Abstract
Molecular networks represent the interactions and relations of genes/proteins, and also encode molecular mechanisms of biological processes, development and diseases. Among the molecular networks, protein-protein Interaction Networks (PINs) have become effective platforms for uncovering the molecular mechanisms of diseases and drug discovery. PINs have been constructed for various organisms and utilized to solve many biological problems. In human, most proteins present their complex functions by interactions with other proteins, and the sum of these interactions represents the human protein interactome. Especially in the research on human disease and drugs, as an emerging tool, the PIN provides a platform to systematically explore the molecular complexities of specific diseases and the references for drug design. In this review, we summarized the commonly used approaches to aid disease research and drug discovery with PINs, including the network topological analysis, identification of novel pathways, drug targets and sub-network biomarkers for diseases. With the development of bioinformatic techniques and biological networks, PINs will play an increasingly important role in human disease research and drug discovery.
Keywords: Protein-protein interaction network, Drug discovery, Network analysis, Sub-network biomarkers, Alzheimer’s disease, Multiple sclerosis
1. INTRODUCTION
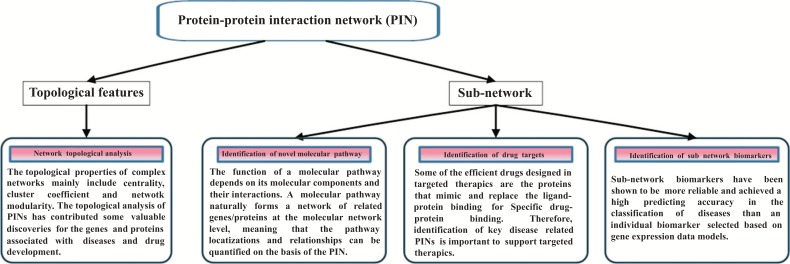
Proteins are the biological molecules that build the microscopic mechanisms of the biological systems. Traditionally, biochemical methods, such as knock-out experiments, targeted mutations, or functional assays, have been widely applied to identify the functions of individual protein [1]. As a result, many basic biological problems are still unanswered. For example, proteins undertake their functions in biological systems by teaming up into macromolecular complexes or transferring signals by interacting with other proteins rather than from single molecule, but traditional methods cannot capture most of these intracellular biochemical activities [2]. Therefore, studies on protein-protein interactions have been proven to be more important in the studies of various mechanisms of cellular activities and diseases. Protein-protein Interaction Network (PIN) presents physical interactions between gene products (i.e., proteins) to accomplish particular cellular functions, such as metabolism, cell cycle regulation and signal transduction. PINs have been constructed for various organisms, including bacteriophages, bacteria, yeast, plants, animals and human [3]. Analysis of PINs has been recognized as an important way to identify the potential genes related to complex diseases or potential drug targets [4]. It plays an important role in predicting the associations between genotype and phenotype [5]. Focusing on human studies, In particular, the PIN and its applications are largely helpful to recent advances in biomedical research because it is an effective index to evaluate their centrality and provide a reference value for protein as a drug target [6]. Therefore, reconstruction and analysis of PINs have become efficacious methods for surveying complex diseases, such as multiple sclerosis [7], Alzheimer’s disease [8], cancer [9], cancer metastasis [10] and drug discovery [11]. Analysis based on the topological features and sub-network has been proved to be a frequently used approach to investigate potentially important proteins in a PIN for diseases or drugs. Based on the topological features, the network topological analysis provides clues for the identification of disease-related genes. Based on the systematic features of sub-networks, PINs can be used for the identification of novel molecular pathways, drug targets and sub-network biomarkers (Fig. 1).
Fig. (1).
PIN and its applications.
2. IDENTIFICATION OF NOVEL MOLECULAR PATHWAYS THROUGH SUB-NETWORK FROM PIN
Sub-network is a part of a global PIN with specific biological functions, which has similar characteristic with a molecular pathway (e.g., signaling pathway). The function of a molecular pathway depends on its molecular components and their interactions. Therefore, a molecular pathway naturally forms a network of related genes/proteins at the molecular level, meaning that the pathway localizations and relationships can be quantified on the basis of PINs. Therefore, sub-network analysis becomes a useful approach for identifying novel molecular pathways. In this process, the human PIN [12] provides a crucial source as a basic global network for the further bioinformatic analysis. For example, based on the molecular pathway distribution in the human PIN, Hu et al. [13] proposed a method that took each pathway as a module and analyzed the relationship between pathways by calculating the distances between different pathway modules. They identified the pathways enriched with Parkinson’s Disease (PD) genes and analyzed the relationships among these pathways with this method. Based on the relationship analysis of the sub-network comprised of the enriched pathways, they discovered some potential pathway targets crucial for the pathology of PD, including apoptosis, focal adhesion pathways and several signaling pathways such as T cell receptor, HIF-1, MAPK and NF-kappa B signaling pathways. In the study of another disease - multiple sclerosis (MS), Baranzini et al. proposed a PIN-based pathway analysis (PINBPA) for two Genome-wide association studies including all the SNPs associated with MS and filtered with Gene-wise P-values < 0.05. This method was applied on a human PIN and identified several MS associated sub-networks with genes from several immunological pathways. Most importantly, they reported for the first time that neural pathways might be involved in the susceptibility of MS [14]. The international multiple sclerosis consortium further used PINBPA combined with functional analysis to identify five highly confident candidate genes with MS susceptibility (BCL10, CD48, REL, TRAF3, and TEC). PINBPA was thus demonstrated as a powerful method to investigate the novel pathways and candidate genes for subsequent genetic studies of complex disease traits [15].
