Abstract
We aimed to validate the prognostic association of p16 expression in ovarian high‐grade serous carcinomas (HGSC) and to explore it in other ovarian carcinoma histotypes. p16 protein expression was assessed by clinical‐grade immunohistochemistry in 6525 ovarian carcinomas including 4334 HGSC using tissue microarrays from 24 studies participating in the Ovarian Tumor Tissue Analysis consortium. p16 expression patterns were interpreted as abnormal (either overexpression referred to as block expression or absence) or normal (heterogeneous). CDKN2A (which encodes p16) mRNA expression was also analyzed in a subset (n = 2280) mostly representing HGSC (n = 2010). Association of p16 expression with overall survival (OS) was determined within histotypes as was CDKN2A expression for HGSC only. p16 block expression was most frequent in HGSC (56%) but neither protein nor mRNA expression was associated with OS. However, relative to heterogeneous expression, block expression was associated with shorter OS in endometriosis‐associated carcinomas, clear cell [hazard ratio (HR): 2.02, 95% confidence (CI) 1.47–2.77, p < 0.001] and endometrioid (HR: 1.88, 95% CI 1.30–2.75, p = 0.004), while absence was associated with shorter OS in low‐grade serous carcinomas (HR: 2.95, 95% CI 1.61–5.38, p = 0.001). Absence was most frequent in mucinous carcinoma (50%), and was not associated with OS in this histotype. The prognostic value of p16 expression is histotype‐specific and pattern dependent. We provide definitive evidence against an association of p16 expression with survival in ovarian HGSC as previously suggested. Block expression of p16 in clear cell and endometrioid carcinoma should be further validated as a prognostic marker, and absence in low‐grade serous carcinoma justifies CDK4 inhibition.
Keywords: ovary, immunocytochemistry, RT‐QPCR
Introduction
CDKN2A (cyclin‐dependent kinase inhibitor 2A) is located on chromosome 9p21.3 and encodes two proteins, p16 and p14ARF, that have different reading frames 1. p14ARF inhibits p53 function and p16 inhibits the CDK4/6 complex acting as a negative cell cycle regulator suppressing the transition from the Gap1 to DNA synthesis (G1/S) phase and arresting the cell cycle in the G1 phase 2. Normal cells express variable amounts of p16 protein that can be detected by immunohistochemistry (IHC) in both nuclear and cytoplasmic localizations (heterogeneous p16 expression pattern) 3. There are two abnormal p16 expression patterns: absent and overexpressed, the latter also referred to as block expression as recommended by the Lower Anogenital Squamous Terminology Standardization Project for HPV‐Associated Lesions (LAST) 4. In keeping with its role as a tumor suppressor, absence of p16 expression can occur due to various mechanisms including homozygous deletion, loss of function mutations, promoter hypermethylation and translational suppression 5. In ovarian carcinoma, homozygous deletion of CDKN2A has been detected in only 3% of high‐grade serous carcinomas (HGSC) 6, 15% of low‐grade serous carcinomas (LGSC) 7, and in 30% of mucinous carcinomas (MC) 8. In contrast, p16 block expression results from a variety of alterations in G1/S cell cycle transition as a compensatory effort to inhibit G1/S transition. p16 block expression is classically observed in human papillomavirus (HPV)‐associated uterine cervical neoplasms, in which viral proteins (E7) inactivate pRB and promote G1/S transition 9, 10. IHC overexpression of p16 is routinely used in clinical diagnostics for identification of HPV‐related neoplasms. Ovarian carcinomas are not associated with HPV infections, but alterations promoting G1/S transition are common, e.g. RB1, CCNE1, CCND1, or MYC 6.
Ovarian carcinoma is a biologically heterogeneous disease 11 composed of five main histotypes: HGSC, LGSC, clear cell carcinoma (CCC), endometrioid carcinoma (EC), and MC, which should be studied separately 12. Older studies combining all histotypes showed that either overexpression or complete absence of p16 were associated with unfavorable outcomes 13, 14, 15. Recently, histotype‐specific studies also reported that normal heterogeneous p16 expression was significantly associated with longer progression‐free and overall survival (OS) in two series of 334 and 115 women with HGSC 16, 17. Therefore, we hypothesized that heterogeneous p16 expression reflecting the normal G1/S transition status is associated with a favorable outcome in HGSC compared to absent or block expression reflecting abnormalities of the G1/S cell cycle checkpoint complex. The purpose of this study was to validate whether abnormal p16 expression is associated with an unfavorable OS in HGSC, and to explore prognostic associations in other histotypes using tissue microarrays (TMAs) from the Ovarian Tumor Tissue Analysis (OTTA) consortium 18, 19.
Methods
Immunohistochemistry
The study investigators obtained tissue from 7492 patients with a diagnosis of primary ovarian, fallopian tube, or peritoneal carcinoma from 24 study sites (Supporting Information, Table S1). Most of these patients also participated in previous OTTA studies 18, 19, 20, and all studies received ethics board approval for tumor profiling. TMAs were constructed containing 1–6 cores of 0.6–1.0 mm in diameter from formalin‐fixed paraffin embedded tissue representing tumor from each patient. p16 IHC was performed centrally at two institutions: Genetic Pathology Evaluation Centre, University of British Columbia, and Calgary Laboratory Services, University of Calgary, Canada. TMAs were stained in five batches with three different protocols (Table S2) using the same antibody (clone E6H4, CINtec, mtm laboratories). Three staining patterns were recorded: absent, heterogeneous and block (Figure S1). Block expression was distinguished from heterogeneous staining by using the recommendation for p16 interpretation from LAST 4; that is, block expression is characterized by diffuse staining of tumor cells in nuclear and/or cytoplasmic compartment with at least moderate intensity with virtually no negative tumor cell clusters. Interobserver reproducibility between two observers (PR and MK) was assessed for a subset of 120 cases. Seventeen studies were scored by PR and the remainder by MK. Cases represented by more than one core and discordant cores were consolidated as heterogeneous if any of a given case score was heterogeneous.
