncol 2016; 62: 101–108. [DOI] [PubMed] [Google Scholar]
- 40. Morbini P, Alberizzi P, Tinelli C, et al Identification of transcriptionally active HPV infection in formalin‐fixed, paraffin‐embedded biopsies of oropharyngeal carcinoma. Hum Pathol 2015; 46: 681–689. [DOI] [PubMed] [Google Scholar]
- 41. Pettus JR, Wilson TL, Steinmetz HB, et al Utility of the Roche Cobas 4800 for detection of high‐risk human papillomavirus in formalin‐fixed paraffin‐embedded oropharyngeal squamous cell carcinoma. Exp Mol Pathol 2017; 102: 47–49. [DOI] [PubMed] [Google Scholar]
- 42. Rooper LM, Gandhi M, Bishop JA, et al RNA in‐situ hybridization is a practical and effective method for determining HPV status of oropharyngeal squamous cell carcinoma including discordant cases that are p16 positive by immunohistochemistry but HPV negative by DNA in‐situ hybridization. Oral Oncol 2016; 55: 11–16. [DOI] [PubMed] [Google Scholar]
- 43. Shelton J, Purgina BM, Cipriani NA, et al p16 immunohistochemistry in oropharyngeal squamous cell carcinoma: a comparison of antibody clones using patient outcomes and high‐risk human papillomavirus RNA status. Mod Pathol 2017; 30: 1194–1203. [DOI] [PubMed] [Google Scholar]
- 44. Volpi CC, Ciniselli CM, Gualeni AV, et al In situ hybridization detection methods for HPV16 E6/E7 mRNA in identifying transcriptionally active HPV infection of oropharyngeal carcinoma: an updating. Hum Pathol 2018; 74: 32–42. [DOI] [PubMed] [Google Scholar]
- 45. Giarre M, Caldeira S, Malanchi I, et al Induction of pRb degradation by the human papillomavirus type 16 E7 protein is essential to efficiently overcome p16INK4a‐imposed G1 cell cycle arrest. J Virol 2001; 75: 4705–4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Munger K, Jones DL. Human papillomavirus carcinogenesis: an identity crisis in the retinoblastoma tumor suppressor pathway. J Virol 2015; 89: 4708–4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schache AG, Liloglou T, Risk JM, et al Evaluation of human papilloma virus diagnostic testing in oropharyngeal squamous cell carcinoma: sensitivity, specificity, and prognostic discrimination. Clin Cancer Res 2011; 17: 6262–6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fischer CA, Zlobec I, Green E, et al Is the improved prognosis of p16 positive oropharyngeal squamous cell carcinoma dependent of the treatment modality? Int J Cancer 2010; 126: 1256–1262. [DOI] [PubMed] [Google Scholar]
- 49. Jordan RC, Lingen MW, Perez‐Ordonez B, et al Validation of methods for oropharyngeal cancer HPV status determination in US cooperative group trials. Am J Surg Pathol 2012; 36: 945–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lassen P, Eriksen JG, Hamilton‐Dutoit S, et al Effect of HPV‐associated p16INK4A expression on response to radiotherapy and survival in squamous cell carcinoma of the head and neck. J Clin Oncol 2009; 27: 1992–1998. [DOI] [PubMed] [Google Scholar]
- 51. Lewis JS Jr, Thorstad WL, Chernock RD, et al p16 positive oropharyngeal squamous cell carcinoma:an entity with a favorable prognosis regardless of tumor HPV status. Am J Surg Pathol 2010; 34: 1088–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Singhi AD, Westra WH. Comparison of human papillomavirus in situ hybridization and p16 immunohistochemistry in the detection of human papillomavirus‐associated head and neck cancer based on a prospective clinical experience. Cancer 2010; 116: 2166–2173. [DOI] [PubMed] [Google Scholar]
- 53. Al‐Swiahb JN, Huang CC, Fang FM, et al Prognostic impact of p16, p53, epidermal growth factor receptor, and human papillomavirus in oropharyngeal cancer in a betel nut‐chewing area. Arch Otolaryngol Head Neck Surg 2010; 136: 502–508. [DOI] [PubMed] [Google Scholar]