The typical experimental approach of inferring the components constituting a pathway is perturbing the cells by molecular interventions [16]. Many wet experiments are needed to determine the molecular mechanisms and regulatory relationships among a set of proteins and metabolites, which is high-cost, time-consuming and error-prone. However, construction the valuable signaling pathways through PINs can not only be time-saving but also contribute to understand the molecular mechanisms of human diseases [5]. Therefore, an important investigation idea about sub-network analysis in PINs is to discover the potential signaling pathways for understanding their roles in signaling transduction, gene expression regulation and diseases [17]. Kiel et al. [18] constructed a PIN to represent the signal transduction process in the visual G protein-coupled receptor (GPCR) rhodopsin and predicted the novel signaling routes (Rac1/RhoA–PDEδ−CRMP-2) functioning in the vesicular transport and cytoskeleton dynamics. Sun et al. [19] proposed a node-weighted Steiner tree approach to detect the important components in a large-scale cancer-related PIN. With this method, 8 and 9 core interactions were identified from the network related to PI3K/Akt and MAPK signaling pathways, respectively. The important function of most of the core interactions were confirmed with literatures. Some new findings were also implied in the results, such as the relationship between protein p53 and NF- κB. In 2010, Bandyopadhyay et al. [20] constructed a large-scale MAPK-related PINs, based on the two-stage yeast two-hybrid (Y2H) method. The kinase sub-network of Filamin protein FLNA, Na-H exchanger NHE1, RAN binding protein RANBP9 and kinesin family member KIF26A were extracted from the global PIN to identify the MAPK scaffolds. In addition to the known function of these scaffolds proteins, NHE1 was found to be a novel plasma-membrane scaffold. Furthermore, some novel interactions with RANBP9 as the core protein were also identified, which indicated that RANBP9 might be a scaffold of MAPK and function as an activator of some transcription factors.
In recent years, the identification of novel pathways based on PINs had made a great process in cancer study. In order to better understand the molecular mechanisms of six neurodegenerative diseases (Parkinson’s disease, dentatorubropallidoluysian atrophy and prion disease, Alzheimer’s disease, Huntington’s disease, amyotrophic lateral sclerosis), Vachiranee et al. [21] constructed the PINs of these neurodegenerative disorders by extracting protein-protein interactions from 80 articles, and then expanding the networks by PubMed and HPRD. With the comparison of these networks, they found that there were 19 common proteins in these diseases, mainly related to cell apoptosis process and mitogen-activated-protein kinase signaling pathways. Stites et al. [22] established a differential equation model of the Ras signaling pathway in normal and cancer cells including heterotrimeric GTP binding protein and small GTPase signaling networks. This model presents that a drug is preferred to be combined with GDP-bound Ras over GTP-bound Ras, in order to affect Ras activity in cancer cells, rather than in normal cells. In the exploration of the value of PINs in the cancer research, the human signaling network may play an import role as that a large part of signaling network is composed of protein-protein interactions and it has been successfully applied in the drug targets prediction of breast cancer [23].
3. NETWORK TOPOLOGICAL ANALYSIS
The topological properties of complex networks mainly include centrality, cluster coefficient and network modularity. The most commonly used centrality feature in the application of PINs is degree centrality. The degree centrality of a node is the number of nodes directly connected to the target node. Degree centrality can describe the interaction between macromolecules and other macromolecules, and reflect the functional diversity and biological importance of macromolecules. In biological networks, the greater the degree a node has, the more network functions or reactions the node is involved in, and thus, it will lead to more serious damage to the biological network if a high degree node (e.g., gene or protein) has been disrupted. Therefore, degree centrality is the most commonly used topological property in biological network analysis [2, 24]. In the PINs, the highest-degree nodes are often called “hubs”. Hubs can be divided into two types: “party” hubs and “date” hubs. “Party” hubs usually bound to the protein adjacent to it, whereas “date” hubs can interact with each other at different space and time [25]. Some studies have found that hub proteins are more essential for cell survival. For example, the differences among the degree of disease proteins, essential proteins and other proteins in PINs are usually compared and analyzed to understand the functional differences of different types of proteins. The proteins with larger degree are often considered to be more important in topological status [26]. Connectivity studies showed that PINs have a property of “small-world”, which means the average distance between proteins is low. In PINs, most proteins are not directly connected, but are linked by a small number of other proteins [6a]. Some disease related studies have shown that highly connected proteins interacting with disease-related proteins are likely to be involved in the same disease based on the observation of strong correlation between protein connectivity and disease associations [27]. Cluster coefficient is used to find the cohesion of proteins and identify the molecular complexes or related functional network modules. Network module refers to a network complex composed of many molecules in the network, which have stable structures and functions. The molecular function of the network module is closely connected. The network module can be divided into functional modules, network topological modules and disease modules. A network topological module is usually a dense area in the network, where nodes have a higher degree of connectivity than the external nodes. A functional module is composed of adjacent nodes with similar biological functions. A disease module is defined as a set of biologically functional nodes, which will lead to diseases when the components in the module is perturbed [28]. In network biology, an important hypothesis points out that proteins involved in the same disease tend to interact each other and form disease modules [2]. Therefore, study of disease modules is helpful to understand the pathogenesis of diseases, and explain the penetrance and expressivity [29],which provides clues to the drug target of gene therapy [30] and identifies or prioritizes new disease and drug proteins [31]. Therefore,through analysis of the disease modules, when some disease proteins have been identified, other diseases associated with the protein will be easily found in the network nearby.