CDKN2A mRNA analysis
A subset of 2280 cases had CDKN2A mRNA expression data from NanoString n‐counter analysis. RNA was extracted from 10 μm sections from formalin fixed paraffin embedded (FFPE) tissue blocks, which were macrodissected to avoid adjacent benign tissue but included tumor stroma using the Qiagen miRNeasy (Qiagen Inc. Toronto, Ontario, Canada) FFPE protocol and quantitated on a Nanodrop spectrophotometer (Thermo‐Fisher Scientific, Waltham, MA, USA). After mixing 500 ng of total RNA per sample with a custom codeset (NanoString Technologies Inc, Seattle, WA, USA) and hybridization buffer (NanoString), hybridization was performed using a Tetrad 2 thermal cycler (Bio Rad Laboratories Inc, Hercules, CA, USA) for 16 or 20 h and then analyzed on a nCounter Digital Analyzer (NanoString). Data was normalized to housekeeping genes (RPL19, ACTB, PGK1, SDHA, and POLR1B) and pre‐processed to a reference of 3 pooled ovarian cancer specimens as described previously 21. We interrogated the cBioportal for associations of CDKN2A alterations with OS in HGSC from TCGA 22, 23.
Statistical tests
Morphology‐based histotype was derived from pathology reports with or without review of reports or slides (Table S1). Because some HGSC were mistakenly classified as other histotypes in the past, we used the highly specific WT1(+)/p53(mutant) IHC combination to reclassify those to HGSC 24. We excluded 409 cases owing to diagnosis other than the five major histotypes, 393 cases being uninterpretable, 31 cases with a combination of absence and block staining, and 134 cases with missing clinical follow‐up data. The final sample size was 6525 (Table 1). The median time from diagnosis to enrollment was 0 days (interquartile range 0–182 days). Patients (n = 331) with missing data for either age or time from diagnosis to enrolment were not part of the multivariate survival analysis.
Table 1.
Clinical characteristics
| High‐grade serous carcinoma | Low‐grade serous carcinoma | Clear cell carcinoma | Endometrioid carcinoma | Mucinous carcinoma | |
|---|---|---|---|---|---|
| Number of cases, n (%) | 4334 (66.4) | 205 (3.1) | 717 (11.0) | 882 (13.5) | 387 (5.9) |
| Age at diagnosis, years, mean ± SD | 59.7 ± 10.7 | 53.8 ± 12.7 | 56.0 ± 11.4 | 54.8 ± 12.0 | 54.5 ± 14.8 |
| Stage, n (%) | |||||
| I/II | 822 (19.5) | 62 (32.3) | 550 (78.2) | 703 (83.5) | 283 (81.9) |
| III/IV | 3402 (80.5) | 130 (67.7) | 154 (21.8) | 139 (16.5) | 67 (19.1) |
| Unknown | 110 | 13 | 13 | 40 | 37 |
| Macroscopic residual disease, n (%) | |||||
| Absent | 1028 (43.7) | 64 (49.6) | 349 (81.0) | 393 (88.3) | 163 (77.2) |
| Present | 1323 (54.3) | 65 (50.4) | 82 (19.0) | 52 (11.7) | 51 (23.8) |
| Unknown | 1983 | 76 | 286 | 437 | 173 |
| Outcome | |||||
| Five year survival, % ± SE1 | 40.7 ± 0.8 | 61.9 ±3.7 | 63.4 ± 1.9 | 81.0 ± 1.5 | 65.3 ± 2.7 |
| Total months followed for censored patients, months, mean ± SD1 | 87 ± 41 | 80 ± 43 | 104 ± 39 | 101 ± 39 | 97 ± 41 |
| p16 expression, n (%) | |||||
| Heterogeneous | 1627 (37.5) | 167 (81.5) | 471 (65.7) | 676 (76.7) | 171 (44.2) |
| Absent | 267 (6.2) | 25 (12.2) | 146 (20.4) | 127 (14.4) | 194 (50.1) |
| Block | 2440 (56.3) | 13 (6.3) | 100 (13.9) | 79 (8.9) | 22 (5.7) |
Follow‐up is right‐censored at 12 years post‐diagnosis.
Associations of p16 IHC expression and CDKN2A mRNA expression with demographic and clinical variables were examined using the chi‐square test and Kruskal–Wallis test, as appropriate. We examined interobserver heterogeneity of p16 interpretation using Kappa coefficients. For individuals with multiple tumor cores, we examined intratumoral heterogeneity of p16 expression using percent discordance. We assessed correlations between (the ordinally scaled) p16 staining values and CDKN2A mRNA expression using Pearson correlation coefficients.
The primary end point for survival analysis was death from any cause. We chose right censoring of follow‐up at 12 years post‐diagnosis to mitigate against deaths from noncancer‐related causes. Kaplan–Meier survival curves and corresponding log‐rank tests were generated to visually assess associations of p16 expression with survival. Cox proportional hazards regression was used for multivariable assessment of hazard ratios (HRs). Models were adjusted for the following confounding factors: study site, age (continuous), FIGO stage (categorized into I/II, III/IV, and missing variable), and residual disease (categorized as absent, i.e. no residual disease, present, and missing). We used left truncation to account for the enrollment of prevalent cases in some studies. We tested whether histotype modified the association between p16 IHC expression and OS by fitting and testing corresponding interaction terms. We assessed the functional form of the association between CDKN2A mRNA expression levels and OS in HGSC using penalized B‐splines 25, adjusting for the same potential confounding variables as described above. All statistical tests were two‐sided, and analyses were carried out using RStudio (Boston, MA, USA) or JMP 13.0.0 (SAS, Cary, NC, USA). This study adhered to the REMARK criteria 26.
Results
p16 protein and CDKN2A mRNA expression across histological types
Table 1 shows the characteristics of the study sample. The histotype distribution is similar to population‐based cohorts except for a slightly higher frequency of MC 27. Inter‐observer agreement for interpretation of p16 IHC was excellent (Cohen's kappa of 0.92). Of the 6525 women, 4046 (62.0%) had more than one interpretable tissue core and, of those, 12.9% had a discordant interpretation between cores. This moderate degree of intratumoral heterogeneity was not statistically different between histotypes (p = 0.11), ranging from 7.4% for LGSC to 16.3% for CCCs. As expected, the distribution of p16 expression categories was significantly different across histotypes (Table 2, p < 0.0001) 28.
Table 2.