- 54. Gao G, Chernock RD, Gay HA, et al A novel RT‐PCR method for quantification of human papillomavirus transcripts in archived tissues and its application in oropharyngeal cancer prognosis. Int J Cancer 2013; 132: 882–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Marklund L, Nasman A, Ramqvist T, et al Prevalence of human papillomavirus and survival in oropharyngeal cancer other than tonsil or base of tongue cancer. Cancer Med 2012; 1: 82–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Reimers N, Kasper HU, Weissenborn SJ, et al Combined analysis of HPV‐DNA, p16 and EGFR expression to predict prognosis in oropharyngeal cancer. Int J Cancer 2007; 120: 1731–1738. [DOI] [PubMed] [Google Scholar]
- 57. Shi W, Kato H, Perez‐Ordonez B, et al Comparative prognostic value of HPV16 E6 mRNA compared with in situ hybridization for human oropharyngeal squamous carcinoma. J Clin Oncol 2009; 27: 6213–6221. [DOI] [PubMed] [Google Scholar]
- 58. Weinberger PM, Yu Z, Kountourakis P, et al Defining molecular phenotypes of human papillomavirus‐associated oropharyngeal squamous cell carcinoma: validation of three‐class hypothesis. Otolaryngol Head Neck Surg 2009; 141: 382–389. [DOI] [PubMed] [Google Scholar]
- 59. Cao D, Begum S, Ali SZ, et al Expression of p16 in benign and malignant cystic squamous lesions of the neck. Hum Pathol 2010; 41: 535–539. [DOI] [PubMed] [Google Scholar]
- 60. Nauta IH, Rietbergen MM, van Bokhoven A, et al Evaluation of the eighth TNM classification on p16‐positive oropharyngeal squamous cell carcinomas in the Netherlands and the importance of additional HPV DNA testing. Ann Oncol 2018; 29: 1273–1279. [DOI] [PubMed] [Google Scholar]
- 61. Boland JM, McPhail ED, Garcia JJ, et al Detection of human papilloma virus and p16 expression in high‐grade adenoid cystic carcinoma of the head and neck. Mod Pathol 2012; 25: 529–536. [DOI] [PubMed] [Google Scholar]
- 62. Bishop JA, Andreasen S, Hang JF, et al HPV‐related multiphenotypic Sinonasal carcinoma: an expanded series of 49 cases of the tumor formerly known as HPV‐related carcinoma with adenoid cystic carcinoma‐like features. Am J Surg Pathol 2017; 41: 1690–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bishop JA, Ogawa T, Stelow EB, et al Human papillomavirus‐related carcinoma with adenoid cystic‐like features: a peculiar variant of head and neck cancer restricted to the sinonasal tract. Am J Surg Pathol 2013; 37: 836–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. McDowell LJ, Young RJ, Johnston ML, et al p16‐positive lymph node metastases from cutaneous head and neck squamous cell carcinoma: no association with high‐risk human papillomavirus or prognosis and implications for the workup of the unknown primary. Cancer 2016; 122: 1201–1208. [DOI] [PubMed] [Google Scholar]
- 65. Westra W, Boy S, El‐Mofty SK, et al Squamous cell carcinoma, HPV‐positive In: WHO Classification of Head and Neck Tumours (4th edn). El‐Naggar A, Chan JKC, Grandis JR, et al (Eds). IARC Press: Lyon, France, 2017. [Google Scholar]
- 66. Ahn SM, Chan JY, Zhang Z, et al Saliva and plasma quantitative polymerase chain reaction‐based detection and surveillance of human papillomavirus‐related head and neck cancer. JAMA Otolaryngol Head Neck Surg 2014; 140: 846–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Chera BS, Amdur RJ. Tepper JE, et al Mature results of a prospective study of deintensified chemoradiotherapy for low‐risk human papillomavirus‐associated oropharyngeal squamous cell carcinoma; Cancer: 2018; 124: 2347–2354. [DOI] [PubMed] [Google Scholar]
- 68. Marur S, Li S, Cmelak AJ, et al E1308: phase II trial of induction chemotherapy followed by reduced‐dose radiation and weekly cetuximab in patients with HPV‐associated resectable squamous cell carcinoma of the oropharynx‐ ECOG‐ACRIN cancer research group. J Clin Oncol 2017; 35: 490–497. [DOI] [PMC free article] [PubMed] [Google Scholar]