The topological analysis of PINs has contributed some valuable discoveries for the diseases/drug related genes/proteins. For example, Taylor et al. [32] collected protein interactions from the literature and high-throughput sequencing to construct a PIN and identified the hub proteins in the network. The expression information in 79 human tissues was then used to measure the extent that a hub protein in the PIN co-expressed in the same tissue with its neighbors. The hubs with low correlation of co-expression protein pairs were identified as intermodular hubs, whereas those with high correlation of co-expression were identified as intramodular hubs. In the analysis of cancer proteins they found that intermodular hubs have greater influence to cancers than intramodular hubs, which indicates that the modularity change of PIN may associates with cancer phenotype because intermodular hubs provide topological linkage to the intramodule hubs. In the further examination of this speculation in the breast cancer patients, pearson correlation coefficient of expression between good outcome group (i.e., surviving longer than 5 years) and poor outcome group (died due to the disease) was used to evaluate the change of the network modularity. They confirmed that the module structure of the network with good outcomes is significantly different from that with poor outcomes. In 2006, Pall et al. [33] found that the network topological characteristics of human proteins encoded by known cancer genes are different from the proteins that are not recorded as mutation proteins in cancer. Their studies have shown that cancer proteins are particularly prone to interacting with more proteins, and they also prefer to involve in the central network hub proteins rather than the network periphery proteins, which presents their higher centrality and participation in the networks as the skeleton of the PINs. However, Goh et al. [26b] presented different views in this issue. They pointed out that the reason for the high topological status of the pathogenic proteins is that the pathogenic genes contain a small number of essential genes, and the topological status of essential proteins is very important. Therefore, Pall et al. probably has overestimated the topological status of pathogenic protein. Furthermore, they studied the topological status of non-essential pathogenic proteins by using the mouse lethal genes as the necessary genes, and showed that the topological status of non-essential pathogenic proteins was in the periphery of the network. They also analyzed the co-expression of non-essential pathogenic genes vs other genes and the co-expression of essential genes vs other genes. They found that there was almost no coordinated expression between non-essential pathogenic genes and other genes, which indicated that the function of non-essential pathogenic genes can be considered to be in the periphery of the network. For these incredible new discoveries, they gave an explanation from the evolutionary point of view. If the degree of the pathogenic proteins is large, then when the disease-causing gene mutation is likely to have an impact on more neighbors, they will endanger cell survival. Thus, in the course of evolution, the population with the high-quality protein interaction network structure of the pathogenic proteins with high topological status was gradually eliminated. For the discovery and explanation of Goh et al. some people think that it is better to answer the question “whether the topological status of pathogenic proteins is higher”[2, 34].
In recent years, a lot of researches have also tried to classify genes, but the conclusion is different from Goh’s work. Barrenas et al. [35] divided the pathogenic genes into single-gene-disease pathogenic genes and complex disease pathogenic genes. Jin et al. [26c] considered the overlap of gene function and divided the pathogenic genes into the genes of both single-gene-diseases and complex diseases, genes of only single-gene-diseases, and genes of only complex diseases. These studies have shown that the pathogenic protein topological status is higher than non-pathogenic proteins. Jingchun et al. [36] explored the topological characteristics of the global and local networks of cancer proteins translated from cancer genes in the human PIN. They further confirmed the results that the network topological properties of cancer proteins are not the same as non-cancer proteins. The topological properties of the proteins translated from the essential genes or control genes are different from that of the cancer proteins. Therefore, they concluded that the coefficient in cancer proteins tended to have higher degrees, higher betweenness, shorter shortest-path distance, and weaker clustering in the human PIN than non-cancer proteins. In 2012, Zhang et al. [37] built a combination classifier based on the topological differences between disease and non-disease proteins in the human PIN. They predicted the candidate genes of coronary artery diseases using this method and obtained 276 candidate genes which showed similar functions to the known disease genes.
Admittedly, the lack of data and data noise become one of the important reasons for the inconsistent conclusions mentioned above. However, there are many improvements in the analyzing strategies. Dickerson et al. [26d] pointed out the big loopholes in the work of Goh et al. At present, the knockout mice gene accounts for only 10% of the total genes, nearly 60% of the human pathogenic genes of the homologous mouse genes do not have corresponding knockout results. Therefore, there is a problem of large data completeness in the work of Goh et al. Moreover, the results of the empirical analysis of Dickerson’s suggest that the topological status of pathogenic proteins is very high. On the other hand, in addition to the degree centrality, it also needs to define the importance of the network topological definition from a multi-angle consideration, and conduct a more detailed discussion [38].
In the drug discovery, network topological analysis showed several important graph properties, including higher degrees and betweenness for the drug targets and drug-regulated genes, though possibly due to network biases. For example, Kotlyar et al. [39] performed the first network topological analysis of proteins encoded by genes that are differentially regulated in response to drugs. Their study showed that these genes and known drug targets had higher centrality in PINs. Compared to these genes, the betweenness and degree of unaffected genes were not that big. The topological structure of the network can also provide a certain reference value for predicting the toxicity of drugs. Toxicity is one of the main reasons for the failure of drug tests. Kotlyar et al. also proposed that topological features of drug-affected genes (e.g., betweenness and centrality in PINs) may be used as an indicator for additional predictive variables. Drug toxicity is proportional to the centrality of proteins translated from regulated genes. More importantly, the system-wide perspective of complex diseases has a great impact on drug discovery processes, because it is helpful to shift from focusing on a single target and single drug to more network-driven methods [27].
4. IDENTIFICATION OF DURG TARGETS USING DISEASE-RELATED PINS
Protein-protein interactions are central to the biological system and have become more and more important to identify targets in drug design. In fact, some of the efficient drugs designed in targeted therapies are the proteins that mimic and replace the ligand-protein binding for specific drug-protein binding. Therefore, identification of key disease related PINs is important to support targeted therapies [6a]. The disease related PIN was usually constructed according to the disease related gene/proteins derived from databases or high throughput experiments, and takes the human interactome as a background dataset. Therefore, Strictly speaking, disease related PIN is a sub-network of the global human PIN. The size of a disease related PIN of some certain disease might be quite small (containing several or tens of proteins) [40], whereas the network for some general disease (such as cancer) or all the human diseases can be quite large. For example, De Las Rivas et al. [41] constructed a cancer related PIN which included 582 cancer proteins and 4968 interactions. Carson et al. [42] constructed a PIN related to all the known human diseases which is composed of the products of 3104 genes, that is, 32% of HPRD proteins with a disease association. The inherent properties of a PIN have a profound influence on drug discovery, which should be discovery of disease-causing networks rather than disease-causing genes.