Association of p16 expression and OS by histotype
| Histotype | Expression | N | HR (95% CI) | P value |
|---|---|---|---|---|
| High‐grade serous | Heterogeneous | 1550 | ref | 0.68 |
| Absent | 244 | 1.06 (0.90–1.25) | ||
| Block | 2292 | 1.03 (0.95–1.11) | ||
| Low‐grade serous | Heterogeneous | 166 | ref | 0.001 |
| Absent | 25 | 2.95 (1.61–5.38) | ||
| Block | 13 | 1.54 (0.72–3.29) | ||
| Clear cell | Heterogeneous | 463 | ref | <0.001 |
| Absent | 138 | 0.67 (0.47–0.96) | ||
| Block | 92 | 2.02 (1.47–2.77) | ||
| Endometrioid | Heterogeneous | 650 | ref | 0.004 |
| Absent | 117 | 0.98 (0.66–1.45) | ||
| Block | 73 | 1.88 (1.30–2.75) | ||
| Mucinous | Heterogeneous | 163 | ref | 0.80 |
| Absent | 187 | 1.05 (0.72–1.55) | ||
| Block | 21 | 1.28 (0.61–2.64) |
Adjusted for study, age, time interval, stage and residual tumor; ref, reference.
Smaller sample size is due to availability of age and time interval information.
A subset of 2280 cases (2010 HGSC, 22 LGSC, 139 EC, 82 CCC, and 27 MC) had corresponding CDKN2A mRNA expression data. CDKN2A mRNA expression was significantly higher in HGSC [normalized mean −2.98 (95% CI ‐3.06 to −2.90)] compared to LGSC [normalized mean −4.90 (95% CI −5.39 to −3.19)], EC [normalized mean −4.82 (95% CI −5.31 to −4.33)], CCC [normalized mean −4.16 (95% CI −4.51 to −3.81)] and MC [normalized mean −4.50 (95% CI −5.07 to −3.93), for all p < 0.0001]. We confirmed the bimodal distribution of CDKN2A mRNA expression in HGSC as previously observed in the TCGA data set 22, 23 (Figure S2). CDKN2A mRNA expression correlated with p16 IHC scores (r = 0.69) overall, and for the specific histotypes (r = 0.69 for HGSC, 0.57 for LGSC, 0.62 for CCC, 0.80 for EC and 0.69 for MC, Figure 1A).
Figure 1.
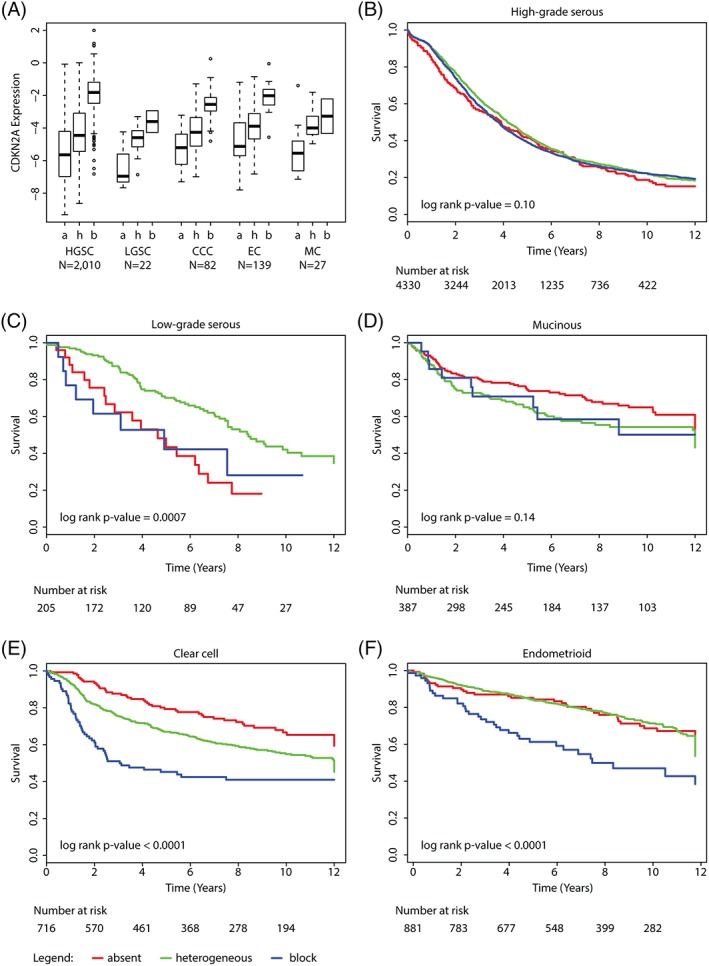
Associations of p16 protein expression with CDKN2A mRNA expression and survival by histotype. (A) Comparison of CDKN2A mRNA values (y‐axis) with p16 scoring categories (x‐axis), by the five major histotypes. a = p16 absence, h = p16 heterogeneous, b = p16 block score, respectively. Kaplan–Meier OS curves of p16 expression within (B) high‐grade serous, (C) low‐grade serous, (D) mucinous, (E) clear cell and (F) endometrioid carcinoma.
Association of p16 protein and CDKN2A mRNA expression and OS in HGSC and LGSC
For HGSC patients, the Kaplan–Meier survival curve showed no difference in OS for the three p16 immunohistochemical expression patterns (Figure 1B, p = 0.32), which was supported by HRs near 1.0 after controlling for the study, age, time to enrollment, stage and residual disease (Table 2).
For mRNA expression in HGSC, we used several different groupings (median split, tertiles) as well as different cut‐offs for dichotomization (mean = −3.03, visual inspection to separate bimodal peaks = −3.7) but there was no association of CDKN2A mRNA levels with OS. Alternatively, we examined the functional form of the association between CDKN2A expression and OS in HGSC using penalized B splines 25. Analyses revealed relatively flat HRs across the entire spectrum of mRNA values with a 95% confidence band that always included an HR of 1.0 (Figure S3). By interrogating 489 HGSC from TCGA via the cBioPortal 22, 23, 92 (19%) showed downregulation of CDKN2A, which was also not associated with survival (p = 0.27).