With the development of disease network, a new discipline called “Network Pharmacology” has emerged. Network analysis related to diseases provides new approaches for discovering drugs, because each protein-target is not working alone but in a framework containing its connectivity with other proteins that may have important alternative targets for drug treatments. Analysis and prediction of biological networks suggest that removing of a single node will have little impact on the disease network. For perturbing a robust phenotype it may be required to modulate multiple interacted proteins in the network [43]. Therefore, network pharmacology is a novel method that can not only improve clinical curative effect of the drugs but also contribute to understand toxicity and potential side effects of drugs, which are one of the most important causes for the failure of new target-driven drug therapies [11a]. The drug target detection with network strategy usually supplies more information than conventional individual target development. Chung et al. [44] calculated the enrichment of 60 published cancer susceptibility genes in the predictions from network-based strategy and traditional expression-alone analysis, respectively. They found that network-based predictions were obviously enriched with cancer susceptibility genes compared to traditional analysis, with some targets (PIK3CA, TP53, ERBB2, HRAS and KRAS) can only be found by network. Some proteins without significantly differential expression in cancer metastasis actually play an important role in the network by interacting with some significantly differentially expressed proteins, and thus were detected as biomarkers, including some well-prognostic biomarkers for breast cancer, such as ERBB2, Myc, and cyclin D1, which were not found in the traditional analysis based only on the differential expression.
Disease related PINs can improve drugs design by determining key protein nodes as potential drug targets. If the target protein is a hub, its inhibition can affect many of the activities which are important to the normal function of the cell, and therefore it is not suitable as a drug target. On the contrary, poorly connected proteins may be sensitive to disease-related networks, and they are more likely to be candidates for disease targets [5]. Construction of signaling sub-networks of the luminal and basal subtypes of the breast cancer revealed the subtype-specific drug targets, most (80%) of which have been experimentally validated using breast cancer cell lines [4]. Furthermore, analysis of sub-networks of the PI3K mutated luminal breast tumors showed that a positive regulatory network loop containing PDGF-D/FLT1/SHC1 could be drug-targeted for improving patient survivals [45]. In the past, a couple of the studies have shown that network motifs/loops could be targeted to treat cancer [46].
In principle, when a drug target is selected in a set of proteins affecting the associated disease, the proteins with lower degrees in the network are more preferably to be selected as drug targets. In consequence, the ideal drug target is a protein whose effects on the interactions in the disease network is essential, but not essential in the network of normal cells [6a]. With this principle, Bergholdt et al. [40] constructed a biological network of type I diabetes by integrating type I diabetes data and protein interactions. In this study, 17 PINs were successfully identified, and these results illustrate the mechanism of type I diabetes pathogenesis and provide the basis for designing new treatment strategies. Some other studies showed that the neighbors of disease-related proteins are prone to interact with the proteins involving in the same disease [47]. This is very important because the investigation of disease related PINs can identify new proteins in the process of disease. Because disease related PINs can change from health state to disease state, the identification of the associations between disease related PINs will help to discover novel drug targets.
In some cases, using of PINs to identify the pathological mechanisms of diseases may help for the development of the diagnosis and treatment strategies for the symptoms and pathogenesis of the diseases [27]. PINs have also become a necessary tool for associating proteins between different phenotypes and diseases, [26b, 48], as well as for studying the relationships between pathological drugs-targets [11b, 49]. It can also help to diagnose and detect sensitive genes in certain patients [50].
Currently, many network-based computational methods were proposed to identify the drug-targets [51] and potential drug combinations [52]. Chen et al. [51] reviewed the main databases, web servers and computational methods for the identification of potential drug-targets. They pointed out that the full utilization of different data sources could contribute to the computational-model-based detection of novel drug–target interactions. Based on this theory, they put forward a serial of novel computational methods working on different data sources for the prediction of drug combinations [52] and miRNA-disease interactions [53]. They further found that the RNA information (such as miRNA, LncRNAs) is also an important source for the identification of the drug targets [54]. It might be a supplement and future direction for the applications of PINs.
5. IDENTIFICATION OF SUB-NETWORK BIOMARKERS
In some diseases, it is necessary to recognize multiple network targets because the disease mechanism could affect multiple genes rather than single genes [55]. For example, cancer is not caused by an individual mutated gene. It could start there, but it enters an ongoing evolutionary battle to combat the biological host defense. In some human cancers, mutations in more than 100 genes have been described [56]. This finding suggests that a single drug target is not effective in treating most common cancers, and that the treatment of cancer needs the understanding of complex pathological mechanisms using drug cocktails [57]. Furthermore,some researchers have concluded that a complete suppression of a single target is less effective than a partial suppression of a small number of targets [43]. Biological network analysis is quite effective in this area, as it seeks to study the participation of multiple drug actions of distinct targets. In fact, the treatment of diseases by regulating more than one drug target is a clear goal of modern systems biology to support the new drug discovery in complex diseases [58] .
The highly interconnected property of the human protein interactome suggests that it is hard to consider diseases as being independent of one another at the molecular level. Traditional methods for identifying biomarkers include expression-data-based and pathway-based approaches. In recent years, sub-network biomarkers have been shown to be more reliable and achieved a high predicting accuracy in the classification of diseases than an individual biomarker selected based on gene expression data models [59]. Currently, the identification of sub-network biomarkers from PINs has been made a great progress. For example, Chuang et al. [44] extracted sub-networks from the human PIN to identify sub-network biomarkers instead of individual genes using a network-based approach. They found that sub-network biomarkers get much higher overlap between predictions and breast cancer cohorts than individual genes (12.7% vs. 1.3%). Besides, the success on the identification of discriminative sub-networks shows the module characteristics of the cancer metastasis mechanism. Compared to individual genes identified by non-network analysis, sub-networks were obvious enriched with cancer susceptibility genes. Furthermore, in the classification test of metastasis and non-metastatic, sub-network biomarkers achieved 48.8% accuracy rate in a single dataset and 55.8% accuracy rate in the reciprocal test of two different datasets, comparatively, the single gene markers got 45.3% and 41.5% accuracy rates. Therefore, they concluded that the identification of sub-network biomarkers using the network-based approach has more advantages than previous methods. First, sub-network biomarkers are more reproducible and robust than individual genes. Second, sub-networks provide more information for disease mechanisms. Third, the disease classification based on network has achieved a high accuracy in prediction. Sub-network biomarkers have been shown to be more predictive for tumor recurrence in prostate and breast cancers [60] .
CONCLUSION
PINs have been proven to be invaluable for the prevention and diagnosis of human diseases and drug target discovery [61], from novel pathways identification, network topological analysis, disease network analysis, to the selection of drug targets. Network based approaches have been widely applied in precision medicine [62]. The identification of specific signaling pathways from the sub-networks can help to understanding the molecular mechanisms of human diseases. Analysis on the property of the network topology contributes to understand the status of disease genes and proteins in the network, further, provides a reference for the designing of drugs. Disease related PINs are the platforms for the investigation of drug targets for diseases. For the treatment of complex diseases, single gene drug target may have side effects and toxicity, and sub-network biomarkers can largely overcome this shortcoming, and become a powerful method for the treatment of human diseases. Although lots of defects still exist in the application of PINs, such as lack of kinetic parameters for calculation or interaction gaps in network itself, PINs provide a systematic platform and bioinformatic tools for the intensive research of human diseases and drug discovery. With the emergence of integrated network in recent years, the integration of PIN with different types of networks, such as metabolic network, transcriptional regulation network and signaling transduction network, will be trends for the functional supplementary of PINs. Besides, the detection of phosphorylation process and RNA targets, which is not included in most current PINs, will also provide more information for the network and improve the accuracy of the prediction on the regulation process. Most importantly, with the increasing understanding of gene/protein-causing diseases, the disease related PIN will be much more perfect in the future for the better performance in the assistant of drug design and disease treatment.
Consent for Publication
Not applicable.
ACKNOWLEDGEMENTS
This work was supported by Grants of the Major State Basic Research Development Program of China (973 programs, 2012CB114405), National Natural Science Foundation of China (31770904), Tianjin Development Program for Innovation and Entrepreneurship, and Innovation Team of Tianjin Fisheries Research System (ITTFRS2017007).
conflict of interest
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Whisstock J.C., Lesk A.M. Prediction of protein function from protein sequence and structure. Q. Rev. Biophys. 2004;36(3):307–340. doi: 10.1017/s0033583503003901. [DOI] [PubMed] [Google Scholar]
- 2.Barabasi A.L., Gulbahce N., Loscalzo J. Network medicine: A network-based approach to human disease. Nat. Rev. Genet. 2011;12(1):56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hao T., Peng W., Wang Q., Wang B., Sun J. Reconstruction and application of protein-protein interaction network. Int. J. Mol. Sci. 2016;17(6):E907. doi: 10.3390/ijms17060907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zaman N., Li L., Jaramillo M.L., Sun Z., Tibiche C., Banville M., Collins C., Trifiro M., Paliouras M., Nantel A., O’Connor-McCourt M., Wang E. Signaling network assessment of mutations and copy number variations predict breast cancer subtype-specific drug targets. Cell Reports. 2013;5(1):216–223. doi: 10.1016/j.celrep.2013.08.028. [DOI] [PubMed] [Google Scholar]
- 5.Safari-Alighiarloo N., Taghizadeh M., Rezaei-Tavirani M., Goliaei B., Peyvandi A.A. Protein-protein interaction networks(PPI) and complex diseases. Gastroenterol. Hepatol. Bed Bench. 2014;7(1):17–31. [PMC free article] [PubMed] [Google Scholar]
- 6.a De Las Rivas J., Prieto C. Protein interactions: mapping interactome networks to support drug target discovery and selection. Methods Mol. Biol. 2012;910:279–296. doi: 10.1007/978-1-61779-965-5_12. [DOI] [PubMed] [Google Scholar]; b Liu A.A., Tang J., Nie W., Su Y. Multi-grained random fields for mitosis identification in time-lapse phase contrast microscopy image sequences. IEEE Trans. Med. Imaging. 2017;36(8):1699–1710. doi: 10.1109/TMI.2017.2686705. [DOI] [PubMed] [Google Scholar]; c Liu A.A., Su Y.T., Nie W.Z., Kankanhalli M. Hierarchical clustering multi-task learning for joint human action grouping and recognition. . IEEE Trans. Pattern Anal. Mach. Intell. 2017;39(1):102–114. doi: 10.1109/TPAMI.2016.2537337. [DOI] [PubMed] [Google Scholar]
- 7.Satoh J.I., Tabunoki H., Yamamura T. Molecular network of the comprehensive multiple sclerosis brain-lesion proteome. Mult. Scler. 2009;15(5):531–541. doi: 10.1177/1352458508101943. [DOI] [PubMed] [Google Scholar]