In contrast, for patients with LGSC, the 5‐year survival rate was significantly lower in tumors with absent p16 expression (Figure 1C, 43.4%, SE 10.7%) and in tumors with block p16 expression (42.2%, SE 14.7%) compared to heterogeneous expression (70.1%, SE 3.8%, p = 0.0005). This was also significant in multivariate analysis for the absence of p16 (Table 2). The limited number of CDKN2A mRNA expression values for non‐HGSC precluded us from examining associations within those histotypes.
No association of p16 protein expression and OS in MC
Figure 1D shows no differences in 5‐year survival for MC with heterogeneous (63.7%, SE 3.8%), absent (73.3%, SE 3.2%) or block expression (72.2%, SE 9.7%, p = 0.12). There was no significant association in multivariate analysis (Table 2).
Association of p16 protein expression and OS in endometriosis related ovarian carcinomas
For CCC, 5‐year survival was more than 20% lower for women with tumor block expression of p16 (45.2%, SE 5.0%) compared to heterogeneous staining (67.0%, SE 2.2%, p < 0.0001, Figure 1E). Similarly, for EC, 5‐year survival was more than 20% lower for women with tumor block expression of p16 (63.0%, SE 5.8%) compared to heterogeneous staining (85.0%, SE 1.4%, p < 0.0001, Figure 1F) or absent staining (85.2%, SE 3.3). In multivariate analysis, there was a significantly increased risk of death for patients with CCC or EC block staining, with HRs of 2.02 (95% CI: 1.47–2.77) and 1.88 (95% CI: 1.30–2.75), respectively (Table 2).
Pooled association of p16 protein expression and OS, and tests of effect modification by histotype
In analyses that combined all five major histological subtypes, women whose tumors exhibited with block expression had poorer OS (HR 1.13, 95% CI 1.06–1.22) than those with absent or heterogeneous expression (Figure S4). The associations between p16 expression and survival were strikingly different across histotypes (Cox regression test for interaction p = 1.4 × 10−8).
Association of p16 protein expression with clinicopathological parameters within histotypes
In EC, a greater proportion of cases with p16 block expression were diagnosed at stage III/IV (33.8% compared to 14.5% for heterogeneous staining, p < 0.0001) and grade 3 (45.7% compared to 16.0% for heterogeneous staining, p < 0.0001, Table S3). CCC cases with p16 block expression were more likely to have residual disease at initial surgery (34.6% compared to 17.1% for heterogeneous staining, p = 0.0067). In LGSC, p16 expression was not associated with age, stage or presence of residual tumor. Associations for HGSC and MC are shown in Table S3. Notably, there was no association for p16 expression status with BRCA1 or BRCA2 mutation status for the subset of HGSCs with available mutation data (n = 1370, p = 0.43).
Discussion
Our investigation showed that associations of p16 staining pattern with OS differ across ovarian carcinoma histotypes. Block p16 expression was significantly associated with shorter survival for endometriosis‐related ovarian carcinomas. In contrast, absence of p16 expression predicted shorter survival in LGSC while no survival associations are observed for MC and HGSC.
In contrast to previous studies 16, 17, we provide strong evidence against a clinically or biologically relevant survival association of p16 expression in HGSC. Our large sample size greatly reduces the potential of this lack of association being a false negative finding. Using our observed sample size and a two‐sided test of hypothesis with Type I error rate of 0.05, we would have 80% power to detect a HR as low as 1.21 comparing absence and 1.10 comparing block to heterogeneous expression, respectively. This null conclusion is supported by a lack of association between OS and CDKN2A mRNA expression data, which correlated positively with p16 IHC‐based protein expression. In keeping with the recommendations from the Institute of Medicine for validation of biomarker studies 29, we used the same antibody and the same scoring system as previous studies. We found excellent interobserver reproducibility regarding the IHC interpretation and similar frequencies of the three staining patterns in HGSC compared to previous studies; and it is, therefore, unlikely that technical or interpretational differences can explain differences in our results from those published previously. We think that the large sample size used in the current study compared to prior studies of HGSC avoided a false positive finding 16, 17. Since p16 block staining is a surrogate for various retinoblastoma pathway alterations, we speculate that different underlying alteration might explain the lack of outcome associations for p16. For example, prognostically opposing underlying alterations (e.g. favorable pRB loss versus unfavorable CCNE1 amplifications), which result in the same p16 block staining, may neutralize each other 30, 31.
Exploring other histotypes, we demonstrate for the first time that block p16 expression is significantly associated with OS in both endometriosis‐associated histotypes: EC and CCC. Overall, those histotype‐specific differences would not have been revealed in a combined histotype analysis and corroborate that biomarker analyses should be done stratified by histotype 12. Yet the subsets of p16 block expression that were significantly associated with unfavorable prognosis were small: 9 and 14% of EC and CCC, respectively. EC is usually associated with a favorable outcome and some patients do not require chemotherapy or could be considered for hormonal therapy if hormone receptors (ER, PGR) are expressed 18, 32. However, estrogen receptor positive Luminal B breast cancers with loss of pRB function and high p16 expression are unresponsive to hormonal therapy 33. Our data suggest that further study of p16 as part of a biomarker panel that identifies EC with unfavorable prognosis would help triage patients to earlier aggressive therapy in the low stage setting, which may not be amenable to hormonal therapy.
We observed a similar negative association between block p16 expression and OS for CCC. Women diagnosed with CCC have a relatively unfavorable prognosis, in part because these tumors are chemotherapy‐resistant and alternative therapeutic options are sparse 34. Among those, radiation has been suggested for CCC 35 and perhaps p16 expression can be assessed for prediction of response to radiation as suggested from other cancer sites 3, 36. The survival associations specifically observed for p16 block staining in the two endometrioisis‐associated histotypes somewhat serves as a cross‐validation. Yet it does not preclude differences in the underlying mechanisms, e.g. CCNE1 copy number gain and overexpression have been reported for CCC but not EC 37. As a limitation, we observed a moderate degree of intratumoral heterogeneity, which was highest in CCC, using TMAs in size akin to pretreatment omental core biopsies. However, CCCs are usually treated by upfront surgery and the p16 assessment on a full histological section should mitigate against intratumoral heterogeneity.