- 8.a Goni J., Esteban F.J., de Mendizabal N.V., Sepulcre J., Ardanza-Trevijano S., Agirrezabal I., Villoslada P. A computational analysis of protein-protein interaction networks in neurodegenerative diseases. BMC Syst. Biol. 2008;2:52. doi: 10.1186/1752-0509-2-52. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Soler-Lopez M., Zanzoni A., Lluis R., Stelzl U., Aloy P. Interactome mapping suggests new mechanistic details underlying Alzheimer’s disease. Genome Res. 2011;21(3):364–376. doi: 10.1101/gr.114280.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.a Pujana M.A., Han J.D., Starita L.M., Stevens K.N., Tewari M., Ahn J.S., Rennert G., Moreno V., Kirchhoff T., Gold B., Assmann V., Elshamy W.M., Rual J.F., Levine D., Rozek L.S., Gelman R.S., Gunsalus K.C., Greenberg R.A., Sobhian B., Bertin N., Venkatesan K., Ayivi-Guedehoussou N., Sole X., Hernandez P., Lazaro C., Nathanson K.L., Weber B.L., Cusick M.E., Hill D.E., Offit K., Livingston D.M., Gruber S.B., Parvin J.D., Vidal M. Network modeling links breast cancer susceptibility and centrosome dysfunction. Nat. Genet. 2007;39(11):1338–1349. doi: 10.1038/ng.2007.2. [DOI] [PubMed] [Google Scholar]; b Su J., Yoon B.J., Dougherty E.R. Identification of diagnostic subnetwork markers for cancer in human protein-protein interaction network. BMC Bioinformatics. 2010;11(Suppl. 6):S8. doi: 10.1186/1471-2105-11-S6-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Wu Z-J., Zhu Y., Huang D-R., Wang Z-Q. Constructing the HBV-human protein interaction network to understand the relationship between HBV and hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2010;29:146. doi: 10.1186/1756-9966-29-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yao C., Li H., Zhou C., Zhang L., Zou J., Guo Z. Multi-level reproducibility of signature hubs in human interactome for breast cancer metastasis. BMC Syst. Biol. 2010;4:151. doi: 10.1186/1752-0509-4-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.a Hopkins A.L. Network pharmacology: The next paradigm in drug discovery. Nat. Chem. Biol. 2008;•••:4. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]; b Pujol A., Mosca R., Farrés J., Aloy P. Unveiling the role of network and systems biology in drug discovery. Trends Pharmacol. Sci. •••;31(3):115–123. doi: 10.1016/j.tips.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 12.Stelzl U., Worm U., Lalowski M., Haenig C., Brembeck F.H., Goehler H., Stroedicke M., Zenkner M., Schoenherr A., Koeppen S., Timm J., Mintzlaff S., Abraham C., Bock N., Kietzmann S., Goedde A., Toksoz E., Droege A., Krobitsch S., Korn B., Birchmeier W., Lehrach H., Wanker E.E. A human protein-protein interaction network: A resource for annotating the proteome. Cell. 2005;122(6):957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 13.Hu Y., Yang Y., Fang Z., Hu Y.S., Zhang L., Wang J. Detecting pathway relationship in the context of human protein-protein interaction network and its application to Parkinson’s disease. Methods. 2017;131:93–103. doi: 10.1016/j.ymeth.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 14.Baranzini S.E., Galwey N.W., Wang J., Khankhanian P., Lindberg R., Pelletier D., Wu W., Uitdehaag B.M., Kappos L., Gene M.S.A.C., Polman C.H., Matthews P.M., Hauser S.L., Gibson R.A., Oksenberg J.R., Barnes M.R. Pathway and network-based analysis of genome-wide association studies in multiple sclerosis. Hum. Mol. Genet. 2009;18(11):2078–2090. doi: 10.1093/hmg/ddp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.International Multiple Sclerosis Genetics Consortium Network-based multiple sclerosis pathway analysis with GWAS data from 15,000 cases and 30,000 controls. Am. J. Hum. Genet. 2013;92(6):854–865. doi: 10.1016/j.ajhg.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.a Sachs K., Perez O., Pe’er D., Lauffenburger D.A., Nolan G.P. Causal protein-signaling networks derived from multiparameter single-cell data. Science. 2005;308(5721):523–529. doi: 10.1126/science.1105809. [DOI] [PubMed] [Google Scholar]; b Silverbush D., Elberfeld M., Sharan R. Optimally orienting physical networks. J. Comput. Biol. 2011;18(11):1437–1448. doi: 10.1089/cmb.2011.0163. [DOI] [PubMed] [Google Scholar]
- 17.Li L., Tibiche C., Fu C., Kaneko T., Moran M.F., Schiller M.R., Li S.S., Wang E. The human phosphotyrosine signaling network: evolution and hotspots of hijacking in cancer. Genome Res. 2012;22(7):1222–1230. doi: 10.1101/gr.128819.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiel C., Vogt A., Campagna A., Chatr-aryamontri A., Swiatek-de Lange M., Beer M., Bolz S., Mack A.F., Kinkl N., Cesareni G., Serrano L., Ueffing M. Structural and functional protein network analyses predict novel signaling functions for rhodopsin. Mol. Syst. Biol. 2011;7:551. doi: 10.1038/msb.2011.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun Y., Ma C., Halgamuge S. The node-weighted Steiner tree approach to identify elements of cancer-related signaling pathways. BMC Bioinformatics. 2017;18(Suppl. 16):551. doi: 10.1186/s12859-017-1958-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bandyopadhyay S., Chiang C-y., Srivastava J., Gersten M., White S., Bell R., Kurschner C., Martin C.H., Smoot M., Sahasrabudhe S., Barber D.L., Chanda S.K., Ideker T. A human MAP kinase interactome. Nat. Methods. 2010;7(10):801–805. doi: 10.1038/nmeth.1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vachiranee L., Seigo T., Susumu G., Kunihiro U., Minoru K. 2007.
- 22.Stites E.C., Trampont P.C., Ma Z., Ravichandran K.S. Network analysis of oncogenic Ras activation in cancer. Science. 2007;318(5849):463–467. doi: 10.1126/science.1144642. [DOI] [PubMed] [Google Scholar]
- 23.Cui Q., Ma Y., Jaramillo M., Bari H., Awan A., Yang S., Zhang S., Liu L., Lu M., O’Connor-McCourt M., Purisima E.O., Wang E. A map of human cancer signaling. Mol. Syst. Biol. 2007;3:152. doi: 10.1038/msb4100200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zanzoni A., Soler-Lopez M., Aloy P. A network medicine approach to human disease. FEBS Lett. 2009;583(11):1759–1765. doi: 10.1016/j.febslet.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 25.Han J-D.J., Bertin N., Hao T., Goldberg D.S., Berriz G.F., Zhang L.V., Dupuy D., Walhout A.J.M., Cusick M.E., Roth F.P., Vidal M. Evidence for dynamically organized modularity in the yeast protein-protein interaction network. Nature. 2004;430(6995):88–93. doi: 10.1038/nature02555. [DOI] [PubMed] [Google Scholar]
- 26.a Tu Z., Wang L., Xu M., Zhou X., Chen T., Sun F. Further understanding human disease genes by comparing with housekeeping genes and other genes. BMC Genomics. 2006;7:31. doi: 10.1186/1471-2164-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Goh K.I., Cusick M.E., Valle D., Childs B., Vidal M., Barabasi A.L. The human disease network. Proc. Natl. Acad. Sci. USA. 2007;104(21):8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Jin W., Qin P., Lou H., Jin L., Xu S. A systematic characterization of genes underlying both complex and Mendelian diseases. Hum. Mol. Genet. 2012;21(7):1611–1624. doi: 10.1093/hmg/ddr599. [DOI] [PubMed] [Google Scholar]; d Dickerson J., Zhu A., Robertson D., Hentges K. Defining the role of essential genes in human disease. PLoS One. 2011;6:e27368. doi: 10.1371/journal.pone.0027368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jaeger S., Aloy P. From protein interaction networks to novel therapeutic strategies. IUBMB Life. 2012;64(6):529–537. doi: 10.1002/iub.1040. [DOI] [PubMed] [Google Scholar]
- 28.Oti; Brunner, The modular nature of genetic diseases. Clin. Genet. 2007;71(1):1–11. doi: 10.1111/j.1399-0004.2006.00708.x. [DOI] [PubMed] [Google Scholar]
- 29.Zaghloul N.A., Katsanis N. Functional modules, mutational load and human genetic disease. Trends Genet. •••;26(4):168–176. doi: 10.1016/j.tig.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao S., Li S. Network-based relating pharmacological and genomic spaces for drug target identification. PLoS One. 2010;5(7):e11764. doi: 10.1371/journal.pone.0011764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Navlakha S., Kingsford C. The power of protein interaction networks for associating genes with diseases. Bioinformatics. 2010;26(8):1057–1063. doi: 10.1093/bioinformatics/btq076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taylor I.W., Linding R., Warde-Farley D., Liu Y., Pesquita C., Faria D., Bull S., Pawson T., Morris Q., Wrana J.L. Dynamic modularity in protein interaction networks predicts breast cancer outcome. Nat. Biotechnol. 2009;27(2):199–204. doi: 10.1038/nbt.1522. [DOI] [PubMed] [Google Scholar]
- 33.Jonsson P.F., Bates P.A. Global topological features of cancer proteins in the human interactome. Bioinformatics. 2006;22(18):2291–2297. doi: 10.1093/bioinformatics/btl390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vidal M., Cusick M.E., Barabasi A.L. Interactome networks and human disease. Cell. 2011;144(6):986–998. doi: 10.1016/j.cell.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barrenas F., Chavali S., Holme P., Mobini R., Benson M. Network properties of complex human disease genes identified through genome-wide association studies. PLoS One. 2009;4(11):e8090. doi: 10.1371/journal.pone.0008090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun J., Zhao Z. A comparative study of cancer proteins in the human protein-protein interaction network. BMC Genomics. 2010;11(Suppl. 3):S5. doi: 10.1186/1471-2164-11-S3-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang L., Li X., Tai J., Li W., Chen L. Predicting candidate genes based on combined network topological features: A case study in coronary artery disease. PLoS One. 2012;7(6):e39542. doi: 10.1371/journal.pone.0039542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Furlong L.I. Human diseases through the lens of network biology. Trends Genet. 2013;29(3):150–159. doi: 10.1016/j.tig.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 39.Kotlyar M., Fortney K., Jurisica I. Network-based characterization of drug-regulated genes, drug targets, and toxicity. Methods. 2012;57(4):499–507. doi: 10.1016/j.ymeth.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 40.Bergholdt R., Brorsson C., Palleja A., Berchtold L.A., Fløyel T., Bang-Berthelsen C.H., Frederiksen K.S., Jensen L.J., Størling J., Pociot F. Identification of Novel Type 1 diabetes candidate genes by integrating genome-wide association data, protein-protein interactions, and human pancreatic islet gene expression. Diabetes. 2012;61(4):954. doi: 10.2337/db11-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.De Las Rivas J., Alonso-Lopez D., Arroyo M.M. Human Interactomics: Comparative analysis of different protein interaction resources and construction of a cancer protein-drug bipartite network. Adv. Protein Chem. Struct. Biol. 2018;111:263–282. doi: 10.1016/bs.apcsb.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 42.Carson M.B., Lu H. Network-based prediction and knowledge mining of disease genes. BMC Med. Genomics. 2015;8(Suppl. 2):S9. doi: 10.1186/1755-8794-8-S2-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Csermely P., Ágoston V., Pongor S. The efficiency of multi-target drugs: The network approach might help drug design. Trends Pharmacol. Sci. 2005;26(4):178–182. doi: 10.1016/j.tips.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 44.Chuang H.Y., Lee E., Liu Y.T., Lee D., Ideker T. Network-based classification of breast cancer metastasis. Mol. Syst. Biol. 2007;3:140. doi: 10.1038/msb4100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McGee S.R., Tibiche C., Trifiro M., Wang E. Network analysis reveals a signaling regulatory loop in the pik3ca-mutated breast cancer predicting survival outcome. Genomics Proteomics Bioinformatics. 2017;15(2):121–129. doi: 10.1016/j.gpb.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cloutier M., Wang E. Dynamic modeling and analysis of cancer cellular network motifs. Integr. Biol. 2011;3(7):724–732. doi: 10.1039/c0ib00145g. [DOI] [PubMed] [Google Scholar]