We also observed a significant association with OS in patients with LGSC. In contrast to the block staining in endometriosis‐associated carcinomas, complete absence was associated with unfavorable outcome in LGSC in keeping with the tumor suppressor function. Although investigating the underlying mechanism of absence of p16 expression is beyond the scope of the current study, the 12% of LGSC showing complete absence of p16 by IHC is strikingly similar to the 15% frequency of the homozygous deletion of the CDKN2A locus reported by Hunter et al 7. We have previously shown that progesterone receptor (PGR) expression is a favorable prognostic factor in LGSC 18. Perhaps PGR and p16 status could help to stratify LGSC regarding prognosis 38. Another consideration is the possible predictive utility of absent p16 expression for CDK4 inhibitors as suggested in clinical trials for breast and other cancers 39. Konecny et al demonstrated that low p16 expression in pRB‐proficient tumors was correlated with in vitro response to CDK4 inhibitors 40. Since other treatment options are limited for LGSC, this may represent an interesting option for recurrent LGSC, a disease often affecting younger women. There was a non‐significant trend for the few LGSC with p16 block expression to have an unfavorable outcome but CKD4 inhibitors will be ineffective in tumors with p16 block expression because p16 block expression already indicates futile intrinsic CDK4 inhibition.
There was no significant prognostic association of p16 expression within MCs despite their having the highest frequency of complete absence (50%) across histotypes. This frequency is slightly higher than the 39% (n = 12/31) frequency of the combined corresponding molecular alterations (homozygous CDKN2A deletion or loss of functional mutations) reported by Ryland et al 8. Absence of p16 expression was most frequently observed in low stage MC and we speculate that a portion of those may be from low transcriptional activity.
A strength of this study is the large sample size providing excellent power to examine differences in protein expression patterns for the most common histotype (HGSC) as well as reasonable power to discern differences within EC and CCC. The lower sample sizes for MC and LGSC reduced our ability to detect differences. We used a diagnostic biomarker panel of WT1/p53 to limit the number of misclassified HGSC into other histotype categories. Within the OTTA consortium, we had the opportunity to analyze protein and mRNA data. Survival analyses were adjusted for confounding factors such as age, stage and residual disease. As a limitation, some of the study sites did not collect detailed treatment data; as such adjustment for treatment was not feasible. However, cases were collected throughout an era of relatively homogeneous standard adjuvant therapy consisting of platinum‐taxane chemotherapy before the introduction of neoadjuvant chemotherapy or PARP inhibitors.
This large‐scale collaborative study did not validate p16 expression as a prognostic marker in HGSC. The frequent block expression as a surrogate for abnormal retinoblastoma pathway activation warrants further study of individual pathway members. The intriguing prognostic associations in endometriosis‐associated EC and CCC make p16 a promising prognostic biomarker that requires further independent validation. The absence of p16 in a subset of LGSC calls for independent validation as a prognostic marker as well as investigation as a predictive marker for CDK4 inhibitors.
Author contributions statement
DGH and MK conceived the study design. PFR, CC, MSA, SMA, CW, AT and MK carried out experiments and interpreted results. RAV and MK analyzed the data. All authors collected data. PFR, RAV and MK wrote the first draft. All authors were involved in writing the paper and had final approval of the submitted and published versions.
Supporting information
SUPPLEMENTARY MATERIAL ONLINE
Figure S1. p16 immunohistochemistry. (A) Heterogeneous staining showing variable staining in tumor cells. (B) Complete absence in tumor with some normal cells staining. (C) Block staining with cytoplasmic and nuclear p16 expression in all tumor cells
Figure S2. Normalized CDKN2A mRNA expression values for HGSC showing bimodal distribution
Figure S3. Assessment of the Functional Form of CDKN2A mRNA values with overall survival in a subset of 1882 women with HGSOC. Vertical dotted lines indicate the mean values of mRNA expression for tumors with absence of p16 staining (left‐most line), heterogeneous staining (middle line) and block staining (right‐most line)
Figure S4. Kaplan–Meier overall survival curves of p16 expression in pooled analysis combining all histotypes
Table S1. Participating studies 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63
Table S2. Immunohistochemical staining protocols
Table S3. Association of p16 expression with clinicopathological parameters
Acknowledgements
We would like to thank all of the women who participated in these research programs. We thank Marjorie J. Riggan (OCAC) for coordination. Acknowledgements for individual studies: The AOCS gratefully acknowledges additional support from S. Boldeman, the Agar family, Ovarian Cancer Action (UK), Ovarian Cancer Australia and the Peter MacCallum Foundation. The AOCS also acknowledges the cooperation of the participating institutions in Australia and acknowledges the contribution of the study nurses, research assistants and all clinical and scientific collaborators to the study. The complete AOCS Study Group can be found at http://www.aocstudy.org. AOV: We thank Mie Konno, Michelle Darago, Faye Chambers. BGS: We thank Breast Cancer Now and the Institute of Cancer Research for support and funding of the Generations Study, Ovarian Cancer Action for funding of work on ovarian cancer within the Study, and the study participants, study staff, and the doctors, nurses and other health care providers and health information sources who have contributed to the study and Professor Robert Brown for advice and help. We acknowledge NHS funding to the Royal Marsden/ICR NIHR Biomedical Research Centre. CAL: We thank Taryn Burghardt for study coordination and Thomas Kryton (BFA, digital imaging specialist for Calgary Laboratory Services) for creating figures. GER: tissue samples were provided by the tissue bank of the National Center for Tumor Diseases (NCT, Heidelberg, Germany) in accordance with the regulations of the tissue bank and the approval of the ethics committee of the University of Heidelberg. SEA: SEARCH team, Craig Luccarini, Caroline Baynes, Don Conroy. UKO: We particularly thank I. Jacobs, E. Wozniak, A. Ryan, J. Ford and N. Balogun. WMH: We thank the Gynaecological Oncology Biobank at Westmead, a member of the Australasian Biospecimen Network‐Oncology group. Peter F. Rambau was supported by a Thomas E. Feasby PhD student fund. NCI/NIH R01CA172404 to Susan J Ramus. Funding for individual studies: AOC: The Australian Ovarian Cancer Study was supported by the U.S. Army Medical Research and Materiel Command under DAMD17‐01‐1‐0729, The Cancer Council Victoria, Queensland Cancer Fund, The Cancer Council New South Wales, The Cancer Council South Australia, The Cancer Foundation of Western Australia, The Cancer Council Tasmania and the NHMRC (ID400413 and ID400281). Professor David Bowtell is supported by the National Health and Medical Research Council of Australia (NHMRC; APP1092856) and the United States Department of Defence (DoD) Ovarian Cancer Transitional Leverage Award (W81XWH‐12‐1‐0104); AOV: Canadian Institutes of Health Research (MOP‐86727).; BAV: ELAN Funds of the University of Erlangen‐Nuremberg; CAL: Cancer Research Society (19319) and CLS Internal Research support RS11‐508; CNI: Instituto de Salud Carlos III (PI 12/01319); Ministerio de Economía y Competitividad (SAF2012); DOV: NCI/NIH R01CA168758, and NCI/NIH R01CA168758. Huntsman Cancer Foundation and the National Cancer Institute of the National Institutes of Health under Award Number P30CA042014. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH; GER: German Federal Ministry of Education and Research, Programme of Clinical Biomedical Research (01GB 9401) and the German Cancer Research Center (DKFZ); HAW: US National Institutes of Health (R01‐CA58598, N01‐CN‐55424 and N01‐PC‐67001); HOP: Department of Defense (DAMD17‐02‐1‐0669) and NCI (K07‐CA080668, R01‐CA95023, P50‐CA159981, MO1‐RR000056, R01‐CA126841); LAX: American Cancer Society Early Detection Professorship (SIOP‐06‐258‐01‐COUN) and the National Center for Advancing Translational Sciences (NCATS), Grant UL1TR000124; MAL: NCI R01‐CA61107, research grant 9422252 from the Danish Cancer Society, Copenhagen, Denmark; and the Mermaid I project; MAY: National Institutes of Health (R01‐CA122443, P30‐CA15083, P50‐CA136393); Mayo Foundation; Minnesota Ovarian Cancer Alliance; Fred C. and Katherine B. Andersen Foundation; SEA: Cancer Research UK C490/A16561, the UK National Institute for Health Research Biomedical Research Centres at the University of Cambridge, Cambridge Cancer Centre. The University of Cambridge has received salary support for PDPP from the NHS in the East of England through the Clinical Academic Reserve. POC: Pomeranian Medical University; TVA: This work was supported by Canadian Institutes of Health Research grant (MOP‐86727) and by NIH/NCI 1 R01CA160669‐01A1; UKO: The UKOPS study was funded by The Eve Appeal (The Oak Foundation) and supported by the National Institute for Health Research University College London Hospitals Biomedical Research Centre; VAN: The Janet D. Cottrelle Foundation Scholars fund provides support to M.S. Anglesio. OVCARE receives core funding from The BC Cancer Foundation and the VGH and UBC Hospital Foundation. WMH: National Health and Medical Research Council of Australia, Enabling Grants ID 310670 and ID 628903. Cancer Institute NSW Grants 12/RIG/1‐17 and 15/RIG/1‐16.
No conflicts of interest were declared.
References
*References cited only in supplementary material, Table S1.
- 1. Quelle DE, Zindy F, Ashmun RA, et al Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell 1995; 83: 993–1000. [DOI] [PubMed] [Google Scholar]
- 2. Lukas J, Parry D, Aagaard L, et al Retinoblastoma‐protein‐dependent cell‐cycle inhibition by the tumour suppressor p16. Nature 1995; 375: 503–506. [DOI] [PubMed] [Google Scholar]
- 3. Witkiewicz AK, Knudsen KE, Dicker AP, et al The meaning of p16(ink4a) expression in tumors: functional significance, clinical associations and future developments. Cell Cycle 2011; 10: 2497–2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Clinton LK, Miyazaki K, Ayabe A, et al The LAST guidelines in clinical practice: implementing recommendations for p16 use. Am J Clin Pathol 2015; 144: 844–849. [DOI] [PubMed] [Google Scholar]
- 5. Merlo A, Herman JG, Mao L, et al 5' CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat Med 1995; 1: 686–692. [DOI] [PubMed] [Google Scholar]
- 6. Cancer Genome Atlas Research N . Integrated genomic analyses of ovarian carcinoma. Nature 2011; 474: 609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hunter SM, Anglesio MS, Ryland GL, et al Molecular profiling of low grade serous ovarian tumours identifies novel candidate driver genes. Oncotarget 2015; 6: 37663–37677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ryland GL, Hunter SM, Doyle MA, et al Mutational landscape of mucinous ovarian carcinoma and its neoplastic precursors. Genome Med 2015; 7: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Imai Y, Matsushima Y, Sugimura T, et al Purification and characterization of human papillomavirus type 16 E7 protein with preferential binding capacity to the underphosphorylated form of retinoblastoma gene product. J Virol 1991; 65: 4966–4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Klaes R, Friedrich T, Spitkovsky D, et al Overexpression of p16(INK4A) as a specific marker for dysplastic and neoplastic epithelial cells of the cervix uteri. Int J Cancer 2001; 92: 276–284. [DOI] [PubMed] [Google Scholar]
- 11. Kurman RJ, Shih IM. The dualistic model of ovarian carcinogenesis: revisited, revised, and expanded. Am J Pathol 2016; 186: 733–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kobel M, Kalloger SE, Boyd N, et al Ovarian carcinoma subtypes are different diseases: implications for biomarker studies. PLoS Med 2008; 5: e232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dong Y, Walsh MD, McGuckin MA, et al Increased expression of cyclin‐dependent kinase inhibitor 2 (CDKN2A) gene product P16INK4A in ovarian cancer is associated with progression and unfavourable prognosis. Int J Cancer 1997; 74: 57–63. [DOI] [PubMed] [Google Scholar]