- 47.Kitano H. A robustness-based approach to systems-oriented drug design. Nat. Rev. Drug Discov. 2007;6(3):202–210. doi: 10.1038/nrd2195. [DOI] [PubMed] [Google Scholar]
- 48.Ideker T., Sharan R. Protein networks in disease. Genome Res. 2008;18(4):644–652. doi: 10.1101/gr.071852.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Berger S.I., Iyengar R. Network analyses in systems pharmacology. Bioinformatics. 2009;25(19):2466–2472. doi: 10.1093/bioinformatics/btp465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fliri A.F., Loging W.T., Volkmann R.A. Cause-effect relationships in medicine: A protein network perspective. Trends Pharmacol. Sci. •••;31(11):547–555. doi: 10.1016/j.tips.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 51.Chen X., Yan C.C., Zhang X., Zhang X., Dai F., Yin J., Zhang Y. Drug-target interaction prediction: Databases, web servers and computational models. Brief. Bioinform. 2016;17(4):696–712. doi: 10.1093/bib/bbv066. [DOI] [PubMed] [Google Scholar]
- 52.Chen X., Ren B., Chen M., Wang Q., Zhang L., Yan G. NLLSS: Predicting synergistic drug combinations based on semi-supervised learning. PLOS Comput. Biol. 2016;12(7):e1004975. doi: 10.1371/journal.pcbi.1004975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.a You Z.H., Huang Z.A., Zhu Z., Yan G.Y., Li Z.W., Wen Z., Chen X. PBMDA: A novel and effective path-based computational model for miRNA-disease association prediction. PLOS Comput. Biol. 2017;13(3):e1005455. doi: 10.1371/journal.pcbi.1005455. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Chen X., Huang L. LRSSLMDA: Laplacian regularized sparse subspace learning for miRNA-disease association prediction. PLOS Comput. Biol. 2017;13(12):e1005912. doi: 10.1371/journal.pcbi.1005912. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Chen X., Niu Y.W., Wang G.H., Yan G.Y. MKRMDA: Multiple kernel learning-based Kronecker regularized least squares for MiRNA-disease association prediction. J. Transl. Med. 2017;15(1):251. doi: 10.1186/s12967-017-1340-3. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Chen X., Huang L., Xie D., Zhao Q. EGBMMDA: Extreme gradient boosting machine for mirna-disease association prediction. Cell Death Dis. 2018;9(1):3. doi: 10.1038/s41419-017-0003-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.a Chen X., Xie D., Zhao Q., You Z.H. MicroRNAs and complex diseases: from experimental results to computational models. Brief. Bioinform. 2017 doi: 10.1093/bib/bbx130. [DOI] [PubMed] [Google Scholar]; b Chen X., Yan C.C., Zhang X., You Z.H. Long non-coding RNAs and complex diseases: from experimental results to computational models. Brief. Bioinform. 2017;18(4):558–576. doi: 10.1093/bib/bbw060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schrattenholz A., Groebe K., Soskic V. Systems biology approaches and tools for analysis of interactomes and multi-target drugs. In: Yan Q., editor. Systems Biology in Drug Discovery and Development: Methods and Protocols. Totowa, NJ: Humana Press; 2010. pp. 29–58. [DOI] [PubMed] [Google Scholar]
- 56.Meyerson M., Gabriel S., Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nat. Rev. Genet. 2010;11(10):685–696. doi: 10.1038/nrg2841. [DOI] [PubMed] [Google Scholar]
- 57.Schrattenholz A., Soskic V. 2008.
- 58.Liu H., Wang J., Zhou W., Wang Y., Yang L. Systems approaches and polypharmacology for drug discovery from herbal medicines: An example using licorice. J. Ethnopharmacol. 2013;146(3):773–793. doi: 10.1016/j.jep.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 59.Al-Harazi O., Al Insaif S., Al-Ajlan M.A., Kaya N., Dzimiri N., Colak D. Integrated genomic and network-based analyses of complex diseases and human disease network. J. Genet. Genomics. 2016;43(6):349–367. doi: 10.1016/j.jgg.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 60.a Fu C., Li J., Wang E. Signaling network analysis of ubiquitin-mediated proteins suggests correlations between the 26S proteasome and tumor progression. Mol. Biosyst. 2009;5(12):1809–1816. doi: 10.1039/B905382D. [DOI] [PubMed] [Google Scholar]; b Paliouras M., Zaman N., Lumbroso R., Kapogeorgakis L., Beitel L.K., Wang E., Trifiro M. Dynamic rewiring of the androgen receptor protein interaction network correlates with prostate cancer clinical outcomes. Integr. Biol. 2011;3(10):1020–1032. doi: 10.1039/c1ib00038a. [DOI] [PubMed] [Google Scholar]; c Wang E. Understanding genomic alterations in cancer genomes using an integrative network approach. Cancer Lett. 2013;340(2):261–269. doi: 10.1016/j.canlet.2012.11.050. [DOI] [PubMed] [Google Scholar]
- 61.a Masoudi-Nejad A., Wang E. Cancer modeling and network biology: Accelerating toward personalized medicine. Semin. Cancer Biol. 2015;30:1–3. doi: 10.1016/j.semcancer.2014.06.005. [DOI] [PubMed] [Google Scholar]; b Wang E., Zaman N., McGee S., Milanese J.S., Masoudi-Nejad A., O’Connor-McCourt M. Predictive genomics: A cancer hallmark network framework for predicting tumor clinical phenotypes using genome sequencing data. Semin. Cancer Biol. 2015;30:4–12. doi: 10.1016/j.semcancer.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 62.a Wang E., Cho W.C.S., Wong S.C.C., Liu S. Disease biomarkers for precision medicine: Challenges and future opportunities. Genomics Proteomics Bioinformatics. 2017;15(2):57–58. doi: 10.1016/j.gpb.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Wang E., Zou J., Zaman N., Beitel L.K., Trifiro M., Paliouras M. Cancer systems biology in the genome sequencing era: part 2, evolutionary dynamics of tumor clonal networks and drug resistance. Semin. Cancer Biol. 2013;23(4):286–292. doi: 10.1016/j.semcancer.2013.06.001. [DOI] [PubMed] [Google Scholar]; c Wang E., Zou J., Zaman N., Beitel L.K., Trifiro M., Paliouras M. Cancer systems biology in the genome sequencing era: part 1, dissecting and modeling of tumor clones and their networks. Semin. Cancer Biol. 2013;23(4):279–285. doi: 10.1016/j.semcancer.2013.06.002. [DOI] [PubMed] [Google Scholar]