- 14. Kommoss S, du Bois A, Ridder R, et al Independent prognostic significance of cell cycle regulator proteins p16(INK4a) and pRb in advanced‐stage ovarian carcinoma including optimally debulked patients: a translational research subprotocol of a randomised study of the Arbeitsgemeinschaft Gynaekologische Onkologie ovarian cancer study group. Br J Cancer 2007; 96: 306–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kudoh K, Ichikawa Y, Yoshida S, et al Inactivation of p16/CDKN2 and p15/MTS2 is associated with prognosis and response to chemotherapy in ovarian cancer. Int J Cancer 2002; 99: 579–582. [DOI] [PubMed] [Google Scholar]
- 16. Milea A, George SH, Matevski D, et al Retinoblastoma pathway deregulatory mechanisms determine clinical outcome in high‐grade serous ovarian carcinoma. Mod Pathol 2014; 27: 991–1001. [DOI] [PubMed] [Google Scholar]
- 17. Beirne JP, McArt DG, James JA, et al p16 as a prognostic indicator in ovarian/tubal high‐grade serous carcinoma. Histopathology 2016; 68: 615–618. [DOI] [PubMed] [Google Scholar]
- 18. Sieh W, Kobel M, Longacre TA, et al Hormone‐receptor expression and ovarian cancer survival: an ovarian tumor tissue analysis consortium study. Lancet Oncol 2013; 14: 853–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kobel M, Madore J, Ramus SJ, et al Evidence for a time‐dependent association between FOLR1 expression and survival from ovarian carcinoma: implications for clinical testing. An ovarian tumour tissue analysis consortium study. Br J Cancer 2014; 111: 2297–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ovarian Tumor Tissue Analysis Consortium, Goode EL, Block MS, et al Dose‐response association of CD8+tumor‐infiltrating lymphocytes and survival time in high‐grade serous ovarian cancer. JAMA Oncol 2017; 3: e173290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Talhouk A, Kommoss S, Mackenzie R, et al Single‐patient molecular testing with NanoString nCounter data using a reference‐based strategy for batch effect correction. PLoS One 2016; 11: e0153844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gao J, Aksoy BA, Dogrusoz U, et al Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013; 6: pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cerami E, Gao J, Dogrusoz U, et al The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012; 2: 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kobel M, Rahimi K, Rambau PF, et al An Immunohistochemical algorithm for ovarian carcinoma typing. Int J Gynecol Pathol 2016; 35: 430–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Paul H. C. Eilers BDM. Flexible smoothing with B‐splines and penalties. Statistical Science 1996; 11: 89–121. [Google Scholar]
- 26. Altman DG, McShane LM, Sauerbrei W, et al Reporting recommendations for tumor marker prognostic studies (REMARK): explanation and elaboration. PLoS Med 2012; 9: e1001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kobel M, Kalloger SE, Huntsman DG, et al Differences in tumor type in low‐stage versus high‐stage ovarian carcinomas. Int J Gynecol Pathol 2010; 29: 203–211. [DOI] [PubMed] [Google Scholar]
- 28. Ferguson DC, Long DJ, Smith MC, et al Comparative analysis of Rb1, P16 and ER as diagnostic, prognostic and potential targets for therapeutic agents in ovarian epithelial tumors: an immunohistochemical study of 130 ovarian carcinomas. J Ovarian Res 2015; 8: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Christine M, Micheel SJN, Omenn GS. Best practices for omics‐based test validation prior to use for patient management decisions in a clinical trial setting In: Evolution of Translational Omics: Lessons Learned and the Path Forward Patient CotRoO‐BTfP, on OiCTBoHCSB, Medicine HSPIo. National Academies Press: Washington, DC, 2012; 65–78. [Google Scholar]
- 30. Garsed DW, Alsop K, Fereday S, et al Homologous recombination DNA repair pathway disruption and retinoblastoma protein loss are associated with exceptional survival in high‐grade serous ovarian cancer. Clin Cancer Res 2017; 24: 569–580. [DOI] [PubMed] [Google Scholar]
- 31. Au‐Yeung G, Lang F, Azar WJ, et al Selective targeting of cyclin E1‐amplified high‐grade serous ovarian cancer by cyclin‐dependent kinase 2 and AKT inhibition. Clin Cancer Res 2017; 23: 1862–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rambau P, Kelemen LE, Steed H, et al Association of hormone receptor expression with survival in ovarian endometrioid carcinoma: biological validation and clinical implications. Int J Mol Sci 2017; 18: E515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Herschkowitz JI, He X, Fan C, et al The functional loss of the retinoblastoma tumour suppressor is a common event in basal‐like and luminal B breast carcinomas. Breast Cancer Res 2008; 10: R75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Anglesio MS, Carey MS, Kobel M, et al Clear cell carcinoma of the ovary: a report from the first ovarian clear cell symposium, June 24th, 2010. Gynecol Oncol 2011; 121: 407–415. [DOI] [PubMed] [Google Scholar]
- 35. Swenerton KD, Santos JL, Gilks CB, et al Histotype predicts the curative potential of radiotherapy: the example of ovarian cancers. Ann Oncol 2011; 22: 341–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chakravarti A, DeSilvio M, Zhang M, et al Prognostic value of p16 in locally advanced prostate cancer: a study based on radiation therapy oncology group protocol 9202. J Clin Oncol 2007; 25: 3082–3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ayhan A, Kuhn E, Wu RC, et al CCNE1 copy‐number gain and overexpression identify ovarian clear cell carcinoma with a poor prognosis. Mod Pathol 2017; 30: 297–303. [DOI] [PubMed] [Google Scholar]
- 38. McIntyre JB, Rambau PF, Chan A, et al Molecular alterations in indolent, aggressive and recurrent ovarian low‐grade serous carcinoma. Histopathology 2017; 70: 347–358. [DOI] [PubMed] [Google Scholar]
- 39. Dickson MA. Molecular pathways: CDK4 inhibitors for cancer therapy. Clin Cancer Res 2014; 20: 3379–3383. [DOI] [PubMed] [Google Scholar]
- 40. Konecny GE, Winterhoff B, Kolarova T, et al Expression of p16 and retinoblastoma determines response to CDK4/6 inhibition in ovarian cancer. Clin Cancer Res 2011; 17: 1591–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. * Merritt MA, Green AC, Nagle CM, et al Talcum powder, chronic pelvic inflammation and NSAIDs in relation to risk of epithelial ovarian cancer. Int J Cancer 2008; 122: 170–1766. [DOI] [PubMed] [Google Scholar]
- 42. * Kelemen LE, Warren GW, Koziak JM, et al Smoking may modify the association between neoadjuvant chemotherapy and survival from ovarian cancer. Gynecol Oncol 2016; 140: 124–130. [DOI] [PubMed] [Google Scholar]
- 43. * Hein A, Thiel FC, Bayer CM, et al Hormone replacement therapy and prognosis in ovarian cancer patients. Eur J Cancer Prev 2013; 22: 52–58. [DOI] [PubMed] [Google Scholar]
- 44. * Bromley AB, Altman AD, Chu P, et al Architectural patterns of ovarian/pelvic high‐grade serous carcinoma. Int J Gynecol Pathol 2012; 31: 397–404. [DOI] [PubMed] [Google Scholar]
- 45. * Kamieniak MM, Rico D, Milne RL, et al Deletion at 6q24.2‐26 predicts longer survival of high‐grade serous epithelial ovarian cancer patients. Mol Oncol 2015; 9: 422–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. * Peterlongo P, Chang‐Claude J, Moysich KB, et al Candidate genetic modifiers for breast and ovarian cancer risk in BRCA1 and BRCA2 mutation carriers. Cancer Epidemiol Biomarkers Prev 2015; 24: 308–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. * Goodman MT, Lurie G, Thompson PJ, et al Association of two common single‐nucleotide polymorphisms in the CYP19A1 locus and ovarian cancer risk. Endocr Relat Cancer 2008; 15: 1055–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. * Lurie G, Wilkens LR, Thompson PJ, et al Genetic polymorphisms in the Paraoxonase 1 gene and risk of ovarian epithelial carcinoma. Cancer Epidemiol Biomarkers Prev 2008; 17: 2070–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. * Lo‐Ciganic WH, Zgibor JC, Bunker CH, et al Aspirin, nonaspirin nonsteroidal anti‐inflammatory drugs, or acetaminophen and risk of ovarian cancer. Epidemiology 2012; 23: 311–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. * Ramus SJ, Antoniou AC, Kuchenbaecker KB, et al Ovarian cancer susceptibility alleles and risk of ovarian cancer in BRCA1 and BRCA2 mutation carriers. Hum Mutat 2012; 33: 690–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. * Glud E, Kjaer SK, Thomsen BL, et al Hormone therapy and the impact of estrogen intake on the risk of ovarian cancer. Arch Intern Med 2004; 164: 2253–2259. [DOI] [PubMed] [Google Scholar]
- 52. * Soegaard M, Jensen A, Hogdall E, et al Different risk factor profiles for mucinous and nonmucinous ovarian cancer: results from the Danish MALOVA study. Cancer Epidemiol Biomarkers Prev 2007; 16: 1160–1166. [DOI] [PubMed] [Google Scholar]
- 53. * Goode EL, Chenevix‐Trench G, Hartmann LC, et al Assessment of hepatocyte growth factor in ovarian cancer mortality. Cancer Epidemiol Biomarkers Prev 2011; 20: 1638–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. * Williams E, Martin S, Moss R, et al Co‐expression of VEGF and CA9 in ovarian high‐grade serous carcinoma and relationship to survival. Virchows Arch 2012; 461: 33–39. [DOI] [PubMed] [Google Scholar]
- 55. * Garcia‐Closas M, Brinton LA, Lissowska J, et al Ovarian cancer risk and common variation in the sex hormone‐binding globulin gene: a population‐based case‐control study. BMC Cancer 2007; 7: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. * Song H, Ramus SJ, Quaye L, et al Common variants in mismatch repair genes and risk of invasive ovarian cancer. Carcinogenesis 2006; 27: 2235–2242. [DOI] [PubMed] [Google Scholar]
- 57. * McGuire V, Felberg A, Mills M, et al Relation of contraceptive and reproductive history to ovarian cancer risk in carriers and noncarriers of BRCA1 gene mutations. Am J Epidemiol 2004; 160: 613–618. [DOI] [PubMed] [Google Scholar]
- 58. * Baxter SW, Choong DY, Eccles DM, et al Transforming growth factor beta receptor 1 polyalanine polymorphism and exon 5 mutation analysis in breast and ovarian cancer. Cancer Epidemiol Biomarkers Prev 2002; 11: 211–214. [PubMed] [Google Scholar]
- 59. * Cook LS, Leung AC, Swenerton K, et al Adult lifetime alcohol consumption and invasive epithelial ovarian cancer risk in a population‐based case‐control study. Gynecol Oncol 2016; 140: 277–284. [DOI] [PubMed] [Google Scholar]
- 60. * Balogun N, Gentry‐Maharaj A, Wozniak EL, et al Recruitment of newly diagnosed ovarian cancer patients proved challenging in a multicentre biobanking study. J Clin Epidemiol 2011; 64: 525–530. [DOI] [PubMed] [Google Scholar]
- 61. * Prentice LM, Klausen C, Kalloger S, et al Kisspeptin and GPR54 immunoreactivity in a cohort of 518 patients defines favourable prognosis and clear cell subtype in ovarian carcinoma. BMC Med 2007; 5: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. * Kobel M, Reuss A, Bois A, et al The biological and clinical value of p53 expression in pelvic high‐grade serous carcinomas. J Pathol 2010; 222: 191–198. [DOI] [PubMed] [Google Scholar]
- 63. * Emmanuel C, Chiew YE, George J, et al Genomic classification of serous ovarian cancer with adjacent borderline differentiates RAS pathway and TP53‐mutant tumors and identifies NRAS as an oncogenic driver. Clin Cancer Res 2014; 20: 6618–6630. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPLEMENTARY MATERIAL ONLINE
Figure S1. p16 immunohistochemistry. (A) Heterogeneous staining showing variable staining in tumor cells. (B) Complete absence in tumor with some normal cells staining. (C) Block staining with cytoplasmic and nuclear p16 expression in all tumor cells
Figure S2. Normalized CDKN2A mRNA expression values for HGSC showing bimodal distribution
Figure S3. Assessment of the Functional Form of CDKN2A mRNA values with overall survival in a subset of 1882 women with HGSOC. Vertical dotted lines indicate the mean values of mRNA expression for tumors with absence of p16 staining (left‐most line), heterogeneous staining (middle line) and block staining (right‐most line)
Figure S4. Kaplan–Meier overall survival curves of p16 expression in pooled analysis combining all histotypes
Table S1. Participating studies 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63
Table S2. Immunohistochemical staining protocols
Table S3. Association of p16 expression with clinicopathological parameters


